Abstract
Over the last approximately 2.6 Myr, Earth's climate has been dominated by cyclical ice ages that have profoundly affected species' population sizes, but the impact of impending anthropogenic climate change on species’ extinction potential remains a worrying problem. We investigated 11 bat species from different taxonomic, ecological and geographical backgrounds using combined information from palaeoclimatic habitat reconstructions and genomes to analyse biotic impacts of historic climate change. We discover tightly correlated fluctuations between species' historic distribution and effective population size, identify frugivores as particularly susceptible to global warming, pinpoint large insectivores as having overall low effective population size and flag the onset of the Holocene (approx. 10–12 000 years ago) as the period with the generally lowest effective population sizes across the last approximately 1 Myr. Our study shows that combining genomic and palaeoclimatological approaches reveals effects of climatic shifts on genetic diversity and may help predict impacts of future climate change.
Keywords: climate change, global warming, pairwise sequentially Markovian coalescent, ecological niche model, demographic history, Chiroptera
1. Introduction
Quaternary climatic fluctuations (i.e. those over the last approx. 2.6 Myr) have produced recurrent glacial periods resulting in global habitat shifts that affect a multitude of organisms [1–3]. Colder climatic episodes have generally led to significant drops in global temperatures and increased glaciation, shrinking habitat space for many taxa (especially at higher latitudes) and forcing them into isolated pockets of local refugia. However, these periods have also produced reductions in global sea levels, thereby connecting isolated landmasses in shelf areas and expanding habitat space for many other organisms [1,4]. Such cyclical periods of isolation and connectivity have alternately triggered fluctuations in population size, with significant implications for speciation and survival potential in animals and plants [1–3,5–9]. If we could characterize these Quaternary fluctuations across a wide panel of species with different ecological backgrounds, it would allow us to pinpoint species groups with specific ecological requirements that will be particularly extinction-prone with impending anthropogenic climate change [10].
Bats (order Chiroptera) are excellent bioindicators as they are highly sensitive to climate change, habitat change, food availability and other environmental aspects [11,12]. For many chiropteran species, climate change has been an agent of isolation, population fluctuations and gradual divergence [13–16]. However, while it is well established that these volant mammals are susceptible to short-term habitat shifts [17,18], the impact of long-term climatic and environmental fluctuations on their population sizes remains relatively unexplored (for exceptions, see [18,19]).
The genetic diversity of a species is positively correlated with its effective population size (Ne) and is an important underlying parameter defining the evolutionary and long-term survival potential of a species [20,21]. A recently developed method, pairwise sequentially Markovian coalescent (PSMC) [22], makes use of heterozygosity and recombination information from a single genome to provide deep insights into a species’s fluctuations in population genetic diversity by estimating fluctuations in effective population size over time [2,23–27]. While the Ne of a single representative genome does not always perfectly reflect the genetic diversity of the whole species, especially in cases of populations that have undergone an atypical population development, PSMC has proven to be a powerful tool to infer temporal trends in genetic diversity [2,27]. In conjunction with ecological niche models that reconstruct changes in distribution on the basis of palaeoclimatic data, such methods can provide much needed information on the effects of episodic climatic fluctuations on the population genetic history of a species and help us understand its vulnerability to future climate change. On a comparative scale, such methods have the power to reveal differences in evolutionary trajectories across species and may help us understand how species biology shapes fluctuations in genomic diversity [24,26,27].
Comparative research to link differences in long-term fluctuations in genetic diversity to species biology remains in its infancy, with few systematic studies reported for major mammalian groups (for exceptions, see [27]). In the present study, we analysed 11 genomes of both megabats and microbats, representing species across most major continents, size classes and ecological requirements, and evaluated fluctuations in Ne across their evolutionary history. We also reconstructed fluctuations in species distribution during the most recent glacial cycles. This new approach of directly comparing palaeo-distributions with fluctuations in Ne allowed us to make inferences about the long-term survival potential of species across different niches and ecological requirements. We predicted that fluctuations in species distribution would correlate with fluctuations in population Ne, and that species biology would affect long-term fluctuations in Ne depending on diet, body size and other ecological factors.
2. Methods
(a). Experimental design
Out of 13 bat genomes on GenBank available at the time of analysis, we used publicly available raw reads from 11 species comprising both megabats and microbats from five out of 18 extant bat families covering a diverse range of size classes, ecological preferences, habitats and geographical regions (electronic supplementary material, table S1). Where possible, we obtained the collection locality for the sampled individual of each species (electronic supplementary material, table S1). Based on our distribution models (see below) as well as the available literature (electronic supplementary material, table S1), we termed each genome as belonging to a refugial or non-refugial population by placing sampling locations (whenever available) or region of occurrence into the context of our reconstructions of species distribution across the late Quaternary.
(b). PSMC analyses
We obtained raw reads of the 11 bat genomes either from the Sequence Read Archive (SRA) or the European Bioinformatics Institute (electronic supplementary material, table S1). We first checked the quality of the raw reads of each species in FastQC [28], and then used the repair.sh script from BBMap 35.51 [29] to repair disordered paired end reads and remove orphan reads. For each species, we performed multiple steps of filtering to remove reads aligning to the mitogenome as well as sex chromosomes and then proceeded to map the remaining reads to the respective genome. Whenever available, we used the mitogenome of the species of interest to map against genomic reads for filtering, or otherwise used the mitogenome of a closely related species within the same genus (except Megaderma lyra and Eidolon helvum; electronic supplementary material, table S1). As the sex chromosomes of bats are not yet characterized, we used the sequences of mouse sex chromosomes (CM001013 and CM001014) for filter mapping. We used the ‘view’ command within SAMtools 0.1.19 [30,31] to obtain unmapped reads (in bam format) and then converted the bam files to paired-end Fastq reads using the bamToFastq tool in the HYDRA 0.5.3 package [32]. These filtered reads (after removing reads mapping to the mitogenome and sex chromosomes) were then mapped onto the genome using the BWA-MEM 0.7.7-r441 [33] algorithm. We only kept reads with a mapping score greater than 20, and used PICARDTOOLS 1.95 (http://broadinstitute.github.io/picard) to sort bam files. We implemented SAMtools mpileup in conjunction with bcftools to identify variable sites, adjusting the mapping quality score (−C50) to reduce the effect of excessive mismatch and setting the minimum depth to ten and maximum depth to 100 for SNP calling. Based on previous studies, a minimum depth coverage of 10 is recommended for SNP calling for PSMC analysis (PSMC instructions; https://github.com/lh3/psmc).
We used the following parameters for PSMC analysis: −t 15 −r 5 −p 4 + 25 * 2 + 4 + 6 (where p is the number of free atomic time intervals, t is the upper limit of time to most recent common ancestor and r is the ratio of the scaled mutation rate and the recombination rate). We performed 30 iterations for optimization of parameters, and ran 100 bootstrap replicates for each genome to determine the uncertainty in our estimates. For bootstrap replicates, long chromosome segments were split into smaller segments and then randomly sampled with replacement.
To obtain estimates of effective population size, we used a previously proposed mammalian mutation rate of 2.2 × 10−9 per base pair per year [34] and three different estimates of generation time (1 year, 2 years and 8 years) as all three estimates are quite commonly used in studies on bats [14,35–42].
(c). Species and climatic data
Species occurrence records were collected from GBIF (accessed August 2017) using the rgbif package [43] as well as from the scientific literature whenever possible (electronic supplementary material, table S2).
We employed ecological niche modelling (ENM) to predict the distribution of each species during four periods: at the present time, Mid-Holocene (approx. 6000 years ago), Last Glacial Maximum (LGM, approx. 20 000 years ago) and Last Interglacial period (LIG, approx. 110 000–130 000 years ago). Environmental variables were extracted from the WORLDCLIM database (version 1.4) [44] at a resolution of 30 arc seconds (for current, Holocene and LIG) and 2.5 min (for LGM). Layers were resized in DIVA GIS (version 7.5) [45] according to the individual species’ study area. From the 19 available bioclimatic variables, we removed non-independent ones and further chose a subset of variables which maximized variability within our dataset (electronic supplementary material) through correlation analyses and principal component analyses to select a total of five variables for ENM: annual mean temperature (Bio1), mean diurnal range (Bio2), isothermality (Bio3), precipitation of driest quarter (Bio17) and precipitation of warmest quarter (Bio18) (electronic supplementary material, table S3). Most of these variables have been previously used for ENM of bats [18]. We additionally followed two alternative approaches in which climatic variables were extracted from distribution points of species subgroups, rather than the global set of distribution points of all 11 species (see electronic supplementary material).
(d). Ecological niche modelling
We applied the MaxEnt algorithm (version 3.3.3 k) [46] for the modeling of species distributions. MaxEnt is based on a probabilistic framework and generates predictions about distributions from an incomplete set of information. The main assumption is based on the probability distribution of maximum entropy, which is subject to certain environmental constraints that along with species occurrence information ultimately generate a species’ potential distribution pattern [46]. MaxEnt consistently outperforms other programs and provides simple predictions for habitat suitability, which are useful for qualitative analyses [47,48]. We mostly applied default parameters, used 25% of data for testing purposes and selected feature classes based on species occurrence records. We set up the following parameters for niche modelling analyses in MaxEnt: 10 000 background points (selected by MaxEnt), 10 runs of cross validations (reduced to one run in analyses under subgroup partitioning schemes, see the electronic supplementary material), 500 iterations and setting the regularization multiplier at 1. Feature selection was in accordance with occurrence records and we generated output in logistic format with probability of presence (electronic supplementary material). We selected the mean representation of all 10 runs for further analysis of each species across each time period.
(e). Model evaluation and area analysis
Prediction accuracy of the modelling exercise was tested using a receiver operating characteristics (ROC) plot analysis, a widely applied measure for model performance evaluation [49,50]. Taking into account the sensitivity and commission error (1-specificity) of the ROC curve plots, we observed that all possible thresholds fell between 0 and 1. A model was considered better than random if the curve was above the diagonal, as indicated by the area under the curve (AUC) being greater than 0.5. Furthermore, a Jackknife test of variable importance was conducted for each species to identify the variable with maximum contributions.
The model maps were transferred to DIVA GIS for further analysis. We opted for a continuous distribution map rather than a binary one as the former map depicts all probable areas (low to high probabilities) and is free of threshold-related uncertainties [51]. Changes across different time periods were assessed only for medium- to high-probability regions (i.e. approx. 0.36–1; blue regions in figure 1) and grid areas were calculated after accounting for latitudinal effects (see the electronic supplementary material).
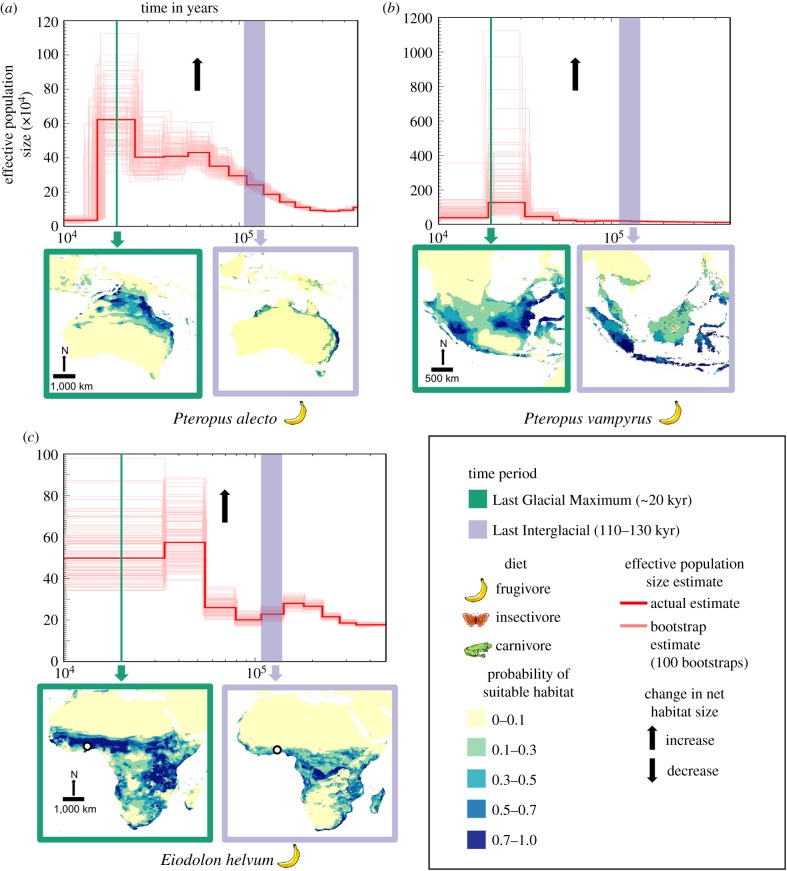
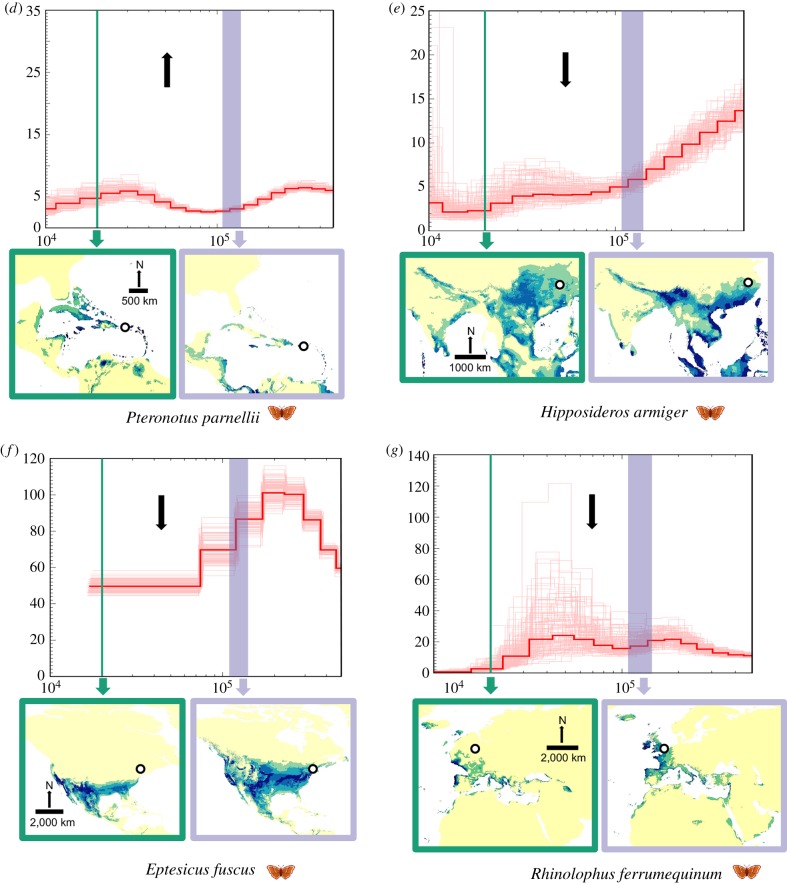
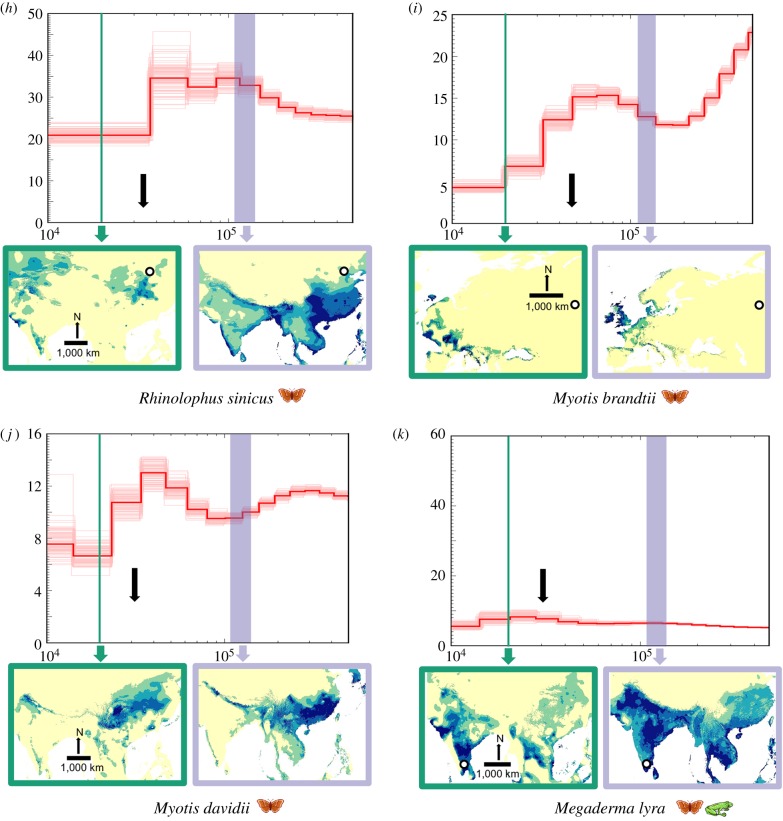
Figure 1.
Temporal fluctuations in effective population size (Ne) between approximately 400 000 and 10 000 years before present, with extent of suitable habitat mapped for the Last Interglacial (LIG) and the Last Glacial Maximum (LGM), respectively. Black arrows indicate estimated net change in suitable habitat availability based on the ecological niche models from the LIG to the LGM (see table 2 for more details). We assumed a generation time of 2 years and a mutation rate of 2.2 × 10−9 per base pair per year for all PSMC plots. (a–d) Species with a relatively high Ne leading to the LGM along with concomitant increase in habitat. (e–j) Species with a relatively low Ne and decrease in habitat leading to the LGM. Megaderma lyra (k) shows a slight increase in Ne and decrease in habitat leading to the LGM. Collection localities for bat genomes are mapped using a white circle with black margin whenever available (see the electronic supplementary material, table S1 for more information). (Online version in colour.)
(f). Correlation between historical fluctuations in Ne and habitat
To assess the correlation between habitat fluctuations and Ne fluctuations across study species, we specifically concentrated on two periods (LIG to LGM, and LGM to Holocene), approximating two key Earth historic events in the last 150 000 years for which we were able to obtain comparative estimates of change in Ne and change in suitable habitat. For each period (LIG to LGM, and LGM to Holocene), we coded changes in Ne and habitat as increasing or decreasing, except for three species (Eidolon helvum, Eptesicus fuscus and Rhinolophus sinicus) in which Ne remained identical between the LGM and Holocene and was hence classified as ‘stable’ (figure 1 and table 1). From a contingency table thus created for each period, we checked for significance of association between habitat and Ne fluctuation using a χ2-test. We further performed a phi-test to determine the extent of significance of the correlation. Values of phi vary between −1 and +1, with 0 meaning no association, values approaching +1 suggesting a strong positive association and values approaching −1 suggesting a strong negative association.
Table 1.
Effective population sizes estimated for approximately 10 000 years ago, the LGM and the LIG. Effective population sizes are approximate values extracted from the PSMC graphs (figure 1; electronic supplementary material, figures S2 and S3). LGM, Last Glacial Maximum; LIG, Last Interglacial.
| effective population size approximately 10 000 years ago |
effective population size during the Last Glacial Maximum |
effective population size during the Last Interglacial |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| generation time (in years) |
generation time (in years) |
generation time (in years) |
|||||||
| species | 1 | 2 | 8 | 1 | 2 | 8 | 1 | 2 | 8 |
| Pteropus alecto | 70 000 | 35 000 | 8000 | 1 260 000 | 620 000 | 156 000 | 600 000 | 300 000 | 70 000 |
| Pteropus vampyrus | 1 300 000 | 640 000 | 160 000 | 2 250 000 | 1 100 000 | 280 000 | 400 000 | 200 000 | 50 000 |
| Eidolon helvum | 1 000 000 | 500 000 | 125 000 | 1 000 000 | 500 000 | 125 000 | 460 000 | 220 000 | 58 000 |
| Pteronotus parnellii | 60 000 | 30 000 | 7500 | 95 000 | 48 000 | 12 000 | 60 000 | 30 000 | 7500 |
| Megaderma lyra | 110 000 | 55 000 | 14 000 | 155 000 | 75 000 | 19 000 | 130 000 | 75 000 | 15 000 |
| Hipposideros armiger | 65 000 | 32 000 | 5600 | 45 000 | 22 000 | 5200 | 345 000 | 170 000 | 43 000 |
| Eptesicus fuscus | n.a. | n.a. | n.a. | 1 000 000 | 500 000 | 125 000 | 1 800 000 | 860 000 | 215 000 |
| Rhinolophus ferrumequinum | 15 000 | 7500 | 2000 | 55 000 | 25 000 | 7500 | 340 000 | 170 000 | 43 000 |
| Rhinolophus sinicus | 420 000 | 210 000 | 52 000 | 420 000 | 210 000 | 52 000 | 660 000 | 330 000 | 82 000 |
| Myotis davidii | 150 000 | 76 000 | 19 000 | 135 000 | 66 000 | 16 500 | 190 000 | 94 000 | 23 500 |
| Myotis brandtii | 85 000 | 45 000 | 11 000 | 85 000 | 45 000 | 11 000 | 255 000 | 115 000 | 31 000 |
3. Results
(a). Strong Quaternary fluctuations in genetic diversity
Our results revealed genomic signatures of drastic fluctuations in Ne correlated with Quaternary climatic oscillations in virtually all 11 bat species under study (figure 1; electronic supplementary material, figure S1 and table S1). We reconstructed the demographic history dating back to at least 1 Myr, and in some cases significantly beyond (electronic supplementary material, figures S1–S3). For all species, whole-genome based estimates of Ne fell within the distribution of bootstrap estimates (figure 1). Although in many species of bat female age of first reproduction varies between 1 and 2 years, the average age of reproduction will be much higher as bats generally live longer than expected based on body size [14,35–42,52]. To account for a wide range of possible generation times, including first and average age of reproduction, we have consequently used three different values for generation time, namely 1, 2 and 8 years, revealing identical temporal patterns of change, with Ne negatively correlated to generation time (electronic supplementary material, figures S1–S3; akin to previous studies [2,26]).
Large-bodied insectivores (including carnivores) experienced comparatively lesser fluctuations in their Ne during the LIG to the LGM compared to small insectivores and large frugivores (figure 1; electronic supplementary material, figures S1–S3). Furthermore, we observed that the overall Ne for most bats was low as they entered the Holocene, especially when assuming a generation time of 8 years (table 1).
(b). Ecological niche modelling reveals fluctuations in geographical distribution
When bioclimatic variables were extracted from the global set of all 11 species' distribution points, MaxEnt generated statistically validated models with AUC > 0.9 (both training and testing AUCs; electronic supplementary material, table S4). Environmental parameters varied in their impact on reconstructed distributions depending on species and time scales (electronic supplementary material, table S5). Isothermality was the most important climatic variable for all three frugivorous bats (electronic supplementary material, table S5). Among insectivores, the importance of climatic variables differed according to time scales (electronic supplementary material, table S5).
The ENM suggested that all bats experienced periodic fluctuations of suitable habitat during the LIG, LIG to LGM, as well as throughout the Holocene (figure 1 and table 2). We used these habitat reconstructions from regions of sample collection as additional information to categorize each genome as part of refugial or non-refugial populations (electronic supplementary material, table S1).
Table 2.
Fluctuations in suitable habitat reconstructed across major climatic periods expressed as percentage of maximum value. LGM, Last Glacial Maximum; LIG, Last Interglacial.
| species | current | Mid-Holocene | LGM | LIG |
|---|---|---|---|---|
| Pteropus alecto | 35.08 | 18.9 | 100 | 4.69 |
| Pteropus vampyrus | 100 | 84.46 | 60.05 | 53.82 |
| Eidolon helvum | 44.31 | 36.67 | 100 | 27.34 |
| Pteronotus parnellii | 74.56 | 100 | 19.06 | 12 |
| Megaderma lyra | 100 | 69.29 | 7.27 | 83.02 |
| Hipposideros armiger | 100 | 84.34 | 21.76 | 57.24 |
| Eptesicus fuscus | 95.72 | 100 | 49.09 | 88.42 |
| Rhinolophus ferrumequinum | 100 | 68.72 | 28.05 | 41.36 |
| Rhinolophus sinicus | 100 | 87.33 | 0.36 | 64.51 |
| Myotis davidii | 99.88 | 94.12 | 14.2 | 100 |
| Myotis brandtii | 100 | 91.64 | 19.38 | 27.53 |
Extracting bioclimatic variables from subgroups of the total set of 11 species (either divided into individual species or into continental groups) led to the selection of sets of variables that exhibited an approximately 17–50% overlap with those from the global species dataset. In the same vein, temporal population trends from the LIG to the LGM and from the LGM to the Holocene (not shown) agreed with those of the global set only 50% (individual species) and 62.5% (continental groups) of the time, confirming previous results that such reconstructions should optimally be based on a large set of species to avoid idiosyncratic biases [53].
(c). Correlation between historical fluctuations in Ne and distribution
We performed a phi-test to assess potential associations between palaeo-habitat fluctuations and Ne fluctuations within our study species during the time intervals between the LIG and the LGM, and between the LGM and the Holocene. During the period of overall climatic cooling from the LIG to the LGM, we observed a significant and strong positive correlation (p-value 0.006; phi-value 0.828) between change in habitat and change in Ne. However, for the period from the LGM to the Holocene, when the most recent climate warming commenced, no significant correlation (p-value 0.632; phi-value 0.289) was observed.
4. Discussion
Quaternary climatic fluctuations have played a major role in shaping evolutionary trajectories of populations and species across all major biomes and taxonomic groups [1,7,8,54,55]. These climatic cycles have caused episodes of population isolation and demographic fluctuations, and remain an important driver of biotic diversification and speciation [1]. Critically, each individual genome carries with it the record of a species’s response to Quaternary warming and cooling episodes [2,22,26,56], teaching us important insights on the impact of climate change on genetic diversity and extinction potential of a species.
In the present era of the sixth mass extinction, knowledge of the standing genetic variation of extant natural populations and their vulnerability to future climate change is imperative for conservation management [57,58]. An effective way to predict vulnerability of species to such future threats is to understand their response to historic climatic change and concomitant habitat fluctuations. In the present study, we have used whole genomes to estimate late Quaternary fluctuations in Ne as a proxy for genetic diversity across a representative panel of bats spanning all major taxonomic groups, size classes and ecological lifestyles (electronic supplementary material, table S1). We also reconstructed historical fluctuations in their distribution and compared these to understand their evolutionary trajectories in response to late Quaternary climatic change.
(a). Climate change leaves a genomic imprint of genetic diversity fluctuations
We show that bats have undergone periods of pronounced fluctuations in Ne and geographical distribution throughout the late Quaternary (figure 1 and table 2). Our comparative analysis focuses on the period between approximately 200 kyr and 10 kyr for two reasons: (1) palaeoclimatic habitat layers are only available until the LIG (http://www.worldclim.org/paleo-climate1) and (2) PSMC inferences based on single genomes are known to be less reliable at very recent timescales, in our case the Holocene [22].
Overall, during the period of global cooling between the LIG and the LGM, most species (10 out of 11) showed a strongly positive correlation between extent of suitable habitat and genetic diversity (figure 1 and table 2). Specifically, five species (figure 1a–d and k) exhibited a relative increase in Ne from the LIG to the LGM, whereas six species (figure 1e–j) displayed a relative decrease in Ne (= drop in diversity).
The only species in which habitat availability did not track Ne fluctuations was Megaderma lyra, the sole carnivorous bat in the panel, whose geographical distribution decreased (figure 1k and table 2) towards the LGM despite a slight increase in net-population genetic diversity (figure 1k). However, the genome-sequenced individual of Megaderma lyra is derived from a population in southern India, well inside the stable, refugial range of this species, suggesting that this population may not reflect the demographic changes that non-refugial populations (i.e. those impacted by climate change) have undergone (figure 1; electronic supplementary material, table S1).
This result is consequential in that our increasing knowledge of palaeo-habitats allows us to make direct inferences on genetic diversity fluctuations, provided we understand the habitat preferences of a species. More importantly, we are able to predict whether future warming trends and concomitant changes in habitat availability will be likely to impact the genetic diversity and survival potential of specific target species.
(b). Species biology predicts response to climate change
Signatures of Ne across eleven bat genomes suggest an important role for species biology and life history in determining the course of genetic diversity fluctuations during the Late Quaternary. Large-bodied frugivores (Pteropus alecto, Pteropus vampyrus and Eidolon helvum), characterized by massive colonies and a great dispersal capability [59,60], showed an overall increase in Ne and distribution range during the most recent major period of climatic cooling (LIG to LGM) (figure 1a–c), perhaps reflecting reduced aridity favouring expansions of fruit-dependent species. Pteropus alecto and Pteropus vampyrus have an archipelagic distribution and have benefitted from Quaternary land expansion for gene flow when sea levels recede [61]. Eidolon helvum, a generalist African open-country dweller, would also have taken advantage of decreased aridity from the LIG to the LGM [62] to increase in Ne (figure 1c).
Across insectivores (including carnivores), larger bats (Megaderma lyra, Hipposideros armiger, Pteronotus parnellii) were characterized by overall low effective population sizes, with often limited fluctuations in population genetic diversity from the LIG to the LGM (figure 1; electronic supplementary material, table S1), whereas smaller bats (Rhinolophus ferrumequinum, Rhinolophus sinicus, Eptesicus fuscus, Myotis davidii and Myotis brandtii) generally tended to exhibit higher values of Ne that often fluctuated more substantially (figure 1), invariably declining from the warmer LIG towards the cooler LGM.
(c). Most bats have entered the Holocene with a low Ne
Most bat species analysed in this study have been characterized by an average vertebrate Ne across the Late Quaternary (table 1). Comparative PSMC analyses across vertebrates remain scant (e.g. six felids and 38 bird species [2,27]): interestingly, the Ne of many bat species in this study roughly equals that of many birds [2] but generally exceeds that of Carnivora [27].
One species, Rhinolophus ferrumequinum, a fairly large-sized insectivore, exhibited an exceptionally low Ne (table 1). Its genetic diversity rivalled that of endangered birds and mega-mammals [2,27,56]. However, its genome was derived from a non-refugial population in Britain, so its exceptionally low current Ne might reflect the great bottlenecks that this peripheral and periodically isolated island population has gone through.
Our comparisons of Ne are based on three plausible generation times according to estimates of first and median age of reproduction (table 1) [35,38,41,63]. If a generation time of 8 years is accepted, many bats in our panel would have entered the Holocene with an alarmingly low population genetic diversity (table 1; electronic supplementary material, figure S2). Regardless of generation time, 7 out of 11 (approx. 64%) species in our panel displayed the absolute lowest levels of genetic diversity during their entry into the Holocene as measured across the entire Late Quaternary (figure 1), attesting that the Early Holocene has not been a time of great population genetic stability in many bat species. Depending on the magnitude of imminent anthropogenic climate change [10], bats may be particularly extinction-prone, given that extreme aridification and warming episodes have been implicated in calamitous bat mortality events [17,41]
(d). Limitations and future challenges
Ancient range reconstruction is a promising method to infer the impact of climate change on natural populations but can be fraught with limitations and imprecision [64,65]. One of the technical challenges of ENM relates to the choice of bioclimatic variables, and whether—in multi-species datasets—these variables should be extracted from the set of distribution points of all species, or subgroups, or individual species. Júnior & Nóbrega [53] showed that inferences on the basis of smaller datasets are subject to strong biases of idiosyncrasy, and that larger datasets combining multiple species should be used as the basis of ENM analyses if present. Our analyses confirm that idiosyncratic biases may operate in our bat dataset, leading us to adopt a global dataset approach for the extraction of bioclimatic variables. Fortunately, our dataset of 11 worldwide species covering approximately 1700 distribution points should render our analysis robust to idiosyncratic biases, but future research into the lower thresholds of distribution data is a worthwhile avenue of inquiry.
A well-established limitation of PSMC analysis is that population subdivision and migration may contribute to genealogies that mimic signals of expansion and contraction [66–70]. Changes in PSMC plots may therefore not always reflect changes in Ne. Yet these changes are still important in terms of a species's extinction potential and susceptibility to environmental change [71]. Similarly, single genome comparisons may often track local population history rather than global species history (this study and [26]). Hence inferences made from PSMC analysis need to be interpreted while keeping in mind the complex demographic history of species in question. Additionally, the choice of mutation rates is known to affect PSMC calculations considerably [22]. For our present study, we used a generic mammalian mutation rate [34] as is commonly employed in the literature [15,72,73] due to the lack of bat specific mutation rates. Regardless, PSMC remains ideal to track changes in population genetic diversity across a species’s evolutionary history.
Future applications should focus on the incorporation of genomic information of multiple individuals to improve Ne estimates across a wider time scale, including into more recent millennia [74]. Given the sharp rise in the availability of genomes over the last few years, we expect that future analyses will be able to incorporate more species, expanding on the vast ecological breadth of bats and other vertebrates, to allow for more detailed insights and conclusions.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Data accessibility
All data generated or analysed during this study are included in this published article and electronic supplementary material. All data points for ENM in this study are provided in electronic supplementary material, table S2.
Authors' contributions
B.C. and F.E.R. conceived the study; B.C., K.M.G. and R.R. performed all data analysis with critical inputs from F.E.R.; B.C., K.M.G. and F.E.R. wrote the manuscript with significant contributions from R.R.
Competing interests
The authors declare no conflict of interest.
Funding
This work was supported by South East Asian Biodiversity Genomics (SEABIG) grant nos. (WBS R-154-000-648-646 and WBS R-154-000-648-733).
References
- 1.Hewitt G. 2000. The genetic legacy of the Quaternary ice ages. Nature 405, 907–913. ( 10.1038/35016000) [DOI] [PubMed] [Google Scholar]
- 2.Nadachowska-Brzyska K, Li C, Smeds L, Zhang G, Ellegren H. 2015. Temporal dynamics of avian populations during Pleistocene revealed by whole-genome sequences. Curr. Biol. 25, 1375–1380. ( 10.1016/j.cub.2015.03.047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Toews DP. 2015. Evolution: a genomic guide to bird population history. Curr. Biol. 25, R465–R467. ( 10.1016/j.cub.2015.04.008) [DOI] [PubMed] [Google Scholar]
- 4.Bintanja R, van de Wal RSW, Oerlemans J.. 2005. Modelled atmospheric temperatures and global sea levels over the past million years. Nature 437, 125–128. ( 10.1038/nature03975) [DOI] [PubMed] [Google Scholar]
- 5.Heaney LR. 1986. Biogeography of mammals in SE Asia: estimates of rates of colonization, extinction and speciation. Biol. J. Linnean Soc. 28, 127–165. ( 10.1111/j.1095-8312.1986.tb01752.x) [DOI] [Google Scholar]
- 6.Ng NS, Wilton PR, Prawiradilaga DM, Tay YC, Indrawan M, Garg KM, Rheindt FE. 2017. The effects of Pleistocene climate change on biotic differentiation in a montane songbird clade from Wallacea. Mol. Phylogenet. Evol. 114, 353–366. ( 10.1016/j.ympev.2017.05.007) [DOI] [PubMed] [Google Scholar]
- 7.Garg KM, Chattopadhyay B, Wilton PR, Prawiradilaga DM, Rheindt FE. 2018. Pleistocene land bridges act as semipermeable agents of avian gene flow in Wallacea. Mol. Phylogenet. Evol. 125, 196–203. ( 10.1016/j.ympev.2018.03.032) [DOI] [PubMed] [Google Scholar]
- 8.Chattopadhyay B, Garg KM, Gwee CY, Edwards SV, Rheindt FE. 2017. Gene flow during glacial habitat shifts facilitates character displacement in a Neotropical flycatcher radiation. BMC Evol. Biol. 17, 210 ( 10.1186/s12862-017-1047-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rebernig CA, Schneeweiss GM, Bardy KE, Schoenswetter P, Villasenor JL, Obermayer R, Stuessy TF, Weiss-Schneeweiss H. 2010. Multiple Pleistocene refugia and Holocene range expansion of an abundant southwestern American desert plant species (Melampodium leucanthum, Asteraceae). Mol. Ecol. 19, 3421–3443. ( 10.1111/j.1365-294X.2010.04754.x) [DOI] [PubMed] [Google Scholar]
- 10.Masson-Delmotte V, et al. 2018. 2018: Global warming of 1.5°C. An IPCC Special Report on the impacts of global warming of 1.5°C above pre-industrial levels and related global greenhouse gas emission pathways, in the context of strengthening the global response to the threat of climate change, sustainable development, and efforts to eradicate poverty See https://www.ipcc.ch/sr15/.
- 11.Jones G, Jacobs DS, Kunz TH, Willig MR, Racey PA. 2009. Carpe noctem: the importance of bats as bioindicators. Endanger. Species Res. 8, 93–115. ( 10.3354/esr00182) [DOI] [Google Scholar]
- 12.Jones G, Rebelo H. 2013. Responses of bats to climate change: learning from the past and predicting the future. In Bat evolution, ecology, and conservation (eds Adams RA, Pedersen SC), pp. 457–478. Berlin, Germany: Springer. [Google Scholar]
- 13.Heaney LR, Walsh JS, Townsend PA. 2005. The roles of geological history and colonization abilities in genetic differentiation between mammalian populations in the Philippine archipelago. J. Biogeogr. 32, 229–247. ( 10.1111/j.1365-2699.2004.01120.x) [DOI] [Google Scholar]
- 14.Flanders J, Jones G, Benda P, Dietz C, Zhang S, Li G, Sharifi M, Rossiter SJ. 2009. Phylogeography of the greater horseshoe bat, Rhinolophus ferrumequinum: contrasting results from mitochondrial and microsatellite data. Mol. Ecol. 18, 306–318. ( 10.1111/j.1365-294x.2008.04021.x) [DOI] [PubMed] [Google Scholar]
- 15.Bhak Y, et al. 2017. Myotis rufoniger genome sequence and analyses: M. rufoniger's genomic feature and the decreasing effective population size of Myotis bats. PLoS ONE 12, e0180418 ( 10.1371/journal.pone.0180418) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin AQ, Csorba G, Li LF, Jiang TL, Lu GJ, Thong VD, Soisook P, Sun KP, Feng J. 2014. Phylogeography of Hipposideros armiger (Chiroptera: Hipposideridae) in the Oriental Region: the contribution of multiple Pleistocene glacial refugia and intrinsic factors to contemporary population genetic structure. J. Biogeogr. 41, 317–327. ( 10.1111/jbi.12163) [DOI] [Google Scholar]
- 17.Welbergen JA, Klose SM, Markus N, Eby P. 2008. Climate change and the effects of temperature extremes on Australian flying-foxes. Proc. R. Soc. B 275, 419–425. ( 10.1098/rspb.2007.1385) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soto-Centeno JA, Steadman DW. 2015. Fossils reject climate change as the cause of extinction of Caribbean bats. Sci. Rep. 5, 7971 ( 10.1038/srep07971) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Razgour O, et al. 2013. The shaping of genetic variation in edge-of-range populations under past and future climate change. Ecol. Lett. 16, 1258–1266. ( 10.1111/ele.12158) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reed DH, Frankham R. 2003. Correlation between fitness and genetic diversity. Conserv. Biol. 17, 230–237. ( 10.1046/j.1523-1739.2003.01236.x) [DOI] [Google Scholar]
- 21.Franklin I, Frankham R. 1998. How large must populations be to retain evolutionary potential? Anim. Conserv. 1, 69–70. ( 10.1111/j.1469-1795.1998.tb00228.x) [DOI] [Google Scholar]
- 22.Li H, Durbin R. 2011. Inference of human population history from individual whole-genome sequences. Nature 475, 493–496. ( 10.1038/nature10231) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nadachowska-Brzyska K, Burri R, Olason PI, Kawakami T, Smeds L, Ellegren H. 2013. Demographic divergence history of pied flycatcher and collared flycatcher inferred from whole-genome re-sequencing data. PLoS Genet. 9, e1003942 ( 10.1371/journal.pgen.1003942) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozma R, Melsted P, Magnússon KP, Höglund J. 2016. Looking into the past–the reaction of three grouse species to climate change over the last million years using whole genome sequences. Mol. Ecol. 25, 570–580. ( 10.1111/mec.13496) [DOI] [PubMed] [Google Scholar]
- 25.Murray GG, et al. 2017. Natural selection shaped the rise and fall of passenger pigeon genomic diversity. Science 358, 951–954. ( 10.1126/science.aao0960) [DOI] [PubMed] [Google Scholar]
- 26.Nadachowska-Brzyska K, Burri R, Smeds L, Ellegren H. 2016. PSMC analysis of effective population sizes in molecular ecology and its application to black-and-white Ficedula flycatchers. Mol. Ecol. 25, 1058–1072. ( 10.1111/mec.13540)27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim S, et al. 2016. Comparison of carnivore, omnivore, and herbivore mammalian genomes with a new leopard assembly. Genome Biol. 17, 211 ( 10.1186/s13059-016-1071-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andrews S.2010. FastQC: a quality control analysis tool for high throughput sequence data. See https://github.com/s-andrews/FastQC .
- 29.Bushnell B. 2016. BBMap short read aligner. Berkeley, CA: University of California; See http://sourceforge.net/projects/bbmap. [Google Scholar]
- 30.Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993. ( 10.1093/bioinformatics/btr509) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. ( 10.1093/bioinformatics/btp352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quinlan AR, Clark RA, Sokolova S, Leibowitz ML, Zhang Y, Hurles ME, Mell JC, Hall IM. 2010. Genome-wide mapping and assembly of structural variant breakpoints in the mouse genome. Genome Res. 20, 623–635. ( 10.1101/gr.102970.109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26, 589–595. ( 10.1093/bioinformatics/btp698) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar S, Subramanian S. 2002. Mutation rates in mammalian genomes. Proc. Natl Acad. Sci. USA 99, 803–808. ( 10.1073/pnas.022629899) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barclay RM, Harder LD. 2003. Life histories of bats: life in the slow lane. In Bat ecology (eds Kunz TH, Fenton M), pp. 209–253. Chicago, IL: University of Chicago Press. [Google Scholar]
- 36.Fleischer T, Gampe J, Scheuerlein A, Kerth G. 2017. Rare catastrophic events drive population dynamics in a bat species with negligible senescence. Sci. Rep. 7, 7370 ( 10.1038/s41598-017-06392-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaillard J-M, Yoccoz NG, Lebreton J-D, Bonenfant C, Devillard S, Loison A, Pontier D, Allaine D. 2005. Generation time: a reliable metric to measure life-history variation among mammalian populations. Am. Nat. 166, 119–123. ( 10.1086/430330) [DOI] [PubMed] [Google Scholar]
- 38.Garg KM, Ramakrishnan U. 2017. Variance in female reproductive success differentially impacts effective population size in the short-nosed fruit bat, Cynopterus sphinx. Evol. Biol. 44, 366–373. ( 10.1007/s11692-017-9414-y) [DOI] [Google Scholar]
- 39.Hammerson G, Kling M, Harkness M, Ormes M, Young B. 2017. Strong geographic and temporal patterns in conservation status of North American bats. Biol. Conserv. 212, 144–152. ( 10.1016/j.biocon.2017.05.025) [DOI] [Google Scholar]
- 40.Korstian JM, Hale AM, Williams DA. 2015. Genetic diversity, historic population size, and population structure in 2 North American tree bats. J. Mammal. 96, 972–980. ( 10.1093/jmammal/gyv101) [DOI] [Google Scholar]
- 41.Storz JF, Beaumont MA. 2002. Testing for genetic evidence of population expansion and contraction: an empirical analysis of microsatellite DNA variation using a hierarchical Bayesian model. Evolution 56, 154–166. ( 10.1111/j.0014-3820.2002.tb00857.x) [DOI] [PubMed] [Google Scholar]
- 42.Sun K, Kimball RT, Liu T, Wei X, Jin L, Jiang T, Lin A, Feng J. 2016. The complex evolutionary history of big-eared horseshoe bats (Rhinolophus macrotis complex): insights from genetic, morphological and acoustic data. Sci. Rep. 6, 35417 ( 10.1038/srep35417) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chamberlain S, Boettiger C, Ram K, Barve V, Mcglinn D. 2016. rgbif: Interface to the global ‘biodiversity'information facility API. R package version 0.9. 5.
- 44.Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. 2005. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25, 1965–1978. ( 10.1002/joc.1276) [DOI] [Google Scholar]
- 45.Hijmans R. 2013. DIVA-GIS, a geographic information system for the analysis of biodiversity data. Version 7.5. See https://diva-gis.org.
- 46.Phillips SJ, Anderson RP, Schapire RE. 2006. Maximum entropy modeling of species geographic distributions. Ecol. Model. 190, 231–259. ( 10.1016/j.ecolmodel.2005.03.026) [DOI] [Google Scholar]
- 47.Elith J, Phillips SJ, Hastie T, Dudík M, Chee YE, Yates CJ. 2011. A statistical explanation of MaxEnt for ecologists. Divers. Distrib. 17, 43–57. ( 10.1111/j.1472-4642.2010.00725.x) [DOI] [Google Scholar]
- 48.Merow C, Smith MJ, Silander JA. 2013. A practical guide to MaxEnt for modeling species' distributions: what it does, and why inputs and settings matter. Ecography 36, 1058–1069. ( 10.1111/j.1600-0587.2013.07872.x) [DOI] [Google Scholar]
- 49.Elith JH, et al. 2006. Novel methods improve prediction of species’ distributions from occurrence data. Ecography 29, 129–151. ( 10.1111/j.2006.0906-7590.04596.x) [DOI] [Google Scholar]
- 50.Peterson AT, Papeş M, Soberón J. 2008. Rethinking receiver operating characteristic analysis applications in ecological niche modeling. Ecol. Modell. 213, 63–72. ( 10.1016/j.ecolmodel.2007.11.008) [DOI] [Google Scholar]
- 51.Lobo JM, Jiménez-Valverde A, Real R. 2008. AUC: a misleading measure of the performance of predictive distribution models. Glob. Ecol. Biogeogr. 17, 145–151. ( 10.1111/j.1466-8238.2007.00358.x) [DOI] [Google Scholar]
- 52.Caswell H. 2001. Matrix population models. Sunderland, MA: Sinauer Associates. [Google Scholar]
- 53.Júnior PDM, Nóbrega CC. 2018. Evaluating collinearity effects on species distribution models: an approach based on virtual species simulation. PLoS ONE 13, e0202403 ( 10.1371/journal.pone.0202403) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taberlet P, Fumagalli L, Wust-Saucy A-G, Cosson JF. 1998. Comparative phylogeography and postglacial colonization routes in Europe. Mol. Ecol. 7, 453–464. ( 10.1046/j.1365-294x.1998.00289.x) [DOI] [PubMed] [Google Scholar]
- 55.Seddon J, Santucci F, Reeve N, Hewitt G. 2001. DNA footprints of European hedgehogs, Erinaceus europaeus and E. concolor: Pleistocene refugia, postglacial expansion and colonization routes. Mol. Ecol. 10, 2187–2198. ( 10.1046/j.0962-1083.2001.01357.x) [DOI] [PubMed] [Google Scholar]
- 56.Mays HL, Jr, et al. 2018. Genomic analysis of demographic history and ecological niche modeling in the endangered Sumatran rhinoceros Dicerorhinus sumatrensis. Curr. Biol. 28, 70–76.e74. ( 10.1016/j.cub.2017.11.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ceballos G, Ehrlich PR, Barnosky AD, García A, Pringle RM, Palmer TM. 2015. Accelerated modern human-induced species losses: entering the sixth mass extinction. Sci. Adv. 1, e1400253 ( 10.1126/sciadv.1400253) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dirzo R, Young HS, Galetti M, Ceballos G, Isaac NJ, Collen B. 2014. Defaunation in the Anthropocene. Science 345, 401–406. ( 10.1126/science.1251817) [DOI] [PubMed] [Google Scholar]
- 59.Kunz TH, Lumsden LF, Fenton M. 2003. Ecology of cavity and foliage roosting bats. In Bat ecology (eds Kunz TH, Fenton M), pp. 3–89. Chicago, IL: University of Chicago Press. [Google Scholar]
- 60.Roberts BJ, Catterall CP, Eby P, Kanowski J. 2012. Long-distance and frequent movements of the flying-fox Pteropus poliocephalus: implications for management. PLoS ONE 7, e42532 ( 10.1371/journal.pone.0042532) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fleming TH, Racey PA. 2010. Island bats: evolution, ecology, and conservation. Chicago, IL: University of Chicago Press. [Google Scholar]
- 62.Abedi-Lartey M, Dechmann DK, Wikelski M, Scharf AK, Fahr J. 2016. Long-distance seed dispersal by straw-coloured fruit bats varies by season and landscape. Glob. Ecol. Conserv. 7, 12–24. ( 10.1016/j.gecco.2016.03.005) [DOI] [Google Scholar]
- 63.Boyles JG, Cryan PM, McCracken GF, Kunz TH. 2011. Economic importance of bats in agriculture. Science 332, 41–42. ( 10.1126/science.1201366) [DOI] [PubMed] [Google Scholar]
- 64.Nogués-Bravo D. 2009. Predicting the past distribution of species climatic niches. Glob. Ecol. Biogeogr. 18, 521–531. ( 10.1111/j.1466-8238.2009.00476.x) [DOI] [Google Scholar]
- 65.Warren DL. 2012. In defense of ‘niche modeling’. Trends Ecol. Evol. 27, 497–500. ( 10.1016/j.tree.2012.03.010) [DOI] [PubMed] [Google Scholar]
- 66.Ptak SE, Przeworski M. 2002. Evidence for population growth in humans is confounded by fine-scale population structure. Trends Genet. 18, 559–563. ( 10.1016/s0168-9525(02)02781-6) [DOI] [PubMed] [Google Scholar]
- 67.Chikhi L, Sousa VC, Luisi P, Goossens B, Beaumont MA. 2010. The confounding effects of population structure, genetic diversity and the sampling scheme on the detection and quantification of population size changes. Genetics 186, 983–995. ( 10.1534/genetics.110.118661) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gattepaille LM, Jakobsson M, Blum MG. 2013. Inferring population size changes with sequence and SNP data: lessons from human bottlenecks. Heredity 110, 409–419. ( 10.1038/hdy.2012.120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Heller R, Chikhi L, Siegismund HR. 2013. The confounding effect of population structure on Bayesian skyline plot inferences of demographic history. PLoS ONE 8, e62992 ( 10.1371/journal.pone.0062992) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mazet O, Rodríguez W, Grusea S, Boitard S, Chikhi L. 2016. On the importance of being structured: instantaneous coalescence rates and human evolution—lessons for ancestral population size inference? Heredity 116, 362–371. ( 10.1038/hdy.2015.104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mazet O, Rodríguez W, Chikhi L. 2015. Demographic inference using genetic data from a single individual: separating population size variation from population structure. Theor. Popul. Biol. 104, 46–58. ( 10.1016/j.tpb.2015.06.003) [DOI] [PubMed] [Google Scholar]
- 72.Freedman AH, et al. 2014. Genome sequencing highlights the dynamic early history of dogs. PLoS Genet. 10, e1004016 ( 10.1371/journal.pgen.1004016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Colella JP, Lan T, Schuster SC, Talbot SL, Cook JA, Lindqvist C. 2018. Whole-genome analysis of Mustela erminea finds that pulsed hybridization impacts evolution at high latitudes. Commun. Biol. 1, 51 ( 10.1038/s42003-018-0058-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schiffels S, Durbin R. 2014. Inferring human population size and separation history from multiple genome sequences. Nat. Genet. 46, 919 –925. ( 10.1038/ng.3015) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article and electronic supplementary material. All data points for ENM in this study are provided in electronic supplementary material, table S2.