Abstract
Electrical lysis of mammalian cells has been a preferred method in microfluidic platforms, because of its simple implementation and rapid recovery of lysates without additional reagents. However, bacterial lysis typically requires at least 10-times higher electric fields (~10 kV/cm), resulting in various technical difficulties. Here, we present a novel, low-field-enabled electromechanical lysis mechanism of bacterial cells using electroconvective vortices near ion selective materials. The vortex-assisted lysis only requires field strength of ~ 100 V/cm, yet efficiently recovering proteins and nucleic acids from a variety of pathogenic bacteria, and operates in a continuous, and ultra-high-throughput (> 1 mL/min) manner. Therefore, we believe that the electromechanical lysis will facilitate microfluidic bacterial sensing and analysis, but also various high-volume applications such as energy-efficient recovery of valuable metabolites in biorefinery pharmaceutical industries, and disinfection of large-volume fluid for water and food industries.
Keywords: bacterial lysis, electromechanical lysis, ion concentration polarization, electroconvective vortex, disinfection
Graphical Abstract
INRODUCTION
Lysis is the disruption of the cell membrane, which is necessary for not only eliminating pathogens but also accessing intracellular contents such as nucleic acids, proteins, metabolites, and other organelles. In particular, the extracted biomolecules from mammalian or microbial cells provide essential information about genetic or disease characteristics1. Thus, the cell lysis is a crucial step for various biological and clinical studies, including genomics, proteomics and metabolomics, with a wide range of applications in medicine and pharmacy, water-food-energy industry, agriculture, and for recovering of valuable intracellular products from recombinant cells2.
Conventionally, many techniques have been developed to secure the highest yield of the intracellular materials from various organisms; among them, chemical, mechanical and other physical methods were commonly employed. Chemical (detergents) or enzymatic permeation of the cell membrane was an attractive way of recovering lysates due to simple operation and high lysis efficiency3–5. However, the added reagents and proteins often interfere with particular biochemical reactions, or damage lysate components, resulting in narrow choices for downstream assays6. In addition, the chemical composition and concentration needed to be specifically optimized according to organisms, so that it was difficult to be applied for lysing various species of bacteria or cells in a complex cell mixture. By contrast, mechanical methods such as bead beating7,8 and sonication9,10, can lyse most cell types without any chemical addictives11. The bead beating uses tiny particles made of glass, zirconia/silica, ceramic, or steel to collide with cells samples and cracks cell membranes for releasing intercellular materials. This method works well for most types of cells. Sonication-based cell lysis results in cell membrane breakdown using an ultrasound frequency (> 20 kHz). Ultrasonic intensity can be adjusted easily with amplitude and operation time settings and appropriate equipment selection. However, both conventional methods appear to be incompatible with very small sample volumes; a minimum of 500 μL is required for stable operation. In addition, the lysis efficiency and recovery rate of lysates are varied due to rather uncontrolled mechanical shearing of released intracellular biomolecules, sensitive to the apparatus and operational conditions9,12. For this reason, the conventional mechanical lysis techniques often require large-volume samples to operate (> 1 mL), causing volume-mismatch challenges for handling limited amount of precious biosamples, or for interfacing with modern analytic tools that only utilized a small quantity of samples (10~100 μL)13. More recently, micro and nanofabrication techniques have been developed for mechanical cell lysis on a chip by using rigid silicon membranes with mechanical sieving structures and/or nanofabricated sharp tips14–17. However, these microfluidic mechanical lysis techniques seem to be applicable only to relatively large cells such as HaCaT cells (DHaCaT= 20 μm)15. Bacterial cells may need at least ten times smaller sieving structures that require even more complex and sophisticated nanofabrication processes.
Electrical cell lysis would be a preferred method for microfluidic systems because of simple operational setting, no lytic additives requirements, and prompt lysis using a sub-microliter solution with a wide range of cellular density (1–107 cells/mL)2,6. Furthermore, the miniaturized electrical lysis module can be directly integrated with post-processing elements, resulting in on-line, all-in-one, in-situ, and accurate analysis of the lysates18. However, for small cells such as bacteria (approximately 1μm long and 0.5 μm thick), the required electric field was extremely high (> 15 kV/cm)19 in order to achieve the necessary transmembrane potential for lysis (~1.5 V)20. Such high electric fields may induce various technical challenges, including biomolecule degradation, Joule heating, and reactive electrochemical byproducts from water dissociation. To alleviate the issues, bacterial lysis by an electric field was performed either in a low salinity solution (e.g., distilled water)21 for minimizing the current density or using a pinched microchannel (e.g., 25 μm width)22 and small electrode gap (e.g., 10–20 μm)23–25 to reduce the electric potential required. This eventually led to the requirement of additional steps to exchange buffers, or resulted in extremely low lysis throughput (< 1 μL/min)22,24,26,27. Therefore, although the on-chip based electrical lysis technique may be beneficial for in-situ analysis and detection, it appears to be generally difficult to produce enough quantity of lysates to implement off-chip post processing and analysis, such as mass spectrometry or capillary electrophoresis, which requires at least 100 μL solutions for handling and processing.
In this work, we developed a novel and effective bacterial lysis mechanism to combine both mechanical shearing and electrical permeation, so called ‘electromechanical lysis’, enabling a rapid, continuous, versatile, and high-throughput lysis of hard-to-lysis bacterial pathogens yet only requiring substantially lower electric field (~100V/cm). First, we demonstrated that ion concentration polarization (ICP) generated near ion selective membranes (ISMs)28,29 facilitates electrical lysis of bacterial cells, owing to formation of fast electroconvective vortices29–31. In addition, the electroconvective vortices concentrated and agitated bacterial cells toward the ISM walls where electric field is significantly enhanced due to significant ion depletion30–32, inducing additional agitation. Second, we characterized the bacterial lysis in various electric field and salinity conditions, and demonstrated that a low electric field (100–300 V/cm) enabled bacterial lysis, therefore minimizing field-associated negative effects and enabling the lysis even in physiological buffer salinity (e.g., 150 mM). Third, the electromechanical lysis operated in a continuous manner to achieve the higher lysis throughput and lysate yield. Fourth, we successfully recovered intracellular biomaterials such as proteins and ribo-nucleic acids (RNAs) from both easy-to-lysis (Escherichia coli, E. coli) and hard-to-lysis (Mycobacterium smegmatis, M. smeg) bacterial pathogens that typically requires extremely high electric potentials (e.g., 10–20 kV/cm)19,21. Last, we also demonstrated that the microfluidic lysis device can be scaled-up toward a large-volume processing platform, resulting in high-throughput (> 1 mL/min) lysis of bacterial pathogens in a power-efficient manner, opening up new application potentials.
RESULTS AND DISCUSSION
Working principle of electromechanical lysis.
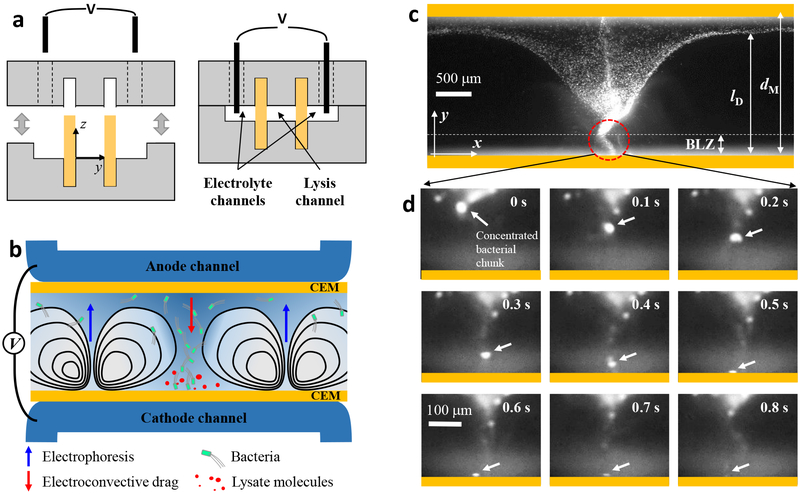
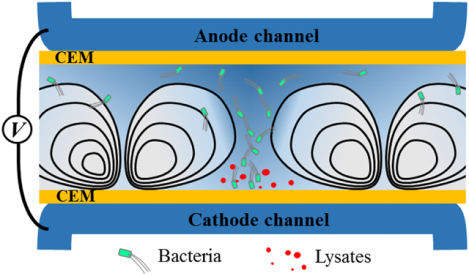
Figure 1a illustrates the fabrication procedure of the ISM-integrated microfluidic devices. First, we integrated two cation exchange membranes (CEMs) that defined a bacterial loading channel for lysis (lysis channel) and two electrolyte rinsing channels to apply electric fields (electrolyte channels), according to our previous ISM-integration technique29,33. We observed no fluid leakage through the gap between CEMs and membrane slots because the CEMs swelled and tightly filled the gap after hydration29. Figure 1b shows experimental setup and working principle of the electromechanical lysis of bacterial cells. The electrical potential applied between the electrodes is predominantly dropped between the two CEMs (lysis channel), because high concentration electrolyte (phosphate buffered saline, 10× PBS 1.5 M) was flown in the electrolyte channels to minimize electrode polarization within the electrolyte channels. In this unipolar ion conduction setting, both ion depletion and ion enrichment regions were generated within the lysis channel near cathodic and anodic sides of CEMs, respectively33.
Figure 1.
Working principle of electromechanical lysis of bacterial cells. a, Fabrication of membrane-integrated microfluidic devices. The top and bottom PDMS blocks were prepared with microchannels (height, h= 250 μm) and high-aspect-ratio membrane slots (thickness, tslot= 0.45 mm, hslot= 3.5 mm) where two CEMs (tMem= 0.45 mm, hMem= 7 mm) were inserted, followed by an irreversible bonding process. b, Schematic illustration to describe principle of the electromechanical lysis for bacterial cells using electroconvective vortices. The ICP phenomena forced bacterial cells to be concentrated into a small spot between vortices and delivered the cells toward the cathodic CEM where the cells underwent electrical permeation and mechanical shearing, resulting in the release of intracellular molecules. c,d, Microscopic observation of the electromechanical lysis at bacterial lysis zone (BLZ). Bacteria cells were first aggregated each other and then experienced a rapid and unstable electro-convectional shearing and mechanical bombardment by the CEM, leading to bacterial lysis within a second.
Meanwhile, bacterial cells (with typical negative zeta potentials) in the lysis channel initially drawn to the anodic CEM due to electrophoresis, then concentrated into a small area between two fully-expanded ion depletion (vortex) regions, forming an inverted-triangular-shaped bacterial plug (Fig. 1b,c). At this relatively high operating voltage / current, ion concentration profile in the channel is largely determined by strong electroconvective vortices, which tend to develop across the entire membrane-to-membrane distance (dM= 2 mm). At this condition, electroconvective vortices continuously delivered concentrated bacterial cells toward the immediate vicinity of the cathodic CEM, namely a bacterial lysis zone (BLZ). Bacterial motion at the BLZ was extremely fast and chaotic due to electrokinetic instabilities at the region, exposing bacterial cells to spatiotemporally unstable electric and fluidic velocity fields, resulting in bacterial lysis. Figure 1d shows microscopic observation of the bacterial lysis at the BLZ; first, bacterial cells were concentrated and formed bacterial chunks at the sharp tip of the bacterial plug, and then lysed within a few seconds.
Visualization and quantification of the lysis performance.
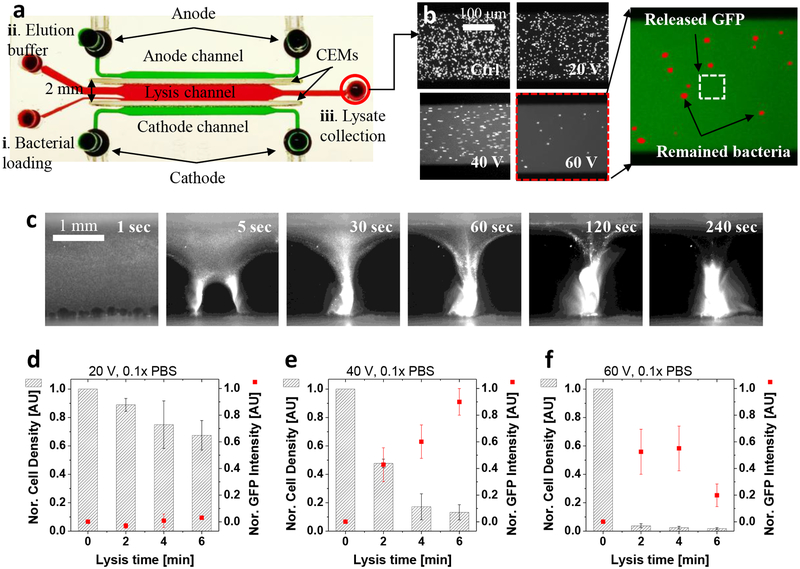
Next, we quantified the lysis performance metrics such as lysis efficiency (rate of bacterial removal) and lysate yield (rate of lysate recovery) using green fluorescent proteins (GFP)-expressing E. coli cells (~108–109 cfu/mL) in 0.1× or 1× PBS buffers. Figure 2a,b and Video S1 describe the batch-mode lysis operation and quantification methods. First, we introduced a bacterial solution (i, ~5 μL) to the lysis channel and applied a DC electric potential (20–60 V) across the electrolyte and lysis channels. After lysis, we flushed the lysis channel with an elution buffer (ii, 100 μL, 1× PBS), followed by collection of lysates and remaining bacterial cells at the outlet reservoirs for quantification (iii, ~105 μL). We measured both bacterial population and background GFP intensity before and after the lysis process by simple image processing (refer Experimental section). The un-lysed bacterial cells showed higher GFP intensity than the background (red spots in Fig. 2b), while the GFP intensity in the background fluid represented the amount of intracellular GFP released by the lysis (green color in Fig. 2b). During the lysis process, the bacterial cells made concentrated bacterial plugs between the vortices and lysed at the BLZ in the cathodic CEM within 6 min (Fig. 2c). Figure 2d–f and Fig. S1 show results of the electromechanical lysis that revealed effects of the electric fields intensity, operational time, and buffer concentration. First, the higher field strength enabled the faster formation of the concentrated bacterial plug and stronger electroconvective vortices, leading to the more rapid bacterial lysis (Fig. S2). In addition, the longer operation time showed the higher bacterial lysis efficiency (removal rates), which was quantified by measuring remaining cell numbers after the lysis. However, the long exposures to the relatively high electric potential (60 V) resulted in reduction of the GFP yields although the lysis efficiency was maintained over 95 %, presumably due to the GFP degradation at such over-processing conditions. This degradation of GFP under the high electric potential can result from a combination effect caused by the excessive shearing of vortices, electro-thermal energy, and local chemical property changes (e.g., ionic strength and pH). Thus, there was a trade-off between the lysis efficiency and lysate yields, and the optimal operating point was found in the moderate potential (40 V) and 4–6 min of operation time.
Figure 2.
Quantification of the lysis efficiency (bacterial removal rate) and lysate yield (GFP recovery rate). a, Experimental setup for the quantification. The batch-mode electromechanical lysis followed three steps: i) bacterial loading and lysis, ii) elution of lysates and un-lysed remaining cells, and iii) collection and quantification by fluorescent signals and image processing. b, Fluorescent microscopic images after the electromechanical lysis in various operational conditions. The lysis efficiency was quantified by comparing bacterial number before and after the lysis (red dots) while the lysate yield was calculated by measuring released GFP intensity (green fluorescent signals in background fluids). The lysis results in various electric potentials obtained in the same operational time (4 min). c, The time-laps images were in-situ observation of the lysis, starting from homogeneously distributed bacterial cells, and leading to fully-lysed concentrated GFP plugs (refer Video S1). d–f, Quantitative lysis results in various electric potentials and operational time, which exhibited three dominant ranges: insufficient (d), appropriate (e), and excessive (f). The excessive lysis condition showed high lysis efficiency but the GFP yield was reduced, while the GFP yield was the maximal at an appropriate condition (40 V, 6 min). The cell density was normalized by the input (0 min) cell number while the GFP intensity was normalized by the maximal GFP recovery value in the lysis experiments. The error bars in Fig. 2d–f were obtained by triplicated experiments.
Interestingly, we enabled bacterial lysis by applying only 20–60 volts across the electrolyte and lysis channels within a few minutes, which is at least 100 fold smaller electric field strength (~100V/cm) compared to the one required for previous electrical lysis techniques for bacterial cells (10–20 kV/cm)19. Furthermore, the lysis operated even using a highly salted solution such as 150 mM buffer (Fig. S1). This was previously challenging because the high current density can cause water dissociation, bubble generation, and pH shift, all of which can cause significant lysate biomolecule degradation. The capability to lyse the bacterial cells in a high salinity buffer is highly desirable because a majority of biofluid samples are of physiological buffer strength. It would be worthwhile to discuss how the bacterial lysis was enabled using the low electric field. This may be attributed to the fact that the additional mechanical shearing applied to bacterial cells by the electroconvective vortices, in addition to the fact that applied electric field was narrowly focused (therefore significantly enhanced) around the tip of concentrated bacterial plug by the ICP phenomena. In this context, our electromechanical approach would efficiently minimize side effects caused by the high current density such as Joule heating and high-salt-related issues by enabling bacterial lysis using a hundred times lower field strength than previous electrical methods. In addition, due to ion exchange membranes, some electric energy seemed to be used for generating vortices and other chaotic flows that are generally difficult to observe in a system without the ion exchanging elements. This extra energy consumption seemed to compromise the power for Joule heating and enabled bacterial lysis in highly salted solution.
Continuous and programmed electromechanical lysis.
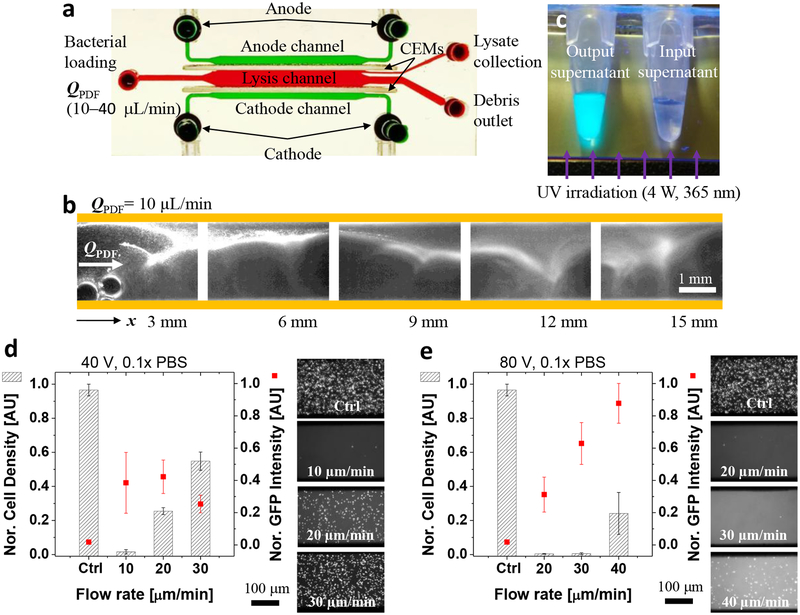
The previous batch-type lysis operation required an elution step, leading to unavoidable lysate dilution and lower processing throughput. Thus, we developed a continuous electromechanical lysis by maintaining a balance between applied electric fields and bacterial flow rates; the flow rate controlled the time that bacterial cells were exposed to chaotic electric and fluidic fields, while the applied electric potential manipulated the electric and vortex strengths. Figure 3a shows the experimental condition for the continuous operation. A pressure driven flow (Q= 10–40 μL/min) continuously delivered bacterial cells to the lysis channel where a DC electric field applied for generating complex and chaotic vortex patterns by the over-limiting ICP phenomena29. As soon as entering the lysis channel, the bacterial cells were transported toward the anodic CEM by electrophoresis, and then gradually lysed by being trapped to the electroconvective vortices along the 15-mm-long channel, resulting in continuous collection of lysates (GFP) at the downstream outlet (Fig. 3b,c and Video S2). The negatively charged GFP tended to be concentrated toward the upper outlet due to electrophoresis and repulsion of ion depletion zones of the cathode CEM, while water and other non-charged debris evenly distributed. In addition, the relatively high flow rate decreased the time for bacteria cells to experience the vortices and electric field, reducing the GFP yield and lysis efficiency (Fig. 3d). However, the GFP yield was also reduced when the flow rate was too low than an optimal value (20 μL/min at 40 V) because of GFP degradation caused by long exposure to the vortices / field (Fig. 3d). At the higher field (80 V), as shown in Fig. 3e, the flow rate to achieve the maximum GFP yield was shifted to a higher flow rate (> 40 μL/min). At the optimal condition (20 μL/min at 40 V), estimated GFP recovery was approximately 75%, which was measured by comparing fluorescent intensity before and after the lysis channel.
Figure 3.
Continuous electromechanical lysis of bacterial cells. a, Experimental set-up for the continuous lysis operation. A constant pressure driven flow (10–40 μL/min) was applied to deliver a fresh bacterial solution and flushed lysates toward downstream, while an electric field generated ICP phenomena at the lysis channel for continuous bacterial lysis. b, Fluorescent microscopic images along the lysis channel. The GFP-expressing bacterial cells underwent chaotic electroconvective vortices and electric fields along the lysis channel, leading to gradual bacterial lysis and recovery of GFP. c, Collected GFP and other lysates using the continuous lysis. The supernatant solution of the lysis output exhibited strong fluorescent intensity under a UV irradiation (4 W, 365 nm) while the supernatant of the input solution showed no distinguishable fluorescent signals. Both supernatant solutions were obtained after centrifugation at 5000 rpm for 3 min to eliminate the GFP signal from live bacterial cells. d,e, Quantification of the continuous lysis performance. The stronger electric potential (e, 80 V) allowed higher flow rates of a bacterial solution than the lower one (d, 40 V), resulting in higher lysis throughput and GFP recovery rate. The cell density was normalized by the input (Ctrl) cell number while the GFP intensity was normalized by the maximal GFP recovery value in the lysis experiments. The error bars in Fig. 3d,e were obtained by triplicated experiments.
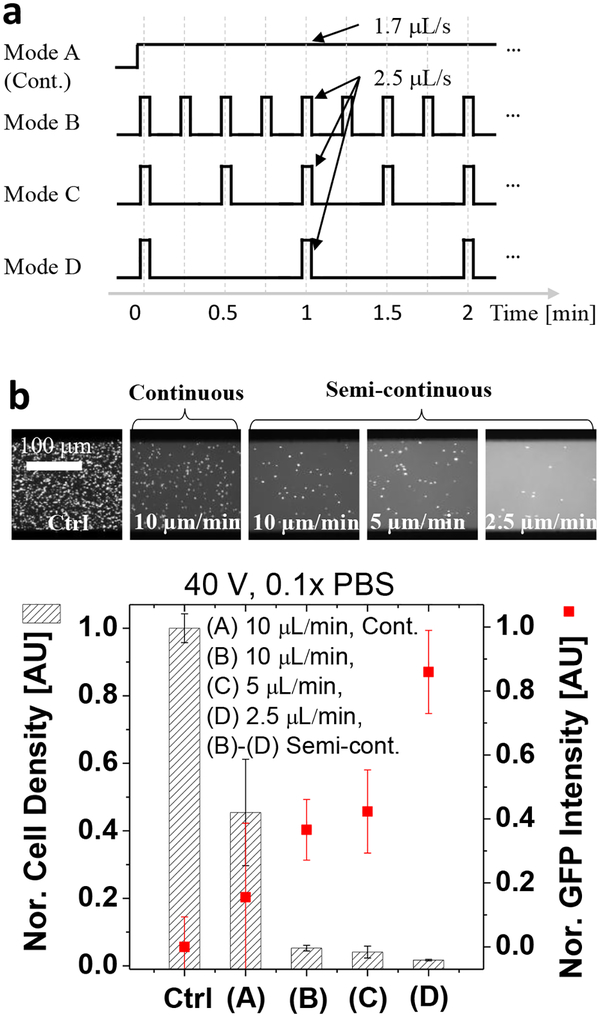
Compared to batch operation, the continuous lysis is enjoying much higher throughput, but some bacterial cells may not be trapped by the electroconvective vortices for lysis in the continuous lysis process. Therefore, it was challenging to simultaneously achieve both high lysate yield (GFP recovery rate) and high lysis efficiency (bacterial removal rate) using the continuous one. Figure 4 shows a semi-continuous lysis mode that can take advantage of both batch and continuous operations, which enjoys both high lysis efficiency and high lysate yield at the same time. That is, the semi-continuous operation is the continuous and automated repetition of the batch electromechanical lysis by programming flow rates of bacterial solutions, without the use of an elution buffer. We fixed a unit volume rate (2.5 μL/s) for the sample loading, and manipulated the bacterial loading frequency using a programmed syringe pump, resulting in flow rates ranged 2.5–10 μL/min (Fig. 4a). Consequently, the semi-continuous method allowed a time for which the bacterial cells located near the anodic CEM moved toward the BLZ by the electroconvective drag, leading to improved performances compared to the continuous method. As shown in Fig. 4b, we achieved high GFP yield (up to 88%) as well as high lysis efficiency (> 97%) from GFP-expressing E. coli (~5×108 cfu/mL) using the semi-continuous modes, while achieving the overall processing flow rate of 2.5 μl/min per single channel.
Figure 4.
Programmed electromechanical lysis of bacterial cells. a, Control of bacterial loading by programming a syringe pump. Mode (A)–(D) exhibited different bacterial loading frequency and flow rates. b, Quantitative results in various programming modes. The semi-continuous lysis enabled high lysis efficiency and high lysate yields compared to the continuous method (A) by allowing more time to experience vortices and electric field. The cell density was normalized by the input (Ctrl) cell number while the GFP intensity was normalized by the maximal GFP recovery value in the lysis experiments. The error bars in Fig. 4b were obtained by triplicated experiments.
RNA recovery from various bacterial strains.
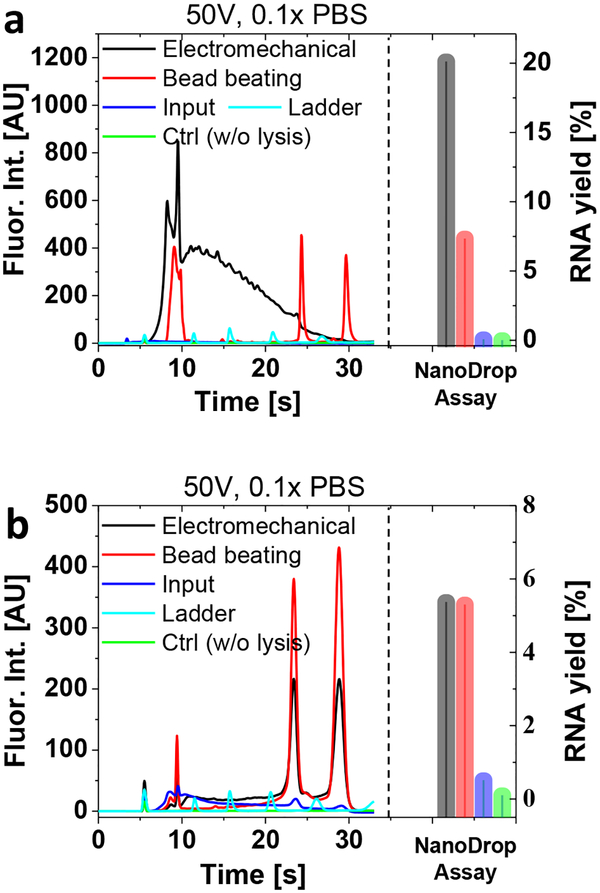
After achieving the recovery of readily detectable lysates such as GFP, we analyzed the lysis recovery for more challenging and fragile biomolecules such as RNAs, which are playing an increasingly important role in clinical and cellular studies1. Figure 5a shows RNA profile and yield obtained from pathogenic E. coli samples (~4×108 cfu/mL) by using on-chip gel electrophoresis and NanoDrop assay, respectively. First, we separately performed both the electromechanical lysis and the gold-standard lysis protocol (bead beating) to individually generate 100 μL lysis samples in 0.1× PBS solutions, and then analyzed the lysed samples to compare the RNA profiles and yields. Interestingly, the RNA gel profile obtained by the electromechanical lysis showed the higher first peak (short RNA including 5S rRNA) than the one recovered by the bead beating although the second and third peaks (longer RNA including 16S and 23S rRNA) were unclear. We also confirmed that the electromechanical lysis exhibited the higher RNA yield than the bead beating by using NanoDrop-based absorbance tests. Figure 5b shows extraction of RNAs from a mycobacterium strain (M. smeg), one of the hard-to-lyse microbes, as they have a heavy and thick outer layer that can be resilient to mechanical, chemical and enzymatic lysis34. The RNA profile showed clear three peaks, comparable to the bead beating results with almost similar RNA recovery rates, approximately 5% of the theoretical maximum. Therefore, the electromechanical lysis enabled extraction of RNAs at the similar efficiency as the gold standard method, from both easy-to-lyse and hard-to-lyse bacterial species by only using a small electric potential (50 V). We note that the positive control using the bead beating was also assisted by other lysis mechanism such as freeze-thaw35 and chemical lysis (RLT buffer)36, while our electromechanical methods performed without any pretreatments. More detailed comparison protocols are available in Experimental section.
Figure 5.
RNA recovery from easy-to-lyse (a, E. coli) and hard-to-lyse (b, M. Smeg) bacterial species. The first fluorescent peak in about 10 seconds represents a bacterial 5s ribosomal RNA followed by two subsequent peaks linked to 16 and 23s ribosomal RNAs. The RNA profiles and yields were obtained by using Bioanalyzer on-chip gel electrophoresis and Nanodrop assays, which revealed comparable RNA yields to conventional bead milling. The RNA yield was calculated by dividing the experimental values with the theoretical maximal.
Parallelization for high-throughput operation.
Another unique advantage of the electromechanical lysis is the scalability, by laterally parallelizing the single lysis unit, CEM and lysis channel. This is possible because our method only employing a unipolar ISM (cation exchange membrane) so that the system is symmetrical and has low complexity, enabling simple stacking and/or parallelization. To demonstrate the scalability, we developed a laterally-arrayed electromechanical lysis device, which consisted with six CEMs and five individual lysis channels connected by a common inlet and outlet (Fig. S3). The five lateral parallelization directly increased the lysis throughput without the need for additional electrode sets, although a higher electric potential was required to maintain the same field strengths in each lysis channel. In addition, the CEM we employed for massive parallelization can minimize non-specific cellular / biomolecular adhesion and clogging due to the same electrical polarity with most biological matters (negatively charged)37, showing efficient lysate isolation and high sustainability for long-term operation33. Although the GFP lysates could not be quantified along individual lysis channels because of the interconnected common inlet and outlet, we observed microscopically that there was no significant channel-to-channel variation on the vortex patterns and GFP signals.
High-throughput lysis device for water disinfection.
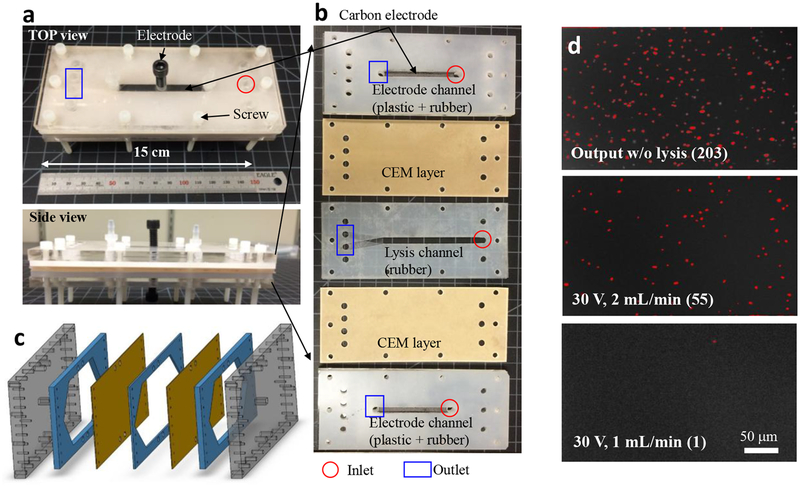
Figure 6a shows another strategy to enhance the lysis throughput. We produced a large-scale plastic device (15 cm × 5 cm) by using a layer-by-layer stacking technique (refer Experimental section). The large-scale device did not allow in situ observation of the process enjoyed by the PDMS-based microchips, but retained the same electromechanical lysis mechanism due to the same CEM-CEM distance (~2 mm). Recently, three-dimensional helical vortex formation near ISMs was demonstrated in both theoretical and empirical manners38, which should enable the electromechanical lysis in the large-scale device in principle. As shown in Fig. 6b, we grooved the lysis channel (12 cm (l) × 5 cm (h)) in a 3-mm-thick conformal rubber sheet and sandwiched the lysis layer with two CEM layers where another rubber and plastic composite layers were covered for the electrolyte channels (8 cm (l) × 5 cm (h)) and electrode interfaces. Figure 6c shows the assembly process of each layer, followed by mechanical clamping by tightening screws to prevent fluid leakages, compressing the compliant lysis layer from 3 mm to 2 mm. During the clamping process, the thickness of the lysis channel layer in the large-scale platform was matched with the lysis channel width in the microfluidic device. That is, we maintained the same CEM-CEM distance (~2 mm), but the height and length of the lysis channel were scaled up to ~10 cm, with the possibility to be scaled up further. Figure 6d shows bacterial lysis results obtained by applying 30 V with significantly higher flow rates (few mL/min) than the microfluidic electromechanical device (~100 μL/min) and other previously reported electrical lysis methods (typically less than μL/min), resulting more than 99 % removal of bacterial cells. We note that the large-scale device may require further engineering for more reliable scaled-up operations. Yet, this would open up new possibilities of carrying out large-volume cell lysis, such as in releasing non-sensitive intracellular metabolites and/or sterilize large-volume solutions in a cost-effective, energy-efficient, portable, and ultra-high-throughput manner. The throughput (over mL/min) and power consumption (~0.5 Wh/L in 0.1× PBS) obtained in this work is the highly competitive to not only other electrical lysis techniques that required extremely high electric fields (~10–20 kV/cm), but also other sterilization techniques such as ultraviolet light irradiation (~0.2 Wh/L)39. While following studies are required to further investigate fundamentals of the lysis mechanism owing to inherent complexity of the electromechanical lysis system, it is worthwhile to discuss the forces that contribute to bacterial lysis. First, the electric permeation of cell membrane would take the largest contribution. In our particular system, the applied electric field was enhanced into small spots (the tip of concentrated bacterial plug) in the BLZ due to ion concentration effects, so that the local electric field strength would be higher than the nominal strength we applied. Second, the rapid electroconvective vortices would facilitate the lysis of bacteria by generating additional mechanical shearing by a viscous vortex flow and colliding with nanoscopically unsmooth membrane surfaces. Third, the cells were rapidly translocated and agitated by the electroconvection that resulted in the exposure of the cells to spatiotemporal changes of many physiochemical factors. For example, the local ionic strength, pH, and temperature would be different according to the location and time line, particularly when the ion concentration polarization is under developing with over-limiting potentials. This temporal change of cellular microenvironments induces much stress to the cells, including the osmotic shock. Fourth, electrothermal effects, which is proportional to the resistance and the square of the current (P ∝ I2∙R) can help the bacteria lysis. Given that the local enhancement of electric field, the Joule effect may increase local temperatures at the concentrated filed regions, although the local temperature would be not enough to boil the solution or damage lysates due to the rapid thermal dissipation in microscale, it might contribute to accelerate kinetics of chemical reaction regarding cell lysis. Unfortunately, all aforementioned factors are mutually coupled in action such that it was almost impossible to in-situ measure the spatiotemporal changes of the electric and fluidic field distribution orthogonally using an empirical way. Therefore, a following-up research based on numerical simulation will unveil the distribution of electric and fluidic fields. This will help to identify whether the potential drop across the cell body is the main cause of lysis or the contribution is made of combination between electric-field-induced forces and the mechanical shearing to the membrane. In addition, it would be interested to understand any correlation between the lysis efficiency and cell properties such as size, stiffness, or osmolality, for potential application to other cell types.
Figure 6.
A large-scale plastic device for ultra-high-throughput electromechanical lysis of bacterial cells for water disinfection. a–c, The large-scale device was made of ISMs, rubber sheets, and plastic plates by layer-by-layer stacking and mechanical clamping. Before the assembly, the rubber layers were grooved to have fluidic channels for electrolytes and bacterial solutions, which was divided by CEMs. The CEM-CEM distance were conserved to be same with the microchip device d, Bacterial density with or without lysis. The output without electric field seems to be the same with the input, but the most bacterial cells was removed after electromechanical lysis (over 99%). The bacterial images were obtained by collecting lysis solutions for ~10 min in each condition, and introduced the lysed solution into a straight microchannel having 25 μm height.
CONCLUSIONS
Herein, we presented a novel electromechanical lysis mechanism that can be used for various bacterial cells, providing highly efficient collection of lysates in a rapid, continuous, and energy efficient manner by only applying a moderate electrical field (< 100 V/cm). In this work, we demonstrated, for the first time, that the ICP phenomena near ISMs facilitated electrical lysis of bacterial cells due to formation of anomalously strong electroconvective vortices. The vortices can be spontaneously generated by applying an overlimiting potential, and contributed to concentrate and agitate bacterial cells toward the ISM walls where the cell underwent additional mechanical shearing and bombardment by the membrane. We successfully performed lysis of bacterial cells by only applying few tens volts in a highly salted buffer (e.g., 150 mM), enabling high recovery rates of intracellular biomaterials such as proteins (> 88 % yield) and RNAs (> 5 % yield). Another unique feature of this technique is that microfluidic electrochemical lysis device can be scaled-up toward milli- or macroscale fluidic platforms, using similar assembly of fluidic channels and ISMs as in electrodialysis, resulting in ultra-high-throughput electromechanical water disinfection (> 99 % bacterial removal rate) in a power-efficient and portable manner. Hence, we believe that the proposed novel lysis mechanism will facilitate not only fundamental studies in microbiology but also various industrial disinfection applications such as wastewater treatment, aquarium sanitation, and food / water sterilization.
EXPERIMENTAL
Experimental setting and procedures.
For the batch-type operation (Fig. 2), E. coli cells (~108 cfu/mL) in a 0.1× or 1× PBS solution were introduced to the lysis device, followed by applying an electrical potential (20–60 V) using Pt wires for generating electric fields and vortices in the lysis channel between membranes. The same bacterial solution with a flow rate (10–40 μL/min) was applied by an electric potential (40 V or 80 V) for obtaining results of the continuous operation (Fig. 3). For UV irradiation results in Fig 3., we used a compact and handheld UV Lamp (UVL-21, Analytik Jena, Jena, Germany) that generates 4W UV with a wavelength of 365 nm. Next, we fixed the electric potential (40 V) and buffer strength (0.1× PBS), and programmed flow rates (2.5–10 μL/min) for obtaining the results of the semi-continuous operation (Fig. 4). For RNA recovery test, we employed the continuous operation mode by applying 50 V to both E. coli and M. smeg in a 0.1× PBS solution with a flow rate of 40 μL/min. The large-scale experiments were conducted by applying 30 V to E. coli cells (~104 cfu/mL) in a 0.1× PBS solution with a flow rate of 1–2 mL/min.
Fabrication and preparation of microfluidic devices.
The ISM-integrated microfluidic device was fabricated and prepared according to our previous protocols29,33. Briefly, a 3-dimensional (3D) master mold was prepared using a stereolithographic technique (3D Systems Inc., Rock Hill, SC, USA), followed by a standard soft lithography process using PDMS elastomer kits (Dow Corning, Midland, MI, USA). The PDMS prepolymer mixed with the curing agent in 10:1 (w/w) ratio, which was poured onto the 3D printed master mold and cured in a 65 °C convection oven over 4 h. After curing, the top PDMS replica was integrated with two CEMs (Fumasep FTCM-E, Fuma-Tech GmbH, Germany) by inserting the membrane into the membrane slots, followed by irreversible bonding with the bottom PDMS block by an oxygen plasma (Harrick Plasma, Ithaca, NY, USA). Then, the electrolyte and lysis channels were filled with deionized water over 48 h at room temperature. This allowed volume expansion of the ISMs by swelling, forming a tight seal between ISMs and the membrane slots. All lysis channels were coated with 0.01% Pluronic surfactant (F-127, Sigma-Aldrich, Natick, MA, USA) to minimize non-specific binding between the cells and PDMS surfaces. Then, the channels were flushed again with 1× PBS to remove the residual chemicals and impurity before loading the cells. We used 10× PBS stock (Thermo Fisher Scientific, Waltham, MA, USA) that was ready to use upon dilution to the desired concentration (e.g., 1×, 0.1×). The PBS is a pH-adjusted blend (pH 7.4) which, when diluted to a 1× working concentration, contains 137 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, and 2 mM KH2PO4. The PBS includes no magnesium and calcium ions.
Fabrication of the large-scale device.
The large-scale plastic-based platform was made of the CEMs, carbon electrodes (Fuel Cell Store, Inc., Boulder, CO, USA), silicon rubber (3.8 mm thickness) and acrylic sheets (5 mm thickness, McMaster-Carr, Elmhurst, IL, USA). First, the electrolyte and lysis channels and holes were formed in the plastic and rubber sheets by a laser and manual cutting, respectively. Then each layer including CEMs was assembled by a mechanical clamping. After fabrication, all membranes were filled with demineralized water over 48 h and coated with Pluronic surfactant in the same manner with experiments on the microdevices.
Preparation of bacterial cells.
We used constitutively GFP-expressing recombinant E. coli, K12 strain to characterize the lysis performances by fluorescent measurement. The wild-type pathogenic strains, E. coli and M. smeg, were used for the RNA recovery experiments. The same culture and preparation protocols were used for all the cells40. Shortly, a single colony grown on a Lysogeny Broth (LB) agar plate was inoculated in a 5 ml LB medium or 7H9 medium (Sigma-Aldrich, Natick, MA, USA) for E. coli and M. smeg, respectively, and grown to a mid-log phase in a rotary-shacking incubator (200 rpm, 37 °C). The culture was centrifuged at 5000× g for 5 min and diluted into 1× or 0.1× PBS solutions at the appropriate concentration based on optical density measurements of the mid-log cultures at 600nm wavelength. We note that all the cells were carefully handled and incubated to protect cellular contamination.
RNA Extraction by Bead Beating and RNA analysis.
The cell culture was centrifuged at 13,000×g for 10 min, were then resuspended in 800 μL of Buffer RLT with 1 % Beta-mercaptoethanol (Sigma-Aldrich, Natick, MA, USA) and incubated at room temperature for 5min, followed by 15 min incubation at −80 °C. After thawing the frozen cell sample on ice, the mixture was then transferred into a lysing matrix, which consist of 400 μL zirconia/silica beads (100 μm diameter, Bio Spec Products, Inc., Bartlesville, OK, USA) in a bead beating tube on ice and shaken vigorously. Bead beating using Mini-beadbeater-16 (BioSpec Products Inc., Bartlesville, OK, USA) was conducted 3 times (3× 60 s) for E. coli and 10 times (10 × 60 s) for M. smeg, with a 1 min rest time in between on ice. Notably, the number of bead beating cycles was chosen to maximize the RNA yield for each bacteria species, respectively. The mixture was centrifuged at 13,000×g for 15 min at 4 °C, and the top aqueous layer above bead matrix was collected. 80 μL aqueous layer was transfer to new tube and mixed with 160 μL Ampure RNAclean SPRI bead solution (Beckman Coulter Inc, Indianapolis, IN, USA). Per the manufacture’s protocol, the RNA sample was washed with 70 % ethanol twice and finally eluded in 40 μL Nuclease-free water. The eluate of RNA sample from SPRI beads was analyzed using on-chip gel electrophoresis with RNA Pico Chip (2100 Bioanalyzer, Agilent technologies, Santa Clara, CA, USA) and NanoDrop assay (ND-1000 Spectrophotometer, NanoDrop Technologies Inc, Wilmington, DE, USA). The eluate from the microfluidic lysis channel was purified and analyzed by the same RNA handling protocol without the RLT treatemnt and bead beating process.
Data acquisition and analysis.
An inverted fluorescence microscope (IX71, Olympus, Tokyo, Japan) equipped with a CCD camera (ORCA-ER, Hamamatsu Photonics, Shizuoka, Japan) was used to obtain the optical microscopic and fluorescent images using an open source software Micromanager (NIH, Bethesda, MD, USA). A pressure-driven flow was generated by a syringe pump (PHD Ultra, Harvard apparatus, Holliston, MA, USA) and constant current and voltage were applied and measured by current-voltage source measurement unit (Keithley 236, Keithley Instruments, OH, USA). Platinum electrodes (Sigma-Aldrich, Natick, MA, USA) were used to exclude electrode and reaction overpotential occurred in the electrolyte rinsing channels. For data analysis and post processing of microscopic images, Image J (NIH, Bethesda, MD, USA) and OriginPro 8 (OriginLab, Wheeling, IL, USA) were used.
Supplementary Material
Video S1: Microscopic visualization of batch-mode electromechanical lysis
Video S2: Collection of lysates (GFP) during continuous mode electromechanical lysis
Figure S1. Characterization of electromechanical lysis at high salinity buffers (1× PBS)
Figure S2. Time-sequential development of vortex patterns at various electric fields
Figure S3. Parallelized and multiplexed electromechanical lysis on a chip
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health (NIH, R01 AI117043), as well as BroadNext10 gift from the Broad Institute.
Footnotes
Supporting Information
Supporting Information is available free of charge on the ACS Publications website as follows.
REFERENCES
- (1).Barczak AK; Gomez JE; Kaufmann BB; Hinson ER; Cosimi L; Borowsky ML; Onderdonk AB; Stanley SA; Kaur D; Bryant KF; Knipe DM; Sloutsky A; Hung DT P Natl Acad Sci USA 2012, 109, 6217–6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Brown RB; Audet J Journal of the Royal Society Interface 2008, 5, S131–S138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Kotlowski R; Martin A; Ablordey A; Chemlal K; Fonteyne PA; Portaels F J Med Microbiol 2004, 53, 927–933. [DOI] [PubMed] [Google Scholar]
- (4).Marcus JS; Anderson WF; Quake SR Anal Chem 2006, 78, 3084–3089. [DOI] [PubMed] [Google Scholar]
- (5).Sarkar A; Kolitz S; Lauffenburger DA; Han J Nat Commun 2014, 5, 3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Nan L; Jiang ZD; Wei XY Lab Chip 2014, 14, 1060–1073. [DOI] [PubMed] [Google Scholar]
- (7).Odumeru J; Gao A; Chen S; Raymond M; Mutharia L Can J Vet Res 2001, 65, 201–205. [PMC free article] [PubMed] [Google Scholar]
- (8).Berasaluce A; Matthys L; Mujika J; Antonana-Diez M; Valero A; Agirregabiria M Rsc Adv 2015, 5, 22350–22355. [Google Scholar]
- (9).Zhang H; Jin WR Electrophoresis 2004, 25, 1090–1095. [DOI] [PubMed] [Google Scholar]
- (10).Taylor MT; Belgrader P; Furman BJ; Pourahmadi F; Kovacs GTA; Northrup MA Anal Chem 2001, 73, 492–496. [DOI] [PubMed] [Google Scholar]
- (11).Di Carlo D; Jeong KH; Lee LP Lab Chip 2003, 3, 287–291. [DOI] [PubMed] [Google Scholar]
- (12).Vandeventer PE; Weigel KM; Salazar J; Erwin B; Irvine B; Doebler R; Nadim A; Cangelosi GA; Niemz A J Clin Microbiol 2011, 49, 2533–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Hou HW; Bhattacharyya RP; Hung DT; Han J Lab Chip 2015, 15, 2297–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).So H; Lee K; Seo YH; Murthy N; Pisano AP ACS Appl Mater Interfaces 2014, 6, 6993–6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Yun SS; Yoon SY; Song MK; Im SH; Kim S; Lee JH; Yang S Lab Chip 2010, 10, 1442–1446. [DOI] [PubMed] [Google Scholar]
- (16).So H; Lee K; Murthy N; Pisano AP ACS Appl Mater Interfaces 2014, 6, 20693–20699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Kim J; Hong JW; Kim DP; Shin JH; Park I Lab Chip 2012, 12, 2914–2921. [DOI] [PubMed] [Google Scholar]
- (18).Cheng J; Sheldon EL; Wu L; Uribe A; Gerrue LO; Carrino J; Heller MJ; O’Connell JP Nature Biotechnology 1998, 16, 541–546. [DOI] [PubMed] [Google Scholar]
- (19).Lee SW; Tai YC Sensor Actuat a-Phys 1999, 73, 74–79. [Google Scholar]
- (20).Gabriel B; Teissie J Biophys J 1999, 76, 2158–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Ma S; Bryson BD; Sun C; Fortune SM; Lu C Anal Chem 2016, 88, 5053–5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Wang HY; Bhunia AK; Lu C Biosens Bioelectron 2006, 22, 582–588. [DOI] [PubMed] [Google Scholar]
- (23).Lu KY; Wo AM; Lo YJ; Chen KC; Lin CM; Yang CR Biosens Bioelectron 2006, 22, 568–574. [DOI] [PubMed] [Google Scholar]
- (24).Lee DW; Cho YH Sensor Actuat B-Chem 2007, 124, 84–89. [Google Scholar]
- (25).Besant JD; Das J; Sargent EH; Kelley SO ACS Nano 2013, 7, 8183–8189. [DOI] [PubMed] [Google Scholar]
- (26).Lu H; Schmidt MA; Jensen KF Lab Chip 2005, 5, 23–29. [DOI] [PubMed] [Google Scholar]
- (27).Mernier G; Piacentini N; Braschler T; Demierre N; Renaud P Lab Chip 2010, 10, 2077–2082. [DOI] [PubMed] [Google Scholar]
- (28).Kim M; Jia M; Kim T Analyst 2013, 138, 1370–1378. [DOI] [PubMed] [Google Scholar]
- (29).Kwak R; Pham VS; Lim KM; Han JY Phys Rev Lett 2013, 110, 114501. [DOI] [PubMed] [Google Scholar]
- (30).Davidson SM; Wessling M; Mani A Sci Rep-Uk 2016, 6, 22505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).de Valenca JC; Wagterveld RM; Lammertink RGH; Tsai PA Physical Review E 2015, 92, 031003. [DOI] [PubMed] [Google Scholar]
- (32).Khair AS Phys Fluids 2011, 23, 072003. [Google Scholar]
- (33).Kim B; Kwak R; Kwon HJ; Pham VS; Kim M; Al-Anzi B; Lim G; Han J Sci Rep-Uk 2016, 6, 31850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Hoffmann C; Leis A; Niederweis M; Plitzko JM; Engelhardt H P Natl Acad Sci USA 2008, 105, 3963–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Johnson BH; Hecht MH Bio-Technol 1994, 12, 1357–1360. [DOI] [PubMed] [Google Scholar]
- (36).Chen YT; Sonnaert M; Roberts SJ; Luyten FP; Schrooten J Tissue Eng Part C-Me 2012, 18, 444–452. [DOI] [PubMed] [Google Scholar]
- (37).Schott H J Pharm Sci 1974, 63, 48–53. [DOI] [PubMed] [Google Scholar]
- (38).Pham SV; Kwon H; Kim B; White JK; Lim G; Han J Physical Review E 2016, 93, 033114. [DOI] [PubMed] [Google Scholar]
- (39).Song K; Mohseni M; Taghipour F Water Res 2016, 94, 341–349. [DOI] [PubMed] [Google Scholar]
- (40).Kim M; Lim JW; Kim HJ; Lee SK; Lee SJ; Kim T Biosens Bioelectron 2015, 65, 257–264. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1: Microscopic visualization of batch-mode electromechanical lysis
Video S2: Collection of lysates (GFP) during continuous mode electromechanical lysis
Figure S1. Characterization of electromechanical lysis at high salinity buffers (1× PBS)
Figure S2. Time-sequential development of vortex patterns at various electric fields
Figure S3. Parallelized and multiplexed electromechanical lysis on a chip