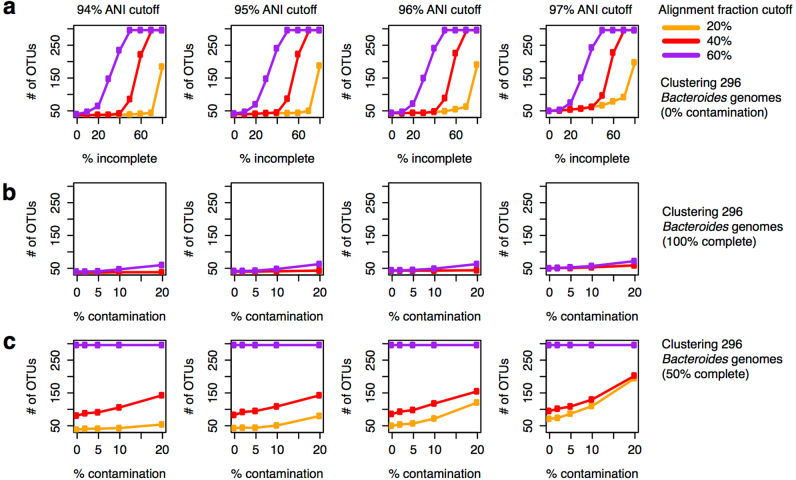
Extended Data Fig. 5. Effect of completeness and contamination on the identification of OTUs from whole genomes.
a–c, OTUs were identified for 296 genomes from the Bacteroides genus on the basis of average-linkage clustering of whole-genome ANI, using the ANIcalculator (v.1.0). The ANI cut-offs used for forming OTUs are indicated in the panel titles (94–97% ANI). The alignment fraction cut-offs, defined as the required percentage of genome length aligned between genome pairs (20–60%), is indicated by line colour. In each panel, the vertical axis indicates the number of OTUs identified from genomes on the basis of the ANI cut-off, alignment fraction cut-off and the degree of incompleteness and/or amount of contamination present in the 296 genomes. a, OTUs were identified for the 296 Bacteroides genomes with up to 80% of genes randomly removed. The number of OTUs is inflated when genomes are incomplete and the alignment fraction is >20%. b, OTUs were identified for the 296 Bacteroides genomes with up to 20% of genes from a different one of the 296 genomes. The number of OTUs is not affected by contamination when genomes are complete. c, OTUs were identified for the 296 Bacteroides genomes with 50% of genes randomly removed and up to 20% of genes from a different one of the 296 genomes, representing a worst-case scenario. The number of OTUs is inflated by contamination when genomes are 50% complete. Using a lower ANI threshold (for example, 94 or 95% versus 96 or 97%) reduces the negative effect of contamination. On the basis of these experiments, we chose an alignment fraction cut-off of 20% and an ANI cut-off of 95% for identifying OTUs from MAGs and reference genomes in the current study.