Abstract
Banana cultivars (Musa ssp.) are diploid, triploid and tetraploid hybrids derived from Musa acuminata and Musa balbisiana. We presented a high-quality draft genome assembly of M. balbisiana with 430 Mb (87%) assembled into 11 chromosomes. We identified that the recent divergence of M. acuminata (A-genome) and M. balbisiana (B-genome) occurred after lineage-specific whole-genome duplication, and that the B-genome may be more sensitive to the fractionation process compared to the A-genome. Homoeologous exchanges occurred frequently between A- and B-subgenomes in allopolyploids. Genomic variation within progenitors resulted in functional divergence of subgenomes. Global homoeologue expression dominance occurred between subgenomes of the allotriploid. Gene families related to ethylene biosynthesis and starch metabolism exhibited significant expansion at the pathway level and wide homoeologue expression dominance in the B-subgenome of the allotriploid. The independent origin of 1-aminocyclopropane-1-carboxylic acid oxidase (ACO) homoeologue gene pairs and tandem duplication-driven expansion of ACO genes in the B-subgenome contributed to rapid and major ethylene production post-harvest in allotriploid banana fruits. The findings of this study provide greater context for understanding fruit biology, and aid the development of tools for breeding optimal banana cultivars.
Subject terms: Next-generation sequencing, Phylogenetics, Genome duplication
A high-quality draft genome assembly of Musa balbisiana (the banana B-genome) is reported. Comparative genomic and gene functional analyses provide insights into the subgenome evolution and functional divergence between the A- and B-subgenomes.
Main
Bananas (Musa ssp.) are large herbaceous plants that are perennial but monocarpic. They originated in Southeast Asia and the Western Pacific and were one of the first crops to be domesticated, about 7,000 years ago, in Southeast Asia1,2. Bananas are widely distributed throughout the tropics and subtropics, where they are a staple food and fruit for millions of people2,3. Moreover, bananas are one of the major export commodities of several developing countries and represent the largest international trade in fruit3,4. Thus, bananas are an essential food resource and have important socioeconomic and ecological roles.
The genus Musa belongs to the monocotyledon Musaceae family along with the genus Ensete. Its wild species have traditionally been subdivided into four sections: Eumusa (x = 11; x represents the number of chromosomes), Rhodochlamys (x = 11), Australimusa (x = 10) and Callimusa (x = 9 or 10)5–7, and refined recently to two sections in which the Rhodochlamys and Australimusa were merged into the Eumusa and Callimusa, respectively8. Most edible bananas belong to the Eumusa (or Musa) section, and are categorized into the dessert or cooking group based on their usage. Furthermore, bananas of this section are distinguished based on their genetic background as Musa acuminata (A-genome, 2n = 2x = 22; n represents the haploid chromosome number), Musa balbisiana (B-genome, 2n = 2x = 22), Musa schizocarpa (S-genome, 2n = 2x = 22) and Australimusa species (T-genome, 2n = 2x = 20)2. The majority of edible cultivated bananas originated from intraspecific or interspecific hybridization between wild diploid M. acuminata (A-genome) and M. balbisiana (B-genome) species. Combinations of these A- and B-genomes have resulted in various genotypes of cultivated edible bananas, including diploid (AA, BB and AB), triploid (AAA, AAB and ABB) and tetraploid (AAAB, AABB, ABBB) variants6. The triploid genotype variants constitute the predominant cultivated varieties that are planted worldwide.
Genome sequencing of the A-genome banana has provided insights into the evolution of monocotyledonous plants1,9. Although the A-genome represents a crucial step in the genetic improvement of banana, the lack of a high-quality B-genome sequence greatly hinders germplasm characterization and the molecular breeding of banana. A draft B-genome has previously been reported, but exhibited low quality, based on assembly and annotation via mapping reads to the A-genome2. Here, we sequenced the genome of the double haploid of the wild diploid genotype Pisang Klutuk Wulung (DH-PKW, 2n = 2x = 22), belonging to the species M. balbisiana that contributed the B-subgenome to cultivated allotriploid bananas. We further compared the B- and A-genomes to investigate subgenome evolution, genetic diversity and the functional divergence of subgenomes in polyploid bananas. Our analyses provide insights into the evolution and regulation of fruit-ripening processes in bananas. In particular, the results highlight a significant contribution of the B-genome towards ethylene biosynthesis and starch metabolism during fruit ripening.
Results
Genome assembly and annotation
To reduce heterozygosity, we used the DH-PKW genotype for our genome sequencing and assembly10. A total of 58.99 gigabases (Gb) (113×) of PacBio single-molecule long reads and 86.34 Gb (166×) Illumina paired-end and mate-pair reads were used for assembly (Supplementary Table 1), producing 492.77 megabases (Mb) of scaffolds. The contig N50 and scaffold N50 of the final assembly were 1.83 and 5.05 Mb, respectively (Supplementary Table 2). K-mer analysis suggested that the draft assembly covers approximately 95% of the genome size of DH-PKW (Supplementary Fig. 1). We further evaluated the completeness of the scaffold assembly using the BUSCO (v.3) plants datasets11. Precise exon placement of 91.3% of the total 1,440 single-copy orthologue groups in the embryophyta dataset was identified in the B-genome assembly. To anchor the scaffolds to chromosomes, we constructed high-throughput chromosome conformation capture (Hi-C) libraries of DH-PKW, generating 72 Gb (138×) Hi-C pair-end reads (Supplementary Table 1). Duplicate removal, sorting and quality assessment were performed with HiC-Pro12, and uniquely mapped valid reads were used for Hi-C scaffolding by LACHESIS software13 (Supplementary Fig. 2). As a result, 430 Mb (87.27%) of the assembly and 94.0% of the genes were placed on 11 chromosome groups (Fig. 1 and Supplementary Table 3). The 11 pseudo-molecules were named in accordance with the M. acuminata (A-genome) reference sequence1,9.
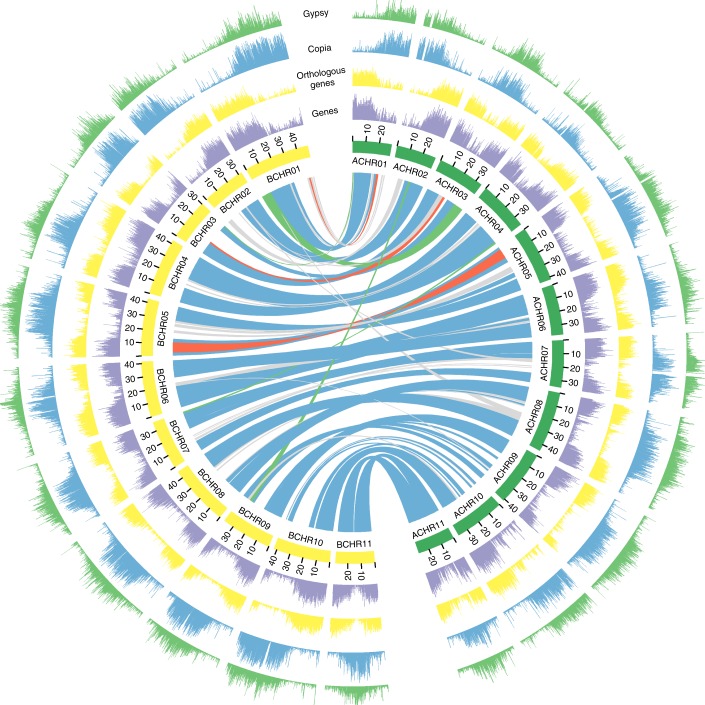
Fig. 1. Characterization of M. balbisiana (B-genome) and M. acuminata (A-genome) chromosomes.
Elements are arranged in the following scheme (from outer to inner). (1) Distribution of Gypsy elements (non-overlapping, window size, 50 kb); (2) distribution of Copia elements (non-overlapping, window size, 50 kb); (3) distribution of orthologous gene pairs between two genomes (non-overlapping, window size, 50 kb); (4) gene density (non-overlapping, window size, 50 kb); (5) syntenic relationships between A- and B-genomes. The connecting blue lines represent alignment blocks, red lines represent inversions, green lines represent translocations and grey lines show small blocks with<30 gene pairs.
About 55.75% of the B-genome assembly was composed of repetitive sequences, which is higher than the 41.85% of the A-genome assembly (Supplementary Table 4). This may be due to the spanning of repetitive regions by long reads14. Long terminal-repeat (LTR) retrotransposons represented the most abundant fraction of transposable elements in the A- and B-genomes, among which the families Gypsy and Copia accounted for 12.88 and 28.04% of the B-genome, respectively. DNA transposons comprised 2.12% of the B-genome and 2.03% of the A-genome (Supplementary Table 4). LTR retrotransposons tended to accumulate near the centromeric and pericentromeric regions (Fig. 1). Active insertions of LTR retrotransposons occurred more recently in the B-genome (peak at 0–0.5 million years ago (MYA)) relative to the A-genome (peak at 1.5–2.0 MYA) after their divergence (Supplementary Fig. 3). Both genomes experienced a wave of LTR retrotransposon amplification but differed in insertion histories.
We annotated the genome using the MAKER pipeline15 incorporating ab initio predictions, homologous proteins and transcriptome data from six samples, resulting in 35,148 protein-coding genes in the B-genome (Supplementary Table 5). Of these, 33,137 (94.27%) were located on the 11 pseudo-chromosomes (Supplementary Table 3). Overall, 86% of the genes transcribed in our RNA sequence (RNA-Seq) analysis. Additionally, we identified 3,329 transcription factors among 88 families in the B-genome using the iTAK programme16 (Supplementary Table 6).
The B-genome is more sensitive to fractionation than the A-genome
Compared to other monocots, the Musa lineage exhibits a relatively slower evolutionary rate as demonstrated previously17 (Supplementary Fig. 4). Phylogenetic analyses based on 519 single-copy orthologous genes indicated a very recent divergence time of about 5.4 MYA for the A- and B-genomes, with their common ancestor having diverged from Poaceae ~134 MYA (Supplementary Fig. 5). This estimate is consistent with a divergence time of 4.6 MYA between the A- and B-genomes, which was estimated based on 17 bacterial artificial chromosome (BAC) clones that contained 23.5 Kb of coding sequence18. Our estimate is more recent than the 20.9 MYA (estimated by three genes and one internal transcribed spacer region) or the 27.9 MYA (estimated by 19F genes)19,20. However, the increased sampling of informative characters in our genome-wide study for phylogenetic analyses should contribute to a more accurate divergence time estimation.
Three rounds of whole-genome duplication (WGD) events (α-, β- and γ-WGD) have occurred in the Musa lineage1, which was validated by our four-fold synonymous third-codon transversion (4dTv) analysis (Supplementary Fig. 6). WGD is frequently, and almost always, followed by diploidization and fractionation, which includes chromosome rearrangement, gene loss and biased retention21. After diploidization and divergence, A- and B-genomes start to evolve independently. To investigate the evolutionary differences between the A- and B-genomes after divergence, we performed three types of comparative analysis: (1) assessment of gene family expansion/contraction between A- and B-genomes by comparison to 14 other plant genomes; (2) comparison of structural variation in the A- and B-genomes by synteny analysis; and (3) comparison of synteny between the ancestral syntenic block and the A/B-genomes.
We analysed the gene family clustering and expansion/contraction of banana genomes of M. acuminata (A-genome) and M. balbisiana (B-genome), compared to 14 other plant genomes using OrthoMCL and CAFE22,23 (Supplementary Table 7 and Supplementary Fig. 7). We found 9,038 gene families that were conserved in M. balbisiana, M. acuminata, Oryza sativa, Brachypodium distachyon and Vitis vinifera. In contrast, 348 and 639 gene families were specific to the A- and B-genome, respectively (Supplementary Fig. 8). After their divergence, 1,761 gene families were expanded and 203 were contracted in the A-genome, while 392 gene families were expanded and 1,008 contracted in the B-genome (Supplementary Fig. 9 and Supplementary Tables 8 and 9). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of the significantly expanded gene families (P < 0.05) in the B-genome suggested that it was enriched in the photosynthesis and biosynthesis of secondary metabolite pathways, including those associated with the metabolism of inositol, starch and sucrose, linoleic acid and arachidonic acid (Supplementary Fig. 10 and Supplementary Table 10). Plants produce a high diversity of secondary metabolites with prominent functions in defence against a variety of herbivores and pathogens, in addition to the mitigation of various types of abiotic stresses24. Thus, these observations are consistent with the association of the B-genome with improved vigour and tolerance to both biotic and abiotic stresses2.
Synteny analysis indicated a high level of genomic colinearity and sequence similarity between the A- and B-genomes (Supplementary Fig. 11). We identified 72 large syntenic blocks between the A- and B-genomes, including 15 large blocks each containing over 900 gene pairs. These 72 syntenic blocks comprised 75.02% of A-genome space (containing 23% transposable elements) and 68.01% of B-genome space (containing 22% transposable elements) (Supplementary Table 11). We also identified two large translocations and two inversions between the A- and B-genome after their divergence. One large reciprocal translocation comprises 7.09 Mb on chromosome (chr)1 of the B-genome and 7.03 Mb on chr:3 of the A-genome, and one large inversion of 9.39 Mb on chr 5 of the B-genome and 8.83 Mb on chr 5 of the A-genome (Fig. 1 and Supplementary Fig. 11). These translocations and inversions were also supported by the rearrangements based on genetic mapping25, and can serve to introduce novel genetic diversity into the A- and B-genomes26.
Previously, 12 Musa ancestral blocks were assembled that represented the ancestral genome before α/β-WGD1,27,28. We identified 97 syntenic blocks resulting from α/β-WGD events in the B-genome by comparison to the 12 Musa ancestral blocks. These blocks contained 24,639 genes and represented 70.10% of all gene models involved in WGD-driven regions. We also identified 100 syntenic blocks that contained 26,780 genes (75.61%) in the A-genome (Supplementary Table 12). Of these ancestral Musa α/β blocks, 56.89% (15,236) and 60.59% (14,930) were singletons in the A- and B-genome, respectively, suggesting that genome fractionation (gene loss) and diploidization processes had occurred extensively after WGD events in the Musa lineage (Supplementary Table 12).
Taken together, our results indicated that the B-genome exhibited less expansion and more contraction of gene families, less syntenic coverage ratio and a higher singleton ratio in the ancestral blocks compared to the A-genome after divergence. Cycles of WGD followed by diploidization and fractionation have occurred across land plants, and are important in determining chromosome structure and gene content. Consequently, these processes have significantly contributed to the evolutionary success of plants21,29. The diploidization and fractionation processes involve a series of evolutionary events, including repetitive DNA loss, chromosome rearrangements and complex patterns of gene loss29,30. The above evidence supports the hypothesis that the B-genome was more sensitive to fractionation than the A-genome after their divergence.
Genetic diversity in polyploid bananas and functional divergence of subgenomes
Polyploid species usually exhibit vigorous growth, including high-quality production and high fitness31. Most banana cultivars are polyploid and exhibit various levels of ploidy and genomic background32. The genetic classification of some bananas is discordant, as is the case for Pelipita. Previous studies have shown that its karyotype comprises eight A and 25 B chromosomes as opposed to the predicted 11 A and 22 B chromosome distribution33. Understanding the genetic diversity and genomic constitution of Musa accessions would inform genomic group classifications, in addition to conservation and breeding strategies. Therefore, we re-sequenced five triploid bananas and four diploid bananas to investigate their genetic diversity (Supplementary Table 13).
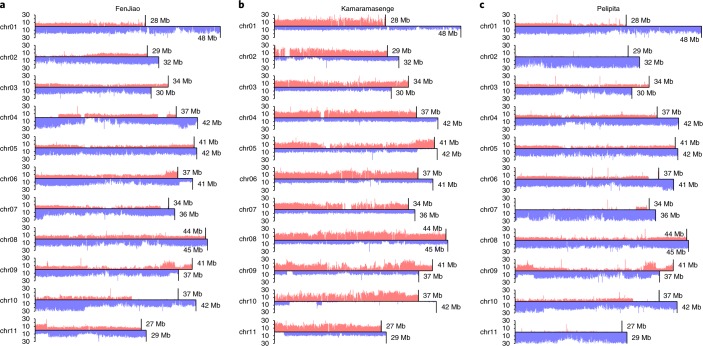
Simultaneous alignment of re-sequencing data to the A- and B-subgenomes identified the uniquely mapped reads that were used to analyse coverage depth, variations calling and homoeologous exchanges on each chromosome (Supplementary Table 14). Homoeologous exchanges were characterized by read coverage that showed a chromosomal region with a duplicated copy from the corresponding homoeologous subgenome34. These analyses confirmed that genome constitutions for the banana accessions were, in most cases, consistent with previous genome group classifications based on morphological traits. We identified 48 segmental homoeologous exchanges in the accession FenJiao, including nine from the B- to the A-subgenome and 39 in the reverse direction (Fig. 2a and Supplementary Table 15). We also found four segmental homoeologous exchanges from the B- to the A-subgenome in the accession Kamaramasenge, and replacement of chromosome 10 of the B-subgenome by the A-subgenome. (Fig. 2b and Supplementary Table 15). For the accession Pelipita, chromosomes 2, 7 and 11 of the A-subgenome were replaced by the B-subgenome and there were 18 segmental homoeologous exchanges on chromosomes 6, 9 and 10 (Fig. 2c and Supplementary Table 15). This indicates the eight A and 25 B chromosome constitution of Pelipita, consistent with previous genomic in situ hybridization studies33. This classification is further supported by phylogenetic analyses based on genotyping data (Supplementary Fig. 12). A total of 18,475,661 single-nucleotide polymorphisms (SNPs), 1,425,391 small insertions and deletions and 220,452 structural variations were identified in the samples (Supplementary Tables 16–18). Analysis of gene and SNP density on the chromosomes indicated that SNPs were preferentially located on non-gene-rich regions (Supplementary Fig. 13). There were ~2.5-fold SNPs on the A-genome of Pisang_Mas and Pisang_Kra compared to the B-genome of Balbisiana (Supplementary Table 16). The nucleotide diversity (π,0.0059) of A-subgenomes was higher than that of the B-subgenomes (0.0031) in accessions Fenjiao, Pelipita and Kamaramasenge.
Fig. 2. Coverage depth and genome structure summary for three allotriploid banana accessions.
a–c, Chromosome coverage and structure for accessions FenJiao (genome group, ABB) (a), Kamaramasenge (genome group, AAB) (b) and Pelipita (genome group, ABB) (c) with 100 kb non-overlapping sliding windows. The upper red bar and lower blue bar represent coverage depth of the A- and B-subgenome, respectively.
Gene family expansion and contraction analysis of M. acuminata and M. balbisiana in comparison to other sequenced genomes indicated that there are 83 gene families significantly expanded in the A-genome (and conversely contracted in the B-genome). These families included plant–pathogen interactions, glycosphingolipid biosynthesis-ganglio series and glycosaminoglycan degradation pathways among others (Supplementary Table 19). Conversely, 33 gene families were significantly expanded in the B-genome (and contracted in the A-genome). These families included those involved in photosynthesis, metabolic pathways and ribosome, among others (Supplementary Table 20). This indicates that the A- and B-genomes may have functionally diverged at the genome level during their respective genome evolution.
To explore the transcription of allopolyploid subgenomes, we assessed the expression of homoeologue genes from the A- and B-subgenomes of the triploid FenJiao. Expression levels were measured within different tissues, at different stages of fruit development and ripening and in banana seedlings after abiotic stress treatments (Supplementary Table 21). A total of 25,717 homoeologous gene pairs were identified between the A- and B-subgenomes based on the gene alignment method with cumulative identity percentage (CIP) ≥ 60% and cumulative alignment length percentage (CALP) ≥ 60% (refs. 35,36). Among the homoeologue gene pairs, 81.83% were further supported by syntenic analysis (Supplementary Table 22). Expression of all homoeologue gene pairs was assessed to determine the distribution of expression fold change of B/A in the triploid ‘FJ’. The log2(RPKM B/RPKM A) was 1.2/1, where RPKM is reads per kilobase million, which differed from the genomic constitution value of 2/1 (Supplementary Fig. 14). This result could be explained by dosage compensation, wherein the triploid expression levels are reduced to a diploid state37,38. We further characterized 1,075 and 4,032 homoeologue gene pairs that showed expression dominance in the A- and B-subgenome, respectively (Supplementary Table 23). KEGG enrichment analysis indicated that genes with expression dominance in the B-subgenome were associated with 2-oxocarboxylic acid metabolism and the arginine biosynthesis pathway (q-value < 0.05) (Supplementary Table 24), whereas those showing expression dominance in the A-subgenome were not significantly enriched in KEGG pathways.
Non-synonymous/synonymous substitution (Ka/Ks) ratios were calculated for all homoeologue gene pairs between the A- and B-subgenomes. The Ka/Ks ratios of genes with expression dominance in the A-subgenome (median, 0.157) were slightly lower than those in the B-subgenome (median, 0.196) and non-dominant genes (median, 0.186) (Supplementary Fig. 15).
We then constructed a gene co-expression network for those genes with expression dominance using weighted gene co-expression network analysis (WGCNA)39. The results indicated that 87 and 295 genes with dominance expression interacted with 4,302 and 4,612 genes in the A- and B-subgenome, respectively. KEGG pathway enrichment analysis suggested that genes in the co-expression network of the A- and B-subgenomes were commonly associated with starch and sucrose metabolism (ko00500) and other metabolic pathways. In particular, ubiquinone and other terpenoid-quinone biosynthesis, photosynthesis–antenna proteins, carotenoid biosynthesis and other glycan degradation pathways were specially enriched in the A-subgenome (q-value < 0.05) (Supplementary Fig. 16 and Supplementary Table 25). In contrast, selenocompound metabolism and cyanoamino acid metabolism pathways were particularly enriched in the B-subgenome (q-value < 0.05) (Supplementary Fig. 17 and Supplementary Table 26). Overall, these results further support the hypothesis of functional divergence between the A- and B-genomes at the transcriptional level.
Expression dominance of homoeologue gene pairs in the ethylene biosynthesis pathway and expansion of the ACO family affect fruit ripening
Ethylene plays a key role in the regulation of climacteric fruit ripening post-harvest40. The core enzymatic steps in the ethylene biosynthesis pathways are well characterized, and include S-adenosyl-l-methionine synthase (SAMS), 1-aminocyclopropane-1-carboxylic acid synthase (ACS) and ACO41,42 (Fig. 3a). However, the expansion and expression dominance of these homoeologue gene pairs during polyploid fruit ripening remains largely unknown.
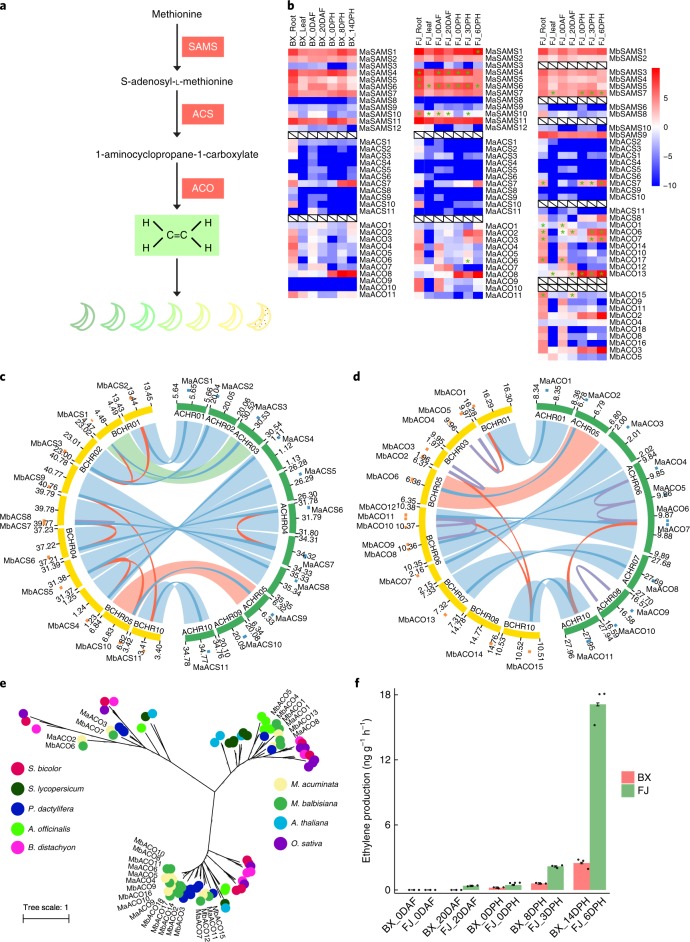
Fig. 3. Phylogeny and expression patterns of ethylene biosynthesis genes between M. acuminata (A-genome) and M. balbisiana (B-genome).
a, Overview of the ethylene biosynthesis pathway. b, Expression patterns of SAMS, ACS and ACO family genes in the root and leaf, and at different stages of fruit development and ripening in BX, the A-subgenome of FJ and the B-subgenome of FJ. Genes aligned horizontally in the heat map indicate homoeologue gene pairs between the A- and B-genomes. White boxes with diagonals indicate the lack of homoeologue gene pairs between the A- and B-subgenomes. Asterisks indicate expression dominance of homoeologue gene pairs between the A- and B-subgenomes of FJ. c,d, Synteny analysis of ACS (c) and ACO (d) families between the A- and B-genomes. Red lines indicate paralogous gene pairs resulting from WGD, blue lines indicate homoeologous gene pairs, purple lines indicate tandem duplication, light blue strips indicate aligned syntenic blocks, light green strip indicates translocation block and light red strips indicate inversion blocks.The blocks in outer ring represent location and length of genes; blue blocks represent genes from A-genome and orange blocks represent genes from B-genome. e, Phylogenetic analysis of ACO family genes among nine species: M. acuminata, M. balbisiana, A. thaliana, O. sativa, Sorghum bicolor, Solanum lycopersicum, Phoenix dactylifera, Asparagus officinalis and B. distachyon. f, Ethylene production at different stages of fruit development and ripening in BX and FJ. Error bars show standard error of the mean from three independent experiments (n = 3).
We identified 12 SAMS, 11 ACS and 11 ACO genes from the A-genome and 10 SAMS, 11 ACS and 18 ACO genes in the B-genome, which represents a significant expansion compared to the seven other sequenced plant species among the monocots and eudicots22 (Supplementary Tables 27 and 28). We further characterized 28 homoeologue gene pairs from the A- and B-genomes (Supplementary Table 29). These gene pairs displayed similar expression profiles in the BaXiJiao (Musa AAA group, cv. Cavendish, BX), the A-subgenome of FenJiao (Musa ABB group, cv Pisang Awak, FJ) and the B-subgenome of FJ (Fig. 3b and Supplementary Tables 30–32). Interestingly, eight gene pairs exhibited homoeologue expression dominance in the B-subgenome and five gene pairs were dominantly expressed in the A-subgenome of FJ (Fig. 3b and Supplementary Tables 33 and 34). The dominant expression of these genes in various tissues may be related to the fundamental role of ethylene biosynthesis.
Both ACS and ACO have previously been demonstrated as limiting enzymes within ethylene biosynthesis41. The expression abundance of MA-ACS1 (AB021906) and MA-ACO1 (X91076) is consistent with ethylene production during the post-harvest banana-ripening period43. Of the ten ACS gene pairs, MaACS7/MbACS7, which is a homologue of MA-ACS1, exhibited high expression levels during fruit ripening and was dominantly expressed in the B-genome (Fig. 3b). MbACS6 and MbACS7 are paralogous in a large syntenic block (the block contains 19 gene pairs), and maintain synteny and close evolutionary relationships to MaACS6 and MaACS7, respectively, suggesting that these genes duplicated from WGD (Fig. 3c and Supplementary Fig. 18). Of the nine ACO gene pairs, three (MaACO2/MbACO6, MaACO3/MbACO7 and MaACO8/MbACO13) exhibited high expression levels during fruit ripening and were dominantly expressed in the B-subgenome (Fig. 3b). MaACO2/MbACO6 and MaACO3/MbACO7 are in the syntenic block of chr:5 and chr:6 between the A- and B-genome, respectively, and belong to the same phylogenetic clade (Fig. 3d,e and Supplementary Table 35). These results indicated that MaACO2/MbACO6 and MaACO3/MbACO7 originated independently and developed crucial functions during fruit ripening.
We also observed high expression levels (log2RPKM > 4 in at least one stage of fruit ripening in the B-genome) of homoeologue gene pairs, probably related to fruit softening (pectin methylesterases, galactosidases, expansions and pectatelyase)44, cell wall modification (xyloglucan endotransglucosylase/hydrolases, fasciclin-like arabinogalactan proteins and β-d-xylosidase)45,46 and aroma production (alcohol dehydrogenases)40. These genes are closely involved in fruit ripening and are regulated by ethylene44. Almost all of these gene pairs showed expression dominance in the B-subgenome of FJ during fruit ripening, which is the same as the dominance expression of gene pairs related to ethylene biosynthesis (Supplementary Fig. 19 and Supplementary Tables 36 and 37). This co-dominance of homoeologue gene pairs in the B-subgenome further supports the significant contribution of the B-genome to ethylene biosynthesis and fruit ripening.
Gene duplication is a major mechanism that generates new genetic diversity as a basis for evolutionary innovation in eukaryotes47. Compared to the 11 ACO genes within the A-genome, the ACO genes in the B-genome expanded significantly to 18 members. The expansion of ACO genes, including MbACO2, -3, -4, -5 in chr 3, MbACO8, -9, -11 in chr 6 and MbACO16, -18 in scaffolds, was driven by tandem duplications in the B-genome (Fig. 3b,d). Of the ACO genes that were expanded in the B-genome, MbACO2 and MbACO3 showed strong expression levels during the fruit-ripening stages with log2RPKM > 11 at 6 days post-harvest (DPH) in FJ, which is coincident with the ethylene climacteric period (Fig. 3b,f and Supplementary Table 38). In addition, MbACO8, -9, -11, -16, -18 exhibited high expression levels in roots and fruits at 0 days after flowering (DAF) (Fig. 3b and Supplementary Table 38). These genes belonged to the same cluster and their expression patterns were highly concordant with their duplication (Fig. 3d,e), suggesting that the expansion and evolution of ACO genes in the B-genome contributed to tissue development and fruit ripening.
Ripening of FJ was more rapid than BX during post-harvest ripening. BX required 8 and 14 DPH to reach the more-green-than-yellow and full-yellow stages, respectively, whereas FJ required 3 and 6 DPH to reach these stages, respectively (Supplementary Fig. 20). The dominant expression of ethylene biosynthesis and fruit ripening-related gene pairs and the expansion of the ACO family in the B-genome could have contributed to increased ethylene production and faster fruit ripening in FJ compared to BX.
The active starch metabolic pathway in the B-genome during fruit development and the ripening process
Starch is the most widespread and abundant storage carbohydrate in plants. It is also a major component of cultivated banana, accumulating to high levels (~60–75% of dry weight) and leading to the presence of large starch granules (~8–30 m) during banana fruit development, along with near-complete conversion to soluble sugars during post-harvest ripening48–51. Thus, banana could serve as an excellent model for the investigatation of starch metabolism in fresh starchy fruits. The major enzymes that are responsible for starch biosynthesis (sugars will eventually be exported transporter: SWEET; sucrose transporter: SUT; sucrose synthase: SuSy; UDP-glucose pyrophosphorylase: UGPase; ADP-glucose pyrophosphorylase: AGPase; granule-bound starch synthase: GBSS; soluble starch synthase: SSS; starch branching enzyme: SBE; and starch debranching enzyme: DBE) and degradation (α-amylase: AMY; β-amylase: BMY; and starch phosphorylase: DPE) are encoded by multigenic families51–56 (Fig. 4a). We identified 101 starch metabolism-related genes in the A-genome, including 77 in the starch synthesis pathway and 24 in the starch degradation pathway. Ninety-six such genes were identified in the B-genome, including 68 in the starch synthesis pathway and 28 in the starch degradation pathway (Supplementary Table 39).
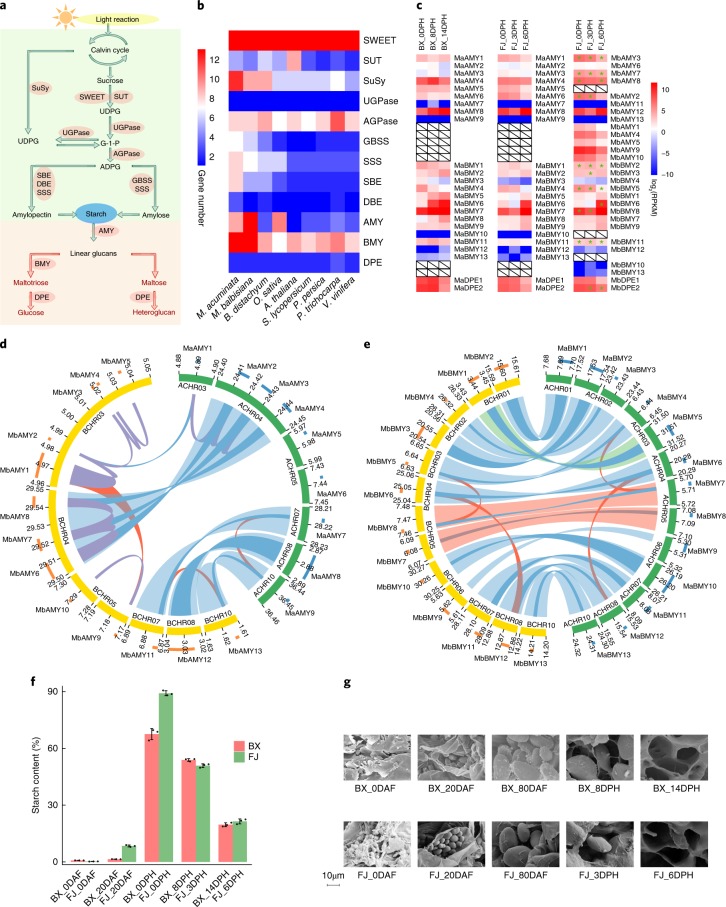
Fig. 4. Comparison of genomic expansion, evolutionary history and differential expression patterns of the starch metabolic pathway between M. acuminata (A-genome) and M. balbisiana (B-genome).
a, Overview of the starch biosynthesis and degradation pathway. b, Gene families in the starch metabolic pathway that are expanded in M. acuminata and M. balbisiana. c, Expression patterns of families AMY, BMY and DPE in the starch degradation pathway in BX, the A-subgenome of FJ and the B-subgenome of FJ during fruit-ripening stages. Horizontally oriented genes in the heat map indicate homoeologue gene pairs between the A- and B-genomes. White boxes with diagonals indicate that no homoeologue gene pairs were identified between the A- and B-genomes. Asterisks indicate expression dominance of homoeologue gene pairs between the A-subgenome of FJ and the B-subgenome of FJ. d,e, Synteny analyses of AMYs (d) and BMYs (e) between the A- and B-genomes. Red lines indicate paralogous gene pairs resulting from segmental/WGD-driven duplication, blue lines indicate homoeologous gene pairs, purple lines indicate tandem duplication, light blue strips indicate aligned syntenic blocks, light green strip indicates translocation block and light red strips indicate inversion blocks. The blocks in the outer ring represent location and length of genes; blue blocks represent genes from A-genome and orange blocks represent genes from B-genome. f, Starch contents at different stages of fruit development and ripening in BX and FJ. Error bars show standard error of the mean from three independent experiments (n = 3). g, Scanning electron microscopy of starch granules at different stages of fruit development and ripening in BX and FJ. The experiment was repeated three times independently with similar results.
In the starch synthesis pathway, five (SuSy, GBSS, SSS, SBE and DBE) out of nine families showed obvious expansion in the A- and B-genomes of banana, compared to seven other plant species that included two fruit plants, tomato and grape. These expansions suggest a potential significance in banana (Fig. 4b and Supplementary Table 40). We characterized 54 homoeologue gene pairs from these families in the A- and B-genomes. Of these, 27 homoeologue gene pairs had expression dominance in root, leaf and fruit tissues with seven dominant in the A-subgenome and 20 in the B-subgenome (Supplementary Fig. 21 and Supplementary Tables 41–45). Consequently, the starch synthesis pathway is more active in different tissues within the B-subgenome than in the A-subgenome. Of those gene pairs with dominant expression in the B-subgenome, MbSWEET17, MbSuSy1, MbSuSy2 and MbSuSy9 had high expression levels (log2-based RPKM > 6) at 0 DAF and 20 DAF, suggesting an important role in starch synthesis during fruit development (Supplementary Fig. 21 and Supplementary Table 43). In contrast, most of the starch synthesis-related genes that were unique to the A- or B-genome exhibited low expression levels (Supplementary Fig. 22).
Within the starch degradation pathway, genome annotation indicated that families AMY and BMY had an obvious expansion in the banana A- and B-genomes compared to seven other plant species (Fig. 4b and Supplementary Table 40). We characterized 21 homoeologue gene pairs within this pathway from the A- and B-genomes. Among these gene pairs, 11 had dominant expression and were associated with the B-subgenome during fruit ripening (Fig. 4c and Supplementary Tables 46–50). Among the dominant genes, MbAMY-2, MbAMY-3, MbAMY-8, MbBMY-6, MbBMY-8 and MbDPE-2 exhibited high expression (log2-based RPKM > 6 in at least one stage) during fruit ripening (Fig. 4c and Supplementary Table 48). MbAMY-1, -2, -3, -4, -5 and MbAMY-6, -7, -8 showed close proximity and an evolutionary relationship, suggesting that these genes duplicated from a tandem copy (Fig. 4d and Supplementary Fig. 23). MbBMY-5, -8 and MbBMY-6, -12 are two paralogous pairs in syntenic blocks of the B-genome and showed close evolutionary relationships, suggesting that these genes duplicated from WGD (Fig. 4e and Supplementary Fig. 24). Taken together, these results suggest that tandem-driven AMY duplication and WGD-driven BMY duplication in the B-genome contribute to starch degradation.
The dominant expression of genes involved in starch biosynthesis in the B-subgenome could have led to increased starch accumulation during fruit development in FJ relative to BX (Fig. 4f,g and Supplementary Fig. 21). In addition, the dominant expression of genes related to starch degradation in the B-subgenome also probably led to elevated starch degradation in FJ (Fig. 4c,f,g). Therefore, the active starch metabolic pathway in the B-genome leads to marked starch accumulation and degradation during the fruit development and ripening processes, respectively.
Discussion
Most cultivated bananas are triploid, having evolved from two wild diploid species, M. acuminata and M. balbisiana6. Our previous M. acuminata genome sequencing efforts provided genomic resources to inform banana breeding1. However, there is also an urgent need to develop the genomic resources of M. balbisiana, which is a crucial contributor to cultivated varieties. Improved understanding of the B-genome structure, subgenome evolution, homoeologous exchange, genetic diversity in polyploidy bananas and gene expansion and expression patterns will help in the design and application of breeding strategies for novel banana cultivars with improved traits.
Cycles of WGD followed by diploidization events have occurred across land plants, and have significantly contributed to their evolutionary success24,25. Our results indicate that the B-genome may be more sensitive to fractionation than the A-genome after WGD, although the A- and B-subgenomes have diverged very recently. Variation in genomic structure between the A- and B-genomes consists of chromosome rearrangements and gene loss during diploidization, which has resulted in the functional divergence of subgenomes in polyploidy bananas. This divergence is supported by differential enrichment of expanded/contracted gene families between the A- and B-genomes and the expression dominance of homoeologue genes from A- and B-subgenomes in triploids. Although homoeologue expression dominance has been identified in certain polyploid species57–60, the relationship between homoeologue expression dominance and functional divergence (especially in regard to ethylene biosynthesis/starch metabolism) of subgenomes in triploids remains to be elucidated. Thus, these results provide an important basis for the improvement of agriculturally important traits by focusing selection on transcriptionally dominant genes. It is worth noting that homoeologous exchanges may obscure the signal of expression dominance in subgenomes of allopolyploids61. The extensive homoeologue exchanges in allopolyploid bananas may remove many progenitor genome conflicts that result in subgenome biases in gene content and expression. Thus, homoeologous exchanges may contribute to the diversity of homoeologue expression dominance and induce a series of rapid genetic and epigenetic modifications for agronomic traits61.
Previous studies have suggested an important role of ethylene production in fruit ripening and starch metabolism with regard to fruit quality post-harvest27,43,55. However, the genetic mechanisms underlying polyploid fruit ripening are less well known. Here, we analysed biological processes related to these two pathways in triploid bananas. We identified significant genomic expansions and dominant expression of homoeologue genes in the B-genome at the pathway level of gene families and, most notably, in the ACO family, known to play a critical role in ethylene production. Our analysis revealed the origin and evolution of crucial gene families in these pathways, particularly for the independent origin of the MaACO2/MbACO6 and MaACO3/MbACO7 gene pairs and their specific function in fruit ripening. Moreover, we identified that this tandem event has led to expansion of ACO genes in the B-genome and to strong expression of these genes during fruit ripening. Our analysis also indicated a potential link between the dominant expression of homoeologue genes and the expansion of gene families with fruit ripening and starch metabolism. Previous studies have demonstrated that the B-genome is associated with improved vigour and tolerance to both biotic and abiotic stresses. Consequently, the B-genome is a target for banana breeding programmes2. Here, we highlight that the B-genome is of great importance in ethylene biosynthesis and post-harvest banana ripening, which will further our understanding of ripening mechanisms in polyploid fruit.
The M. balbisiana genome assembly, along with our previously acquired M. acuminata genome data, may aid in the discovery of cultivar-specific sequences that are related to important cultivar-specific traits, including shelf life, quality and stress tolerance. Thus, these resources can be used to build molecular characterization strategies for various cultivars to assist in rapid quality control and the conservation of biodiversity. The data from this study also pave the way for whole-genome association studies, germplasm improvement and genetic modification of bananas to meet increasing commercial demands. These genomic resources and results also reinforce the use of the banana as an appropriate model to study subgenome evolution and fruit biology in triploid variants. Due to the sterility and seedlessness of banana cultivars, further efforts will be needed to leverage the key gene resources from precisely characterized germplasms to achieve effective breeding schemes.
Methods
Sample collection
A double haploid of the wild diploid genotype PKW; 2n = 2x = 22 was provided by the Centre de Coopération Internationale en Recherche Agronomique pour le Développement (CIRAD) for genome sequencing. Fresh, unexpanded leaves were harvested and then frozen immediately with liquid nitrogen to preserve genomic DNA for isolation. High-molecular weight genomic DNA was extracted using a standard cetyltrimethyl ammonium bromide (CTAB) method62. DNA integrity was assessed by agarose gel electrophoresis (concentration of agarose gel, 1%; voltage, 150 V; electrophoresis time, 40 min). Finally, DNA was purified from the gel using a QIAquick Gel Extraction kit (QIAGEN).
Library construction and sequencing
One paired-end and eight mate-pair libraries were constructed for short-read sequencing on the Illumina HiSeq 2000 platform, which generated around 86.34 Gb (166× coverage) of high-quality data. For long-read DNA sequencing, 5.79 million SMRT long reads (58.99 Gb data, 113× coverage) were sequenced using the PacBio Sequel system with libraries of 20-kb insert size; sub-reads had a mean length of 10.2 kb and N50 length of 16.6 kb. One Hi-C library was prepared and sequenced on Illumina NovaSeq 6000 to generate 71.96 Gb (138× coverage) of high-quality data63 (Supplementary Table 1). Additional details are available in the Supplementary note.
Genome assembly
De novo assembly of DH-PKW was performed using wtdbg (v.1.2.8; https://github.com/ruanjue/wtdbg) based on PacBio data (only reads longer than 1 kb were used in assembly). The assembled genome was corrected for two rounds using the ‘wtdbg-cns’ programme in the wtdbg package. We then used the algorithm Arrow (https://github.com/PacificBiosciences/GenomicConsensus), which takes into account all of the underlying data and the raw quality values inherent in SMRT sequencing, to polish the assembly again for the final consensus accuracies. The final consensus contigs were scaffolded using the SSPACE-standard programme64 (v.3.0) with meta-pair reads from libraries of insert size 2–20 kb. Based on Hi-C data, 430.02-Mb scaffolds were anchored to 11 pseudo-molecules using LACHESIS software13 (Supplementary Fig. 2). Chromosomes were numbered according to the linkage group nomenclature adopted for M. acuminata. Additional details regarding genome assembly are provided in the Supplementary note.
Evaluation of assembly quality
BUSCO (v.3) was used to assess assembly completeness11. We mapped 29,610 M. acuminata-expressed sequence tags (ESTs) to the assembled genome using BLAT65 (v.35) with default parameters. In total, 93.59% of the ESTs were aligned to the genome with identity >90%. Additionally, BWA66 v.0.7.12 (aln -l 35) was used to map 59× Illumina reads to the assembly, and 96.11% of the reads were mapped to the assembled genome. Additional details are available in the Supplementary note.
Genome annotation
Repetitive sequences within the M. balbisiana genome were identified by a combination of homology-based and de novo approaches (Supplementary Table 1). Gene structures were annotated iteratively using three main approaches (ab initio predictions, homologue proteins and transcriptome data) that were combined using MAKER15 (v.2.31.8) (Supplementary Table 5). Gene functions were annotated according to the best match of the alignments using blastp67 (E-value < 1 × 10–5) against the Swiss-Prot68, TrEMBL68, NR (https://ftp.ncbi.nlm.nih.gov/blast/db/FASTA/), KOG69 and KEGG70 databases. Additional details are available in the Supplementary note.
Transcription factors
We used the iTAK programme16 to identify transcription factors based on the consensus rules that are mainly summarized within PlnTFDB and PlantTFDB71,72, with families from PlantTFcat73 and AtTFDB74 used as supporting evidence. In total, we identified 3,329 transcription factor genes in M. balbisiana and 3,899 in M. acuminata (Supplementary Table 6).
Gene family analysis
A total of 500,142 genes from 16 plant species with available whole-genome sequences were used for gene family clustering analysis. BLAST67 (v.2.2.26; -p blastp) was used to generate pairwise protein sequence alignments with E-values of < 1 × 10–5. Then, OrthoMCL22 was used to cluster similar genes by setting the main inflation value at 1.5 and using default settings for the other parameters. These analyses resulted in 39,358 gene families comprising 393,700 genes from the 16 species (Supplementary Table 7 and Supplementary Fig. 7).
We identified 519 single-copy gene families shared among the 16 species, and constructed a phylogenetic tree using MrBayes (v.3.1.2)75 software with the general time-reversible model (Supplementary Fig. 4). Divergence times for the 16 species were also estimated based on fourfold degenerate sites of all single-copy orthologous genes using the MCMCTree programme in the PAML package (v.4.4)76 (Supplementary Fig. 5).
CAFÉ (v.2.1)23 was used to analyse the expansion and contraction of gene families. A random birth-and-death model was used to assess gene gain or loss in gene families across the specified phylogenetic tree (Supplementary Fig. 9). Families with P < 0.05 were considered as significant expansion or contraction, and pathway enrichment analysis of these families was conducted using the enrichment pipeline77. Additional details are available in the Supplementary note.
Whole-genome alignment analysis
MCSCAN78 (parameters: -a -e 1e-5 -s 5) was used to detect collinearity within M. acuminata (A-genome) and M. balbisiana (B-genome) and among various species. Syntenic blocks containing at least ten gene pairs were obtained. All of the orthologous and paralogous gene pairs were extracted from the syntenic blocks for calculation of 4dTv79 distances using the HKY substitution model80 (Supplementary Fig. 8). Additional details are available in the Supplementary note.
Orthologous gene pair analysis
BLAST67 (v.2.2.26, -p blastp) was used to align M. acuminata proteins to M. balbisiana proteins for identification of orthologous genes. The value 1 × 10–5 was used as a cut-off to define the raw orthologues. We then filtered the BLAST results using two parameters (CIP ≥ 60% and CALP ≥ 60% (ref. 35). We identified 25,717 orthologous gene pairs (81.83% consistency with syntenic blocks) between M. acuminata and M. balbisiana using these two parameters (Supplementary Table 22). The orthologous gene pairs were first aligned using MUSCLE (v.3.8.31)81, then the Ka/Ks ratio of each gene pair was calculated using KaKs_Calculator (v.2.0)82 with model yn00. The significant difference between Ka/Ks values was detected by Student’s t-test.
Re-sequencing analysis
Nine different genotypes of banana were used for re-sequencing, including the triploid plants BaXiJiao (subgroup Cavendish, AAA), Gros_Michel (subgroup Gros Michel, AAA), FenJiao (subgroup Pisang Awak, ABB), Kamaramasenge (AAB) and Pelipita (ABB), in addition to the diploid plants Pisang_Mas (subgroup Sucrier, AA), Pisang_Kra (subsp. malaccensis, AA), DH-PKW (BB) and balbisiana (BB) (Supplementary Table 13). BaXiJiao, Gros_Michel and FenJiao were obtained from the Tissue Culture Centre of the Chinese Academy of Tropical Agricultural Sciences (CATAS). Pelipita, Pisang_Mas, Pisang_Kra, M. balbisiana and Kamaramasenge were provided by the Bioversity International Musa Transit Centre. Genomic DNA was extracted from fresh leaves of seedlings at the five-leaf stage using the CTAB method62.
Paired-end reads with libraries of 500-bp insert size were aligned to the A- and B-genomes simultaneously using BWA (v.0.7.12)66 with the parameters ‘bwa aln -t 20 -l 35’ (Supplementary Table 14), and only uniquely mapped reads were kept. Potential PCR duplicates were marked using Picard (v.1.54, https://broadinstitute.github.io/picard/) and indexed using the SAMtools package83. The Genome Analysis Toolkit84 was then used to infer SNPs and InDels. SNP identifications were filtered based on the following parameters: ‘QD < 2.0 || FS > 60.0 || MQ < 40.0 || MQRankSum < −12.5 || ReadPosRankSum < −8.0’, and InDels were filtered based on the following parameters: ‘QD < 2.0 || FS > 200.0 || ReadPosRankSum < −20.0’ (Supplementary Tables 16 and 17). Breakdancer (http://breakdancer.sourceforge.net/)85 was used to detect structural variations by checking paired-end reads with an abnormal orientation and/or span. The final structural variations were filtered using the following requirements: (1) minimum size of 100 and maximum size of 1,000,000; (2) minimum score of ≥30; and (3) minimum number of required reads supporting each structural variation ≥5 (Supplementary Table 18). Nucleotide diversity (π) was analysed using VCFtools (v.0.1.13; https://vcftools.github.io/man_latest.html).
Analysis of homoeologous exchanges
Assessment of read coverage depth was used to detect homoeologous exchanges between the A- and B-subgenomes. We inferred these based on cases where reads coverage of a given region on one parental genome was significantly high while the orthologous one was low or had no coverage. High coverage indicates duplication, and low or no coverage indicates loss34. The uniquely mapped paired-end reads were used to calculate the coverage depth of each sample on the A- and B-genomes (Supplementary Figs. 25–27). According to coverage depth, we detected homoeologous exchanges in the triploids ‘FenJiao (ABB)’, ‘Pelipita (ABB)’ and ‘Kamaramasenge (AAB)’ (Supplementary Table 15). Additional details are available in the Supplementary note.
Transcriptome analysis
Banana fruits at different stages of development (0, 20 and 80 DAF) and ripening (8 and 14 DPH for BX, 3 and 6 DPH for FJ) were obtained from the banana plantation at the Institute of Tropical Bioscience and Biotechnology (Chengmai, Hainan, 20° N, 110° E). Two-month-old BX and FJ banana seedlings were obtained from the Tissue Culture Centre of CATAS and cultured in Hoagland’s solution. Seedlings at the five-leaf stage were treated with 200 mM mannitol for 7 days, 300 mM NaCl for 7 days and low-temperature conditions (4 °C) for 22 h. Fruit, root and leaf samples were frozen in liquid nitrogen and stored at −80 °C until RNA extraction for transcriptome analysis.
Total RNAs were isolated using a plant RNA extraction kit (TIANGEN). Total RNA (3 μg) from each sample was converted to complementary DNA using a RevertAid First-Strand cDNA Synthesis Kit (Fermentas). cDNA libraries were constructed using TruSeq RNA Library Preparation Kit v.2, and were subsequently sequenced on the Illumina HiSeq 2000 platform using the Illumina RNA-Seq protocol. Two biological replicates were used for each sample. A total of 159.14 Gb (Supplementary Table 21) of high-quality clean data were produced. Gene expression levels were calculated as RPKM86. Differentially expressed genes were identified by methods previously established with the read count of two replicates for each gene (fold change ≥2; false discovery rate ≤0.001)87. Additional details are available in the Supplementary note.
Weighted gene co-expression network analysis
Gene expression patterns for all identified genes were used to construct a co-expression network using WGCNA (v.1.47)39. Genes without expression detected in all tissues were removed before analyses. Soft thresholds were set based on the scale-free topology criterion employed in ref. 88. An adjacency matrix was developed using squared Euclidean distance values, and the topological overlap matrix was calculated for unsigned network detection using the Pearson method. Co-expression coefficients >0.55 between the target genes were then selected. Finally, the network connections were visualized using cytoscape89.
Identification of gene families involved in ethylene biosynthesis and starch metabolism pathways
We compared genes related to ethylene biosynthesis and starch metabolism in the M. balbisiana genome to those annotated in both M. acuminata and other plant genomes, including B. distachyon, O. sativa, Arabidopsis thaliana, Solanum lycopersicum, Prunus persica, Populus trichocarpa and V. vinifera. We retrieved protein sequences of these gene families from 16 species (Supplementary Table 51) for homologue-based searches with the criteria similarity >80% and coverage >80%. We then confirmed the presence of the conserved domain within all protein sequences and removed members without a complete domain.
Determination of total starch content
Banana pulp was immersed in 0.5% sodium bisulfite for 10 min to prevent browning, and then dried at 40 °C for 24 h. Pulp was then ground and centrifuged. The residue was suspended in 5 ml of 80% Ca(NO3)2 in a boiling water bath for 10 min then centrifuged for 4 min at 4,000 r.p.m. The supernatant was transferred to a 20-ml volumetric flask and the residue was extracted twice with 80% Ca(NO3)2, which yielded a combined extract volume of 20 ml. All experiments were repeated three times. The total starch content was assessed following methods described by Yang et al.90.
Scanning electron microscopy
Samples were fixed in stubs using double-faced tape and coated with a 10-nm platinum layer in a Bal-tec MEDo020 coating system (Kettleshulme). The prepared samples were analysed using an FEI Quanta 600 FEG scanning electron microscope (FEI Co.). Observations were performed in secondary electron mode while operating at 15 kV.
Measurement of ethylene production during fruit post-harvest stage
Ethylene production was measured by enclosing fruit samples in an airtight container for 2 h at 25 °C. After incubation, 1 ml of the headspace gas was withdrawn and injected into a gas chromatograph (GC-2010, Shimadzu) fitted with a flame ionization detector and an activated alumina column. Ethylene production measurements were obtained as recommended by the manufacturer.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Supplementary Note and Supplementary Figs. 1–27.
Supplementary Tables 1–52.
Acknowledgements
This work was supported by the earmarked fund for the National Key Research and Development Programme (No. 2018YFD1000200), Modern Agro-industry Technology Research System (No. CARS-32), National Nonprofit Institute Research Grant of CATAS (No. 1630052012012), the National Natural Science Foundation of China (No. 31872161),the Natural Science Foundation of Hainan Province (No. 2019CXTD412), the Central Public-interest Scientific Institution Basal Research Fund for CATAS (Nos. 1630052017018, 1630052019024, 1630052016005, 1630052016006 and 1630052017017), and the CGIAR Research Programme on Roots, Tubers and Bananas.
Author contributions
Z.J., B.X., X.F. and A.D.-H. conceived the experiments. Z.W., H.M., J.H.L., C.J., J.Y.W., J.Z., W.H., W.T., Y.Y., Z.D., J.Y. L., W.M., Y.X., J.S.W., M.P., A.G., J.X., Y.L., C.H., N.L., R.H., F.S. and S.R. participated in various aspects of biological sample collection, preparation and quality control. X.Y., C.X., Z.W., L.Y., Y.L.Y., C.L., Y.G. and S.S. sequenced the genomes. X.Y., C.X., Z.W., M.R., F.-C.B. and G.M. assembled the genomes. X.Y., C.X. and Z.W. annotated the genomes. Z.W., H.M., W.H., X.Y., C.X., S.Z., Z.Y.W., L.Y., Y.L.Y., C.L., Y.G., O.G. and N.Y. analysed the genomes. W.H., Z.W., H.M., C.X., B.X., Z.J., A.D.-H. and M.R. wrote the manuscript.
Data availability
Raw sequence reads for B-genome assembly and transcriptome for all samples were deposited in the CNSA (https://db.cngb.org/search/project/CNP0000292/) of CNGBdb with accession number CNP0000292 and Sequence Read Archive of the National Centre for Biotechnology Information (NCBI) under the BioProject (No. PRJNA432894). Genome assembly and annotation of DH-PKW were submitted to NCBI (No. PYDT00000000). Assembly and gene annotation of the A-genome (DH-Pahang) are available on the Banana Genome Hub (http://banana-genome-hub.southgreen.fr/).
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information: Nature Plants thanks M. Schranz, Robert VanBuren and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zhuo Wang, Hongxia Miao, Juhua Liu, Biyu Xu, Xiaoming Yao, Chunyan Xu.
Contributor Information
Angélique D’Hont, Email: dhont@cirad.fr.
Wei Hu, Email: huwei2010916@126.com.
Zhiqiang Jin, Email: 18689846976@163.com.
Supplementary information
Supplementary information is available for this paper at 10.1038/s41477-019-0452-6.
References
- 1.D’Hont A, et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature. 2012;488:213–217. doi: 10.1038/nature11241. [DOI] [PubMed] [Google Scholar]
- 2.Davey MW, et al. A draft Musa balbisiana genome sequence for molecular genetics in polyploid, inter-and intra-specific Musa hybrids. BMC Genom. 2013;14:683. doi: 10.1186/1471-2164-14-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Langhe E, et al. Why bananas matter: an introduction to the history of banana domestication. Ethnobot. Res. Appl. 2009;7:165–177. doi: 10.17348/era.7.0.165-177. [DOI] [Google Scholar]
- 4.Paul JY, et al. Golden bananas in the field: elevated fruit pro‐vitamin A from the expression of a single banana transgene. Plant Biotechnol. J. 2017;15:520–532. doi: 10.1111/pbi.12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheesman EE. Classification of the bananas. Kew Bull. 1947;2:97–117. doi: 10.2307/4109206. [DOI] [Google Scholar]
- 6.Simmonds NW, Shepherd K. The taxonomy and origins of the cultivated bananas. Bot. J. Linn. Soc. 1955;55:302–312. doi: 10.1111/j.1095-8339.1955.tb00015.x. [DOI] [Google Scholar]
- 7.Daniells, J. et al. Diversity in the GenusMusa (INIBAP, 2001).
- 8.Häkkinen M. Reappraisal of sectional taxonomy in Musa (Musaceae) Taxon. 2013;62:809–813. doi: 10.12705/624.3. [DOI] [Google Scholar]
- 9.Martin G, et al. Improvement of the banana ‘Musa acuminata’ reference sequence using NGS data and semi-automated bioinformatics methods. BMC Genom. 2016;17:243. doi: 10.1186/s12864-016-2579-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Assani A, et al. Production of haploids from anther culture of banana [Musa balbisiana (BB)] Plant Cell Rep. 2003;21:511–516. doi: 10.1007/s00299-002-0557-6. [DOI] [PubMed] [Google Scholar]
- 11.Simao FA, et al. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 12.Servant N, et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015;16:259. doi: 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burton JN, et al. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat. Biotechnol. 2013;31:1119. doi: 10.1038/nbt.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li C, et al. Genome sequencing and assembly by long reads in plants. Genes. 2017;9:6. doi: 10.3390/genes9010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell MS, et al. Genome annotation and curation using MAKER and MAKER‐P. Curr. Protoc. Bioinformatics. 2014;48:4–11. doi: 10.1002/0471250953.bi0411s48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng Y, et al. iTAK: a program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant. 2016;9:1667–1670. doi: 10.1016/j.molp.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 17.Givnish TJ, et al. Monocot plastid phylogenomics, timeline, net rates of species diversification, the power of multi-gene analyses, and a functional model for the origin of monocots. Am. J. Bot. 2018;105:1888–1910. doi: 10.1002/ajb2.1178. [DOI] [PubMed] [Google Scholar]
- 18.Lescot M, et al. Insights into the Musa genome: syntenic relationships to rice and between Musa species. BMC Genomics. 2008;9:58. doi: 10.1186/1471-2164-9-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christelova P, et al. A multi gene sequence-based phylogeny of the Musaceae (banana) family. BMC Evol. Biol. 2011;11:103. doi: 10.1186/1471-2148-11-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janssens SB, et al. Evolutionary dynamics and biogeography of Musaceae reveal a correlation between the diversification of the banana family and the geological and climatic history of Southeast Asia. New Phytol. 2016;210:1453–1465. doi: 10.1111/nph.13856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mandáková T, et al. How diploidization turned a tetraploid into a pseudotriploid. Am. J. Bot. 2016;103:1187–1196. doi: 10.3732/ajb.1500452. [DOI] [PubMed] [Google Scholar]
- 22.Li L, Stoeckert CJ, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Bie T, et al. CAFE: a computational tool for the study of gene family evolution. Bioinformatics. 2006;22:1269–1271. doi: 10.1093/bioinformatics/btl097. [DOI] [PubMed] [Google Scholar]
- 24.Bennett RN, Wallsgrove RM. Secondary metabolites in plant defence mechanisms. New Phytol. 1994;127:617–633. doi: 10.1111/j.1469-8137.1994.tb02968.x. [DOI] [PubMed] [Google Scholar]
- 25.Baurens FC, et al. Recombination and large structural variations shape interspecific edible bananas genomes. Mol. Biol. Evol. 2018;36:97–111. doi: 10.1093/molbev/msy199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saxena RK, Edwards D, Varshney RK. Structural variations in plant genomes. Brief. Funct. Genom. 2014;13:296–307. doi: 10.1093/bfgp/elu016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jourda C, et al. Expansion of banana (Musa acuminata) gene families involved in ethylene biosynthesis and signalling after lineage specific whole genome duplications. New Phytol. 2014;202:986–1000. doi: 10.1111/nph.12710. [DOI] [PubMed] [Google Scholar]
- 28.Garsmeur O, et al. Two evolutionarily distinct classes of paleopolyploidy. Mol. Biol. Evol. 2013;31:448–454. doi: 10.1093/molbev/mst230. [DOI] [PubMed] [Google Scholar]
- 29.Dodsworth S, Chase MW, Leitch AR. Is post‐polyploidization diploidization the key to the evolutionary success of angiosperms? Bot. J. Linn. Soc. 2016;180:1–5. doi: 10.1111/boj.12357. [DOI] [Google Scholar]
- 30.Langham RJ, et al. Genomic duplication, fractionation and the origin of regulatory novelty. Genetics. 2004;166:935–945. doi: 10.1534/genetics.166.2.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ni Z, et al. Altered circadian rhythms regulate growth vigor in hybrids and allopolyploids. Nature. 2009;457:327. doi: 10.1038/nature07523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Jesus ON, et al. Genetic diversity and population structure of Musa accessions in ex situ conservation. BMC Plant Biol. 2013;13:41. doi: 10.1186/1471-2229-13-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.D’Hont A, et al. The interspecific genome structure of cultivated banana, Musa spp. revealed by genomic DNA in situ hybridization. Theo. Appl. Genet. 2000;100:177–183. doi: 10.1007/s001220050024. [DOI] [Google Scholar]
- 34.Chalhoub Boulos, et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 2014;345:950–953. doi: 10.1126/science.1253435. [DOI] [PubMed] [Google Scholar]
- 35.Salse J, et al. Improved criteria and comparative genomics tool provide new insights into grass paleogenomics. Brief. Bioinformatics. 2009;10:619–630. doi: 10.1093/bib/bbp037. [DOI] [PubMed] [Google Scholar]
- 36.Salse J, et al. Reconstruction of monocotelydoneous proto-chromosomes reveals faster evolution in plants than in animals. Proc. Natl Acad. Sci. USA. 2009;106:14908–14913. doi: 10.1073/pnas.0902350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ren L, et al. Determination of dosage compensation and comparison of gene expression in a triploid hybrid fish. BMC Genom. 2017;18:38. doi: 10.1186/s12864-016-3424-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo M, Davis D, Birchler JA. Dosage effects on gene expression in a maize ploidy series. Genetics. 1996;142:1349–1355. doi: 10.1093/genetics/142.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar R, Khurana A, Sharma AK. Role of plant hormones and their interplay in development and ripening of fleshy fruits. J. Exp. Bot. 2014;65:4561–4575. doi: 10.1093/jxb/eru277. [DOI] [PubMed] [Google Scholar]
- 41.Adams DO, Yang SF. Ethylene biosynthesis: identification of 1-aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene. Proc. Natl Acad. Sci. USA. 1979;76:170–174. doi: 10.1073/pnas.76.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang SF, Hoffman NE. Ethylene biosynthesis and its regulation in higher plants. Annu. Rev. Plant Physiol. 1984;35:155–189. doi: 10.1146/annurev.pp.35.060184.001103. [DOI] [Google Scholar]
- 43.Liu X, et al. Characterization of ethylene biosynthesis associated with ripening in banana fruit. Plant Physiol. 1999;121:1257–1265. doi: 10.1104/pp.121.4.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iqbal N, et al. Ethylene role in plant growth, development and senescence: interaction with other phytohormones. Front. Plant. Sci. 2017;8:475. doi: 10.3389/fpls.2017.00475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teh BT, et al. The draft genome of tropical fruit durian (Durio zibethinus) Nat. Genet. 2017;49:1633. doi: 10.1038/ng.3972. [DOI] [PubMed] [Google Scholar]
- 47.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 48.Hubbard NL, Pharr DM, Huber SC. Role of sucrose phosphate synthase in sucrose biosynthesis in ripening bananas and its relationship to the respiratory climacteric. Plant Physiol. 1990;94:201–208. doi: 10.1104/pp.94.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.do Nascimento JRO, et al. Beta-amylase expression and starch degradation during banana ripening. Postharvest Biol. Tec. 2006;40:41–47. doi: 10.1016/j.postharvbio.2005.11.008. [DOI] [Google Scholar]
- 50.Tribess T, et al. Thermal properties and resistant starch content of green banana flour (Musa cavendishii) produced at different drying conditions. LWT-Food Sci. Technol. 2009;42:1022–1025. doi: 10.1016/j.lwt.2008.12.017. [DOI] [Google Scholar]
- 51.Jourda C, et al. Lineage-specific evolutionary histories and regulation of major starch metabolism genes during banana ripening. Front. Plant. Sci. 2016;7:1778. doi: 10.3389/fpls.2016.01778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martin C, Smith AM. Starch biosynthesis. Plant Cell. 1995;73:2141–2145. doi: 10.1105/tpc.7.7.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tiessen A, et al. Starch synthesis in potato tubers is regulated by post-translation redox modification of ADP-glucose pyrophosphorylase: a novel regulatory mechamism linking starch synthesis to the sucrose supply. Plant Cell. 2002;14:2191–2213. doi: 10.1105/tpc.003640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tetlow IJ, et al. Analysis of protein complexes in wheat amyloplasts reveals functional interactions among starch biosynthetic enzymes. Plant Physiol. 2008;146:1878–1891. doi: 10.1104/pp.108.116244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiao YY, et al. A comprehensive investigation of starch degradation process and identification of a transcriptional activator MabHLH6 during banana fruit ripening. Plant Biotechnol. J. 2017;16:151–164. doi: 10.1111/pbi.12756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miao H, et al. Soluble starch synthase III-1 in amylopectin metabolism of banana fruit: characterization, expression, enzyme activity, and functional analyses. Front. Plant. Sci. 2017;8:454. doi: 10.3389/fpls.2017.00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang T, et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015;33:531–537. doi: 10.1038/nbt.3207. [DOI] [PubMed] [Google Scholar]
- 58.International Wheat Genome Sequencing Consortium. A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science. 2014;345:1251788. doi: 10.1126/science.1251788. [DOI] [PubMed] [Google Scholar]
- 59.Grover CE, et al. Homoeolog expression bias and expression level dominance in allopolyploids. New Phytol. 2012;196:966–971. doi: 10.1111/j.1469-8137.2012.04365.x. [DOI] [PubMed] [Google Scholar]
- 60.Yang J, et al. The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat. Genet. 2016;48:1225–1232. doi: 10.1038/ng.3657. [DOI] [PubMed] [Google Scholar]
- 61.Bird KA, et al. The causes and consequences of subgenome dominance in hybrids and recent polyploids. New Phytol. 2018;220:87–93. doi: 10.1111/nph.15256. [DOI] [PubMed] [Google Scholar]
- 62.Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4326. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boetzer M, et al. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2010;27:578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 65.Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mount, D. W. Using the basic local alignment search tool (BLAST). Cold Spring Harb. Protoc.10.1101/pdb.top17 (2007). [DOI] [PubMed]
- 68.Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000;28:45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tatusov RL, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ogata H, et al. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pérez-Rodríguez P, et al. PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Res. 2009;38:D822–D827. doi: 10.1093/nar/gkp805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jin J, et al. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2013;42:D1182–D1187. doi: 10.1093/nar/gkt1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dai X, et al. PlantTFcat: an online plant transcription factor and transcriptional regulator categorization and analysis tool. BMC Bioinformatics. 2013;14:321. doi: 10.1186/1471-2105-14-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Davuluri RV, et al. AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics. 2003;4:25. doi: 10.1186/1471-2105-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 76.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 77.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2008;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tang H, et al. Synteny and collinearity in plant genomes. Science. 2008;320:486–488. doi: 10.1126/science.1153917. [DOI] [PubMed] [Google Scholar]
- 79.Huang S, et al. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 2009;41:1275–1281. doi: 10.1038/ng.475. [DOI] [PubMed] [Google Scholar]
- 80.Hasegawa M, Kishino H, Yano TA. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 81.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang D, et al. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinf. 2010;8:77–80. doi: 10.1016/S1672-0229(10)60008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen K, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat. Methods. 2009;6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mortazavi A, et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 87.Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- 88.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005;4:17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 89.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang J, Bi C. A half-grain method for analyzing the amylose content in rice grains and its application. Acta Agron. Sin. 1992;18:366–372. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Note and Supplementary Figs. 1–27.
Supplementary Tables 1–52.
Data Availability Statement
Raw sequence reads for B-genome assembly and transcriptome for all samples were deposited in the CNSA (https://db.cngb.org/search/project/CNP0000292/) of CNGBdb with accession number CNP0000292 and Sequence Read Archive of the National Centre for Biotechnology Information (NCBI) under the BioProject (No. PRJNA432894). Genome assembly and annotation of DH-PKW were submitted to NCBI (No. PYDT00000000). Assembly and gene annotation of the A-genome (DH-Pahang) are available on the Banana Genome Hub (http://banana-genome-hub.southgreen.fr/).