Abstract
Color discrimination requires the input of different photoreceptor cells that are sensitive to different wavelengths of light. TheDrosophila visual system contains multiple classes of photoreceptor cells that differ in anatomical location, synaptic connections, and spectral sensitivity. The Rh5 and Rh6 opsins are expressed in nonoverlapping sets of R8 cells and are the onlyDrosophila visual pigments that remain uncharacterized. In this study, we ectopically expressed Rh5 and Rh6 in the major class of photoreceptor cells (R1–R6) and show them to be biologically active in their new environment. The expression of either Rh5 or Rh6 in “blind” ninaE17 mutant flies, which lack the gene encoding the visual pigment of the R1–R6 cells, fully rescues the light response. Electrophysiological analysis showed that the maximal spectral sensitivity of the R1–R6 cells is shifted to 437 or 508 nm when Rh5 or Rh6, respectively, is expressed in these cells. These spectral sensitivities are in excellent agreement with intracellular recordings of the R8p and R8y cells measured inCalliphora and Musca. Spectrophotometric analyses of Rh5 and Rh6 in vivo by microspectrophotometry, and of detergent-extracted pigments in vitro, showed that Rh5 is reversibly photoconverted to a stable metarhodopsin (λmax = 494 nm), whereas Rh6 appears to be photoconverted to a metarhodopsin (λmax = 468 nm) that is less thermally stable. Phylogenetically, Rh5 belongs to a group of short-wavelength-absorbing invertebrate visual pigments, whereas Rh6 is related to a group of long-wavelength-absorbing pigments and is the first member of this class to be functionally characterized.
Keywords: Drosophila melanogaster, fruit fly, rhodopsin, visual pigment, spectral tuning, green-absorbing rhodopsin, blue-absorbing rhodopsin, protein expression
Color vision is the ability of an organism to distinguish differences in wavelength independent of differences in light intensity (Jacobs, 1981). With the exception of the use of oil droplets or screening pigments in some photoreceptor cells, color vision is dependent on the expression of spectrally distinct forms of the visual pigment rhodopsin in different photoreceptor cells (Jacobs, 1981; Hardie, 1985; Nathans et al., 1986a,b; Nathans, 1992). This prerequisite for color vision is fulfilled in the Drosophila compound eye, which is composed of ∼750 ommatidia (for review, see Wolff and Ready, 1993). Each ommatidium contains a bundle of 8 photoreceptor cells and 12 auxiliary cells. The photoreceptor cells differ in their position within the ommatidium, their synaptic connections within the optic lobes of the brain, and the opsin genes they express. Each photoreceptor cell contains a rhabdomere, a microvillar structure that contains the visual pigment and serves as the compartment for visual transduction. The rhabdomeres are arranged concentrically with the six peripheral rhabdomeres of the R1–R6 cells surrounding one central rhabdomere formed by the R7 or R8 cells in the distal or proximal portion of the ommatidium, respectively. The rhabdomeres of the R1–R6 photoreceptor cells span the length of the ommatidium and contain the major visual pigment of the Drosophila compound eye, the blue-absorbing Rh1 rhodopsin (ninaE) (O'Tousa et al., 1985;Zuker et al., 1985; Feiler et al., 1988). The R7 cells express either Rh3 or Rh4, UV-absorbing visual pigments, in nonoverlapping subsets of cells (Fryxell and Meyerowitz, 1987; Montell et al., 1987; Zuker et al., 1987; Feiler et al., 1992). The R8 cells express either Rh5 or Rh6 in nonoverlapping subsets of cells (Chou et al., 1996, 1999; Huber et al., 1997; Papatsenko et al., 1997). A minor class of ommatidia, located along the dorsal margin, contains R7 and R8 cells that both express Rh3 (Fortini and Rubin, 1990; Feiler et al., 1992). Rh2 encodes a violet-absorbing visual pigment that is expressed in the ocelli, simple eyes located on the vertex of the head (Cowman et al., 1986;Feiler et al., 1988).
The present studies were undertaken to characterize the spectral and photochemical properties of the R8 photoreceptor cell-specific rhodopsins Rh5 and Rh6. Previous work in larger flies,Calliphora and Musca, suggested that these visual pigments are likely to have unique spectral properties (for review, seeHardie, 1985, 1986). The characterization of these pigments provides insight into the relationship between visual pigment structure and the regulation of spectral tuning. Furthermore, phylogenetic analyses of Rh5 and Rh6 place both of these pigments into newly defined clades of visual pigment genes that have not been well characterized. Thus the characterization of Rh5 and Rh6 provides a basis for examining other cloned invertebrate pigments and completes the spectral characterization of the visual pigments of the Drosophilaeye that is essential for a detailed examination of color vision inDrosophila.
MATERIALS AND METHODS
Ectopic expression of the Rh5 and Rh6 opsin genes.Flies expressing the Rh5 opsin gene in the R1–R6 photoreceptor cells of ninaE17 mutants (y w; ninaE17; P[Rh1 + 5, y+]) have been described previously (Chou et al., 1996). Flies expressing the Rh6 opsin gene (y w; ninaE17; P[Rh1 + 6, y+]) were generated in a similar manner. Briefly, the Rh6 cDNA (a 1.4 kbEcoRI/KpnI fragment from clone 4.1A, containing the entire Rh6-coding region and polyA tail) was inserted immediately 3′ of an expression cassette containing the Rh1 promoter (2.4 kb including 33 bp of the 5′-untranslated region). The transcriptional fusion was subcloned into they+-marked P-element vector “C4” obtained from Pam Geyer (University of Iowa, Ames, IA). The construct was injected into y w; sr ninaE17 mutant embryos, and multiple independent P-element-mediated germline transformants were obtained using standard techniques (Karess and Rubin, 1984).
Histology. The immunohistochemistry and confocal imaging were performed as described previously (Chou et al., 1996). Head sections were incubated overnight at 4°C with rabbit anti-Rh1 (1:250 dilution), monoclonal anti-Rh5 (clone 7F1; IgG1 subclass; 1:20 dilution), and monoclonal anti-Rh6 (clone 14C5; IgM subclass; 1:20 dilution). After washes in PBS containing 0.1% saponin, the sections were incubated for 1 hr at room temperature with three secondary antibodies: Texas Red-conjugated goat anti-rabbit IgG (1:100 dilution), Cy5-conjugated goat anti-mouse IgG1 (1:100 dilution), and FITC-conjugated goat anti-mouse IgM (1:100 dilution). The confocal images were collected using a Bio-Rad MRC-1024 (Hercules, CA) with a Nikon Labphot-2 microscope (Melville, NY). Secondary antibodies and other immunological reagents were obtained from Jackson ImmunoResearch (West Grove, PA) and Southern Biotechnology (Birmingham, AL).
Sequence alignment and phylogenetic analysis. Amino acid sequences were aligned using Clustal X (Thompson et al., 1997). Phylogenetic trees were constructed with PAUP* 4.0b2a (Swofford, 1998) using maximum parsimony (unweighted) and neighbor-joining methods. To ensure robustness of the results and to estimate confidence intervals, we bootstrapped all trees 100 times. For each parsimony bootstrap replicate, five random additions and tree-bisection reconnection branch swapping were performed. In all analyses, bovine rhodopsin was designated an outgroup.
Electrophysiology. Electroretinogram (ERG) recordings were obtained from immobilized white-eyed (w) flies using glass microelectrodes filled with normal saline (0.9% NaCl, w/v) as described previously (Chou et al., 1996; Townson et al., 1998). Flies were stimulated with light from a xenon arc lamp (450 W; Osram; Oriel, Stratford, CT), using interference and neutral density filters to select specific wavelengths and intensities of light. Light intensity was measured using a calibrated silicon photodiode (model 71883; Oriel) and an optical power meter (OPM model 70310; Oriel).
Spectral sensitivity was measured using a modification of the voltage-clamp method of Franceschini (1979, 1984), which we have described in detail elsewhere (Townson et al., 1998). Briefly, the amplitude of the ERG response to a flickering (10 Hz) monochromatic stimulus was maintained at a criterion level by continuously adjusting the light intensity, as the wavelength of stimulating light was varied during a scan. In previous experiments, we have found that directly measuring and “clamping” the area (amplitude) of the ERG response can be problematic. This is because the scans require several minutes to complete and there is a significant problem in defining the baseline from which the ERG area or amplitude is calculated, especially when the baseline drifts. In the measurements in this study, for each ∼0.3 sec window (ERG response to approximately three flickers), we averaged all of the ERG voltages during this period and used the SD as an estimate of the amplitude (width) of the response (Press et al., 1992). The SD is a function of the variance between the individual data points and the mean voltage during the sinusoidal ERG response. Thus, the SD is related to the response amplitude, although we have found that it is much less sensitive to baseline drift and noise.
During an experiment, as the monochrometer was stepped through a scan, the SD of the ERG response to three pulses of light (during 0.3 sec) was calculated and compared with a criterion set point. The ERG response was maintained near the set point during the scan by constantly adjusting the light intensity using a proportional-integral-derivative algorithm (Corripio, 1990). Spectral sensitivity (SS) was defined as the inverse of the light flux required to produce the criterion response, taking into account the wavelength and intensity of the stimulating light [i.e., SS ∝ 1/(light intensity (μW/cm2) × wavelength)]. Raw sensitivity data were normalized to an amplitude of 1.0 at the wavelength of maximum sensitivity, multiple individual measurements were averaged, and values that differed by >10% from the mean value of a 10 nm window were filtered out. The filtered spectra were then smoothed with a window of 12 nm.
Microspectrophotometry. Heads from white-eyed flies were bisected (maintaining the retinas intact) and mounted between quartz coverslips in PBS. A single eye was illuminated antidromically so that light passed through the specimen into the objective lens of the single-beam microspectrophotometer [Leitz (MPV2; Wetzler, Germany) equipped with Zeiss ultrafluar optics and a Products for Research photomultiplier (RCA model C31034A02; Danvers, MA)]. To adapt the specimen, light emitted from a super-pressure mercury arc lamp (HBO 100 W; Osram, Berlin, Germany) and passed through an interference filter was used to photoconvert the visual pigment from its rhodopsin (R) to its metarhodopsin (M) state. The wavelength (λ1) of the adapting light was selected to shift the photosteady state preferentially between R and M. To measure the transmission through the specimen, light from a tungsten halogen source (12 V/100 W; model 64623; Osram) was passed through a 1/8-m monochromator (model 7420; Oriel) and directed through the specimen. Transmission was measured continuously with a photomultiplier as the monochromator scanned from 350 to 700 nm. The measuring light did not significantly alter the steady state between the two photopigment states. A second adapting light at a different wavelength (λ2) was then used to shift the photosteady state of the R and M states back toward the R state, and the transmission of the specimen was measured again. A difference spectrum ΔE (λ) was calculated from these measurements: ΔE(λ) = [log (Iλ2(λ)/Iλ1(λ)], where Iλ1(λ) andIλ2(λ) are the light intensities transmitted through the specimen after adaptation at λ1 and λ2, respectively. The difference spectra were normalized, averaged, and smoothed with a window of 10 nm.
Electrophysiological and spectroscopic data acquisition and instrument control were performed with a Power Macintosh 7600/120 (Apple Computer, Cupertino, CA) equipped with a National Instruments (Austin, TX) PCI-MIO-16XE-50 multifunction input/output board running LabView 5.0.
Preparation of visual pigment extracts and spectrophotometry. Drosophila ninaE17 mutants, white-eyed (w1118) control flies, and transgenic strains expressing different Drosophila visual pigments in aninaE17 mutant background were dark adapted for 48 hr. Between 200 and 1000 eyes were dissected under dim red light (>665 nm) and collected in water. The water was removed, and 200 μl of homogenization buffer (10% sucrose and 0.1m sodium phosphate buffer, pH 6.0) was added. The samples were homogenized and then centrifuged for 20 sec at low speed (1000 × g) to spin down the chitin debris and unhomogenized tissue. The supernatant was removed, and the procedure was repeated twice. The combined supernatants were centrifuged for 10 min at 100,000 × g, and the membrane pellet was extracted with 40 μl of 4% digitonin in sodium phosphate buffer, pH 6.0, for 40 min at 20°C in the dark. Difference spectra were recorded with a Kontron Uvikon 930 spectrophotometer at 10°C (for measuring absorbance changes of extracts containing Rh1, Rh3, Rh4, or Rh5 rhodopsin) or at 1°C (for extracts containing Rh6 rhodopsin or extracts obtained from ninaE17 mutants). The extracts were irradiated in the cuvette with monochromatic light (Schott interference filters) using a 50 W xenon lamp until a steady state was achieved (2–4 min) at the wavelengths indicated in the figure legends. To ensure that there was little or no effect on the photosteady state of the pigment caused by measuring the spectrum, before each experiment we measured and then immediately measured again the spectrum of the dark-adapted pigment. The difference spectrum of these two measurements is shown as the “baseline” in relevant figure panels.
Rhodopsin nomogram modeling. Rhodopsin and metarhodopsin absorption spectra were calculated from the difference spectra recorded from the microspectrophotometry (MSP) and detergent-extracted pigments using the exponential function described by Stavenga et al. (1993). Briefly, the spectral shape of the rhodopsin α-band absorption can be described by the following log normal function: α = Aexp[−a0x2(1 + a1x +a2x2)], where x =10log(λ/λmax),A = 1, a0 = 380,a1 = 6.09, anda2 = 3a12/8. Taking this relationship and assuming that the spectral shape of metarhodopsin is also described by this equation (but with different A and λmax), we fit the measured difference spectra to an equation in which the α-band absorption of rhodopsin was subtracted from that of metarhodopsin. A curve-fitting routine was implemented in KaleidaGraph (version 3.08d; Synergy Software, Reading, PA) using the Levenberg–Marquardt (nonlinear least-squares) algorithm (Press et al., 1992). The computer solved for the λmax and amplitude of both the rhodopsin and metarhodopsin absorption spectra and calculated the SD for each variable and the correlation coefficient (Pearson's r).
Stavenga et al. (1993) have shown that an improved fit can be obtained in the lower wavelength region by incorporating additional terms for the rhodopsin β-band absorption. These terms were not incorporated in the curve fitting because too little is known about the characteristics of M β-band absorption.
RESULTS
Ectopic expression of the Rh5 and Rh6 opsins in transgenic flies
The Drosophila visual system has proven to be an extremely useful tool for studying novel invertebrate opsins in vivo (Feiler et al., 1988, 1992; Britt et al., 1993; Townson et al., 1998). To characterize the spectral and biophysical properties of the Rh5 and Rh6 opsins, we expressed the genes encoding these visual pigments in the R1–R6 photoreceptor cells of a mutant strain of flies (ninaE17). By coupling the structural gene of Rh5 or Rh6 to the promoter region from the Rh1 opsin gene, we targeted the expression of the novel opsin to the R1–R6 photoreceptor cells. These cells comprise the major class of photoreceptor cells in Drosophila. The R1–R6 photoreceptor cells dominate the physiological and photochemical properties of the compound eye and are primarily responsible for the optomotor behavior of fruit flies (Heisenberg and Wolf, 1984). TheninaE17 mutant strain serves as an appropriate host for the expression of the Rh5 and Rh6 opsins because these flies lack the Rh1 visual pigment that is normally expressed in the R1–R6 cells. White-eyed flies (w) were used in these experiments because the removal of the red-screening pigments of the eye increases the animal's sensitivity to light, allows for the measurement of rhodopsin difference spectra by MSP and from visual pigment extracts, and also simplifies the immunofluorescence studies of visual pigment expression patterns.
To characterize the expression pattern of the endogenous and ectopically expressed opsins, we used specific polyclonal and monoclonal antibodies directed against the C termini of the opsin proteins (see Materials and Methods). As shown in Figure1A, the Rh1 opsin is expressed throughout the eye of the control flies (y w; red). Specifically, these opsins are expressed in the rhabdomeres of the R1–R6 cells, which extend the full length of the retina and constitute the majority of the photoreceptor cells found in the compound eye (Fig. 1A). In contrast, the endogenous Rh5 (Fig. 1A, blue) and Rh6 (green) opsins localize to the R8 cells, which comprise only a subset of the total photoreceptor cell population and are located proximally within the retina. Not labeled in these figures are the endogenous opsins that localize to the R7 cells, a subset of photoreceptor cells that are located in the distal retina. The host strain (y w; ninaE17) lacks Rh1 expression in the R1–R6 photoreceptor cells because of a deletion within the Rh1 gene, but this strain does express the endogenous opsins found in the R7 and R8 cells (Fig. 1B). In the transgenic flies, the Rh5 and Rh6 opsins (y w; ninaE17 P[Rh1 + 5, y+] and y w; ninaE17 P[Rh1 + 6, y+], respectively) are expressed throughout the compound eye, in the R1–R6 photoreceptor cells of ninaE mutant flies (Fig. 1C,D). Note that the expression of the endogenous opsins in the R8 cells is not disrupted in the transgenic flies and there is no endogenous Rh1 opsin expressed in the retina.
Fig. 1.
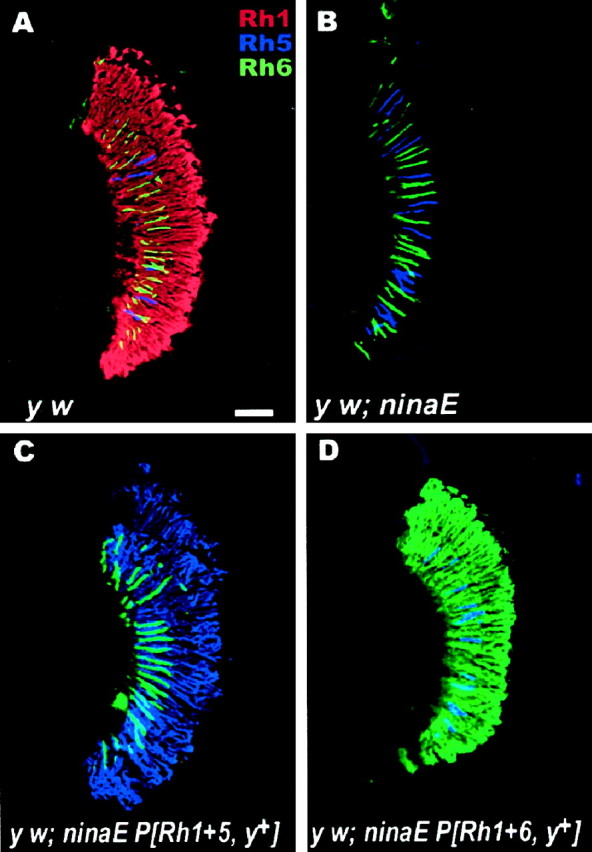
Expression of Rh5 and Rh6 in the compound eye ofDrosophila. Shown are confocal immunofluorescence images of longitudinal sections of retinas from white-eyed flies.A, In control flies (y w), Rh1 (red) is expressed in the rhabdomeres of the R1–R6 photoreceptor cells. Endogenous Rh5 (blue) and Rh6 (green) opsins are expressed in the rhabdomeres of the R8 cells, which occupy the proximal half of the retina. Rh3 and Rh4 are expressed in the rhabdomeres of R7 photoreceptor cells, which occupy the distal half of the retina (not labeled). B,In the mutant host strain (y w; ninaE17), a portion of the Rh1 gene (ninaE) has been deleted, and Rh1 is not expressed in the R1–R6 cells. However, the endogenous minor opsins, localized to the R7 (not labeled) and R8 (Rh5 and Rh6 shown as blueand green, respectively) cells, are still expressed.C, In transgenic flies that ectopically express Rh5 under the control of the Rh1 promoter (y w; ninaE17 P[Rh1 + 5, y+]), Rh5 (blue) is found throughout the retina in the R1–R6 photoreceptor cells. D, Similarly, in transgenic flies that ectopically express Rh6 (y w; ninaE17 P[Rh1 + 6, y+]), Rh6 (green) is expressed in the R1–R6 cells. Because the host strain for the expression experiment is mutant forninaE17 (as in B), there is no detectable Rh1 opsin expressed in either of the transgenic strains (C,D). Scale bar inA, 50 μm.
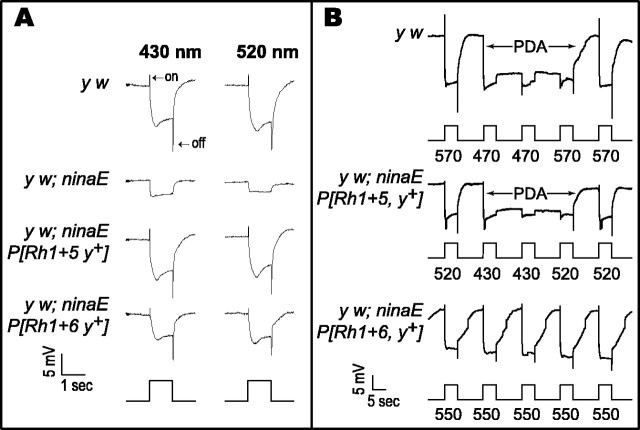
To test whether Rh5 and Rh6 are biologically active in their new cellular environment, ERGs were recorded from several different lines of control and transgenic flies. Figure2 shows wild-type ERGs recorded from white-eyed (y w) flies, which express the Rh1 opsin in the R1–R6 photoreceptor cells (Fig. 2A, top rowof traces). In these ERGs, a hyperpolarizing “on-” and a depolarizing “off-”transient appears at the onset and termination of the light stimulus, respectively. In addition there is a robust depolarization that is maintained for the duration of the stimulus. The on- and off-transients have been shown to originate in the first optic neuropile (the lamina) and are induced only by activation of the R1–R6 photoreceptor cells (Heisenberg, 1971; Heisenberg and Wolf, 1984;Laughlin, 1989). The maintained depolarization is generated directly at the level of the retina by the action of all three classes of photoreceptors contained within the compound eye. In the ERGs recorded from the y w; ninaE17 host strain, no on- or off-transients can be detected, and the magnitude of the depolarization is dramatically reduced (Fig. 2A, second row of traces). The remnant depolarization recorded from these flies arises from the R7 and R8 photoreceptor cells, which still express functional opsins (Fig.1B). When either Rh5 or Rh6 is introduced into theninaE17 mutant host strain, the light response in the ERG is completely restored (Fig. 2A,third, fourth rows oftraces). This indicates that these genes encode functional opsins capable of forming rhodopsins that are activated by light and that couple to the downstream components of the phototransduction cascade within the R1–R6 photoreceptor cells.
Fig. 2.
ERGs recorded from transgenic flies expressing Rh5 and Rh6. A, Responses to different wavelengths are arranged in columns, and responses recorded from specific genetic backgrounds are arranged inrows. Control y w flies (toprow of traces), which express the Rh1 opsin in the R1–R6 photoreceptor cells, have a robust response to light at both 430 and 520 nm. The depolarization is preceded and followed by on- and off-transients, respectively, which originate in the lamina and reflect the activation of the R1–R6 cells (Heisenberg, 1971; Heisenberg and Wolf, 1984; Laughlin, 1989). The mutant host strain that lacks the Rh1 opsin, y w; ninaE17 (secondrow of traces from thetop), has a dramatically reduced receptor potential and lacks both on- and off-transients. The residual response is derived from the R7 and R8 photoreceptor cells that are unaffected by theninaE17 mutation. The transgenic flies that express Rh5 or Rh6, y w; ninaE17P[Rh1 + 5, y+](thirdrow of traces from the top) and y w; ninaE17P[Rh1 + 6, y+](fourthrow oftraces from the top), display robust, wild-type photoresponses at either wavelength of light, demonstrating that both Rh5 and Rh6 are functional when expressed in the R1–R6 photoreceptor cells. Response amplitudes are not directly comparable between strains because of differences in the transgene expression levels. All of the strains were stimulated with the same intensity of light, which was ∼1.09 μW/cm2 at 430 nm and 0.9 μW/cm2 at 520 nm. B,When a control fly (y w; toptrace) is stimulated with intense light at 570 nm, there are a robust depolarization and immediate repolarization at the end of the stimulus. A PDA is induced when the fly is stimulated with light at 470 nm (thus producing a large amount of activated M-form). The depolarization is maintained after the cessation of the stimulus, and the photoreceptor cells are relatively inactivated to further stimuli. When the fly is again stimulated with 570 nm light, the PDA is terminated by the photoconversion of the M-form back to the R-form. Transgenic flies expressing Rh5 (y w; ninaE17 P[Rh1 + 5, y+]; secondtrace from the top) can undergo a PDA when stimulated with light at 430 nm, and this can be terminated by stimulation with light at 520 nm. Transgenic flies expressing Rh6 (y w; ninaE17 P[Rh1 + 6, y+]; thirdtrace from the top) do not show a PDA when stimulated with 550 nm light. This wavelength would be expected to produce maximal conversion of Rh6 from the R- to the M-form with minimal photoconversion back to the R-form (see Fig. 4, for Rh6 extract difference spectra). Stimulation of Rh6 transgenic flies at 350, 430, 470, 520, and 570 nm was also insufficient to induce a PDA (data not shown). Light intensity was unattenuated in these experiments and was ∼0.5 mW/cm2 at each of the wavelengths tested. Thebottomrow in A and therowbelow each trace inB show the stimulus, with the time and voltage scales indicated on the bottomleft of eachpanel.
Spectral and photochemical analysis of Rh5 and Rh6
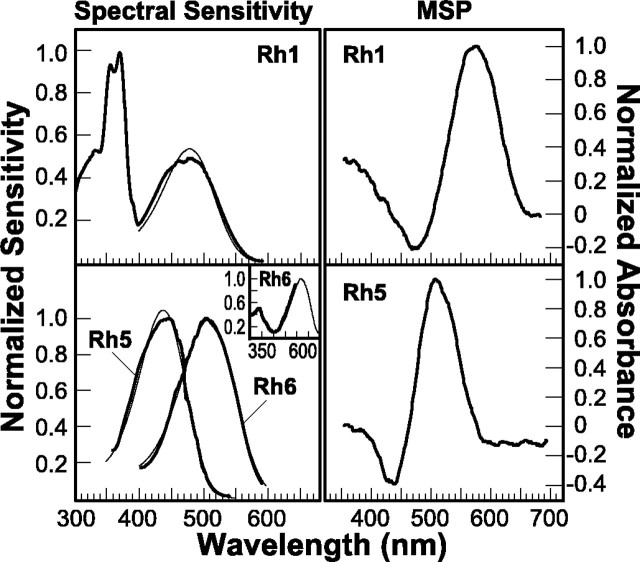
To characterize the spectral properties of the Rh5 and Rh6 opsins, we used both spectrophotometric and electrophysiological techniques. To determine the spectral sensitivity of transgenic flies expressing either the Rh5 or Rh6 opsin, we used a modified version of Franceschini's voltage-clamp technique (Franceschini, 1979, 1984). As shown in Figure 3, top left(Rh1), white-eyed control flies (y w), which express Rh1 in the R1–R6 photoreceptor cells, have a dual sensitivity in the UV and visible region of the spectrum. The sensitivity peak in the visible region has a maximum near 475 nm, which is well fit by a rhodopsin nomogram having an absorption maximum at 478 nm. The sensitivity in the UV region is thought to arise as the result of a sensitizing pigment that transfers the energy of an absorbed photon to the Rh1 chromophore, thereby inducing isomerization and visual pigment activation (Kirschfeld et al., 1977; Minke and Kirschfeld, 1979). In contrast, the spectral sensitivity of flies expressing the Rh5 or Rh6 minor opsins in the R1–R6 photoreceptor cells has been markedly altered (Fig. 3, bottom left). In the case of flies expressing Rh5, there is a peak of sensitivity near 440 nm, which is well fit by a rhodopsin nomogram having an absorption maximum at 437 nm. The spectral sensitivity of flies expressing Rh6 shows a peak in the green region near 510 nm, which is well fit by a rhodopsin nomogram having an absorption maximum at 508 nm.
Fig. 3.
Spectral sensitivity and in vivospectroscopy of Rh5 and Rh6. Spectral sensitivity was measured using a modification of the voltage-clamp method of Franceschini (1979, 1984) (see Materials and Methods for details). TopLeft (Rh1), Recordings averaged from y wcontrol flies (n = 2). These animals express the Rh1 (ninaE) opsin in the R1–R6 photoreceptor cells and have prominent sensitivity peaks in the blue and UV regions of the spectrum (boldtrace). The single peak at ∼475 nm (blue) is attributable to direct absorption by, and activation of, the Rh1 pigment. The doublet at ∼355 and 370 nm (UV) is attributable to a UV-sensitizing pigment. The sensitizing pigment is thought to transfer the energy of absorbed UV quanta to the Rh1 chromophore, thereby inducing isomerization and visual pigment activation (Kirschfeld et al., 1977). The peak of Rh1 sensitivity in the blue region is fit well by a calculated rhodopsin α-band absorption (finetrace) having a maximal absorption at 478 nm (r = 0.983) (Stavenga et al., 1993). The measured spectral sensitivity is somewhat broader than the calculated absorption, and this could be caused by waveguide or self-screening effects (Smakman and Stavenga, 1986). Bottom Left, The averaged spectral sensitivities of transgenic flies expressing either Rh5 (n = 4) or Rh6 (n = 5) in the R1–R6 cells (same genotypes indicated in Fig. 2). Flies expressing Rh5 have a prominent peak of sensitivity at ∼440 nm, whereas flies expressing Rh6 have a prominent peak at ∼510 nm (bold traces). The spectral sensitivities of Rh5 and Rh6 were well fit by calculated rhodopsin α-band absorptions (finetraces) having absorption maxima at 437 nm (r = 0.995) and 508 nm (r = 0.998) for Rh5 and Rh6, respectively. When Rh5 or Rh6 is expressed in a host strain that lacks the response of the R7 and R8 cells (w norpAP24;ninaE17 P[Rh1+norpA cDNA, w+]; see Results for description), we find that Rh5 transgenic flies display no UV sensitivity (data not shown).Inset, In contrast, Rh6 transgenic flies showing UV sensitivity. The finetrace shows the same curve shown in the mainpanel, and the boldcurve is a high-resolution scan through the lower wavelength region. The amplitude of the high-resolution scan was normalized to that of the short-wavelength limb of the visible region scan (finetrace) for comparison. The y w; ninaE17 mutant host strain did not have a detectable spectral sensitivity in this assay, because the amplitude of the ERG response was not high enough to meet the criterion of the recording paradigm (see Materials and Methods). Right, Rhodopsin and metarhodopsin difference spectra measured in vivo by MSP. TopRight, Control flies (y w; Rh1; n = 3).BottomRight, Rh5 transgenic flies (y w; ninaE17 P[Rh1 + 5, y+]; n = 2). Rh1-expressing flies were illuminated with adapting lights (λ1 and λ2) of 475 and then 580 nm, to shift the photosteady state from the Rh1 R-form to the M-form, and then vice versa. Rh5 transgenic flies were illuminated with adapting lights (λ1 and λ2) of 418 and 524 nm. No difference spectrum could be generated from flies expressing the Rh6 opsin construct after adaptation at multiple wavelengths (see Results).
To test whether either the Rh5 or Rh6 rhodopsins are capable of coupling to the sensitizing pigment and thus conferring UV sensitivity to the R1–R6 cells, we examined the spectral sensitivity of Rh5 and Rh6 when expressed in a special host strain. As described above and shown in Figure 2A,ninaE17 flies have functional R7 and R8 cells that are unaffected by the ninaE17mutation and produce a small depolarization in the ERG. The R7 photoreceptor cells have a pronounced UV sensitivity that could potentially interfere with the analysis of Rh5 and Rh6 in the UV region of the spectrum. Therefore, in transgenic flies expressing either Rh5 or Rh6, the norpA-encoded phospholipase C that is required for visual transduction in all photoreceptor cells of the compound eye was removed genetically (by the norpA mutation) (Bloomquist et al., 1988), and then this activity was restored only in the R1–R6 cells by expressing the norpA cDNA under the control of theninaE (Rh1) promoter (Pearn et al., 1996). Because these flies also lack the Rh1 visual pigment gene in the R1–R6 cells (ninaE17 mutant), all of the light-induced response was derived from Rh5 or Rh6 expression in the R1–R6 cells. Flies expressing Rh5 have no detectable sensitivity in the UV region (data not shown). We would expect that even a small peak of UV sensitivity would be detectable because there would be little overlap between the short-wavelength limb of the sensitivity peak of Rh5 transgenics and that attributable to the sensitizing pigment. By contrast, flies expressing Rh6 do show a small sensitivity peak in the UV (Fig. 3, bottom left, inset). This most likely reflects the coupling of Rh6 to the sensitizing pigment in the R1–R6 cells, although the efficiency of coupling appears reduced in Rh6 transgenics in the UV region compared with Rh1, as demonstrated by relative sensitivity ratios of ∼2:1 (UV/visible) for Rh1 versus ∼0.5:1 for Rh6. Furthermore, we did not observe the expected fine structure in the UV region for Rh6-expressing flies that is normally observed for other rhodopsins that couple to the sensitizing pigment and has also been observed in the R8y photoreceptor cells of larger flies (Hardie and Kirschfeld, 1983; Feiler et al., 1988). Our inability to detect any fine structure may be caused by the relatively low UV sensitivity of the Rh6-expressing flies.
We examined the transgenic flies expressing Rh5 and Rh6 using microspectrophotometry (MSP) to determine the absorption profiles of rhodopsin and metarhodopsin for each of the novel pigments. Photoconversion of R produces the activated form of the visual pigment M. For many invertebrate pigments, the M-form of the visual pigment is thermally stable, and its absorbance λmax is dramatically shifted from that of the R-form of the pigment (Stavenga, 1989, 1992). In addition the R- and M-forms are photointerconvertible, so that when the λmax of R and M differ significantly the ratio of the rhodopsin molecules in the R- and M-forms can be manipulated using different illumination conditions. In the photosteady state, the ratio of the R- and M-forms is dependent on the spectral composition of the illuminating light and the absorption profiles of the two states of the pigment (Hamdorf et al., 1973;Hamdorf, 1979). For example, when the Rh1 rhodopsin (expressed iny w control flies) is illuminated with blue light near 480 nm, ∼70% of the Rh1 R-form is converted to the M-form at steady state (Huber et al., 1990). Subsequent illumination with orange light near 580 nm photoconverts 100% of the M-form back to the R-form of rhodopsin. When absorption spectra are recorded before and after an illumination that shifts a substantial amount of the R-form to the M-form and these spectra are subtracted from each other, the resulting difference spectrum reflects the decrease in R-form absorption and the increase in M-form absorption (Fig. 3, top right). We used MSP to measure the difference spectrum of flies expressing the Rh5 opsin and found that Rh5 is photoconverted to a thermally stable metarhodopsin that absorbs maximally near 500 nm (Fig. 3, bottom right). No difference spectrum could be generated for Rh6-expressing flies by MSP, suggesting either (1) that there is not enough Rh6 expressed to be detectable by MSP, (2) that the M-form of Rh6 is not stable, or (3) that there is sufficient overlap in the absorption spectra of the R- and M-forms that illumination at many different wavelengths (402, 418, 430, 442, 456, 475, 499, 524, 563, and 584 nm; data not shown) did not lead to a detectable shift in the ratio of R and M in the photosteady state.
To examine the photoconversion between the R- and M-forms of Rh5 and Rh6 further, we attempted to generate prolonged depolarizing afterpotentials (PDAs) in transgenic flies expressing these pigments. A PDA is only generated in white-eyed flies when a substantial amount of rhodopsin has been photoconverted to metarhodopsin. During a PDA, the depolarization is maintained after the cessation of the light stimulus, and the response is inactivated to further stimuli (Pak, 1979). The PDA is thought to result from the inadequate inactivation of the activated metarhodopsin state. Metarhodopsin is inactivated in part by a stoichiometric interaction with an abundant, cytosolic protein, arrestin. Because arrestin is expressed at a fraction of the concentration of rhodopsin, excessive conversion of the visual pigment to its activated M-form is thought to overcome the ability of arrestin to inactivate it, and this produces a PDA (Dolph et al., 1993). This requires that (1) the visual pigment must be expressed and activated at sufficiently high levels that the concentration barrier to PDA generation is overcome, (2) the absorptions of the R- and M-forms are sufficiently different that illumination creates a photosteady state in which a high percentage of the visual pigment is present in the M-form, and (3) the M-form of the pigment must be stable for the period of time in which the PDA is observed. Figure 2B, top trace, shows that when y w control flies are illuminated with intense orange light at 570 nm, which does not shift a significant amount of the R-form to the M-form, there is a robust depolarization that ends abruptly at the end of the stimulus. When the animals are illuminated with blue light at 470 nm, converting 70% of R to M, there is a robust depolarization that is maintained after the stimulus ends, and the fly eye is relatively inactivated to further stimuli. When orange light is used to photoconvert all of M to R, the PDA is immediately terminated, and the response returns to baseline. When flies expressing Rh5 are stimulated with appropriate wavelengths of light, a similar “retuned” PDA can be generated. The PDA can be initiated with intense light at 430 nm, converting ∼50% of the Rh5 R-form to M, and this response can be terminated by illumination at 520 nm (photoconverting the M-form back to the R-form) (Fig. 2B, second trace from the top). This retuning of the PDA to wavelengths of light that can shift the steady state ratio of the R- and M-forms of ectopically expressed pigment has been demonstrated in previous studies with transgenic flies expressing Rh2, Rh3, and Rh4 (Feiler et al., 1988, 1992). Interestingly, stimulation of Rh6 transgenic flies at many wavelengths (Fig.2B, third trace from the top; 550 nm is shown) was insufficient to induce a PDA. As was the case with the MSP studies, this suggests either that the pigment is not being expressed or activated at high levels, that there is a significant overlap between R- and M-form absorption, or that the M-form is unstable. To resolve these possibilities we examined the absorption profiles of theDrosophila Rh5 and Rh6 opsins in detergent extracts prepared from the retinas of dark-adapted flies (Fig.4). We found that the Rh5 R-form can be reversibly photoconverted to a stable M-form (Fig. 4, top left), whereas photoactivation of Rh6 at a reduced temperature (1°C) produces an M-form that was not photoconverted back to the R-form in detectable amounts, and no stable M-form could be detected in extracts at higher temperatures (10°C) (Fig. 4, top right). This finding suggests that the M-form of Rh6 is not as stable as that of Rh5, at least when the pigments are expressed ectopically in the R1–R6 cells and extracted in digitonin. This finding provides one possible explanation for our inability to detect the Rh6 M-form by MSP and to induce a PDA in Rh6-expressing flies.
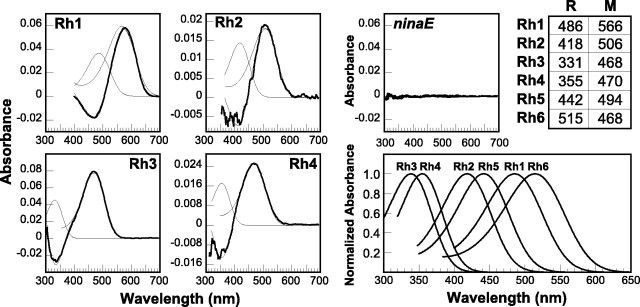
Fig. 4.
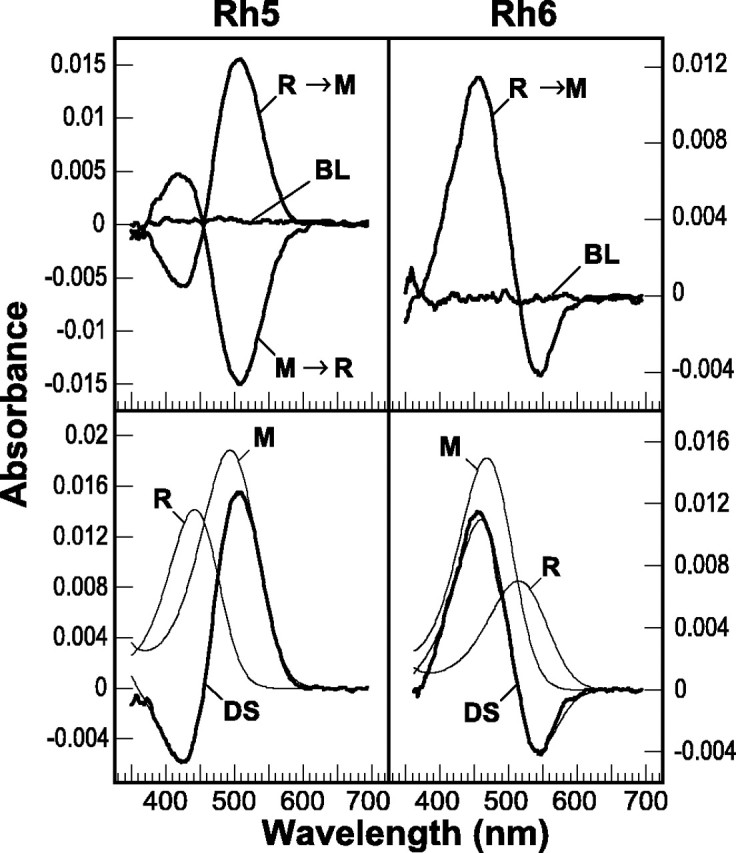
In vitro spectroscopy of Rh5 and Rh6 visual pigment extracts. Top, Difference spectra obtained from digitonin extracts of Rh5 (left) and Rh6 (right) transgenic flies are shown. Extracts were prepared in dim red light from 260 or 700 hand-dissected eyes of flies expressing Rh5 or Rh6, respectively, in the R1–R6 cells. Measurement of the absorption spectrum had no measurable effect on the samples, as shown by the baseline (BL) difference spectra calculated from sequential scans of the sample without other illumination. For both Rh5 and Rh6, when the extracts were illuminated with light selected to convert preferentially the R-form to the M-form (R → M) (4 min at 421 nm and 2 min at 560 nm for Rh5 and Rh6, respectively), a difference spectrum is generated. For Rh5, but not Rh6, the difference spectrum could be generated at 10° C. The Rh6 difference spectrum could only be generated at a reduced temperature (1° C). When the extracts were illuminated with light selected to convert preferentially the M-form back to the R-form (M → R) (4 min at 560 nm for Rh5; not shown for Rh6), a difference spectrum could be generated for Rh5 but not for Rh6. Bottom, The Rh5 and Rh6 difference spectra (DS) were fit with calculated rhodopsin and metarhodopsin absorption profiles as described in Materials and Methods (curve fits for Rh5 and Rh6 are shown asfinetraces overlying theDS in the bottomright andleftpanels, respectively). The Rh5 difference spectrum was best fit with R- and M-form absorptions having maxima at 442 and 494 nm, respectively (r = 0.998). The calculated ratio of R- and M-form extinctions (R/M) at the λmax for each form was 1:1.3. The Rh6 difference spectrum was best fit with R- and M-form absorptions having maxima at 515 and 468 nm, respectively (r = 0.991). The calculated ratio of R/M extinctions at the λmax for each form was 1:2.1.
An alternative explanation of these findings may be that there is too little Rh6 expressed in the transgenic flies to induce a PDA or be detectable by MSP. The evidence that this may be the case is based on the absorbance change, corresponding to the formation of metarhodopsin, observed in visual pigment extracts. We calculated the amount of rhodopsin per eye, assuming that the molar extinction coefficients of the M-forms of the Drosophila rhodopsins are approximately the same as has been determined for the Calliphora Rh1 rhodopsin, i.e., 72,000 m−1cm−1 (Stavenga and Schwemer, 1984). Although wild-type flies contain ∼220 fmol of Rh1 per eye, the amount of photoconvertible Rh5 and Rh6 rhodopsin expressed in the R1–R6 and R8 cells of the transgenic strains is only 45 and 9 fmol/eye, respectively. So although the relatively low amount of Rh5 present may be at the lower limit of the amount of photoconvertible rhodopsin required to induce a PDA (∼10–20% of wild-type Rh1 levels), the even smaller amount of functional Rh6 present may be the major reason for our inability to generate a PDA in these flies.
The theoretical absorption spectra of the R- and M-forms of each pigment were derived from the difference spectra by fitting the data to a model in which the absorption of the R-form was subtracted from the absorption of the M-form. The absorptions of the R- and M-forms were calculated using an exponential function that describes the shape of the rhodopsin absorption curve as a function of wavelength (described in Materials and Methods) (Stavenga et al., 1993). We found that the difference spectrum of the detergent-extracted Rh5 was fit well by R- and M-forms having absorption maxima at 442 and 494 nm, respectively (Fig. 4, bottom left). This is in fairly good agreement with a similar analysis of the MSP data [R (λmax) = 450 nm; M (λmax) = 496 nm] and the spectral sensitivity measurements determined electrophysiologically (λmax = 437 nm). Differences in the calculated absorption spectra of the R-form could be caused by detergent effects, or waveguide effects within the photoreceptor rhabdomere, although they more likely are the result of the derived nature of the difference spectrum and the fact that the extinction coefficient of the R-form is approximately one-half that of the M-form. Similar analysis of the detergent-extracted Rh6 difference spectrum showed that it was fit well by R- and M-forms having absorption maxima at 515 and 468 nm, respectively (Fig. 4, bottom right). Again, the maximal absorption of the R-form differs slightly from the spectral sensitivity determined electrophysiologically (λmax = 508 nm).
Photoactivation of rhodopsin to metarhodopsin is associated with the isomerization of the chromophore from the 11-cis to the all-trans form as well as with conformational changes within the rhodopsin protein that allow it to couple to and activate the G-protein transducin. These structural changes are associated with changes in the absorption of the visual pigment, which are likely to reflect changes in the interaction of the chromophore with amino acid residues within the chromophore-binding pocket. Thus, the study of R- and M-form absorption changes in different visual pigments may potentially provide important insight into the activation process. Analyses of the difference spectra obtained from extracts or MSP of transgenic flies expressing Rh1–Rh4 and the absorption spectra of the calculated R- and M-forms are shown in Figure5. The absorption maxima for both states of each pigment are also indicated in tabular form (Fig. 5, top right). It is interesting to note that all of theDrosophila rhodopsins undergo a bathochromic (red) shift after photoactivation, except for Rh6 that undergoes a hypsochromic (blue) shift. This hypsochromic shift in the metarhodopsin absorption of Rh6 is typical for pigments having a rhodopsin absorption maxima at wavelengths longer than 500 nm (Stavenga, 1989, 1992).
Fig. 5.
Spectroscopy of the DrosophilaRh1–Rh4 visual pigments. Left, Middle, Difference spectra that were measured from digitonin extracts of flies expressing Rh1, Rh3, and Rh4 andninaE17 mutant controls and MSP of flies expressing Rh2 are shown. In each case the difference spectra were calculated from spectra measured after illumination with adapting lights (using a single-adapting light and subtracting from the dark-adapted state or with λ1 and λ2 as described above), which were 461 nm (for Rh1), 418 and 524 nm (for Rh2), 344 nm (for Rh3), 384 nm (for Rh4), and 560 and 442 nm (forninaE). Difference spectra reflecting the R → M conversion were generated from flies expressing Rh1–Rh4; however no difference spectrum was generated from theninaE17 mutant host strain. Each difference spectrum (boldtrace in eachpanel) was fit with calculated rhodopsin and metarhodopsin absorption profiles as described in Materials and Methods (curve fits of the difference spectrum and the R- and M-forms are shown as finetraces in eachpanel). Table, TopRight, The calculated absorption maxima of the R- and M-forms for each pigment are indicated. The r values for the fits and the ratio of the R- and M-form extinctions λmax were as follows: Rh1, r = 0.996 and R/M = 1:1.6; Rh2, r = 0.0.987 and R/M = 1:1.5; Rh3, r = 0.998 and R/M = 1:1.7; and Rh4, r = 0.997 and R/M = 1:1.5.BottomRight, The calculated rhodopsin α-band absorptions based on the absorption maxima listed in thetable are shown. As discussed in the text, the calculated R- and M-form absorption maxima differ somewhat between the different measurement methods. Difference spectroscopy would be expected to yield a fairly accurate estimate of M-form absorption, because its extinction coefficient is greater than that of the R-form. In the absence of waveguide or self-screening effects, the spectral sensitivity measured physiologically would be expected to yield a fairly accurate estimate of the R-form absorption. For reference, the maximal sensitivities of flies expressing Rh1–Rh6 are 478 nm (Rh1), 420 nm (Rh2), 345 nm (Rh3), 375 nm (Rh4), 437 nm (Rh5), and 508 nm (Rh6) (Feiler et al., 1988, 1992; this paper).
Figure 5, bottom right, shows the calculated rhodopsin absorption curves for Rh1-Rh6 based on the R-form curve fits to the MSP and extract data. The Drosophila visual pigments have absorption maxima that differ by almost 200 nm, ranging from the UV to the green region of the spectrum. This remarkable range of spectral sensitivity is shared by many invertebrate organisms; however fruit flies are the first invertebrate organism from which the complete complement of all known visual pigment genes has been expressed and characterized in detail.
Relationship between opsins from Drosophila and other invertebrate species
To examine the relationship between the visual pigments ofDrosophila and other invertebrate species, we aligned the amino acid sequences for many of the known genes and generated a phylogenetic tree to evaluate their relatedness. As shown in Figure6, visual pigments that are thought to have similar spectral properties share a high degree of structural relatedness. The Rh5 visual pigment is most similar to other opsins that are thought to be short-wavelength absorbing, including the locust 2, bee blue, and moth Manop3 opsins, to which Rh5 is ∼46, 49, and 50% identical, respectively (Fig. 6, SW clade). In contrast, Rh5 is only 30–35% identical to theDrosophila Rh1, Rh2, and Rh6 opsins and 40–44% identical to the Rh3 and Rh4 opsins. Interestingly, a subset of the SW-absorbing pigments appears to share a common ancestor with the UV-absorbing pigments. The Rh6 visual pigment belongs to a group of opsins that are proposed to form long-wavelength-absorbing rhodopsins (Fig. 6,LW clade). As with Rh5, Rh6 shares a greater similarity (>55%) with opsins of the LW group than with otherDrosophila opsins. Rh6 shares a 51% identity with Rh1 and Rh2 and only a 30–33% identity with Rh3–Rh5 (Chou et al., 1996;Huber et al., 1997).
Fig. 6.
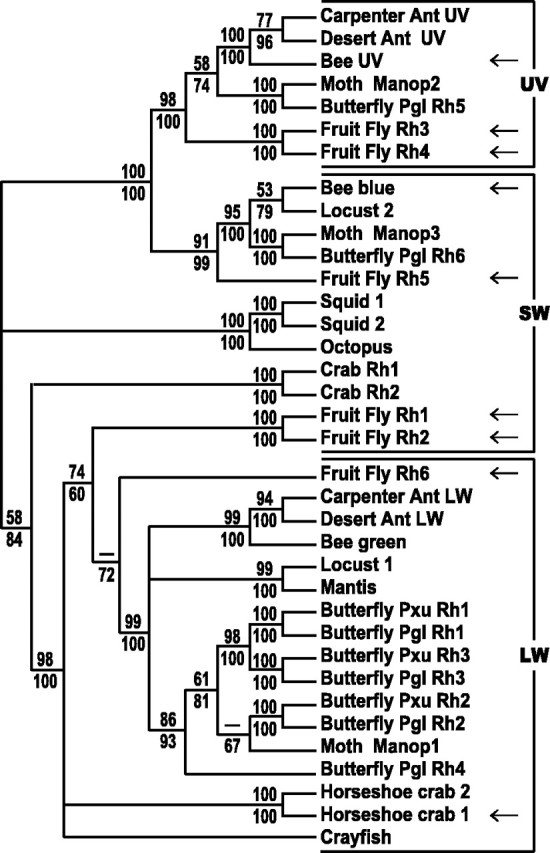
Phylogenetic relationships between selected invertebrate opsins. The deduced amino acid sequences of cloned invertebrate opsin genes were aligned and used to generate a phylogenetic tree, as described in Materials and Methods. Both maximum parsimony (unweighted) and neighbor-joining methods were used, and thevalues at each node represent the percentage of 100 bootstrap replicates that yielded the indicated structure (parsimony/neighbor-joining). Nodes with bootstrap values <60% are indicated as − and if <60% for both analyses were collapsed. Visual pigments were grouped into three families based on their spectral properties. The ultraviolet (UV) pigment family includes those with maximal absorptions below 400 nm. The short-wavelength (SW) family includes those pigments having maximal absorptions between 400 and 500 nm. The long-wavelength (LW) family includes those pigments with maximal absorptions above 500 nm. Inclusion within a family was based on one or more of three criteria: (1) direct characterization of the rhodopsin after ectopic expression inDrosophila, (2) well characterized spectral sensitivity or absorption measured from the native organism that is consistent with the placement within the tree, and (3) position within the phylogenetic tree with respect to other better-characterized visual pigments. The first criterion was met by Drosophila (fruit fly) Rh1–Rh6, bee blue and UV, and the horseshoe crab 1 pigments (indicated with ←) (Feiler et al., 1988, 1992; Townson et al., 1998) (Knox, Salcedo, Smith, Chou, Chadwell, Britt, and Barlow, unpublished results; this paper). The second criterion was met by the following pigments. Squid 1 and 2 are thought to have a λmax of 494 and 480 nm, respectively (Suzuki et al., 1976; Morris et al., 1993). The desert and carpenter ant UV pigments are thought to have a λmax of 360 nm, whereas the desert and carpenter ant LW pigments are thought to have a λmax of 510 nm (Popp et al., 1996; Smith et al., 1997). The bee green pigment is thought to have a λmax of 526 nm (Chang et al., 1996). The octopus pigment is thought to have a λmax of 475 nm (Koutalos et al., 1989). One or both of the crab pigments are thought to have a λmax of ∼480 nm (Sakamoto et al., 1996). The crayfish pigment appears to have a λmax of 533 nm (Hariyama et al., 1993; Zeiger and Goldsmith, 1994). Locust 1 and 2 are thought to have a λmax of 520 and 430 nm, respectively (Towner et al., 1997). The moth Manop1, Manop2, and Manop3 pigments are thought to have a λmax of 520, 357, and 450 nm, respectively (Chase et al., 1997). Butterfly Pxu Rh1 and Rh2 are thought to have a λmax of 515 nm, whereas Pxu Rh3 is thought to have a λmax of 575 nm (Kitamoto et al., 1998; Arikawa et al., 1999). The horseshoe crab 2 pigment is thought to have a λmax of 530 nm (Smith et al., 1993). The remaining pigments were organized on the basis of the third criterion. This group includes the visual pigments cloned from Papilio glaucus(Pgl Rh1–Rh6) that are closely related to the Pxu Rh1–Rh3 or the moth Manop1 and Manop2 opsins (Briscoe, 1998, 1999) (A. D. Briscoe, personal communication) and the Mantis opsin, which is closely related to Locust 1. The phylogenetic tree shows that opsins are typically more closely related to pigments that share similar spectral properties than to pigments from the same species. Rh5 belongs to a group of related visual pigments that appear to be SW absorbing, whereas Rh6 belongs to a class of LW-absorbing pigments. Rh6 is the first opsin of this class to be functionally characterized. GenBank accession numbers: Apis mellifera (Bee UV, AF004169; blue, AF004168; green, U26026), Camponotus abdominalis(Carpenter Ant UV, AF042788; LW, U32502), Cataglyphis bombycina (Desert Ant UV, AF042787; LW, U32501),Drosophila melanogaster (Fruit Fly Rh1, P06002; Rh2,P08099; Rh3, P04950; Rh4, P29404; Rh5, U67905; Rh6, Z86118),Hemigrapsus sanguineus (Crab Rh1, D50583; Rh2, D50584),Limulus polyphemus [Horseshoe crab 1 (lateral eye),L03781; 2 (ventral eye), L03782], Loligo forbesi (Squid 1, X56788), Octopus dofleini (Octopus, X07797),Papilio xuthus (Butterfly Pxu Rh1, AB007423; Pxu Rh2,AB007424; Pxu Rh3 AB007425), Papilioglaucus (Butterfly Pgl Rh1, AF077189; Pgl Rh2, AF077190; Pgl Rh3, AF067080; Pgl Rh4, AF077193; Pgl Rh5, AF077191; Pgl Rh6,AF077192), Procambrus clarkii (Crayfish, S53494),Schistocerca gregaria (Locust 1, X80071; Locust 2,X80072), Sphodromantis sps (Mantis, X71665), andTodarodes pacificus (Squid 2, X70498). Genome Sequence Database (GSDB) accession numbers (www.ncgr.org/cgi-bin/ff):Manduca Sexta (Moth Manop1, 76082; Manop2, 109852; Manop3, 1249561).
It is noteworthy that the only cloned invertebrate pigments that have been functionally expressed and characterized are theDrosophila rhodopsins Rh1–Rh6 (Feiler et al., 1988, 1992) (this paper) and the honeybee blue- and UV-absorbing pigments (Townson et al., 1998) (Fig. 6, indicated with ←). This group includes a large number of the UV- and SW-absorbing pigments shown in the tree; however Rh6 is the first and only member of the LW group to be formally characterized. As such, Rh6 both confirms and defines this group of pigments as likely having absorption maxima at longer wavelengths of light. In other experiments, we have found that the more distantly related Limulus lateral eye opsin (horseshoe crab 1) also confers LW sensitivity when ectopically expressed inDrosophila (B. E. Knox, E. Salcedo, W. C. Smith, W.-H. Chou, L. V. Chadwell, S. G. Britt, and R. Barlow, unpublished results).
The characterization of Rh6 as an LW-absorbing invertebrate opsin is particularly important because there is a large group of visual pigments that are within this clade. Although none of these pigments have been expressed and characterized, some of them are thought to encode red-absorbing visual pigments on the basis of their expression pattern (e.g., Pxu Rh3) (Kitamoto et al., 1998), whereas other long-wavelength-absorbing pigments have been characterized spectrophotometrically but have not yet been cloned (Schwemer and Paulsen, 1973; Langer et al., 1986). In addition, it is interesting to note that many of the SW visual pigments are included within clades that share common ancestors either with the UV visual pigments (in the case of Rh5 and its related pigments) or with the LW visual pigments (in the case of Rh1 and its related pigments). This suggests that there may be specific amino acid differences between the SW pigment group (like Rh5) and the UV pigments that are responsible for their different spectral properties. Thus our analysis of the Drosophila Rh5 and Rh6 opsins provides a framework in which the other invertebrate pigments can be grouped and studied, which is based on both structural and functional information about the pigments.
DISCUSSION
In this paper, we have described the spectral characterization of two Drosophila visual pigments, Rh5 and Rh6. Each protein falls into a structurally related group of rhodopsins that are distinguished from other groups by having absorption maxima at different wavelengths of light. Both Rh5 and Rh6 were ectopically expressed in the R1–R6 photoreceptor cells of blindninaE17 flies, and we found that they are capable of restoring the light response of this mutant. Rh5 encodes a visual pigment that confers a maximal spectral sensitivity at 437 nm to the R1–R6 cells, whereas Rh6-expressing cells are maximally sensitive to light at 508 nm. We examined the absorption properties of the Rh5 opsin in situ using MSP and in visual pigment extracts and found that it can form a thermally stable metarhodopsin (λmax = 494 nm) that can be reversibly photoconverted. After activation, Rh6 is converted to a metarhodopsin (λmax = 468 nm), which appears to be less stable than the M-forms of other Drosophila rhodopsins.
The spectral and physiological properties of the DrosophilaR8 photoreceptor cells have been difficult to examine because of their proximal location in the ommatidium and their small size. An early analysis of Drosophila photoreceptor cell function was undertaken using adapting lights and mutations to eliminate specific classes of photoreceptor cells (Harris et al., 1976). These studies showed that the R8 photoreceptor cells of Drosophila are sensitive to blue-green light with a maximal sensitivity near 500 nm. Physiological studies performed in larger flies, Calliphoraor Musca, have identified four different classes of R8 photoreceptor cells. Two minor classes include the R8marg cells, whose rhabdomeres are located beneath those of the R7marg cells along the dorsal margin of the eye. The R8marg cells express a UV-absorbing pigment identical to that found in the R7marg cells (Hardie, 1984). InDrosophila, the R7marg and R8marg cells express Rh3 (Fortini and Rubin, 1990; Feiler et al., 1992). The R8r cells have only been found in Musca males and express a pigment that is identical to that found in both the R1–R6 cells and the sex-specific R7r cells whose rhabdomeres lie above the R8r rhabdomeres (Hardie, 1983). The two major classes of R8 cells (R8p and R8y) are distinguished on the basis of their paired occurrence within ommatidia that contain the R7p or R7y photoreceptor cells, respectively. Interestingly, we have found that the pairing of different classes of R7 and R8 photoreceptor cells in individual ommatidia also occurs in Drosophila. Specifically, the R7p and R8p photoreceptors correspond to the R7 and R8 cells of Drosophila that express Rh3 and Rh5 in a pairwise manner, and R7y and R8y correspond to theDrosophila R7 and R8 cells that express Rh4 and Rh6 (Chou et al., 1996, 1999). These classes of ommatidia were originally identified on the basis of their appearance under blue illumination and fluorescence (y for yellow; p for pale) (Kirschfeld et al., 1978;Franceschini et al., 1981). The R8p and R8y cells ofCalliphora and Musca have maximum spectral sensitivities at ∼440 and 540 nm, respectively (Hardie et al., 1979;Smola and Meffert, 1979). The sensitivity of the R8y cell is thought to be shifted from the predicted rhodopsin λmax(∼520 nm) to longer wavelengths (540 nm) because of the screening effects of the overlying R7y cell rhabdomere (Hardie et al., 1979). The R8y cells also displayed a small triplet of peaks in the UV region, indicating the presence of a sensitizing pigment similar to that seen in the R1–R6 cells; however no UV sensitivity was found in the R8p cells (Hardie and Kirschfeld, 1983).
The spectral sensitivities measured in the present study from transgenic flies expressing Rh5 and Rh6 are in excellent agreement with the measurements from the R8p and R8y cells of larger flies, respectively, both in terms of their spectral sensitivities and in that Rh6 but not Rh5 appears to couple to the sensitizing pigment. Furthermore, our current results are also in good agreement with previous studies in Drosophila. Spectral sensitivity measurements made from sev; ora flies, which lack the R7 photoreceptor cells and the Rh1 opsin, demonstrated that the remaining R8 cells had blue-green sensitivity with a maximum near 500 nm (Harris et al., 1976; Washburn and O'Tousa, 1989). Although these experiments did not resolve the diversity of R8 cells present in wild-type flies, the spectral sensitivity is consistent with that of the class of R8 cells that express Rh6 (λmax = 508 nm). Interestingly, in other experiments examining the basis for the paired expression of opsin genes in the R7 and R8 cells of individual ommatidia, we have found that the expression of Rh5 versus Rh6 is dependent on the presence of an R7 cell. Indeed, in sevmutant flies (as would be the case in sev; ora), we have found that there is a dramatic decrease in the number of R8 cells that express Rh5, and virtually all of the R8 cells express Rh6 (Chou et al., 1996, 1999). And just as we were unable to measure a difference spectrum by MSP in vivo or induce a PDA in transgenic flies expressing Rh6, no MSP difference spectrum could be elicited fromsev; ora flies, and the spectral sensitivity of sev; ora flies was unchanged by light adaptation (Harris et al., 1976). These results are consistent with the idea that the Rh6 metarhodopsin may be short-lived and that the primary class of R8 photoreceptor cells that is present in sev; ora flies is that expressing Rh6. However, on the basis of the available data we cannot eliminate the possibility that our inability to obtain an MSP spectrum and to induce a PDA in flies expressing Rh6 in R1–R6 cells simply reflects the low level of photoconvertible rhodopsin present in the eyes of these flies. Whether or not a reduced thermal stability is characteristic of all M-forms derived from the LW class of invertebrate rhodopsins remains to be determined. Complete photoreversibility of the extracted pigments, i.e., photoconversion of R to M and vice versa without decay of substantial amounts of rhodopsin, has only been shown at −15°C for the long-wavelength-absorbing (520 nm) visual pigment of the mothDeilephila that forms an M-form with maximal absorbance at 475 nm (Schwemer and Paulsen, 1973).
This work completes the characterization of all of the known, cloned, Drosophila opsin genes. Although more opsin genes may potentially be identified as the Drosophila genome is completed, there are no known classes of photoreceptor cells that would necessarily require additional genes. The characterization of Rh5 and Rh6 is an important addition to the characterization of invertebrate visual pigments, because they have unique spectral properties and functionally define groups of related pigments that are as yet uncharacterized. As described in Results, Rh6 is the first cloned LW-absorbing invertebrate visual pigment to be functionally characterized. Its position within a well supported clade of visual pigments that are thought to be LW absorbing provides the only available evidence that these genes do in fact encode LW pigments.
One of the primary interests behind the study of the invertebrate visual pigments is the study of color vision. Several invertebrate species have been show to use color vision in specific behaviors. Honeybees are well known to use color vision in foraging and in returning to the hive (for review, see Menzel and Muller, 1996). Swallowtail butterflies, blowflies, and the mantis shrimp have also been shown to use color vision (Fukushi, 1985, 1989; Troje, 1993;Marshall et al., 1996; Kinoshita et al., 1999). Limited evidence suggests that Drosophila may also be able to discriminate between different colors (Quinn et al., 1974; Spatz et al., 1974; Menne and Spatz, 1977; Bicker and Reichert, 1978). The visual system ofDrosophila is ideally suited to an analysis of color vision because of the wide range of genetic and molecular approaches that are available to identify new genes, manipulate them, and study the effectsin vivo, in the intact fly. These approaches have provided important insights into the basis of photoreceptor cell recruitment and phototransduction (Dickson and Hafen, 1993; Zuker, 1996; Treisman and Heberlein, 1998) and have the potential to reveal important aspects of color vision in Drosophila. The characterization of Rh1–Rh6 provides a comprehensive view of the visual pigments and photoreceptor cell sensitivities in the Drosophila compound eye and opens up the possibility of conducting molecular and genetic studies of color vision in a well defined, genetically tractable experimental system.
Footnotes
This work was supported by National Eye Institute Grant R01EY10759 to S.G.B. and by funds from the European Union (BMH4-CT97-2341) to R.P. We thank J. O'Tousa for fly stocks and antibodies and A. Briscoe, R. Hardie, and D. Stavenga for comments on this manuscript. We are especially grateful to K. Kirschfeld and R. Feiler (Max-Planck-Institut für Biologische Kybernetik, Tübingen, Germany) for helping us assemble and modify the scanning spectral sensitivity instrument and microspectrophotometer that they developed.
Correspondence should be addressed to Dr. Steven G. Britt, Departments of Cellular and Structural Biology and Ophthalmology, University of Colorado Health Sciences Center, 4200 East Ninth Avenue, Box B111, Denver, CO 80262. E-mail: steve.britt@uchsc.edu.
Mr. Salcedo's present address: Departments of Cellular and Structural Biology and Ophthalmology, University of Colorado Health Sciences Center, 4200 East Ninth Avenue, Box B111, Denver, CO 80262.
REFERENCES
- 1.Arikawa K, Scholten DGW, Kinoshita M, Stavenga DG. Tuning of photoreceptor spectral sensitivities by red and yellow pigments in the butterfly Papilio xuthus. Zool Sci. 1999;16:17–24. [Google Scholar]
- 2.Bicker G, Reichert H. Visual learning in a photoreceptor degeneration mutant of Drosophila melanogaster. J Comp Physiol [A] 1978;127:29–38. [Google Scholar]
- 3.Bloomquist BT, Shortridge RD, Schneuwly S, Perdew M, Montell C, Steller H, Rubin G, Pak WL. Isolation of a putative phospholipase C gene of Drosophila, norpA, and its role in phototransduction. Cell. 1988;54:723–733. doi: 10.1016/s0092-8674(88)80017-5. [DOI] [PubMed] [Google Scholar]
- 4.Briscoe AD. Molecular diversity of visual pigments in the butterfly Papilio glaucus. Naturwissenschaften. 1998;85:33–35. doi: 10.1007/s001140050448. [DOI] [PubMed] [Google Scholar]
- 5.Briscoe AD. Intron splice sites of Papilio glaucus PglRh3 corroborate insect opsin phylogeny. Gene. 1999;230:101–109. doi: 10.1016/s0378-1119(99)00027-x. [DOI] [PubMed] [Google Scholar]
- 6.Britt SG, Feiler R, Kirschfeld K, Zuker CS. Spectral tuning of rhodopsin and metarhodopsin in vivo. Neuron. 1993;11:29–39. doi: 10.1016/0896-6273(93)90268-v. [DOI] [PubMed] [Google Scholar]
- 7.Chang BS, Ayers D, Smith WC, Pierce NE. Cloning of the gene encoding honeybee long-wavelength rhodopsin: a new class of insect visual pigments. Gene. 1996;173:215–219. doi: 10.1016/0378-1119(96)00165-5. [DOI] [PubMed] [Google Scholar]
- 8.Chase MR, Bennett RR, White RH. Three opsin-encoding cDNAS from the compound eye of Manduca sexta. J Exp Biol. 1997;200:2469–2478. doi: 10.1242/jeb.200.18.2469. [DOI] [PubMed] [Google Scholar]
- 9.Chou WH, Hall KJ, Wilson DB, Wideman CL, Townson SM, Chadwell LV, Britt SG. Identification of a novel Drosophila opsin reveals specific patterning of the R7 and R8 photoreceptor cells. Neuron. 1996;17:1101–1115. doi: 10.1016/s0896-6273(00)80243-3. [DOI] [PubMed] [Google Scholar]
- 10.Chou WH, Huber A, Bentrop J, Schulz S, Schwab K, Chadwell LV, Paulsen R, Britt SG. Patterning of the R7 and R8 photoreceptor cells of Drosophila: evidence for induced and default cell-fate specification. Development. 1999;126:607–616. doi: 10.1242/dev.126.4.607. [DOI] [PubMed] [Google Scholar]
- 11.Corripio AB. Tuning of industrial control systems. Instrument Society of America; Research Triangle Park, NC: 1990. [Google Scholar]
- 12.Cowman A, Zuker C, Rubin G. An opsin gene expressed in only one photoreceptor cell type of the Drosophila eye. Cell. 1986;44:705–710. doi: 10.1016/0092-8674(86)90836-6. [DOI] [PubMed] [Google Scholar]
- 13.Dickson B, Hafen E. Genetic dissection of eye development in Drosophila. In: Bate M, Arias AM, editors. The development of Drosophila melanogaster. Cold Spring Harbor Laboratory; Plainview, NY: 1993. pp. 1327–1362. [Google Scholar]
- 14.Dolph PJ, Ranganathan R, Colley NJ, Hardy RW, Socolich M, Zuker CS. Arrestin function in inactivation of G protein-coupled receptor rhodopsin in vivo. Science. 1993;260:1910–1916. doi: 10.1126/science.8316831. [DOI] [PubMed] [Google Scholar]
- 15.Feiler R, Harris WA, Kirschfeld K, Wehrhahn C, Zuker CS. Targeted misexpression of a Drosophila opsin gene leads to altered visual function. Nature. 1988;333:737–741. doi: 10.1038/333737a0. [DOI] [PubMed] [Google Scholar]
- 16.Feiler R, Bjornson R, Kirschfeld K, Mismer D, Rubin GM, Smith DP, Socolich M, Zuker CS. Ectopic expression of ultraviolet-rhodopsins in the blue photoreceptor cells of Drosophila: visual physiology and photochemistry of transgenic animals. J Neurosci. 1992;12:3862–3868. doi: 10.1523/JNEUROSCI.12-10-03862.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fortini ME, Rubin GM. Analysis of cis-acting requirements of the Rh3 and Rh4 genes reveals a bipartite organization to rhodopsin promoters in Drosophila melanogaster. Genes Dev. 1990;4:444–463. doi: 10.1101/gad.4.3.444. [DOI] [PubMed] [Google Scholar]
- 18.Franceschini N (1979) Voltage clamp by light. Invest Ophthalmol [Suppl]:5.
- 19.Franceschini N. Chromatic organization and sexual dimorphism of the fly retinal mosaic. In: Borsellino A, Cervetto L, editors. Photoreceptors. Plenum; New York: 1984. pp. 319–350. [Google Scholar]
- 20.Franceschini N, Kirschfeld K, Minke B. Fluorescence of photoreceptor cells observed in vivo. Science. 1981;213:1264–1267. doi: 10.1126/science.7268434. [DOI] [PubMed] [Google Scholar]
- 21.Fryxell KJ, Meyerowitz EM. An opsin gene that is expressed only in the R7 photoreceptor cell of Drosophila. EMBO J. 1987;6:443–451. doi: 10.1002/j.1460-2075.1987.tb04774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fukushi T. Visual learning in walking blowflies, Lucilia cuprina. J Comp Physiol [A] 1985;157:771–778. doi: 10.1007/BF01350074. [DOI] [PubMed] [Google Scholar]
- 23.Fukushi T. Learning and discrimination of coloured papers in the walking blowfly, Lucilia cuprina. J Comp Physiol [A] 1989;166:57–64. doi: 10.1007/BF00190210. [DOI] [PubMed] [Google Scholar]
- 24.Hamdorf K. The physiology of invertebrate visual pigments. In: Autrum H, editor. Handbook of sensory physiology. Springer; Berlin: 1979. pp. 145–224. [Google Scholar]
- 25.Hamdorf K, Paulsen R, Schwemer J. Photoregeneration and sensitivity control of photoreceptors of invertebrates. In: Langer H, editor. Biochemistry and physiology of visual pigments. Springer; Berlin: 1973. pp. 155–166. [Google Scholar]
- 26.Hardie RC. Projection and connectivity of sex-specific photoreceptors in the compound eye of the male housefly (Musca domestica). Cell Tissue Res. 1983;233:1–21. doi: 10.1007/BF00222228. [DOI] [PubMed] [Google Scholar]
- 27.Hardie RC. Properties of photoreceptors R7 and R8 in the dorsal marginal ommatidia in the compound eyes of Musca and Calliphora. J Comp Physiol [A] 1984;154:157–165. [Google Scholar]
- 28.Hardie RC. Functional organization of the fly retina. In: Autrum H, Ottoson D, Perl ER, Schmidt RF, Shimazu H, Willis WD, editors. Progress in sensory physiology. Springer; Berlin: 1985. pp. 1–79. [Google Scholar]
- 29.Hardie RC. The photoreceptor array of the dipteran retina. Trends Neurosci. 1986;9:419–423. [Google Scholar]
- 30.Hardie RC, Kirschfeld K. Ultraviolet sensitivity of fly photoreceptors R7 and R8: evidence for a sensitising function. Biophys Struct Mech. 1983;9:171–180. [Google Scholar]
- 31.Hardie RC, Franceschini N, McIntyre PD. Electrophysiological analysis of fly retina. II. Spectral and polarisation sensitivity in R7 and R8. J Comp Physiol [A] 1979;133:23–39. [Google Scholar]
- 32.Hariyama T, Ozaki K, Tokunaga F, Tsukahara Y. Primary structure of crayfish visual pigment deduced from cDNA. FEBS Lett. 1993;315:287–292. doi: 10.1016/0014-5793(93)81180-8. [DOI] [PubMed] [Google Scholar]
- 33.Harris WA, Stark WS, Walker JA. Genetic dissection of the photoreceptor system in the compound eye of Drosophila melanogaster. J Physiol (Lond) 1976;256:415–439. doi: 10.1113/jphysiol.1976.sp011331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heisenberg M. Separation of receptor and lamina potentials in the electroretinogram of normal and mutant Drosophila. J Exp Biol. 1971;55:85–100. doi: 10.1242/jeb.55.1.85. [DOI] [PubMed] [Google Scholar]
- 35.Heisenberg M, Wolf R. Vision in Drosophila. Springer; New York: 1984. [Google Scholar]
- 36.Huber A, Smith DP, Zuker CS, Paulsen R. Opsin of Calliphora peripheral photoreceptors R1–6. Homology with Drosophila Rh1 and posttranslational processing. J Biol Chem. 1990;265:17906–17910. [PubMed] [Google Scholar]
- 37.Huber A, Schulz S, Bentrop J, Groell C, Wolfrum U, Paulsen R. Molecular cloning of Drosophila Rh6 rhodopsin: the visual pigment of a subset of R8 photoreceptor cells. FEBS Lett. 1997;406:6–10. doi: 10.1016/s0014-5793(97)00210-x. [DOI] [PubMed] [Google Scholar]
- 38.Jacobs GH. , series, author. Comparative color vision. In: Carterette EC, Friedman MP, editors. Academic Press series in cognition and perception. Academic; New York: 1981. [Google Scholar]
- 39.Karess R, Rubin G. Analysis of P transposable element functions in Drosophila. Cell. 1984;38:135–146. doi: 10.1016/0092-8674(84)90534-8. [DOI] [PubMed] [Google Scholar]
- 40.Kinoshita M, Shimada N, Arikawa K. Colour vision of the foraging swallowtail butterfly Papilio xuthus. J Exp Biol. 1999;202:95–102. doi: 10.1242/jeb.202.2.95. [DOI] [PubMed] [Google Scholar]
- 41.Kirschfeld K, Franceschini N, Minke B. Evidence for a sensitising pigment in fly photoreceptors. Nature. 1977;269:386–390. doi: 10.1038/269386a0. [DOI] [PubMed] [Google Scholar]
- 42.Kirschfeld K, Feiler R, Franceschini N. A photostable pigment within the rhabdomeres of fly photoreceptors no. 7. J Comp Physiol [A] 1978;125:275–284. [Google Scholar]
- 43.Kitamoto J, Sakamoto K, Ozaki K, Mishina Y, Arikawa K. Two visual pigments in a single photoreceptor cell: identification and histological localization of three mRNAs encoding visual pigment opsins in the retina of the butterfly Papilio xuthus. J Exp Biol. 1998;201:1255–1261. doi: 10.1242/jeb.201.9.1255. [DOI] [PubMed] [Google Scholar]
- 44.Koutalos Y, Ebrey TG, Tsuda M, Odashima K, Lien T, Park MH, Shimizu N, Derguini F, Nakanishi K, Gilson HR, Honig B. Regeneration of bovine and octopus opsins in situ with natural and artificial retinals. Biochemistry. 1989;28:2732–2739. doi: 10.1021/bi00432a055. [DOI] [PubMed] [Google Scholar]
- 45.Langer H, Schmeinck G, Anton-Erxleben F. Identification and localization of visual pigments in the retina of the moth, Anteraea polyphemus (Insecta, Saturniidae). Cell Tissue Res. 1986;245:81–89. [Google Scholar]
- 46.Laughlin SB. The role of sensory adaptation in the retina. J Exp Biol. 1989;146:39–62. doi: 10.1242/jeb.146.1.39. [DOI] [PubMed] [Google Scholar]
- 47.Marshall NJ, Jones JP, Cronin TW. Behavioral evidence for colour vision in stomatopod crustaceans. J Comp Physiol [A] 1996;179:473–481. [Google Scholar]
- 48.Menne D, Spatz H-C. Colour vision in Drosophila melanogaster. J Comp Physiol [A] 1977;114:301–312. [Google Scholar]
- 49.Menzel R, Muller U. Learning and memory in honeybees: from behavior to neural substrates. Annu Rev Neurosci. 1996;19:379–404. doi: 10.1146/annurev.ne.19.030196.002115. [DOI] [PubMed] [Google Scholar]
- 50.Minke B, Kirschfeld K. The contribution of a sensitizing pigment to the photosensitivity spectra of fly rhodopsin and metarhodopsin. J Gen Physiol. 1979;73:517–540. doi: 10.1085/jgp.73.5.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Montell C, Jones K, Zuker C, Rubin G. A second opsin gene expressed in the ultraviolet sensitive R7 photoreceptor cells of Drosophila melanogaster. J Neurosci. 1987;7:1558–1566. doi: 10.1523/JNEUROSCI.07-05-01558.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morris A, Bowmaker JK, Hunt DM. The molecular basis of a spectral shift in the rhodopsins of two species of squid from different photic environments. Proc R Soc Lond [Biol] 1993;254:233–240. doi: 10.1098/rspb.1993.0151. [DOI] [PubMed] [Google Scholar]
- 53.Nathans J. Rhodopsin: structure, function, and genetics. Biochemistry. 1992;31:4923–4931. doi: 10.1021/bi00136a001. [DOI] [PubMed] [Google Scholar]
- 54.Nathans J, Thomas D, Hogness DS. Molecular genetics of human color vision: the genes encoding blue, green, and red pigments. Science. 1986a;232:193–202. doi: 10.1126/science.2937147. [DOI] [PubMed] [Google Scholar]
- 55.Nathans J, Piantanida TP, Eddy RL, Shows TB, Hogness DS. Molecular genetics of inherited variation in human color vision. Science. 1986b;232:203–210. doi: 10.1126/science.3485310. [DOI] [PubMed] [Google Scholar]
- 56.O'Tousa JE, Baehr W, Martin RL, Hirsh J, Pak WL, Applebury ML. The Drosophila ninaE gene encodes an opsin. Cell. 1985;40:839–850. doi: 10.1016/0092-8674(85)90343-5. [DOI] [PubMed] [Google Scholar]
- 57.Pak WL. Study of photoreceptor function using Drosophila mutants. In: Breakefield X, editor. Neurogenetics: genetic approaches to the nervous system. Elsevier; North-Holland, NY: 1979. pp. 67–99. [Google Scholar]
- 58.Papatsenko D, Sheng G, Desplan C. A new rhodopsin in R8 photoreceptors of Drosophila: evidence for coordinate expression with Rh3 in R7 cells. Development. 1997;124:1665–1673. doi: 10.1242/dev.124.9.1665. [DOI] [PubMed] [Google Scholar]
- 59.Pearn MT, Randall LL, Shortridge RD, Burg MG, Pak WL. Molecular, biochemical, and electrophysiological characterization of Drosophila norpA mutants. J Biol Chem. 1996;271:4937–4945. doi: 10.1074/jbc.271.9.4937. [DOI] [PubMed] [Google Scholar]
- 60.Popp MP, Grisshammer R, Hargrave PA, Smith WC. Ant opsins: sequences from the Saharan silver ant and the carpenter ant. Invert Neurosci. 1996;1:323–329. doi: 10.1007/BF02211912. [DOI] [PubMed] [Google Scholar]
- 61.Press WH, Teukolsky SA, Vetteringly WT, Flannery BR. Numerical recipes in C, the art of scientific computing, 2nd Edition. Cambridge UP; Cambridge, UK: 1992. [Google Scholar]
- 62.Quinn WG, Harris WA, Benzer S. Conditioned behavior in Drosophila melanogaster. Proc Natl Acad Sci USA. 1974;71:708–712. doi: 10.1073/pnas.71.3.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sakamoto K, Hisatomi O, Tokunaga F, Eguchi E. Two opsins from the compound eye of the crab Hemigrapsus sanguineus. J Exp Biol. 1996;199:441–450. doi: 10.1242/jeb.199.2.441. [DOI] [PubMed] [Google Scholar]
- 64.Schwemer J, Paulsen R. Three visual pigments in Deilephila elpenor (Lepidoptera, Sphingidae). J Comp Physiol [A] 1973;86:215–229. [Google Scholar]
- 65.Smakman JG, Stavenga DG. Spectral sensitivity of blowfly photoreceptors: dependence on waveguide effects and pigment concentration. Vision Res. 1986;26:1019–1025. doi: 10.1016/0042-6989(86)90036-2. [DOI] [PubMed] [Google Scholar]
- 66.Smith WC, Price DA, Greenberg RM, Battelle BA. Opsins from the lateral eyes and ocelli of the horseshoe crab, Limulus polyphemus. Proc Natl Acad Sci USA. 1993;90:6150–6154. doi: 10.1073/pnas.90.13.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smith WC, Ayers DM, Popp MP, Hargrave PA. Short wavelength-sensitive opsins from the Saharan silver and carpenter ants. Invert Neurosci. 1997;3:49–56. doi: 10.1007/BF02481714. [DOI] [PubMed] [Google Scholar]
- 68.Smola U, Meffert P. The spectral sensitivity of the visual cells R7 and R8 in the eye of the blowfly Calliphora erythrocephala. J Comp Physiol [A] 1979;133:41–52. [Google Scholar]
- 69.Spatz HC, Emanns A, Reichert H. Associative learning of Drosophila melanogaster. Nature. 1974;248:359–361. doi: 10.1038/248359a0. [DOI] [PubMed] [Google Scholar]
- 70.Stavenga DG. Pigments in compound eyes. In: Stavenga DG, Hardie RC, editors. Facets of vision. Springer; Berlin: 1989. pp. 152–172. [Google Scholar]
- 71.Stavenga DG. Eye regionalization and spectral tuning of retinal pigments in insects. Trends Neurosci. 1992;15:213–218. doi: 10.1016/0166-2236(92)90038-a. [DOI] [PubMed] [Google Scholar]
- 72.Stavenga DG, Schwemer J. Visual pigments of invertebrates. In: Ali MA, editor. Photoreception and vision in invertebrates. Plenum; New York: 1984. pp. 12–61. [Google Scholar]
- 73.Stavenga DG, Smits RP, Hoenders BJ. Simple exponential functions describing the absorbance bands of visual pigment spectra. Vision Res. 1993;33:1011–1017. doi: 10.1016/0042-6989(93)90237-q. [DOI] [PubMed] [Google Scholar]
- 74.Suzuki T, Uji K, Kito Y. Studies on cephalopod rhodopsin: photoisomerization of the chromophore. Biochim Biophys Acta. 1976;428:321–338. doi: 10.1016/0304-4165(76)90040-4. [DOI] [PubMed] [Google Scholar]
- 75.Swofford DL. PAUP*. Phylogenetic analysis using parsimony (*and other methods), Version 4. Sinauer; Sunderland, MA: 1998. [Google Scholar]
- 76.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Towner P, Harris P, Wolstenholme AJ, Hill C, Worm K, Gartner W. Primary structure of locust opsins: a speculative model which may account for ultraviolet wavelength light detection. Vision Res. 1997;37:495–503. doi: 10.1016/s0042-6989(96)00198-8. [DOI] [PubMed] [Google Scholar]
- 78.Townson SM, Chang BS, Salcedo E, Chadwell LV, Pierce NE, Britt SG. Honeybee blue- and ultraviolet-sensitive opsins: cloning, heterologous expression in Drosophila, and physiological characterization. J Neurosci. 1998;18:2412–2422. doi: 10.1523/JNEUROSCI.18-07-02412.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Treisman JE, Heberlein U. Eye development in Drosophila: formation of the eye field and control of differentiation. Curr Top Dev Biol. 1998;39:119–158. doi: 10.1016/s0070-2153(08)60454-8. [DOI] [PubMed] [Google Scholar]
- 80.Troje N. Spectral categories in the learning behavior of blowflies. Z Naturforsch [C] 1993;48:96–104. [Google Scholar]
- 81.Washburn T, O'Tousa JE. Molecular defects in Drosophila rhodopsin mutants. J Biol Chem. 1989;264:15464–15466. [PubMed] [Google Scholar]
- 82.Wolff T, Ready DF. Pattern formation in the Drosophila retina. In: Bate M, Arias AM, editors. The development of Drosophila melanogaster. Cold Spring Harbor Laboratory; Plainview, NY: 1993. pp. 1277–1325. [Google Scholar]
- 83.Zeiger J, Goldsmith TH. Behavior of crayfish rhodopsin and metarhodopsin in digitonin: the 510 and 562 nm “visual pigments” are artifacts. Vision Res. 1994;34:2679–2688. doi: 10.1016/0042-6989(94)90224-0. [DOI] [PubMed] [Google Scholar]
- 84.Zuker C, Montell C, Jones K, Laverty T, Rubin G. A rhodopsin gene expressed in photoreceptor cell R7 of the Drosophila eye: homologies with other signal-transducing molecules. J Neurosci. 1987;7:1550–1557. doi: 10.1523/JNEUROSCI.07-05-01550.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zuker CS. The biology of vision of Drosophila. Proc Natl Acad Sci USA. 1996;93:571–576. doi: 10.1073/pnas.93.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zuker CS, Cowman AF, Rubin GM. Isolation and structure of a rhodopsin gene from D. melanogaster. Cell. 1985;40:851–858. doi: 10.1016/0092-8674(85)90344-7. [DOI] [PubMed] [Google Scholar]