Abstract
Protein 4.1N is a neuronal selective isoform of the erythrocyte membrane cytoskeleton protein 4.1R. In the present study, we demonstrate an interaction between 4.1N and nuclear mitotic apparatus protein (NuMA), a nuclear protein required for mitosis. The binding involves the C-terminal domain of 4.1N. In PC12 cells treatment with nerve growth factor (NGF) elicits translocation of 4.1N to the nucleus and promotes its association with NuMA. Specific targeting of 4.1N to the nucleus arrests PC12 cells at the G1 phase and produces an aberrant nuclear morphology. Inhibition of 4.1N nuclear translocation prevents the NGF-mediated arrest of cell division, which can be reversed by overexpression of 4.1N. Thus, nuclear 4.1N appears to mediate the antiproliferative actions of NGF by antagonizing the role of NuMA in mitosis.
Keywords: 4.1N, nuclear mitotic apparatus protein (NuMA), nerve growth factor (NGF), nuclear translocation, G1 phase arrest, mitosis
Protein 4.1 is a cytoskeletal protein most extensively studied in red blood cells where it interacts with spectrin, actin, band 3, and glycophorin C (Anderson and Marchesi, 1985; Pasternack et al., 1985; Correas et al., 1986a,b; Lombardo et al., 1992; Gascard and Cohen, 1994; Hemming et al., 1994; Hemming et al., 1995). Patients with hereditary elliptocytosis caused by 4.1 deficiency display major abnormalities in red cell function, emphasizing the importance of 4.1 for cellular integrity (Conboy et al., 1991; Walensky et al., 1998a). Although the great bulk of 4.1 research has been confined to red blood cells, 4.1 has been detected in other tissues, including the brain (Baines and Bennett, 1985; Krebs et al., 1987; Sihag et al., 1994; Walensky et al., 1998b). Recently, we discovered a distinct gene for a novel form of 4.1 expressed selectively in the nervous system. Like the erythroid form 4.1 (4.1R), the nervous system form of 4.1, designated 4.1N, is non-nuclear and predominantly associated with the neuronal plasma membrane (Walensky et al., 1999).
Nuclear mitotic apparatus protein (NuMA) was originally identified as a non-histone protein that leaves the nucleus at mitosis and became associated with the poles of the mitotic spindle (Lydersen and Pettijohn, 1980). Several lines of evidence suggest that NuMA plays a major role in organizing the spindle apparatus during mitosis and nuclear reassembly at the end of mitosis (Price and Pettijohn, 1986;Compton et al., 1992; Yang and Snyder, 1992; Kallajoki et al., 1993; He et al., 1995). In the present study, we demonstrate protein–protein interactions between 4.1N and NuMA. Independently, Mattagajasingh et al. (1999) have observed binding of 4.1R to NuMA.
In PC12 cells, the neurotrophic actions of nerve growth factor (NGF) are accompanied inhibition of proliferation elicited by cell arrest in the G1 phase of the cell cycle (Rudkin et al., 1989; Yan and Ziff, 1995; van Grunsven et al., 1996a,b). Molecular mechanisms accounting for this G1 phase arrest have been obscure. Here we demonstrate that treatment with NGF translocates 4.1N to the nucleus in association with NuMA. Moreover, we show that inhibition of this translocation prevents the NGF-mediated arrest of cell division, which can be reversed by overexpression of 4.1N. Thus, NGF-induced translocation of 4.1N into the nucleus appears to prevent the role of NuMA role in mitosis, indicating that 4.1N mediates the antiproliferative actions of NGF.
MATERIALS AND METHODS
Materials. PC12 cells were maintained in DMEM with 10% fetal bovine serum (FBS), 5% horse serum, and 100 U of penicillin-streptomycin at 37°C with 5% CO2atmosphere in a humidified incubator. For immunofluorescent staining, PC12 cells were anchored onto poly-d-lysine-treated coverslips. Differentiation was initiated by addition of 50 ng/ml NGF with culture medium changed to DMEM with 2% horse serum and 1% FBS. NGF, epidermal growth factor (EGF), 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside, and isopropyl-1-thio-β-d-galactopyranoside were purchased from Boehringer Mannheim (Indianapolis, IN). Mouse monoclonal anti-NuMA, anti-hemagglutinin (HA), anti-Myc, and rabbit polyclonal anti-HA antibodies were supplied by Calbiochem (La Jolla, CA). Protein A-conjugated agarose beads was also from Calbiochem. Mouse monoclonal anti-cyclin D1, cyclin B1, and proliferating cell number antigen (PCNA) were supplied by Santa Cruz Biotechnology (Santa Cruz, CA). Mouse monoclonal T7.Tag antibody was from Novagen (Madison, WI). Protein concentration was determined by the Bradford method. Plasmids were purified using a Maxiprep kit from Qiagen (Hilden, Germany). −Leu DO supplement, −Leu/−Trp/−His DO supplement, minimal SD agar base, and minimal SD base for yeast two-hybrid screen were from Clontech (Cambridge, UK). Glutathione-Sepharose 4B was supplied by Amersham Pharmacia Biotech (Uppsala, Sweden). Vectorshield mounting medium was from Vector Laboratories (Burlingame, CA). All chemicals not included above were purchased from Sigma (St. Louis, MO).
Yeast two-hybrid screen. Two-hybrid screening was conducted using Y190 yeast strain containing the HIS3 and β-galactosidase (β-gal) reporter genes and the pPC97 and pPC86 expression vectors. The C-terminal domain of 4.1N (4.1N CTD) was cloned into yeast expression vector pPC97 (containing the GAL4 DNA-binding domain) as a bait. This was used to screen a rat hippocampal and cortex cDNA library cloned into pPC86 (Li and Snyder, 1995; Lai et al., 1998), containing the GAL4 transcription domain. The 4.1N CTD plasmid was transformed into yeast using the lithium acetate-polyethylene glycol method (Ausubel et al., 1990). The transformation of the hippocampal and cortex cDNA library into yeast expressing the GAL4–4.1N CTD fusion was performed essentially as described (Walensky et al., 1998a). A total of 2 × 106 independent clones were screened, and positive interactive proteins were identified by selecting for His+ growth phenotype. Positive clones were further evaluated for β-gal expression by nitrocellulose filter lift assay as described (Walensky et al., 1998a). The plasmid was isolated from a colony displaying β-gal activity using glass beads, transformed into bacteria by electroporation, and then DNA sequenced.
In vitro binding assays. GlutathioneS-transferase (GST) fusion proteins were prepared according to the manufacturer's recommendations (Amersham Pharmacia Biotech) and coupled to glutathione-Sepharose beads. Twenty-four hours after transfection with 10 μg of human NuMA cDNA, a 10 cm plate of HEK293 cells was washed once in PBS, lysed in 1 ml of lysis buffer A [50 mm Tris, pH 7.4, 40 mmNaCl, 1 mm EDTA, 0.5% Triton X-100, 1.5 mmNa3VO4, 50 mm NaF, 10 mm sodium pyrophosphate, 10 mm sodium β-glycerophosphate, 1 mm phenylmethylsulfonyl fluoride (PMSF), 5 mg/ml aprotinin, 1 mg/ml leupeptin, and 1 mg/ml pepstatin A], was centrifuged for 10 min at 14,000 × g at 4°C. From this, 500 μl of supernatant was added to 50 μl of GST, GST-4.1N-N-terminal domain (NTD), or GST-4.1N CTD agarose, incubated with slow rotation for 1 hr, and washed three times with 500 μl of lysis buffer each time. The agarose then was resuspended in 30 μl of sample buffer separated by SDS-PAGE followed by immunoblot using the rabbit polyclonal anti-NuMA antibody with 1:2000 dilution. Equal loading of GST or GST fusion proteins was confirmed with Coomassie blue staining. His-tagged NuMA and FKBP12 were purified according to the manufacturer's recommendations (Novagen). Purified GST-4.1N CTD was coupled to glutathione-Sepharose beads and respectively incubated with His-NuMA, His-FKBP12, and bacterial lysate at 4°C for 2 hr; after extensive washing, the agarose then was resuspended in 30 μl of sample buffer separated by SDS-PAGE followed by immunoblot using the mouse monoclonal anti-T7-Tag antibody with 1:2000 dilution.
Coimmunoprecipitation of 4.1 and NuMA from transfected 293 cells. Ten centimeter dishes of HEK293 cells were cotransfected with 5 μg each of HA-NuMA (plasmid encoded a 473 amino acid peptide with sequence homology to the human NuMA 1440–1913) and myc-4.1N CTD, HA-NuMA and myc-4.1N NTD, myc-NuMA (amino acids 1440–1913) and HA-4.1N CTD, myc-NuMA and HA-4.1N NTD by the calcium phosphate precipitation method. The supernatant was prepared as above. After normalizing the protein concentration, 2 μl of anti-myc antibody and 40 μl of 50% slurry protein G-agarose (Calbiochem) were added to the supernatant and incubated with rotation at 4°C for 3 hr. The agarose pellet was washed three times with 500 μl of lysis buffer each time. The agarose then was resuspended in 30 μl of sample buffer separated by SDS-PAGE followed by immunoblot using anti-HA antibody with 1:2000 dilution. For the cotransfection of human full-length NuMA cDNA and 4.1N full-length cDNA and human full-length NuMA cDNA and myc-NLS-4.1N full-length cDNA, 2 μl of rabbit polyclonal anti-NuMA antibody was used and immunoblotted with anti-4.1 antibody. The protein expression levels were confirmed by immunoblot with anti-HA and -4.1 antibodies.
Coimmunoprecipitation of 4.1 and NuMA from PC12 cells. PC12 cells were treated with 50 ng/ml NGF in DMEM with 1% FBS, 2% horse serum, and 100 U of penicillin-streptomycin at the indicated times, whereupon cells were washed twice before lysis in 500 μl of lysis buffer A. After centrifugation for 5 min at 13,000 × gin a bench-top centrifuge, the supernatant was mixed with 1 μl of rabbit polyclonal anti-NuMA antibody and 40 μl of 50% slurry protein A-agarose (Calbiochem), followed by incubation with rotation at 4°C for 3 hr. The agarose pellet was washed three times with 500 μl of lysis buffer each time. The agarose then was resuspended in 30 μl of sample buffer and separated by SDS-PAGE followed by immunoblot using anti-4.1 antibody with 1:5000 dilution.
Isolation of nuclear fraction. The PC12 cell nuclei were isolated essentially as previously described (Rosenberg, 1996; Sawa et al., 1997) with minor modifications. Briefly, 1 × 108 cells were rinsed with PBS, removed from the dish with a cell scraper, and collected by centrifugation (1500 rpm for 10 min). The pellet was resuspended and washed twice with cold PBS and then resuspended in a 5 packed cell volume (PCV) of hypotonic buffer (10 mm HEPES, pH 7.9, 10 mmKCl, 1.5 mm MgCl2, 0.5 mmPMSF, 0.5 μg/ml of leupeptin-pepstatin A-aprotinin, and 1 mm DTT), and the cells were pelleted. The cells were swollen by resuspending in 2 PCV of hypotonic buffer and incubated on ice for 10–15 min, followed by homogenization using a Dounce glass homogenizer with a loose-fitting pestle. Cell lysis was monitored microscopically. The homogenates were centrifuged through a 1.5 ml cushion of buffer (10 mm HEPES-KOH, pH 7.6, 2.4m sucrose, 15 mm KCl, 2 mm EDTA, 1 mm DTT, and 0.5 mm PMSF) in a Beckman Instruments Inc. (Palo Alto, CA) SW 70.1 Ti rotor at 25,000 rpm for 30 min at 4°C. The transparent supernatant above the cushion is the non-nuclear fraction. The nuclear pellet was resuspended in lysis buffer A and incubated for 10 min on ice, and the lysate was centrifuged at 13,000 × g for 5 min at 4°C. After normalizing the protein concentration of the supernatant, the lysate was boiled in SDS sample buffer. SDS-PAGE and Western blotting were performed as described above.
Transient transfection. Transfection was performed according to the manufacturer's protocol (Life Technologies, Gaithersburg, MD). Briefly, PC12 cells were plated on a poly-d-lysine-treated coverslip in a six-well tissue culture plate 1 d before the transfection. The cells were incubated until they achieved 50% confluency. For each transfection, 2 μg of plasmid DNA was diluted into 100 μl of serum free OptiMEM-I medium; then 6 μl PLUS reagent was added to the DNA solution, and the mixture was incubated at room temperature for 15 min. For each transfection, LipofectAMINE (4 μl) was mixed with 100 μl of OptiMEM-I, which was added to the DNA-PLUS mixture, mixed gently, and incubated at room temperature for another 15 min. The cells were washed once with the serum-free OptiMEM-I medium, to which 0.8 ml of OptiMEM-I was added and put back in the incubator. After 15 min, the 0.2 ml DNA-PLUS-LipofectAMINE solution was removed to the cells. After 3 hr of incubation, the transfection mixture was replaced with 2 ml of growth medium. After overnight recovery, the cells were either differentiated by 50 ng/ml NGF or left untreated.
Immunocytochemistry. Fixation, permeabilization, blocking, and PBS washes were all performed as described (Ye et al., 1998). Briefly, PC12 cells growing on the poly-d-lysine-treated coverslips were fixed by immersion in 3.7% formaldehyde in PBS for 10 min at room temperature. The fixed cells were then permeabilized for 5 min with 0.5% Triton X-100 in PBS. The cells were then rinsed with PBS three times at room temperature. The cells were blocked by incubation in 10% FBS-PBS for 10 min and rinsed twice with PBS. Subsequent antibody incubations were performed at 37°C in a humid chamber. Cells were incubated in mouse monoclonal anti-NuMA antibody diluted 1:100 in 1% bovine serum albumin (BSA)-PBS for 1 hr and rinsed in BSA-PBS for three times for 10 min each at room temperature. The cells were incubated 40 min with Texas Red-conjugated goat anti-mouse secondary antibody (Jackson ImmunoResearch, West Grove, PA) diluted 1: 125 in BSA-PBS and rinsed in BSA-PBS three times for 10 min each at room temperature. Rabbit polyclonal anti-4.1N antibody diluted 1:400 or rabbit polyclonal anti-HA antibody diluted 1:200 were used to stain the cells for another 1 hr and rinsed in BSA-PBS three times for 10 min each at room temperature. Then the cells were incubated with fluorescein isothiocyanate (FITC) donkey anti-rabbit secondary antibody (Jackson ImmunoResearch) diluted 1:125 for 40 min and rinsed in BSA-PBS three times for 10 min each at room temperature. DNA was detected with 4′,6-diamino-2-phenylindole (DAPI, 0.6 μg/ml; Sigma). Coverslips were mounted with Vectashield mounting medium for fluorescence (Vector Laboratories) and studied under the confocal microscope.
Cyclin D1 staining. Exponentially growing PC12 cells, NGF-treated 2 d PC12 cells, and HA-NLS-4.1N-, HA-NLS-4.1N CTD-, and HA-4.1N NTD-transfected PC12 cells were fixed, permeabilized, and blocked as described above. Mouse monoclonal antibody cyclin D1 was used at 1:200 dilution in BSA-PBS solution, and the transfected cells were also stained with rabbit polyclonal anti-HA antibody at 1:200 dilution.
RESULTS
Binding of 4.1N to NuMA
The specific domains upstream of the COOH domain of 4.1N share high homology with the corresponding regions of 4.1R, which has been well characterized in terms of its interactions with a variety of cytoskeletal proteins (Walensky et al., 1999). By contrast, the C-terminal region of 4.1N differs considerably from 4.1R, and interactions of the C-terminal domain of 4.1N have not been delineated. Accordingly, we conducted yeast two-hybrid analysis using as bait the C-terminal area (679–879 amino acids) of 4.1N (Table1). Of 2 million transformants in a cDNA library of the rat cerebral cortex and hippocampus, we have identified 28 His-positive clones and 13 that are positive for both His+ and β-gal. One of these is NuMA. Direct examination of interactions of 4.1N and NuMA in yeast two-hybrid analysis reveals strongly positive interactions whether 4.1N is used as bait or prey. By contrast, the N-terminal region (1–321 amino acids) of 4.1N fails to interact with NuMA.
Table 1.
Two-hybrid analysis of 4.1N
| Library, rat cortex and hippocampus | No. transformants (2 × 106) | No. His+(28) | No. His+ and β-gal positive (13) |
|---|---|---|---|
| Binding domain fusion protein (pPC97) | Activation domain fusion protein (pPC86) | Histidine prototrophy | β-Gal filter assay |
| 4.1N (aa 679–879) | NuMA (aa 1440–1913) | + | + |
| NuMA (aa 1440–1913) | 4.1N (aa 679–879) | + | + |
| 4.1N (aa 1–321) | NuMA (aa 1440–1913) | − | − |
| NuMA (aa 1440–1913) | 4.1N (aa 1–321) | − | − |
To explore protein–protein interactions directly, we used GST-linked 4.1N (Fig. 1A). The N- and C-terminal domains of GST-4.1N were attached to glutathione-Sepharose beads, which were incubated with HEK293 cell lysates transfected with human NuMA cDNA. After extensive washing, we conducted a Western blot analysis with anti-NuMA antibody. NuMA binds to the C-terminal domain of GST-4.1N but not to the N-terminal domain, which is consistent with our yeast two-hybrid findings. To further investigate whether this interaction between the two proteins is direct, we purified the C-terminal domain of GST-4.1N, His-tagged NuMA fragment and His-tagged FKBP12, and performed an in vitrobinding assay. The C-terminal domain of GST-4.1N selectively binds NuMA but not FKBP12 directly (Fig. 1B).
Fig. 1.
4.1 and NuMA associate in vitro andin vivo. A, In vitro binding of human NuMA to GST-4.1 CTD. Lysates from HEK 293 cells transfected with full-length human NuMA cDNA were incubated with GST, GST-4.1N NTD, or GST-4.1N CTD. Bound proteins were visualized by Western immunoblotting with anti-NuMA antibody. B, In Vitrodirect binding of His-tagged NuMA fragment to GST-4.1N CTD. The purified recombinant His-tagged NuMA fragment, His-tagged FKBP12 proteins, and bacterial lysates were respectively incubated with purified GST-4.1N CTD at 4°C for 2 hr, and the binding proteins were visualized by Western immunoblotting with anti-T7.Tag antibody. C, Coimmunoprecipitation of NuMA (amino acids 1440–1913) with 4.1N CTD. HEK293 cells were respectively cotransfected with Myc- or HA-tagged NuMA and 4.1N CTD or 4.1N NTD. After immunoprecipitation with anti-myc antibody, bound proteins were visualized by Western blotting with anti-HA antibody. D, Similar levels of transfected HA- or Myc-tagged NuMA, 4.1N NTD and 4.1N CTD in 293 cell lysate were confirmed by Western blot with anti-HA antibody.
To ascertain whether NuMA and 4.1N interacts in intact cells, we used immunoprecipitation experiments (Fig. 1C,D). NuMA and the C-terminal or N-terminal domain of 4.1N were tagged with Myc or HA and immunoprecipitated with anti-Myc, and Western blot analysis of SDS-PAGE was conducted with anti-HA antibody. NuMA coprecipitates with the C-terminal but not the N-terminal domain of 4.1. The same results are obtained regardless of whether NuMA or 4.1N is tagged with HA or Myc. Western blot analysis of the HEK-293 cell lysates with antibodies to HA-NuMA, the N-terminal domain of 4.1N, or the C-terminal domain of 4.1N shows that all the transfected proteins are equally expressed.
Nerve growth factor causes 4.1N to translocate to the nucleus
To evaluate the localizations of 4.1N and NuMA in PC12 cells, we conducted immunofluorescent studies using confocal microscopy (Fig.2). In the absence of NGF treatment, although very weak staining of protein 4.1N appears in nuclei, 4.1N localizations are most pronounced at the periphery of cells. By contrast, NuMA occurs in the nucleus, whose morphology and localization are confirmed by DAPI staining (data not shown). In some cells undergoing mitosis, NuMA staining is associated with the spindle pole bodies (Fig. 2B). Overlay analysis reveals no coincidence for 4.1N or NuMA staining in the absence of NGF (Fig.2C).
Fig. 2.
4.1N colocalizes with NuMA in the nucleus of NGF-differentiated PC12 cells but not in untreated PC12 cells. Exponentially growing PC12 cells (A–C) and NGF (50 ng/ml)-treated and differentiated PC12 cells (D–F) were double-labeled with mouse monoclonal anti-NuMA antibody and rabbit polyclonal anti-4.1N antibody. The secondary antibodies were Texas Red-conjugated goat anti-mouse antibody (NuMA, red) and FITC-conjugated donkey anti-rabbit antibody (4.1N, green). 4.1N colocalizes with NuMA in the differentiated PC12 cells (F,yellow-stained nuclei) but not in exponentially growing cells. Scale bar, 20 μm.
Treatment with NGF (50 ng/ml) for 2 d leads to substantial sprouting of neurites. In contrast to the non-nuclear staining of 4.1N in untreated cells, NGF-treated cells manifest green 4.1N staining throughout the processes and their terminals as well as in the cell body including the nucleus (Fig. 2D). After NGF treatment, red NuMA staining remains associated with the nucleus, with an absence of staining in the nucleolus (Fig. 2E). Overlay analysis reveals yellow staining of the nucleus, indicating that NGF has elicited translocation of 4.1N to the nucleus in apparent colocalization with NuMA (Fig. 2F). Furthermore, time course experiments suggest that the 4.1N nuclear translocation persists as long as the cells are maintained in NGF. In addition, 4.1N nuclear translocation was also detected in sympathetic neurons after NGF treatment (data not shown).
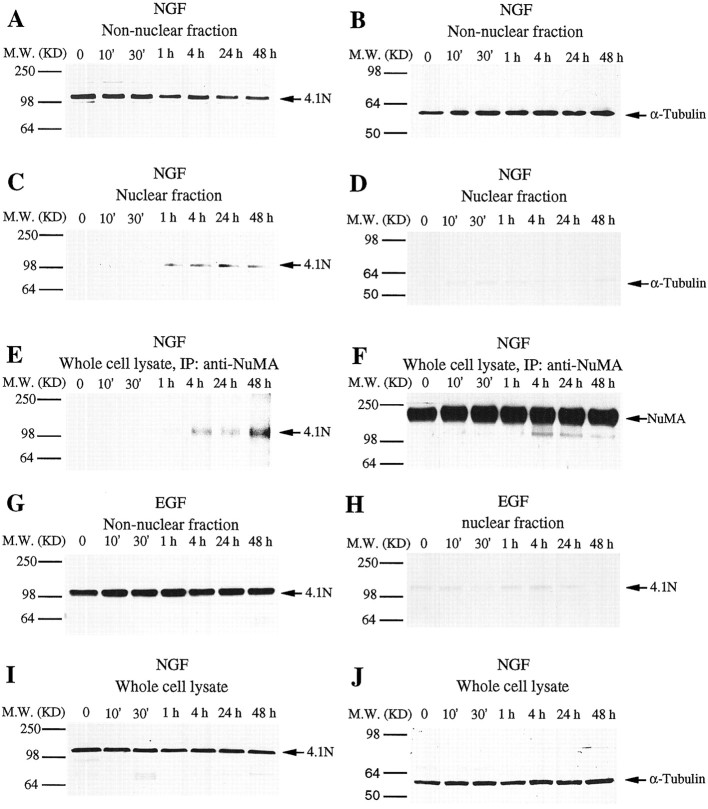
We used biochemical experiments to explore further the NGF-induced translocation of 4.1N to the nucleus (Fig.3). NGF treatment produces a time-dependent decrease in levels of 4.1N in non-nuclear areas accompanied by appearance of 4.1N in nuclear fractions (Fig.3A,C). 4.1N is first demonstrable in the nucleus at 1 hr and remains there at 4, 24, and 48 hr. In the nucleus, 4.1N is associated with NuMA, because immunoprecipitation with rabbit polyclonal anti-NuMA antibody also reveals a progressive increase of coprecipitated 4.1N in nuclear fractions between 4 and 48 hr after NGF treatment (Fig. 3E). At all time points the same amount of NuMA is immunoprecipitated (Fig. 3F). Selectivity for the translocation of 4.1N is evident by the absence of translocation for α-tubulin after NGF treatment (Fig.3B,D). In whole-cell extracts, no change in the total level of 4.1N or α-tubulin is evident after NGF treatment, establishing that NGF elicits translocation of 4.1N rather than a change in overall biosynthesis (Figure, 3I,J).
Fig. 3.
4.1N translocates from plasma membrane to nucleus and binds NuMA in PC12 cells in response to NGF treatment. After NGF (50 ng/ml) treatment, 4.1N concentration decreases gradually in non-nuclear fractions (A) but increases in nuclear fractions (C). As a control, the concentration of α-tubulin is unchanged in non-nuclear fractions (B) and remains undetectable in all nuclear fractions (D). E, 4.1N coimmunoprecipitates with NuMA in response to NGF treatment. PC12 cells were treated with 50 ng/ml NGF, and at the indicated times cells were lysed and immunoprecipitated (IP) with anti-NuMA antibody. Coprecipitated 4.1N was detected by Western blotting as described. F, Western blot of NuMA shows that same amount of the NuMA protein was immunoprecipitated in each lane.G, EGF (50 ng/ml) treatment of PC12 cells does not alter 4.1N levels in non-nuclear fractions. H, 4.1N remains undetectable in nuclear fractions after EGF (50 ng/ml) treatment of PC12 cells. I, NGF treatment does not alter 4.1N expression level in whole-cell lysates. PC12 cells were treated with 50 ng/ml NGF and harvested as previously described. Equal quantities (100 μg) of protein were loaded in each lane, and 4.1N was visualized by Western blotting. As a control, the conentration of α-tubulin is unaltered in response to NGF treatment (J).M.W., Molecular weight.
EGF shares mitogenic and certain signal transduction actions with NGF, but EGF does not display the neurotrophic and antiproliferative effects characteristic of NGF (Cho et al., 1989; Yan et al., 1991; Mothe et al., 1993; Hempstead et al., 1994; Blumberg et al., 1995). EGF fails to elicit translocation of 4.1N to the nucleus (Fig.3G,H), results also obtained by immunofluorescent staining (data not shown).
Nuclear targeting of 4.1N incurs G1 phase arrest
The two principal effects of NGF on PC12 cells are extension of neurites and inhibition of cell proliferation. NuMA plays a major role in the regulation of mitosis (Van Ness and Pettijohn, 1983; Compton et al., 1992; Yang et al., 1992; Yang and Snyder, 1992; Compton and Cleveland, 1993, 1994; Compton and Luo, 1995). Accordingly, we wondered whether the nuclear targeting of 4.1N and its association with NuMA would influence mitotic events. We asked whether specific targeting of 4.1N into the nucleus in the absence of NGF might mimic the antiproliferative actions of NGF. Therefore, we transfected various forms of 4.1N into the nucleus using 4.1N tagged with an HA-labeled nuclear localization signal (NLS) (HA-NLS-4.1N) and evaluated mitotic events in comparison with cells in which full-length 4.1N was transfected without NLS (Table 2). In control PC12 cells not treated with NGF, ∼2.5% of the cells are in mitosis, and 24% are in the G1 phase, illustrated by the nuclear localized G1 phase-specific marker cyclin D1. Transfection of full-length 4.1N without NLS produces no change in these ratios. By contrast, transfection of full-length 4.1N containing NLS increases the percentage of cells in G1 by ∼80% and leads to an absence of any mitotic figures. A similar elimination of mitotic figures and increase of cells in the G1 phase occurs with transfection of the C-terminal domain of NLS-4.1N (HA-NLS-4.1N CTD), consistent with the ability of the C-terminal domain of 4.1N to bind NuMA. However, transfection of the N-terminal domain of NLS-4.1N (HA-NLS-4.1N NTD) fails to inhibit mitosis, which accords with the inability of the N-terminal domain of 4.1N to bind NuMA. Although the N-terminal domain of 4.1 strongly binds to the plasma membrane, the HA-NLS-4.1N NTD is indeed targeted to nucleus (data not shown).
Table 2.
Nuclear targeting of 4.1N elicits G1 phase arrest and aberrant nuclear morphology in PC12 cells
| Plasmid | |||||
|---|---|---|---|---|---|
| HA-NLS-4.1N (FL) | HA-NLS-4.1N CTD | HA-NLS-4.1N NTD | 4.1N (FL) | Control | |
| Metaphase (%) | 0 | 0 | 2 ± 1* | 2 ± 1* | 2.5 ± 1* |
| G1 phase (%) | 42 ± 1.8* | 41 ± 3* | 22 ± 4* | 25 ± 5* | 24 ± 5* |
| Aberrant nuclear morphology (%) | 5 ± 1.5* | 5 ± 1.2* | 0 | 0 | 0 |
More than 500 transfected cells were analyzed in each of three independent experiments. Data are presented as the mean ± SEM. Metaphase staging was based on DAPI staining and microtubule morphology. G1 phase was evaluated by cyclin D1 nuclear translocation. Aberrant nuclear morphology was evident as bipartite nuclei with a dumbbell shape.
*p < 0.001.
The antiproliferative effects of NGF in PC12 cells are associated with alterations in disposition of a number of proteins associated with the cell cycle (Buchkovich and Ziff, 1994; Yan and Ziff, 1995; Movsesyan et al., 1996; van Grunsven et al., 1996a,b). One of the most notable of these changes is an increase in nuclear level of cyclin D1 (van Grunsven et al., 1996a). To further evaluate how nuclear targeting of 4.1N might mimic actions of NGF, we stained cells for cyclin D1 (Fig.4). Transfection with full-length NLS-4.1N or its C-terminal domain produces an 80% increase in the percentage of cells staining for nuclear cyclin D1, closely similar to the augmentation elicited by NGF itself. By contrast, transfection with the N-terminal domain of NLS-4.1N fails to produce such an increase.
Fig. 4.

Nuclear targeting of 4.1N arrests PC12 cells at G1 phase. Exponentially growing PC12 cells (A, B), NGF-treated PC12 cells (C, D), and untreated PC12 cells transfected with HA-NLS-4.1N [full length (FL)] (E–H) were stained with cyclin D1 antibody (A, C, F), whereas their nuclei were labeled with DAPI (B, D, G). The transfected cells were also labeled with anti-HA antibody (E). H, In exponentially growing cultures, ∼24% of untreated cells express cyclin D1 in the nucleus compared with 45% in NGF-treated cells. Transfection with HA-NLS-4.1N (FL) and HA-NLS-4.1N CTD causes G1 arrest in >40% of transfected PC12 cells, as indicated by nuclear cyclin D1 staining. As a control, only ∼20% of HA-4.1N NTD-transfected PC12 cells show G1 arrest (H). *p < 0.001.
BrdU incorporation and PCNA accumulation in nucleus are widely used as S phase markers (Moir et al., 1994; Thomaidou et al., 1997; Gould et al., 1998). To further explore the effects of nuclear targeting of 4.1N on cell cycle profile, we respectively stained cells for BrdU incorporation and PCNA nuclear localization. None of the HA-NLS-4.1N-, HA-NLS-4.1N CTD-, or HA-4.1N NTD-transfected cells shows significant difference from the NGF-treated or the control PC12 cells. Presumably, variations in numbers of cells in different stages preclude detection of possible BrdU staining alterations associated with NLS-4.1N transfection.
In cells transfected with full-length NLS-4.1N (HA-NLS-4.1N) or its C-terminal domain (HA-NLS-4.1N CTD), ∼5% of the cells manifest aberrant nuclear morphology (Fig. 5). The nuclei are bipartite with a dumbbell shape (Fig.5D–F). Moreover, cells with the bipartite nuclei are substantially larger than other cells (Fig. 5G–L).
Fig. 5.
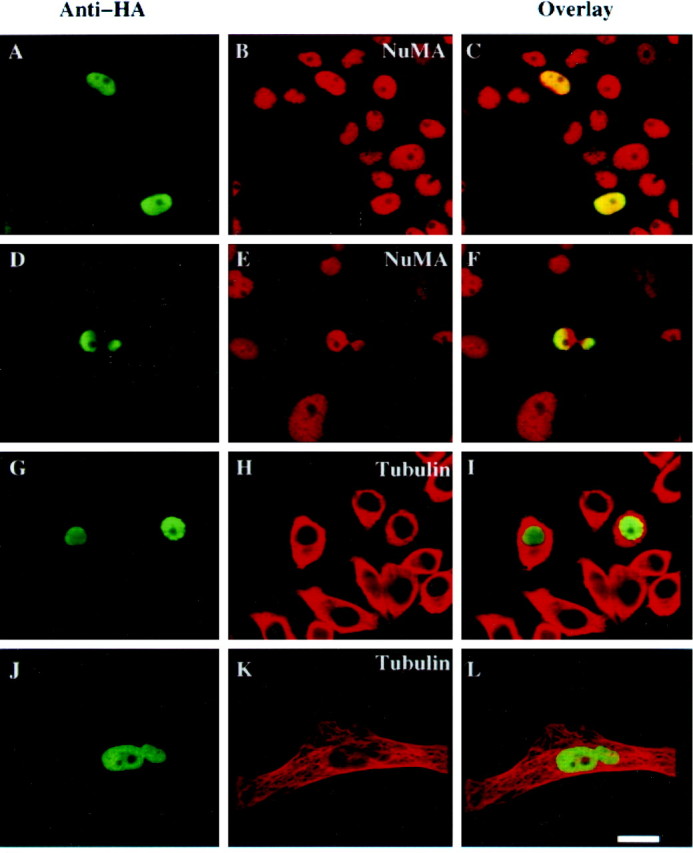
Nuclear targeting of 4.1N alters PC12 nuclear morphology. PC12 cells were transfected with HA-NLS-4.1N (A, D, G, J) and stained with anti-NuMA antibody (B, E) or anti-α-tubulin antibody (H, K), followed by Texas Red-conjugated goat anti-mouse antibody. Transfected cell nuclei were labeled by rabbit polyclonal anti-HA antibody (A, D, G, J). Overlay analysis (C, F, I, L) shows an aberrant, dumbbell-shaped nuclear morphology in ∼5% of the transfected cells. The morphology of microtubules in HA-NLS-4.1N-transfected cells does not differ from the untransfected cells (G–I). Transfected cells with aberrant nuclear morphology (J–L) appear larger than transfected or untransfected cells with normal nuclear morphology (G–I). Scale bar, 10 μm.
Extranuclear NuMA prevents 4.1N translocation to the nucleus and inhibits the antimitogenic effects of NGF
Our findings indicate that NGF triggers translocation of 4.1N to the nucleus associated with the antiproliferative effects of NGF. We reasoned that prevention of 4.1N nuclear translocation should reverse the antiproliferative effect of NGF. We sought to prevent 4.1N nuclear translocation by selectively overexpressing NuMA in the cytoplasm so that it might bind endogenous 4.1N and prevent its nuclear translocation after NGF treatment. To keep NuMA out of the nucleus, we used constructs of NuMA lacking NLS, which has been shown to accumulate in the cytoplasm as a large mass approximately the same size as the cell nucleus during interphase, when it is expressed in BHK-21 cells. When cells enter mitosis, NuMA lacking NLS associates normally with the mitotic spindle without causing any apparent deleterious effects on the progression of mitosis (Saredi et al., 1996). Transfection of cells with NuMA lacking NLS results in intense densities of extranuclear NuMA colocalized with 4.1N (Fig.6A–C). We observe the same extranuclear concentrations of NuMA together with 4.1N in the absence and presence of NGF treatment. Thus, extranuclear NuMA markedly diminishes nuclear localizations of 4.1N (Fig.6D–F).
Fig. 6.

Extranuclear NuMA prevents 4.1N translocation to the nucleus and inhibits the antimitogenic effects of NGF, which can be reversed by overexpression of 4.1N. PC12 cells were transfected with NuMA lacking a nuclear localization signal (NuMA-ΔNLS) and then stained with 4.1N (A, D) and NuMA (B, E) antibodies. In the absence of NGF, NuMA-ΔNLS accumulates in the transfected cell cytoplasm (B, red circleabove rednucleus with unstained nucleoli) and colocalizes with endogenous 4.1N (C, yellow circle). In the presence of NGF, ∼6% of the NuMA-ΔNLS-transfected PC12 cells show two duplicated and separated nuclei (E). Transfected NuMA colocalizes with endogenous 4.1N in the NGF differentiated cells (F). To reverse the effects of NuMA-ΔNLS, PC12 cells were cotransfected with HA-4.1N and NuMA-ΔNLS and then stained with anti-HA (G) and anti-NuMA (H) antibodies. In the presence of NGF, transfected NuMA-ΔNLS accumulates outside the nucleus (H) and colocalizes with transfected 4.1N (I). The transfected HA-4.1N appears in the nucleus (G, I). Duplicated and segregated nuclei in one differentiated cell phenotype are never observed. Scale bar, 15 μm.
In control cells in the absence of NGF, ∼2.5% of cells display mitotic figures, whereas NGF treatment virtually abolishes such figures. Transfection of NuMA without NLS dramatically reverses the NGF effects on mitotic figures (Table 3). Most of the cells transfected with NLS-deficient NuMA display substantial neurite extension with NuMA accumulated in the cytoplasm. Strikingly, ∼6% of the transfected cells contain two duplicated and segregated nuclei after NGF treatment (Fig.6D–F).
Table 3.
Extranuclear NuMA prevents antimitogenic effects of NGF
| Events | Mitotic figures (%) | |
|---|---|---|
| Control − NGF | 2.5 ± 0.8* | |
| Control + NGF | 0.2 ± 0.1* | |
| NuMA-NLS − NGF | 3 ± 0.5* | |
| NuMA-NLS + NGF | 6 ± 1.2* (2 nuclei in 1 cell) | |
| NuMA-NLS + HA-4.1N + NGF | 0 (2 nuclei in 1 cell) | |
More than 500 transfected cells were analyzed in each of the three independent experiments. Data are presented as the mean ± SEM. Mitotic cells were identified by DAPI staining and NuMA translocation to spindle pole bodies. Most of the cells transfected with NLS-deficient NuMA displayed substantial neurite extension with NuMA accumulated in the cytoplasm. Approximately 6% of the transfected cells contained two duplicated and segregated nuclei after NGF treatment. Overexpression of 4.1N reversed these effects and restored the antimitogenic actions of NGF.
*p < 0.001.
We postulate that the extranuclear NuMA stimulates “mitosis” by preventing nuclear translocation of 4.1N. If this is the case, then overexpression of 4.1N might be expected to overcome these effects and restore the antimitogenic actions of NGF. Accordingly, we transfected cells with HA-4.1N in addition to NuMA lacking NLS. This treatment restores the cell cycle-arresting influences of NGF. Immunofluorescent staining shows that a substantial portion of the HA-4.1N enters the nucleus (Fig. 6G–I). To ensure that the actions of HA-4.1N are caused by 4.1N and not HA, we performed similar experiments using the C- and N-terminal domains for HA-4.1N. The C-terminal domain of HA-4.1N, like full-length 4.1N, restores the cell cycle-arresting actions of NGF, whereas the N-terminal domain has no effect. This result also indicates that the HA-4.1 exerts its effects by binding NuMA, because the C-terminal but not the N-terminal domain of 4.1N binds NuMA.
DISCUSSION
In the present study, we have demonstrated an interaction between 4.1N and NuMA using the yeast two-hybrid technique as well as several other procedures. Independently, Mattagajasingh et al. (1999)observed binding of 4.1R to NuMA. In both cases, the binding involves the C-terminal domain of 4.1. Thus, though the C-terminal domain differs ∼50% between 4.1R and 4.1N, both forms of the protein contain the essential elements that permit interactions with NuMA.
We observed that NGF elicits translocation of 4.1 to the nucleus, findings established by both immunofluorescent staining and subcellular fractionation. 4.1N was phosphorylated on tyrosine residue in response to NGF treatment, and the tyrosine phosphorylation was identified 10 min after NGF treatment (data not shown). It is possible that the Trk receptor directly phosphorylates 4.1N on its tyrosine group and mediates its translocation into nucleus.
The nuclear translocation of 4.1N appears to mediate antiproliferative effects of NGF. Thus, transfection of 4.1N with a nuclear localization signal mimics effects of NGF. In these experiments we observe aberrant nuclear morphology, involving bipartite nuclei in ∼5% of cells. Molecular mechanisms to account for these effects are unclear. However, NuMA has been suggested to play a role in nuclear reassembly at the end of mitosis (Price and Pettijohn, 1986; Compton et al., 1992;). In addition, NuMA occurs in a filamentous network in the interphase nucleus and may regulate the morphology of the nuclear cytoskeleton (Zeng et al., 1994; Merdes and Cleveland, 1998). Presumably, overexpression of 4.1N interferes with the normal cytoskeletal actions of NuMA in the nucleus.
We provided further evidence for the role of 4.1N in antiproliferative effects of NGF by showing that extranuclear overexpression of NuMA keeps 4.1N out of the nucleus and prevents the antimitogenic effects of NGF. Interestingly, preventing nuclear translocation of 4.1N does not block the ability of NGF to promote neurite extension. Thus, we observed that many cells contain duplicated and segregated nuclei but with substantial neurite expression, something that is unprecedented for NGF actions. These experiments dissociate the neurotrophic and antiproliferative effects of NGF, consistent with previous findings (Greene and Tischler, 1976; Ignatius et al., 1985; Rudkin et al., 1989;Yan and Ziff, 1995).
To establish that overexpression of extranuclear NuMA blocks the antimitogenic effects of NGF by preventing 4.1 entry into the nucleus, we showed that overexpressing 4.1N can restore its nuclear localization and reestablish the antimitogenic effects of NGF.
Our findings establish a major role for 4.1N in mediating NGF actions. By translocating to the nucleus and binding NuMA, 4.1N prevents the mitotic actions of NuMA, enabling the antiproliferative actions of NGF. Thus, we think that our findings establish 4.1N as a mediator of the antimitogenic influences of NGF.
4.1N is not the only neural form of 4.1. We recently showed that 4.1R also occurs in the brain, where it is discretely localized to granule cells in the cerebellum and the dentate gyrus (Walensky et al., 1998b). Mice with targeted deletion of 4.1R manifest deficits in movement, coordination, balance, and learning consistent with the areas of 4.1R enrichment in the brain. By contrast, 4.1N mRNA and protein occur in virtually all neurons in the brain (Walensky et al., 1999). We have recently cloned another brain-specific form of 4.1, designated 4.1B, which is most enriched in Purkinje cells and thalamic neurons (Parra et al., 1998). We also have cloned 4.1G, so designated because it occurs generally throughout the body in many different tissues (Walensky et al., 1998a), whereas 4.1N and 4.1B are predominantly neuronal. In the brain 4.1G is localized to glia (Walensky et al., 1998a). 4.1N, 4.1R, 4.1G, and 4.1B all arise from distinct genes (von Ruckmann et al., 1997; Walensky et al., 1998a, 1999).
Heretofore, 4.1 has been selectively associated with extranuclear events as a critical structural component of the erythrocyte membrane cytoskeleton. Protein 4.1 binds spectrin and actin and potentiates interactions of spectrin tetramers with F-actin (Ohanian et al., 1984;Correas et al., 1986a,b). The membrane binding domain of 4.1 links the cytoskeletal scaffold to the plasma membrane through interactions with band 3 and glycophorin C (Anderson and Marchesi, 1985; Pasternack et al., 1985; Correas et al., 1986a,b; Hemming et al., 1994, 1995). Consistent with our discovery that 4.1N may translocate into the nucleus of PC12 cells, 4.1R has been detected in the nuclei of fibroblast cells and COS cells (Krauss et al., 1997; Gascard et al., 1999). Our findings and the independent observations of Mattagajasingh et al. (1999) provide the important demonstration of a nuclear role for any form of 4.1. Interactions with NuMA appear to account for the mediation by 4.1N of the anti-proliferative actions of NGF. Whether other proteins of the nuclear cytoskeleton act together with NuMA and 4.1 in mediating these effects remains to be elucidated.
Footnotes
This work is supported by United States Public Health Service Grants MH18501 (S.H.S.), Research Scientist Grant DA-00074 (S.H.S.), Medical Scientist Training Grant GM-07309 (M.M.L.), and Grant GM51542 (D.A.C.). We are indebted to Drs. Christopher Ferris, Masaaki Takahashi, Patrick E. Burnett, and Xiaodong Li for valuable suggestions. We also thank Dr. Harish C. Joshi for thoughtful discussion and advice.
Correspondence should be addressed to Dr. Solomon H. Snyder, Department of Neuroscience, Johns Hopkins University School of Medicine, 725 North Wolfe Street, Baltimore, Maryland 21205.
REFERENCES
- 1.Anderson RA, Marchesi VT. Regulation of the association of membrane skeletal protein 4.1 with glycophorin by a polyphosphoinositide. Nature. 1985;318:295–298. doi: 10.1038/318295a0. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. Wiley; New York: 1990. [Google Scholar]
- 3.Baines AJ, Bennett V. Synapsin I is a spectrin-binding protein immunologically related to erythrocyte protein 4.1. Nature. 1985;315:410–413. doi: 10.1038/315410a0. [DOI] [PubMed] [Google Scholar]
- 4.Blumberg D, Radeke MJ, Feinstein SC. Specificity of nerve growth factor signaling: differential patterns of early tyrosine phosphorylation events induced by NGF, EGF, and bFGF. J Neurosci Res. 1995;41:628–639. doi: 10.1002/jnr.490410509. [DOI] [PubMed] [Google Scholar]
- 5.Buchkovich KJ, Ziff EB. Nerve growth factor regulates the expression and activity of p33cdk2 and p34cdc2 kinases in PC12 pheochromocytoma cells. Mol Biol Cell. 1994;5:1225–1241. doi: 10.1091/mbc.5.11.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho KO, Skarnes WC, Minsk B, Palmieri S, Jackson-Grusby L, Wagner JA. Nerve growth factor regulates gene expression by several distinct mechanisms. Mol Cell Biol. 1989;9:135–143. doi: 10.1128/mcb.9.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Compton DA, Cleveland DW. NuMA is required for the proper completion of mitosis. J Cell Biol. 1993;120:947–957. doi: 10.1083/jcb.120.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Compton DA, Cleveland DW. NuMA, a nuclear protein involved in mitosis and nuclear reformation. Curr Opin Cell Biol. 1994;6:343–346. doi: 10.1016/0955-0674(94)90024-8. [DOI] [PubMed] [Google Scholar]
- 9.Compton DA, Luo C. Mutation of the predicted p34cdc2 phosphorylation sites in NuMA impair the assembly of the mitotic spindle and block mitosis. J Cell Sci. 1995;108:621–633. doi: 10.1242/jcs.108.2.621. [DOI] [PubMed] [Google Scholar]
- 10.Compton DA, Szilak I, Cleveland DW. Primary structure of NuMA, an intranuclear protein that defines a novel pathway for segregation of proteins at mitosis. J Cell Biol. 1992;116:1395–1408. doi: 10.1083/jcb.116.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conboy JG, Shitamoto R, Parra M, Winardi R, Kabra A, Smith J, Mohandas N. Hereditary elliptocytosis due to both qualitative and quantitative defects in membrane skeletal protein 4.1. Blood. 1991;78:2438–2443. [PubMed] [Google Scholar]
- 12.Correas I, Leto TL, Speicher DW, Marchesi VT. Identification of the functional site of erythrocyte protein 4.1 involved in spectrin-actin associations. J Biol Chem. 1986a;261:3310–3315. [PubMed] [Google Scholar]
- 13.Correas I, Speicher DW, Marchesi VT. Structure of the spectrin-actin binding site of erythrocyte protein 4.1. J Biol Chem. 1986b;261:13362–13366. [PubMed] [Google Scholar]
- 14.Gascard P, Cohen CM. Absence of high-affinity band 4.1 binding sites from membranes of glycophorin C- and D-deficient (Leach phenotype) erythrocytes. Blood. 1994;83:1102–1108. [PubMed] [Google Scholar]
- 15.Gascard P, Nunomura W, Lee G, Walensky LD, Krauss SW, Takakuwa Y, Chasis JA, Mohandas N, Conboy JG. Deciphering the nuclear import pathway for the cytoskeletal red cell protein 4.1R. Mol Biol Cell. 1999;10:1783–1798. doi: 10.1091/mbc.10.6.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gould E, Tanapat P, McEwen BS, Flugge G, Fuchs E. Proliferation of granule cell precursors in the dentate gyrus of adult monkeys is diminished by stress. Proc Natl Acad Sci USA. 1998;95:3168–3171. doi: 10.1073/pnas.95.6.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greene LA, Tischler AS. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc Natl Acad Sci USA. 1976;73:2424–2428. doi: 10.1073/pnas.73.7.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He D, Zeng C, Brinkley BR (1995) Nuclear matrix proteins as structural and functional components of the mitotic apparatus. Int Rev Cytol 1–74. [DOI] [PubMed]
- 19. Hemming NJ, Anstee DJ, Mawby WJ, Reid ME, Tanner MJ. Localization of the protein 4.1-binding site on human erythrocyte glycophorins C and D. Biochem J 299 1994. 191 196[erratum (1994) 300:920]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hemming NJ, Anstee DJ, Staricoff MA, Tanner MJ, Mohandas N. Identification of the membrane attachment sites for protein 4.1 in the human erythrocyte. J Biol Chem. 1995;270:5360–5366. doi: 10.1074/jbc.270.10.5360. [DOI] [PubMed] [Google Scholar]
- 21.Hempstead BL, Birge RB, Fajardo JE, Glassman R, Mahadeo D, Kraemer R, Hanafusa H. Expression of the v-crk oncogene product in PC12 cells results in rapid differentiation by both nerve growth factor- and epidermal growth factor-dependent pathways. Mol Cell Biol. 1994;14:1964–1971. doi: 10.1128/mcb.14.3.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ignatius MJ, Chandler CR, Shooter EM. Nerve growth factor-treated, neurite-bearing PC12 cells continue to synthesize DNA. J Neurosci. 1985;5:343–351. doi: 10.1523/JNEUROSCI.05-02-00343.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kallajoki M, Harborth J, Weber K, Osborn M. Microinjection of a monoclonal antibody against SPN antigen, now identified by peptide sequences as the NuMA protein, induces micronuclei in PtK2 cells. J Cell Sci. 1993;104:139–150. doi: 10.1242/jcs.104.1.139. [DOI] [PubMed] [Google Scholar]
- 24.Krauss SW, Larabell CA, Lockett S, Gascard P, Penman S, Mohandas N, Chasis JA. Structural protein 4.1 in the nucleus of human cells: dynamic rearrangements during cell division. J Cell Biol. 1997;137:275–289. doi: 10.1083/jcb.137.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krebs KE, Zagon IS, Sihag R, Goodman SR. Brain protein 4.1 subtypes: a working hypothesis. BioEssays. 1987;6:274–279. doi: 10.1002/bies.950060607. [DOI] [PubMed] [Google Scholar]
- 26.Lai MM, Burnett PE, Wolosker H, Blackshaw S, Snyder SH. Cain, a novel physiologic protein inhibitor of calcineurin. J Biol Chem. 1998;273:18325–18331. doi: 10.1074/jbc.273.29.18325. [DOI] [PubMed] [Google Scholar]
- 27.Li XJ, Snyder SH. Molecular cloning of Ebnerin, a von Ebner's gland protein associated with taste buds. J Biol Chem. 1995;270:17674–17679. doi: 10.1074/jbc.270.30.17674. [DOI] [PubMed] [Google Scholar]
- 28.Lombardo CR, Willardson BM, Low PS. Localization of the protein 4.1-binding site on the cytoplasmic domain of erythrocyte membrane band 3. J Biol Chem. 1992;267:9540–9546. [PubMed] [Google Scholar]
- 29.Lydersen BK, Pettijohn DE. Human-specific nuclear protein that associates with the polar region of the mitotic apparatus: distribution in a human/hamster hybrid cell. Cell. 1980;22:489–499. doi: 10.1016/0092-8674(80)90359-1. [DOI] [PubMed] [Google Scholar]
- 30.Mattagajasingh SN, Huang SC, Hartenstein JS, Snyder M, Marchesi VT, Benz EJ. A nonerythroid isoform of protein 4.1R interacts with the nuclear mitotic apparatus (NuMA) protein [in process citation]. J Cell Biol. 1999;145:29–43. doi: 10.1083/jcb.145.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merdes A, Cleveland DW. The role of NuMA in the interphase nucleus. J Cell Sci. 1998;111:71–79. doi: 10.1242/jcs.111.1.71. [DOI] [PubMed] [Google Scholar]
- 32.Moir RD, Montag-Lowy M, Goldman RD. Dynamic properties of nuclear lamins: lamin B is associated with sites of DNA replication. J Cell Biol. 1994;125:1201–1212. doi: 10.1083/jcb.125.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mothe I, Ballotti R, Tartare S, Kowalski-Chauvel A, Van Obberghen E. Cross talk among tyrosine kinase receptors in PC12 cells: desensitization of mitogenic epidermal growth factor receptors by the neurotrophic factors, nerve growth factor and basic fibroblast growth factor. Mol Biol Cell. 1993;4:737–746. doi: 10.1091/mbc.4.7.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Movsesyan V, Whalin M, Shibutani M, Katagiri Y, Broude E, Guroff G. Down-regulation of cyclin F levels during nerve growth factor-induced differentiation of PC12 cells. Exp Cell Res. 1996;227:203–207. doi: 10.1006/excr.1996.0268. [DOI] [PubMed] [Google Scholar]
- 35.Ohanian V, Wolfe LC, John KM, Pinder JC, Lux SE, Gratzer WB. Analysis of the ternary interaction of the red cell membrane skeletal proteins spectrin, actin, and 4.1. Biochemistry. 1984;23:4416–4420. doi: 10.1021/bi00314a027. [DOI] [PubMed] [Google Scholar]
- 36.Parra M, Walensky L, Chan N, Snyder HS, Mohandas N, Conboy J. Charaterization of protein 4.1B, a novel member of the protein family with high level, focal expression in brain. Mol Biol Cell [Suppl] 1998;9:1535. doi: 10.1074/jbc.275.5.3247. [DOI] [PubMed] [Google Scholar]
- 37.Pasternack GR, Anderson RA, Leto TL, Marchesi VT. Interactions between protein 4.1 and band 3. An alternative binding site for an element of the membrane skeleton. J Biol Chem. 1985;260:3676–3683. [PubMed] [Google Scholar]
- 38.Price CM, Pettijohn DE. Redistribution of the nuclear mitotic apparatus protein (NuMA) during mitosis and nuclear assembly. Properties of purified NuMA protein. Exp Cell Res. 1986;166:295–311. doi: 10.1016/0014-4827(86)90478-7. [DOI] [PubMed] [Google Scholar]
- 39.Rosenberg IM. Protein analysis and purification, benchtop techniques. Birkhauser; Cambridge, MA: 1996. [Google Scholar]
- 40.Rudkin BB, Lazarovici P, Levi BZ, Abe Y, Fujita K, Guroff G. Cell cycle-specific action of nerve growth factor in PC12 cells: differentiation without proliferation. EMBO J. 1989;8:3319–3325. doi: 10.1002/j.1460-2075.1989.tb08493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saredi A, Howard L, Compton DA. NuMA assembles into an extensive filamentous structure when expressed in the cell cytoplasm. J Cell Sci. 1996;109:619–630. doi: 10.1242/jcs.109.3.619. [DOI] [PubMed] [Google Scholar]
- 42.Sawa A, Khan AA, Hester LD, Snyder SH. Glyceraldehyde-3-phosphate dehydrogenase: nuclear translocation participates in neuronal and nonneuronal cell death. Proc Natl Acad Sci USA. 1997;94:11669–11674. doi: 10.1073/pnas.94.21.11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sihag RK, Wang LW, Cataldo AM, Hamlin M, Cohen CM, Nixon RA. Evidence for the association of protein 4.1 immunoreactive forms with neurofibrillary tangles in Alzheimer's disease brains. Brain Res. 1994;656:14–26. doi: 10.1016/0006-8993(94)91361-7. [DOI] [PubMed] [Google Scholar]
- 44.Thomaidou D, Mione MC, Cavanagh JF, Parnavelas JG. Apoptosis and its relation to the cell cycle in the developing cerebral cortex. J Neurosci. 1997;17:1075–1085. doi: 10.1523/JNEUROSCI.17-03-01075.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Grunsven LA, Thomas A, Urdiales JL, Machenaud S, Choler P, Durand I, Rudkin BB. Nerve growth factor-induced accumulation of PC12 cells expressing cyclin D1: evidence for a G1 phase block. Oncogene. 1996a;12:855–862. [PubMed] [Google Scholar]
- 46.van Grunsven LA, Billon N, Savatier P, Thomas A, Urdiales JL, Rudkin BB. Effect of nerve growth factor on the expression of cell cycle regulatory proteins in PC12 cells: dissection of the neurotrophic response from the anti-mitogenic response. Oncogene. 1996b;12:1347–1356. [PubMed] [Google Scholar]
- 47.Van Ness J, Pettijohn DE. Specific attachment of nuclear-mitotic apparatus protein to metaphase chromosomes and mitotic spindle poles: possible function in nuclear reassembly. J Mol Biol. 1983;171:175–205. doi: 10.1016/s0022-2836(83)80352-0. [DOI] [PubMed] [Google Scholar]
- 48.von Ruckmann B, Jons T, Dolle F, Drenckhahn D, Schubert D. Cytoskeleton-membrane connections in the human erythrocyte membrane: band 4.1 binds to tetrameric band 3 protein. Biochim Biophys Acta. 1997;1325:226–234. doi: 10.1016/s0005-2736(96)00261-1. [DOI] [PubMed] [Google Scholar]
- 49.Walensky LD, Gascard P, Fields ME, Blackshaw S, Conboy JG, Mohandas N, Snyder SH. The 13-kD FK506 binding protein, FKBP13, interacts with a novel homologue of the erythrocyte membrane cytoskeletal protein 4.1. J Cell Biol. 1998a;141:143–153. doi: 10.1083/jcb.141.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walensky LD, Shi ZT, Blackshaw S, DeVries AC, Demas GE, Gascard P, Nelson RJ, Conboy JG, Rubin EM, Snyder SH, Mohandas N. Neurobehavioral deficits in mice lacking the erythrocyte membrane cytoskeletal protein 4.1. Curr Biol. 1998b;8:1269–1272. doi: 10.1016/s0960-9822(07)00536-2. [DOI] [PubMed] [Google Scholar]
- 51.Walensky LD, Blackshaw S, Liao D, Watkins CC, Weier HU, Parra M, Huganir RL, Conboy JG, Mohandas N, Snyder SH. A novel neuron-enriched homolog of the erythrocyte membrane cytoskeletal protein 4.1 [in process citation]. J Neurosci. 1999;19:6457–6467. doi: 10.1523/JNEUROSCI.19-15-06457.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yan GZ, Ziff EB. NGF regulates the PC12 cell cycle machinery through specific inhibition of the Cdk kinases and induction of cyclin D1. J Neurosci. 1995;15:6200–6212. doi: 10.1523/JNEUROSCI.15-09-06200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yan H, Schlessinger J, Chao MV. Chimeric NGF-EGF receptors define domains responsible for neuronal differentiation. Science. 1991;252:561–563. doi: 10.1126/science.1850551. [DOI] [PubMed] [Google Scholar]
- 54.Yang CH, Snyder M. The nuclear-mitotic apparatus protein is important in the establishment and maintenance of the bipolar mitotic spindle apparatus. Mol Biol Cell. 1992;3:1259–1267. doi: 10.1091/mbc.3.11.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang CH, Lambie EJ, Snyder M. NuMA: an unusually long coiled-coil related protein in the mammalian nucleus. J Cell Biol. 1992;116:1303–1317. doi: 10.1083/jcb.116.6.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ye K, Ke Y, Keshava N, Shanks J, Kapp JA, Tekmal RR, Petros J, Joshi HC. Opium alkaloid noscapine is an antitumor agent that arrests metaphase and induces apoptosis in dividing cells. Proc Natl Acad Sci USA. 1998;95:1601–1606. doi: 10.1073/pnas.95.4.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeng C, He D, Brinkley BR. Localization of NuMA protein isoforms in the nuclear matrix of mammalian cells. Cell Motil Cytoskeleton. 1994;29:167–176. doi: 10.1002/cm.970290208. [DOI] [PubMed] [Google Scholar]