Abstract
Recent European genome-wide association studies (GWAS) have revealed strong statistical correlations between MDD and numerous zero-to-high linked variants in the genomic region containing major histocompatibility complex (MHC) genes (MHC region), but the underlying biological mechanisms are still unclear. To better understand the roles of this genomic region in the neurobiology of MDD, we applied a convergent functional genomics approach to integrate GWAS data of MDD relevant biological phenotypes, gene-expression analyses results obtained from brain samples, and genetic analyses of independent Chinese MDD samples. We observed that independent MDD risk variants in the MHC region were also significantly associated with the relevant biological phenotypes in the predicted directions, including the emotional and cognitive-related phenotypes. Gene-expression analyses further revealed that mRNA expression levels of several MHC region genes in the human brain were associated with MDD risk SNPs and diagnostic status. For instance, a brain-enriched gene ZNF603P consistently showed lower mRNA levels in the individuals carrying MDD risk alleles and in MDD patients. Remarkably, we further found that independent MDD risk SNPs in the MHC region likely converged to affect the mRNA level(s) of the same gene(s), and Europeans and Han Chinese populations have a substantial shared genetic and molecular basis underlying MDD risk associations in the MHC region. These results highlighted several potential pivotal genes at the MHC region in the pathogenesis of MDD. Their common impacts on multiple psychiatric relevant phenotypes also implicated the neurological processes shared by different psychological processes, such as mood and/or cognition, shedding lights on their potential biological mechanisms.
Subject terms: Functional genomics, Genetic markers, Genetics research, Depression
Introduction
Major depressive disorder (MDD) is a chronic severe neuropsychiatric illness whose worldwide lifetime prevalence was estimated to be up to 13% [1]. Previous studies have revealed a moderate (~37%) heritability of this illness [2], and genetic approaches are thus presumed to be viable to find at least part of the mechanisms for MDD pathogenesis. However, genetic analyses of MDD, including recent genome-wide association study (GWAS) [3–10], have earned less success than expected during the last decade. Generally limited statistical power of small sample sizes, population heterogeneity, and phenotypic complexity were thought to restrict the detecting power of current MDD genetic studies [11], and international collaboration projects by multiple groups were therefore elicited to gain better insights. Indeed, many novel risk loci have been reported through these large collaborative studies, and the knowledge of genetic basis of MDD has been significantly enriched [12–16].
While genetic analysis has provided essential information about MDD pathogenesis, such results still remained difficult to integrate and explain. Thanks to the accumulation of basic and clinical studies of MDD, researchers have found that MDD patients often exhibit a series of pathological alterations in their emotional and cognitive abilities [17, 18]. In addition, impairment and dysfunction of certain brain regions engaged in processes for emotion and cognition (the prefrontal cortex, hippocampus, etc.) [19, 20], as well as dysregulated synaptic function and neuronal development in these brain areas [21–23], have been widely reported in subjects affected by MDD. Despite the complicated neurological mechanisms, phenotypic uncertainties, and heterogeneity of precipitating factors in MDD, these pathological features observed in patients were thought to be at least partially linked to genetic risk factors of this illness. In the past few years, researchers have conducted multiple convergent functional genomic studies [24–27] to test this hypothesis and have thereby proven the involvement of multiple genetic loci in MDD (e.g., PCDH9, CREB1, and SLC6A15) [28–30]. Therefore, one main task for researchers has been to examine whether MDD risk genes/variants also affect the key aspects of the neurobiology of the illness. Beyond the genetic analyses, recent advances suggest that genetic variations (including noncoding variations) usually contribute to complex diseases (e.g., MDD) via altering expression of certain genes [31, 32]. Intriguingly, altered expression levels of specific genes are also found to exert pivotal roles in the pathogenesis of MDD [33–35]. For example, transcriptome analyses of specific human brain regions have identified multiple genes that are differentially expressed between MDD patients and healthy controls [34]. We herein performed the current study to further dissect the genetic and pathological basis of MDD via analyzing the link between top MDD risk loci and/or the genes and relevant pathological phenotypes.
In the latest GWAS of MDD (named PGC2 MDD GWAS) [16], the genomic region containing major histocompatibility complex (MHC) genes (the MHC region) has been one of the most significant genomic regions associated with this illness. The MHC molecules are the tissue–antigens necessary for the function of immune cells, which can be classified as MHC class I (MHC-I), class-II (MHC-II), and class III (MHC-III) molecules [36]. These molecules play vital roles in the immune system and determine the host histocompatibility as well as acquired immune responses [37]. Despite the well-known function of these molecules in the immune system, they have not been extensively implicated in psychiatric diseases until the application of GWAS [38]. Furthermore, in a recent study combining human brain transcriptome analysis and mouse models, scientists reported the crucial roles of C4A (encodes the acidic form of complement factor 4) in MHC class III region in schizophrenia [39]. In addition to the recent finding that the MHC region being one of the most significant genomic regions associated with MDD [16], a recent fine-mapping analysis through imputing HLA alleles and C4 haplotypes also indicated that the observed association signal at MHC in MDD were unlikely arising from HLA alleles or C4 haplotypes, providing other possibilities that the MDD risk signal derived from additional genes at MHC [40].
Nevertheless, there are still several unanswered questions: (1) despite high linkage disequilibrium (LD) and gene density in the MHC region, what is the exact LD structure of risk associations? (2) Are the MDD risk SNPs also associated with any of the relevant biological phenotypes? (3) Are there any functional consequences caused by the MDD risk alleles? (4) Are the mRNA levels of MHC genes altered in MDD patients? (5) Can the MDD risk SNPs in PGC2 GWAS [16] be replicated across different populations? Based on the theory of convergent functional genomics [24–27], gene(s) significantly associated with multiple MDD relevant phenotypes and aberrantly expressed in patients is likely an authentic susceptibility gene. We therefore performed careful examination of the risk loci in MHC regions in diverse GWAS resources and expression datasets, and carried out replication analyses in distinct populations. This integrative analyses highlighted several genes in the MHC region (but not limited to them) deserving further investigations.
Materials and methods
All the protocols and methods were approved by the institutional review board of the Kunming Institute of Zoology, Chinese Academy of Sciences and the ethics committees of all participating hospitals and universities. The full descriptions about the materials and methods were shown in Supplementary Materials. The sequence of this hypothesis-driven integrative analysis is summarized in Fig. 1.
Fig. 1.
Overview of research strategy and study design. The study focused the recently reported genome-wide significant SNPs for MDD in the MHC region, examining their associations with the relevant biological phenotypes in human populations, and independent replications of clinical associations with MDD across populations. The gene mRNA expression in the MHC region were then tested for their associations with the MDD genetic risk SNPs and diagnostic status. This integrative analyses identified a set of MHC SNPs and genes showing consistent associations with the illness and biological phenotypes
Descriptions of PGC2 MDD GWAS sample
The primary MDD statistical data in Europeans were from a recent PGC2 GWAS [16] of 135,458 MDD cases and 344,901 controls, including PGC29 cohort and six expanded samples (deCODE, Generation Scotland, GERA, iPSYCH, UK Biobank, and 23andMe). The 23andMe data (75,607 cases and 231,747 controls) were not publicly released, therefore the statistical results of 59,851 MDD cases and 113,154 controls were utilized in the present study. Detailed information about the cohorts, genotyping, and quality control, as well as statistical methods can be found in the original GWAS report [16]. For the threshold of association significance with MDD, we empirically defined a p-value < 5.00 × 10−8 to be genome-wide statistically significant.
LD analysis of MHC SNPs
A total of 83,279 SNPs in the MHC genomic region (chr6:26Mb-34Mb) were covered in the PGC2 MDD GWAS [16]. We examined the linkage disequilibrium (LD) pattern of MDD risk SNPs in the MHC region using genotype data from European individuals (N = 503) in 1000 Genomes Project [41], and calculated the tagging SNPs based on the tagger procedure implemented in Haploview (pairwise tagging only, r2 threshold = 0.6) [42]. The LD structure of the tagging SNPs was constructed using r2 algorithm implemented in the Haploview program [42] and the online tool SHEsis (http://analysis.bio-x.cn/) [43, 44].
Analyses on MDD-related biological phenotypes
The 83,279 MHC SNPs were aligned to the other GWAS datasets of related biological phenotypes, such as neuroticism [45], depressed affect and worry [45], cognitive performance [46], verbal–numerical reasoning [47], intelligence [48], depressive symptoms [45], and subjective well-being [49]. After alignment, a total of 73,733 SNPs remained for further analyses, of which 581 SNPs were genome-wide significantly associated with MDD in the PGC2 GWAS [16]. For the definitions of association significance threshold with these biological phenotypes, since we are primarily interested in whether the MDD genome-wide risk SNPs (N = 581) affected the biological phenotypes, and a total of eight phenotypes were examined, we therefore set a p-value of 0.05/581/8≈1.08 × 10−5 as region-wide significance (we did not consider LD relationship among the 581 SNPs during the definitions of significance; thus a p-value of 1.08 × 10−5 was more stringent than expected), and a p-value < 0.05 was considered nominally significant. Detailed information about the related biological phenotypes can be found in the Supplementary Materials.
Expression quantitative trait loci (eQTL) analysis in human brains in public datasets and our own Han Chinese sample
To identify the functional consequences associated with the MDD risk alleles within the MHC region, we examined the effects of index SNPs on the variations of primarily MHC gene-expression levels in human brains using two public eQTL datasets: (1) BrainSeq [50] and (2) Genotype-Tissue Expression project (GTEx) [51]. Detailed descriptions of the two brain eQTL datasets can be found in the Supplementary Materials.
We also collected amygdala tissues of 65 non-psychiatric controls from the Chinese Brain Bank Center. The total RNA was extracted from these samples, and the reverse transcription reaction was conducted to synthesize cDNA using a first-strand cDNA synthesis kit (K1612, ThermoFisher Scientific, USA) according to the manufacturer’s protocol. The quantitative real-time PCR was then performed on a 7900HT Fast Real-Time PCR System (Applied Biosystems, USA), with a reaction mixture of 10.0 μL 2 × SYBR master mix (Roche, USA), 2.0 μL of primers (10 μM), 1.0 μL of cDNA, and 7.0 μL of nuclease-free water for each sample. The relative gene expression was presented as the means of –ΔCt for a statistical test against genotypic groups [52]. The sequences of primers used for amplification of RPS13 were 5′-CCCCACTTGGTTGAAGTTGA-3′ (forward) and 5′-CTTGTGCAACACCATGTGAA-3′ (reverse); sequences of primers for BTN3A2 were 5′-GTCCAGTGAGATAGAAAGTGAGCA-3′ (forward) and 5′-TTATTGGTATCGGACGAAGACTC-3′ (reverse); and sequences of primers for ZNF603P were 5′-CCCTGTTGGTCTGAAGAAGATAGTA-3′ (forward) and 5′-CTACGTTCATAATGGTGCTGCTC-3′ (reverse).
Diagnostic analysis of MHC gene expression in brains
We downloaded the raw RNA-seq data (fastq files) from two independent Gene Expression Omnibus (GEO) datasets. The first GEO dataset (GSE102556) included human postmortem brain dorsolateral PFC (BA8/9) tissues from 26 MDD cases and 22 matched controls from the Douglas Bell Canada Brain Bank (DBCBQ; Douglas Mental Health Institute, Verdun, Québec) [34]. The second GEO dataset (GSE101521) consisted of brain tissues from 30 DSM-IV MDD cases and 29 controls collected at The Division of Molecular Imaging and Neuropathology, New York State Psychiatric Institute and Columbia University [35]. In both datasets, quality control, alignment, and gene-expression quantification were conducted using the same standard as described in the Supplementary Materials. We quantified gene-level expression based on known genes of Ensembl build GRCh38.91 using featureCounts [53], and FPKM (fragments per kilobase of exon per million fragments mapped) was calculated to measure gene-level expression.
For each differential expression analysis, a mean gene FPKM value of 0.001 across all samples was used as the minimum threshold for inclusion. Principal components (PCs) were calculated using the transformed FPKM (log2 with an offset of 1) in the genome, and the first 10 PCs were used as covariates in the following diagnostic analyses. Statistical analyses of mRNA expression associated with diagnosis were conducted in R 3.4.2 using the linear regression model, treating log2-transformed expression levels of FPKM as the outcome, and covaried for top 10 expression PCs. In the datasets utilized in the present study, the top 10 PCs were significantly associated with sex, age, pH, RNA integrity number (RIN), and postmortem interval (PMI) (Figure S1).
Descriptions of Han Chinese MDD case-control samples
CONVERGE GWAS sample
Data of 5303 MDD patients and 5337 non-psychiatric controls from Han Chinese population were retrieved from the CONVERGE GWAS dataset [15]. Cases were diagnosed using the Composite International Diagnostic Interview (WHO lifetime version 2.1; Chinese version) according to the DSM-IV criteria. Controls were recruited either from local communities or from the patients who underwent minor surgical procedures at the general hospitals.
Chinese replication sample
This sample contained 1789 MDD cases and 2464 controls of Chinese ancestry. All patients were diagnosed with MDD strictly according to the DSM-IV criteria via standard diagnostic assessments, supplemented with clinical information through thorough review of medical records and interview with family informants. Control subjects were mentally healthy local volunteers. All participants provided written informed consents.
Meta-analysis of MHC SNPs with MDD in Han Chinese
For genetic analyses, we performed logistic regression analyses to test the association between SNP dosages and the risk of MDD in the case-control samples (CONVERGE GWAS and the Chinese MDD replication sample). To conduct the meta-analysis, values of odds ratio (OR) and standard error (SE) of each sample were retrieved to calculate inter-sample heterogeneity, total pooled OR, and the overall 95% confidence interval (CI). A p-value < 0.05 was considered nominally significant.
Results
LD analysis of the MDD risk SNPs in the MHC region
As described in the Materials and Methods, a total of 73,733 SNPs within the MHC region were selected for the analyses. We first plotted the association results of these 73,733 SNPs with MDD using the LocusZoom program in Figure S1 [54]. To depict a more intuitive MDD risk association pattern for the MHC region, we also made the plot using SNPs with p-value lower than 0.001 (Fig. 2). In brief, among the 73,733 SNPs in the MHC region, 581 SNPs showed genome-wide significant associations with MDD in the PGC2 GWAS (p < 5.00 × 10−8) [16]. The LD pattern of these 581 genome-wide risk SNPs varied substantially in Europeans, we thus calculated the tagging SNPs (using pairwise tagging only with r2 threshold = 0.6) in European individuals of 1000 Genomes Project (N = 503) [41] using the Haploview software [42], and chose nine tagging SNPs for subsequent analysis. These nine tagging SNPs have encompassed a large genomic region, and were in weak-to-moderate LD with each other (0.000 ≤ r2 ≤ 0.497, Figure S3 and Table S1).
Fig. 2.
Genetic associations of MHC variants with MDD in the PGC2 GWAS [16], and the associations of MHC tagging SNPs with multiple MDD relevant biological phenotypes. A physical map of the region is given and depicts known genes within the region. DS depressive symptoms, DA depressed affect sub-cluster, SWB subjective well-being, CP cognitive performance, VNR verbal–numerical reasoning, √ in black, nominal significance; √ in gray, genome-wide significance (for MDD) or region-wide significance (for relevant biological phenotypes); × not significant, NA not available. The MDD association figure in PGC2 GWAS [16] was made using the online program LocusZoom (http://locuszoom.sph.umich.edu//), and only SNPs with p-value lower than 0.001 were plotted [54]
The independent MDD risk alleles in the MHC region were associated with worse cognitive functions
Aberrant cognitive functions have been repeatedly observed in MDD patients compared with healthy controls [18], and we thus hypothesized that SNPs associated with the risk of MDD should also affect cognitive abilities. By examining these SNPs in a large-scale GWAS of cognitive performance (N = 257,828), we found that independent MDD risk SNPs were region-wide significantly associated with cognitive performance (Table 1) [46]. We also ensured that such association signals were not due to the LD signal from one particular tagging SNP. For example, rs1936365 and rs9379871, two SNPs in negligible LD in Europeans (r2 = 0.003, locates 1.89 Mb far between them) with genome-wide associations with MDD (rs1936365, p = 3.86 × 10−11; rs9379871, p = 1.24 × 10−8; Table 1), were both significantly associated with cognitive performance (rs1936365, p = 2.68 × 10−11; rs9379871, p = 1.79 × 10−13; Table 1), and their MDD risk alleles both predicted impaired cognitive performance.
Table 1.
Associations of the nine MHC tagging SNPs with MDD and related phenotypes in published GWAS datasets
| SNP | rs9379871 | rs13194053 | rs6904596 | rs6940116 | rs1936365 | rs144447022 | rs9257803 | rs3094067 | rs3094084 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Position | 26375854 | 27143883 | 27491299 | 27708732 | 28268452 | 29244219 | 29344395 | 30299245 | 30947509 | |
| Effect allele/non-effect allele | C/G | T/C | A/G | A/G | C/G | T/G | T/C | T/G | A/T | |
| MDD_OR | (N = 173,005) | 1.072 | 1.061 | 0.931 | 1.060 | 0.929 | 0.924 | 0.932 | 1.072 | 1.061 |
| MDD_P-val | 1.24 × 10−8 | 1.32 × 10−8 | 2.81 × 10−8 | 4.91 × 10−8 | 3.86 × 10−11 | 2.23 × 10−9 | 1.03 × 10−8 | 2.89 × 10−8 | 3.48×10−8 | |
| DS_Z score | (N = 381,455) | 3.888 | 4.524 | −5.276 | 3.601 | −4.336 | −4.449 | −3.793 | 3.894 | 2.623 |
| DS_P-val | 1.01 × 10−4 | 6.08 × 10−6 | 1.32 × 10−7 | 3.17 × 10−4 | 1.45 × 10−5 | 8.61 × 10−6 | 1.49 × 10−4 | 9.87 × 10−5 | 0.00872 | |
| DA_Beta | (N = 357,957) | 0.009445 | 0.009604 | −0.01281 | 0.007579 | −0.00811 | −0.01117 | −0.008315 | 0.008533 | 0.004524 |
| DA_P-val | 0.00563 | 0.00117 | 2.45 × 10−4 | 0.0119 | 0.00963 | 0.00185 | 0.0138 | 0.0128 | 0.13 | |
| Worry_Beta | (N = 348,219) | 0.0159 | 0.01627 | −0.01711 | 0.01307 | −0.01195 | −0.01941 | −0.01585 | 0.01798 | 0.01402 |
| Worry_P-val | 5.98 × 10−6 | 9.42 × 10−8 | 1.93 × 10−6 | 2.51 × 10−5 | 2.10 × 10−4 | 1.48 × 10−7 | 5.02 × 10−6 | 3.45 × 10−7 | 5.25 × 10−6 | |
| SWB_Beta | (N = 298,420) | −0.008b | −0.017 | 0.024 | −0.016a | 0.018 | NA | NA | NA | NA |
| SWB_P-val | 0.102b | 4.15 × 10−5 | 6.31 × 10−6 | 2.46 × 10−4a | 9.72 × 10−5 | NA | NA | NA | NA | |
| Neuroticism_Z score | (N = 390,278) | 3.986 | 4.212 | −5.020 | 4.073 | −4.189 | −4.778 | −4.191 | 4.482 | 4.080 |
| Neuroticism_P-val | 6.73 × 10−5 | 2.53 × 10−5 | 5.17 × 10−7 | 4.65 × 10−5 | 2.80 × 10−5 | 1.77 × 10−6 | 2.78 × 10−5 | 7.39 × 10−6 | 4.51 × 10−5 | |
| CP_Beta | (N = 257,828) | −0.031 | −0.026 | 0.035 | −0.026 | 0.026 | 0.04 | NA | −0.033c | NA |
| CP_P-val | 1.79 × 10−13 | 1.04 × 10−12 | 1.09 × 10−15 | 3.10 × 10−12 | 2.68 × 10−11 | 2.02 × 10−19 | NA | 6.36×10−15c | NA | |
| VNR_Z score | (N = 168,033) | −6.686 | −6.388 | 6.924 | −6.484 | 6.215 | 8.140 | 8.119 | −7.204 | −4.400 |
| VNR_P-val | 2.29 × 10−11 | 1.68 × 10−10 | 4.40 × 10−12 | 8.95 × 10−11 | 5.14 × 10−10 | 3.94 × 10−16 | 4.71 × 10−16 | 5.86 × 10−13 | 1.08 × 10−5 | |
| Intelligence_Beta | (N = 269,867) | −0.0199 | −0.0138 | 0.0172 | −0.0130 | 0.0117 | 0.0281 | 0.0254 | −0.0237 | −0.0069 |
| Intelligence_P-val | 1.10 × 10−5 | 2.43 × 10−4 | 7.35 × 10−5 | 5.23 × 10−4 | 0.00271 | 7.06 × 10−8 | 1.99 × 10−8 | 1.37 × 10−6 | 0.0873 | |
MDD major depressive disorder, OR odds ratio, DS depressive symptoms, DA depressed affect, SWB subjective well-being, CP cognitive performance, VNR verbal–numerical reasoning, NA not available
aIndex SNP, rs2056923, r2 = 1.00 with rs6940116 in Europeans, and the beta effect was shown in accordance with the effect allele of rs6940116 based on LD
bIndex SNP, rs2073531, r2 = 1.00 with rs9379871 in Europeans, and the beta effect was shown in accordance with the effect allele of rs9379871 based on LD
cIndex SNP, rs3130356, r2 = 1.00 with rs3094067 in Europeans, and the beta effect was shown in accordance with the effect allele of rs3094067 based on LD
In the GWAS samples of VNR (N = 168,033) [47], most of the MDD risk MHC SNPs were also significantly associated with VNR in the expected directionality (Table 1). In another GWAS of intelligence (N = 269,867) [48], most of the MDD genome-wide risk SNPs were again nominally or region-wide significantly associated with intelligence (Table 1). These series of analyses are in agreement with a previous study [55], in which the authors identified a schizophrenia risk SNP rs6904071 in the MHC region showing association with cognitive performance (i.e., episodic memory) and hippocampal volume. Notably, rs6904071 was also genome-wide significantly associated with MDD in the PGC2 GWAS (p = 8.21 × 10−9) [16]. Taken together, although we cannot preclude the possibility that there are substantial overlap in the samples of these GWAS on cognition-related phenotypes, the consistent data could still provide strong evidence that MDD risk SNPs in the MHC region likely also affected cognitive functions.
The MHC MDD SNPs were correlated with neuroticism, depressed affect, worry, depressive symptoms, and subjective well-being
Neuroticism is a predictive factor that is strongly associated with the onset of MDD [56, 57]. Several GWAS of neuroticism also reported shared genetic components between neuroticism and various psychiatric illnesses, including MDD [49, 58, 59]. Here, we utilized a recently published GWAS resource (N = 390,278) [45] and observed strong associations between neuroticism and MDD risk SNPs, such as rs6904596 and rs9257803 (p = 5.17 × 10−7 and 2.78 × 10−5, respectively; r2 = 0.001 and locates 1.85 Mb far between them, Table 1). More intriguingly, carriers of the MDD risk alleles at all SNPs tended to show increased vulnerable personality traits compared with the protective allele carriers (Table 1). In addition, we further examined the associations of those MHC tagging SNPs with depressed affect (N = 357,957) and worry (N = 348,219), the two sub-clusters that derived from neuroticism items in a recent GWAS in the UKB cohort [45]. Intriguingly, the MHC tagging SNPs were also significantly associated with both depressed affect and worry sub-clusters in the predicted directions (Table 1), even though the two neuroticism-related sub-clusters were genetically distinguishable [60]. These results suggested that the MHC region might affect the shared neurological basis underlying both psychological traits.
The associations between MDD risk SNPs in the MHC region and depressive symptoms (N = 381,455) [45] further strengthened the link between these SNPs and the illness, as most of the tested SNPs were nominally or region-wide significantly associated with depressive symptoms in the expected directions (Table 1). However, it should be noted that the samples used for depressive symptoms [45] had substantial overlap with that in the PGC2 MDD GWAS [16], although the depressive symptom GWAS used a continuous phenotype rather than a binary status for analyses in part of the samples (i.e., UKB).
We also tested the associations between the MDD risk SNPs in the MHC region and subjective well-being, a phenotype genetically correlated with both MDD and neuroticism [49]. Using the statistics of 298,420 individuals in a recent GWAS of subjective well-being [49], we found that only five of the nine tagging SNPs (or their LD index SNPs) in the MHC region were included in this GWAS, and four of them were nominally or region-wide significantly associated with subjective well-being (Table 1), with the lowest p-value of 6.31 × 10−6 for rs6904596. Also, the MDD risk alleles at these four SNPs predicted worse subjective well-being (Table 1).
Associations of MHC risk SNPs with gene expression in brains
As discussed, there are many low-to-high LD SNPs linked to MDD spanning a large genomic region in the MHC region. Since an effective strategy to understand the underlying molecular mechanisms of these genetic risk signals is to perform the eQTL analyses [61], we examined the associations between the MDD risk SNPs and mRNA expression levels of nearby genes using RNA-seq results of DLPFC from the BrainSeq dataset (N = 273 healthy individuals) [50]. In these series of eQTL analyses, to maximally capture the genes of expression showing associations with genetic risk, we investigated all abovementioned 581 genome-wide MDD risk SNPs. We identified 426 pairs of significant SNP-gene eQTL associations in the BrainSeq sample (FDR-corrected p-value < 0.1, Table S2), and 284 pairs of them were also nominally significant in the GTEx dataset of frontal cortex (BA9) tissues (Table S2) [51] with the same directions of allelic effects (p < 0.05, Table S2). The significant eQTL genes of risk SNPs in both datasets included BTN3A2, HCG17, HCG4P3, HLA-C, PSORS1C1, VARS2, ZKSCAN3, ZNF192P1, ZNF192P2, ZNF602P, ZNF603P, ZSCAN26, and ZSCAN31 (Table S2). Intriguingly, convergent eQTL associations of independent tagging SNPs with the same gene expression were frequently observed (Table S2). For example, rs6940116 and rs1936365, though in low LD (r2 = 0.006, locates 559.72 Kb far between them), both indicated higher expression of ZNF602P corresponding to their MDD risk alleles (Figure S4); moreover, both of their risk alleles pointed to lower mRNA levels of ZNF603P (Figure S4). We also found that many MDD risk SNPs were eQTLs of multiple genes in the brain, for instance, in addition to ZNF602P and ZNF603P, rs1936365 was also significantly associated with the mRNA expression of other genes (ZKSCAN3 and ZSCAN26), and the MDD risk alleles were associated with either higher (ZNF602P, ZKSCAN3, and ZSCAN26) or lower (ZNF603P) gene expression in the brain (Table S2).
The convergent eQTL associations of independent SNPs (e.g., rs6940116 and rs1936365) with the same gene(s) might suggest convergence of the pathway(s) or molecular mechanisms underlying their roles in the genetic risk of MDD. Besides, the opposite directions of eQTL associations of the risk allele at one SNP (e.g., rs1936365) on multiple genes suggested potential pleiotropic effects of this genomic locus and highlighted the existence of potentially complicated regulatory element(s), resulting in such varied eQTL effects. Nevertheless, it is also possible that the eQTL SNPs (e.g., rs1936365) might be in substantial LD with multiple independent causative variants, and the currently observed variable eQTL associations with different genes were the reflections of functional consequences attributed to different causative variants.
European and Han Chinese populations shared risk SNPs and molecular basis at MHC region
Owning to the differences in demographic histories and genomic structures between distinct continental populations, replicative analyses of some authentic disease risk SNPs across different populations don’t always return positive results [62, 63]. Previous analyses have revealed intensive impact of natural selection on the MHC genomic region during human evolution, probably due to their central roles in the immune system [64, 65], we therefore sought to understand if the PGC2 (comprising of Europeans) MDD risk SNPs in the MHC region conferred risk of the illness in common world populations or solely in the Europeans.
We firstly examined the statistical associations of the 581 PGC2 MDD genome-wide risk SNPs in the MHC region in the CONVERGE GWAS of Han Chinese populations (consisting of 5303 cases and 5337 controls) [15]. Not surprisingly, 424 of the tested SNPs were not included in the Han Chinese CONVERGE GWAS (Table S3), likely due to their monomorphic status in this distinct population. For the 157 remaining SNPs, 66 of them were nominally associated with MDD in CONVERGE GWAS in the consistent directions (p < 0.05, Table S3), which primarily included two independent tagging SNPs (rs9379871 and rs9257803, r2 = 0.001 in Europeans and Han Chinese, respectively) and their linked SNPs (e.g., rs9379871, p = 0.0109; rs9257803, p = 0.00387; Table 2). However, one of the leading SNPs (rs1936365) in PGC2 GWAS was not associated with MDD in the CONVERGE GWAS (p = 0.364), likely reflecting the heterogeneity of the MHC loci between continental populations. In addition, we also noticed the SNP rs9262201, which did not reach genome-wide significance in the PGC2 MDD GWAS (p = 8.65 × 10−8) [16], was nominally associated with MDD in the CONVERGE GWAS (p = 0.0357; Table 2) [15], and its directions of allelic effects were the same in both datasets. LD analysis showed that rs9262201 was in low LD with rs9379871 and rs9257803 in either Europeans or Han Chinese (all r2 ≤ 0.005, Table S4). We then performed further replication analyses of the three SNPs (rs9379871, rs9257803, and rs9262201) in an independent Han Chinese sample (including 1789 cases and 2464 controls), and two of the three SNPs were again nominally associated with MDD in consistent allelic directions (rs9257803, p = 0.00231; rs9262201, p = 0.0351; Table 2). Meta-analysis of the two Han Chinese samples also found that all these three SNPs were significantly associated with MDD in this European-divergent population (rs9379871, p = 0.0478; rs9257803, p = 0.000139; rs9262201, p = 0.0101; Table 2).
Table 2.
Associations of independent MHC SNPs with MDD in distinct populations
| SNP | Position | LD with rs9379871 | Allele | Freq in Europeans | Freq in Chinese | PGC2 GWAS [16] (59,851/113,154)a | CONVERGE GWAS [15] (5303/5337)a | Chinese Replicationb (1789/2464)a | Meta-analysis in Chinese | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | P-value | OR | P-value | OR | P-value | OR | P-value | ||||||
| rs9379871 | 26375854 | / | C/G | 0.888 | 0.937 | 1.072 | 1.24×10−8 | 1.127 | 0.0109 | 1.005 | 0.954 | 1.091 | 0.0478 |
| rs9257803 | 29344395 | 0.001 | T/C | 0.095 | 0.049 | 0.932 | 1.03×10−8 | 0.822 | 0.00387 | 0.698 | 0.00231 | 0.784 | 0.000139 |
| rs9262201 | 30760886 | 0.001 | A/G | 0.780 | 0.874 | 1.052 | 8.65×10−8 | 1.085 | 0.0357 | 1.156 | 0.0351 | 1.108 | 0.0101 |
LD linkage disequilibrium, Allele effect allele/non-effect allele, OR odds ratio, Freq frequency of effect allele
aThe sample size in bracket is shown as MDD cases/normal controls
b“Chinese Replication” means our collected further replication samples from Han Chinese population, which is independent from CONVERGE GWAS [15]
The “Meta-analysis in Chinese” is calculated based on the OR and standard error (SE) from CONVERGE GWAS [15] and our collected Chinese replication sample
These three MDD risk SNPs are located in the noncoding regions, and their functionalities are still unknown. We thus applied the plausible approach, eQTL analysis [32], to investigate their potential physiological impacts. Intriguingly, two of the three risk SNPs displayed eQTL effects in brains. The strongest eQTL association appeared between BTN3A2 and rs9379871 (p = 4.79 × 10−16 in the DLPFC of BrainSeq and p = 4.20 × 10−13 in the frontal cortex of GTEx, Figure S5), and subjects carrying the risk allele [C] of rs9379871 showed an increase of BTN3A2 mRNA levels in the peripheral blood, indicating that higher expression levels of this gene might be a risk factor for MDD. Since rs9379871 was also associated with MDD in the Han Chinese population, we then investigated the association between this SNP and BTN3A2 expression in the amygdala tissues (N = 65) collected from Han Chinese donors. Again, the risk allele [C] of rs9379871 predicted a higher mRNA expression level of BTN3A2 (p = 0.0002, Figure S6). There was another SNP rs9262201 showing association with MDD in both Europeans and Han Chinese, and exhibited significant eQTL associations with HCG20 mRNA expression in the BrainSeq and GTEx datasets (p = 1.19 × 10−7 in the DLPFC of BrainSeq and p = 5.10 × 10−4 in the frontal cortex of GTEx, Figure S5). The MDD risk allele [A] at rs9262201 was associated with a higher expression level of HCG20 in both datasets. Since the expression levels of HCG20 in brains were relatively low and thus undetectable using our quantitative real-time PCR (qRT-PCR) method, we were not able to test the associations between rs9262201 and HCG20 expression in our Han Chinese amygdala tissues. Nevertheless, these results suggested potential common molecular genetic basis of MDD associations in the MHC region shared by Europeans and Han Chinese, i.e., increased expression of BTN3A2 are likely risk factors for MDD in both ethnic groups.
The mRNA expression of a brain-enriched gene ZNF603P was associated with MDD diagnosis status and genetic risk
To gain further insights into the potential pathophysiological roles of genes in the MHC region in MDD, we assessed the effects of diagnostic status on mRNA expression patterns in human brains. We re-analyzed the raw RNA-seq data from two independent cohorts of European ancestry [34, 35], and quantified the gene-expression level changes in MDD patients by covaring expression heterogeneity using PCA analysis. Considering the relatively small sizes of these two samples (GSE102556: 26 MDD cases and 22 controls [34]; GSE101521: 30 MDD cases and 29 controls [35]), we conducted a meta-analysis using their summary statistics (Beta and SE). In this meta-analysis, 11 MHC genes showed nominally differential expression levels between cases and controls (p < 0.05, Table S5). Although none of these genes survived multiple corrections according to the number of genes in the MHC regions (which might be due to the limited statistical power of the small sample size), they showed the same direction of diagnostic effects in both datasets. Some of these genes, based on previous findings, worth further attention. For example, ZSCAN31, which has been reported to show lower expression levels in schizophrenia cases compared with controls [66], was also downregulated in MDD patients compared with healthy subjects in our analysis (p = 0.0214, Table S5), and the MDD risk allele was associated with decreased expression of ZSCAN31 in the eQTL analysis of BrainSeq and GTEx datasets (the bottom line of Table S2).
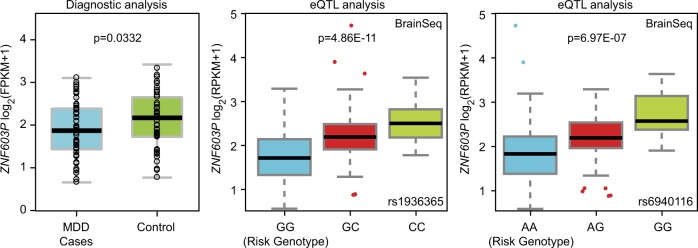
In addition to ZSCAN31, ZNF603P was decreased in MDD cases compared with controls (p = 0.0332). This gene was also highlighted in the earlier eQTL analyses (it showed significant associations with multiple MDD risk alleles), and both the eQTL analysis and diagnosis analysis exhibited the same directions (Fig. 3), i.e., if the gene was downregulated in MDD patients compared with healthy controls (e.g., ZNF603P, p = 0.0332 in Table S5), the linked MDD risk alleles also pointed to a lower mRNA expression level of this gene in the eQTL analysis (e.g., the eQTL association between rs1936365 and ZNF603P was p = 4.86 × 10−11, Table S2). Therefore, ZNF603P is likely a susceptibility gene for MDD at least in Europeans. To identify if this gene also confers risk of MDD in Han Chinese, we measured the mRNA levels of this gene in our Han Chinese amygdala samples using qRT-PCR. Of note, we observed the eQTL associations between rs1936365 and ZNF603P expression in Han Chinese (p = 0.00744, Table S6) in the same direction of allelic effects as that in Europeans, suggesting it is a common eQTL variant in both populations. However, rs1936365 was not associated with MDD in the Han Chinese CONVERGE GWAS (p = 0.364, Table S4). We also conducted genome-wide SNP array analysis using Illumina GSA platforms in a subset of our Han Chinese amygdala samples (N = 59) followed by high-quality imputation. This analysis yielded a total of 57,423 SNPs within the MHC region (chr6:26-34 M) passing quality control (INFO > 0.8, call rate > 0.95). A linear regression analysis of these SNPs against ZNF603P expression identified multiple significant eQTL SNPs, but none of them were associated with MDD in the Han Chinese CONVERGE GWAS (Table S6). Therefore, based on the existing data, we cannot conclude that ZNF603P is a risk gene for MDD in Han Chinese, and further analyses are necessary.
Fig. 3.
Risk genotype and diagnosis predict ZNF603P mRNA expression. (Left) Diagnostic analysis of ZNF603P expression in a meta-analysis of two RNA-seq datasets [34, 35] from frontal cortex (56 MDD cases and 51 healthy controls). (middle) Association of rs1936365 with ZNF603P expression in the 237 non-psychiatric controls from DLPFC in BrainSeq dataset [50]. (Right) Association of rs6940116 with ZNF603P expression in the 237 non-psychiatric controls from DLPFC in BrainSeq dataset [50]. The source of the eQTL plots for rs1936365 and rs6940116 was also utilized in Figure S4. eQTL, expression quantitative trait loci
The function of ZNF603P in brain is still unclear. It was proposed to be a pseudogene belonging to the lncRNA class. However, the mRNA levels of ZNF603P in different brain tissues, as presented in the two MDD RNA-seq datasets [34, 35], are moderate (average FPKM ≥ 3.00). Moreover, using the RNA-seq data from diverse human tissues included in the GTEx dataset [51], we found that ZNF603P was preferably expressed in several brain regions known to participate in cognitive processes and frequently implicated in the neuropathology of MDD (such as the frontal cortex (BA9)), while the mRNA expression levels of ZNF603P in peripheral tissues were relatively low (the expression profile in diverse tissues is shown in Figure S7). Therefore, ZNF603P likely exerts pivotal but unidentified functions in brains and further study characterizing its function is needed.
Discussion
The advancements in genetic techniques and large-scale international collaboration have greatly improved our understanding of the neurobiology of MDD [67]. To delve into the genetic and physiological basis of MDD, we have explored the MHC region genetic variants showing genome-wide significant associations with this illness in several GWAS datasets of MDD-relevant phenotypes, and presented convergent and consistent lines of evidence suggesting that several MHC region genes are risk factors for MDD. Although the results for most of these SNPs in MDD-related phenotypes did not reach genome-wide level of significance, the consistent findings across different analyses that the MDD risk alleles all pointed to abnormalities linked to the disease strongly supported the reliability of the study.
We also attempted to obtain a broader view in the search for potential causal variants via performing the trans-ethnic replication analysis of the European MDD risk SNPs in Han Chinese individuals, as leveraging variations in the LD structure across groups with different genetic backgrounds was proposed to be helpful to achieve this goal [30, 68–70]. Although this method could miss certain risk loci restricted to a particular population, common risk loci in the general world populations are likely detected with greater efficiency. However, as mentioned earlier, the MHC region is a target of natural selection during human evolution, and it is unclear whether human evolution has any impact on the genetic risk associations with MDD. Indeed, among the hundreds of genome-wide risk variants in Europeans, only three independent SNPs were also nominally associated with MDD in two independent Han Chinese samples (Table 2). We then calculated the integrated haplotype score (iHS) of MHC SNPs using genotype data from 1000 Genomes Project [41] with Selscan [71, 72], which could reflect recent positive selection during human evolution. Intriguingly, a substantial portion of the MDD genome-wide risk SNPs underwent positive selection in Africans or Europeans (The criteria for SNP under positive selection should have |iHS| > 1.96, which empirically corresponds to the most extreme 5% of iHS values across the genome with minor allele frequency > 0.05), while few of them showed signature of natural selection in East Asians (Table S7 and Figure S8). These preliminary results suggested distinct effects of natural selection on the MHC variants in divergent populations, which may eventually influence the genetic risk architectures of complex disorders, such as MDD. This observation is also in line with our LD analysis results of the 581 genome-wide risk SNPs in Europeans and in Han Chinese, in which we observed sharp differences in LD structures of the MHC region between these two populations (Figure S9). Therefore, there are likely both common and population-specific genetic risk variants for MDD within the MHC region.
Taken one step further, we utilized the public RNA-seq resources to examine the expression patterns of MHC region genes in MDD patients and controls, as well as their associations with the MDD risk SNPs in this genomic region. These series of analyses have identified a brain-enriched gene ZNF603P showing consistent patterns of allelic and diagnostic associations. For this gene, the MDD risk genotypes and diagnostic status predicted the same direction of gene-expression levels in brains, indicating that aberrant transcriptional regulation of ZNF603P was likely correlated to the mechanisms underlying the genetic risk associations of the MHC region variants. These results corroborate the contention that complex disease GWAS loci usually exert effects via altering gene-expression levels in relevant tissues [31, 32, 61, 73]. While their roles in the pathogenesis of MDD are still unclear, a possible explanation is that those genes contribute to the immune dysfunction in MDD [74, 75]. Nevertheless, the hypothesis that MDD is in part attributed to immune dysfunction still remains to be tested, as Glanville et al. reported that C4 haplotypes or HLA alleles, which play a major role in the susceptibility to autoimmune diseases, were not associated with the risk of MDD [40]. This contention appears to be in line with our results. However, given the substantial overlap between the samples used in their and our studies, further investigation into this and other hypothesized etiologic mechanisms underlying the genetic risk of MDD conferred by the MHC region variants should be performed.
Despite these intriguing findings, there are also limitations calling for precautions and further investigations. For example, if the strength of the present study is the comprehensive integration of publicly available databases, a major weakness will be the lack of functional validations of the risk genes, which should be carried out in the future. In addition, although we have reported a set of susceptibility genes in the MHC region, we believe that there are still other susceptibility genes involved in the risk of MDD yet to be identified given the high density of genes in this genomic region. Third, it is well-characterized that there are many genes belonging to the same family in the MHC region, especially the genes encoding the HLA class I heavy-chain paralogs (e.g., HLA-A, HLA-B, HLA-C). While these genes may share the same DNA sequence elements, they could also result in the misalignment during RNA-seq results analyses, in which event false positive or negative results in identifying those homologous genes may occur [76]. While we are cautious in the interpretations of the currently available data, we believe that the best solution to this problem is to apply the long reads sequencing methods such as PacBio. Additionally, we also realized that the genotyping platforms and quality control standards are different between datasets, which may result in missing of certain SNPs and even the incomplete overall picture of the MHC associations. Meanwhile, we replicated the eQTL associations of both BTN3A2 and ZNF603P expression in our Han Chinese amygdala sample using qRT-PCR, whereas the European eQTL datasets used RNA-seq analysis; also, we were not able to detect HCG20 expression using qRT-PCR in our sample, it is thus necessary to examine the variation resulted from different gene-expression quantification techniques. It is also important to note that MDD is a complex illness with strong phenotypic heterogeneity during clinical diagnosis. Although we have excluded certain individuals affected by known confounding factors, we cannot rule out the possibility that the remaining MDD patients are still heterogeneous. For example, most cases included in the current study were recurrent MDD patients, and thus have received antidepressant therapeutics. We therefore cannot exclude the confounding effects of antidepressant therapeutics on the risk association signals between MDD and MHC region variants, as certain genes at MHC region might be targets of antidepressant therapeutics. Further analyses either considering antidepressant therapeutics as covariates or stratify patients according to their responses to antidepressant therapeutics should be carried out. Finally, although we found the independent MHC SNPs are associated with the same gene(s) expression, and this might suggest convergence of the pathway(s) or molecular mechanisms underlying independent genetic risk; however, it is also possible that the independent MHC SNPs are both in substantial LD with the same causative or regulative variant(s). In such cases, the observed eQTL associations of independent SNPs with the same genes should not be considered as evidence of convergent signal, but were the reflections of functional consequences attributed to the causative variants.
Funding and disclosure
All authors have no conflicts of interest to declare. This work was supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant no. XDB13000000 to ML), National Natural Science Foundation of China (Grant nos. 81722019 to ML, 31701133 to XX, 31701088 to LL, 81871067 to HC, 81671330 to LX-L, 31730037 to Y-GY, 81471358 to CZ, and 81771450 to CZ), Yunnan Applied Basic Research Projects (2018FB051 to XX, 2018FB136 to HC), the Medical and Health Science and Technology project in Zhejiang (2018KY721 to DSZ), Shanghai Municipal Education Commission—Gaofeng Clinical Medicine Grant Support (20152530 to CZ), the Shanghai Municipal Commission of Health and Family Planning Foundation, Key Developing Disciplines (2015ZB0405 to CZ), the High Scientific and Technological Research Fund of Xinxiang Medical University (Grant no. 2017ZDCG-04 to LX-L), the Medical science and technology research project of Henan Province (201702131 to YF-Y), the Fundamental and Advancing Foundation of Henan Province (132300413216 to LX-L), the Training plan for young excellent teachers in Colleges and Universities of Henan (No. 2016GGJS-106 to WQ-L), the Science and technology project of Xinxiang (CXGG17030 to WQ-L), and the support project for the Disciplinary group of Psychiatry and Neuroscience, Xinxiang Medical University. Xiao Xiao was also supported by the Chinese Academy of Sciences Western Light Program, and Youth Innovation Promotion Association, CAS. Ming Li was also supported by CAS Pioneer Hundred Talents Program and the 1000 Young Talents Program.
Supplementary information
Acknowledgements
The authors sincerely acknowledge with appreciation all the individuals with major depressive disorders and healthy controls whose contributions made this work possible. The authors are deeply grateful to all the participants as well as to the physicians working on this project.
Footnotes
Co-first authors: Huijuan Li, Hong Chang, Xueqin Song
Co-senior authors: Xiao Xiao, Luxian Lv, Ming Li
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xiao Xiao, Phone: +86 871 65190612, Email: xiaoxiao2@mail.kiz.ac.cn.
Luxian Lv, Email: lvx928@126.com.
Ming Li, Phone: +86 871 65190612, Email: limingkiz@mail.kiz.ac.cn.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41386-019-0346-3).
References
- 1.Hasin DS, Goodwin RD, Stinson FS, Grant BF. Epidemiology of major depressive disorder: results from the National Epidemiologic Survey on Alcoholism and Related Conditions. Arch Gen Psychiatry. 2005;62:1097–1106. doi: 10.1001/archpsyc.62.10.1097. [DOI] [PubMed] [Google Scholar]
- 2.Sullivan PF, Neale MC, Kendler KS. Genetic epidemiology of major depression: review and meta-analysis. Am J Psychiatry. 2000;157:1552–1562. doi: 10.1176/appi.ajp.157.10.1552. [DOI] [PubMed] [Google Scholar]
- 3.Kohli MA, Lucae S, Saemann PG, Schmidt MV, Demirkan A, Hek K, et al. The neuronal transporter gene SLC6A15 confers risk to major depression. Neuron. 2011;70:252–265. doi: 10.1016/j.neuron.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis CM, Ng MY, Butler AW, Cohen-Woods S, Uher R, Pirlo K, et al. Genome-wide association study of major recurrent depression in the U.K. population. Am J Psychiatry. 2010;167:949–957. doi: 10.1176/appi.ajp.2010.09091380. [DOI] [PubMed] [Google Scholar]
- 5.Major Depressive Disorder Working Group of the Psychiatric Gwas Consortium. Ripke S, Wray NR, Lewis CM, Hamilton SP, Weissman MM, et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2013;18:497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muglia P, Tozzi F, Galwey NW, Francks C, Upmanyu R, Kong XQ, et al. Genome-wide association study of recurrent major depressive disorder in two European case-control cohorts. Mol Psychiatry. 2010;15:589–601. doi: 10.1038/mp.2008.131. [DOI] [PubMed] [Google Scholar]
- 7.Power RA, Tansey KE, Buttenschon HN, Cohen-Woods S, Bigdeli T, Hall LS, et al. Genome-wide association for major depression through age at onset stratification: Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium. Biol Psychiatry. 2017;81:325–335. doi: 10.1016/j.biopsych.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rietschel M, Mattheisen M, Frank J, Treutlein J, Degenhardt F, Breuer R, et al. Genome-wide association-, replication-, and neuroimaging study implicates HOMER1 in the etiology of major depression. Biol Psychiatry. 2010;68:578–585. doi: 10.1016/j.biopsych.2010.05.038. [DOI] [PubMed] [Google Scholar]
- 9.Shi J, Potash JB, Knowles JA, Weissman MM, Coryell W, Scheftner WA, et al. Genome-wide association study of recurrent early-onset major depressive disorder. Mol Psychiatry. 2011;16:193–201. doi: 10.1038/mp.2009.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wray NR, Pergadia ML, Blackwood DH, Penninx BW, Gordon SD, Nyholt DR, et al. Genome-wide association study of major depressive disorder: new results, meta-analysis, and lessons learned. Mol Psychiatry. 2012;17:36–48. doi: 10.1038/mp.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levinson DF, Mostafavi S, Milaneschi Y, Rivera M, Ripke S, Wray NR, et al. Genetic studies of major depressive disorder: why are there no genome-wide association study findings and what can we do about it? Biol Psychiatry. 2014;76:510–512. doi: 10.1016/j.biopsych.2014.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howard David M., Adams Mark J., Clarke Toni-Kim, Hafferty Jonathan D., Gibson Jude, Shirali Masoud, Coleman Jonathan R. I., Hagenaars Saskia P., Ward Joey, Wigmore Eleanor M., Alloza Clara, Shen Xueyi, Barbu Miruna C., Xu Eileen Y., Whalley Heather C., Marioni Riccardo E., Porteous David J., Davies Gail, Deary Ian J., Hemani Gibran, Berger Klaus, Teismann Henning, Rawal Rajesh, Arolt Volker, Baune Bernhard T., Dannlowski Udo, Domschke Katharina, Tian Chao, Hinds David A., Trzaskowski Maciej, Byrne Enda M., Ripke Stephan, Smith Daniel J., Sullivan Patrick F., Wray Naomi R., Breen Gerome, Lewis Cathryn M., McIntosh Andrew M. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nature Neuroscience. 2019;22(3):343–352. doi: 10.1038/s41593-018-0326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howard DM, Adams MJ, Shirali M, Clarke TK, Marioni RE, Davies G, et al. Genome-wide association study of depression phenotypes in UK Biobank identifies variants in excitatory synaptic pathways. Nat Commun. 2018;9:1470. doi: 10.1038/s41467-018-03819-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hyde CL, Nagle MW, Tian C, Chen X, Paciga SA, Wendland JR, et al. Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat Genet. 2016;48:1031–1036. doi: 10.1038/ng.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Converge consortium. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature. 2015;523:588–591. doi: 10.1038/nature14659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50:668–681. doi: 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fales CL, Barch DM, Rundle MM, Mintun MA, Snyder AZ, Cohen JD, et al. Altered emotional interference processing in affective and cognitive-control brain circuitry in major depression. Biol Psychiatry. 2008;63:377–384. doi: 10.1016/j.biopsych.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinkelmann K, Moritz S, Botzenhardt J, Riedesel K, Wiedemann K, Kellner M, et al. Cognitive impairment in major depression: association with salivary cortisol. Biol Psychiatry. 2009;66:879–885. doi: 10.1016/j.biopsych.2009.06.023. [DOI] [PubMed] [Google Scholar]
- 19.Hamilton JP, Siemer M, Gotlib IH. Amygdala volume in major depressive disorder: a meta-analysis of magnetic resonance imaging studies. Mol Psychiatry. 2008;13:993–1000. doi: 10.1038/mp.2008.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Y, Coupland NJ, Lebel RM, Carter R, Seres P, Wilman AH, et al. Structural changes in hippocampal subfields in major depressive disorder: a high-field magnetic resonance imaging study. Biol Psychiatry. 2013;74:62–68. doi: 10.1016/j.biopsych.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 21.Forrest MP, Parnell E, Penzes P. Dendritic structural plasticity and neuropsychiatric disease. Nat Rev Neurosci. 2018;19:215–234. doi: 10.1038/nrn.2018.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci. 2011;14:285–293. doi: 10.1038/nn.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duman RS, Aghajanian GK. Synaptic dysfunction in depression: potential therapeutic targets. Science. 2012;338:68–72. doi: 10.1126/science.1222939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ayalew M, Le-Niculescu H, Levey DF, Jain N, Changala B, Patel SD, et al. Convergent functional genomics of schizophrenia: from comprehensive understanding to genetic risk prediction. Mol Psychiatry. 2012;17:887–905. doi: 10.1038/mp.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurian SM, Le-Niculescu H, Patel SD, Bertram D, Davis J, Dike C, et al. Identification of blood biomarkers for psychosis using convergent functional genomics. Mol Psychiatry. 2011;16:37–58. doi: 10.1038/mp.2009.117. [DOI] [PubMed] [Google Scholar]
- 26.Le-Niculescu H, Kurian SM, Yehyawi N, Dike C, Patel SD, Edenberg HJ, et al. Identifying blood biomarkers for mood disorders using convergent functional genomics. Mol Psychiatry. 2009;14:156–174. doi: 10.1038/mp.2008.11. [DOI] [PubMed] [Google Scholar]
- 27.Niculescu AB, Le-Niculescu H. Convergent Functional Genomics: what we have learned and can learn about genes, pathways, and mechanisms. Neuropsychopharmacology. 2010;35:355–356. doi: 10.1038/npp.2009.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schuhmacher A, Lennertz L, Wagner M, Hofels S, Pfeiffer U, Guttenthaler V, et al. A variant of the neuronal amino acid transporter SLC6A15 is associated with ACTH and cortisol responses and cognitive performance in unipolar depression. Int J Neuropsychopharmacol. 2013;16:83–90. doi: 10.1017/S1461145712000223. [DOI] [PubMed] [Google Scholar]
- 29.Xiao X, Zhang C, Grigoroiu-Serbanescu M, Wang L, Li L, Zhou D, et al. The cAMP responsive element-binding (CREB)-1 gene increases risk of major psychiatric disorders. Mol Psychiatry. 2017;23:1–11. doi: 10.1038/mp.2017.243. [DOI] [PubMed] [Google Scholar]
- 30.Xiao X, Zheng F, Chang H, Ma Y, Yao YG, Luo XJ, et al. The gene encoding protocadherin 9 (PCDH9), a novel risk factor for major depressive disorder. Neuropsychopharmacology. 2018;43:1128–1137. doi: 10.1038/npp.2017.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bray NJ, Hill MJ. Translating genetic risk loci into molecular risk mechanisms for schizophrenia. Schizophr Bull. 2016;42:5–8. doi: 10.1093/schbul/sbv156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edwards SL, Beesley J, French JD, Dunning AM. Beyond GWASs: illuminating the dark road from association to function. Am J Hum Genet. 2013;93:779–797. doi: 10.1016/j.ajhg.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jansen R, Penninx BW, Madar V, Xia K, Milaneschi Y, Hottenga JJ, et al. Gene expression in major depressive disorder. Mol Psychiatry. 2016;21:339–347. doi: 10.1038/mp.2015.57. [DOI] [PubMed] [Google Scholar]
- 34.Labonte B, Engmann O, Purushothaman I, Menard C, Wang J, Tan C, et al. Sex-specific transcriptional signatures in human depression. Nat Med. 2017;23:1102–1111. doi: 10.1038/nm.4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pantazatos SP, Huang YY, Rosoklija GB, Dwork AJ, Arango V, Mann JJ. Whole-transcriptome brain expression and exon-usage profiling in major depression and suicide: evidence for altered glial, endothelial and ATPase activity. Mol Psychiatry. 2017;22:760–773. doi: 10.1038/mp.2016.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.The MHC sequencing consortium. Complete sequence and gene map of a human major histocompatibility complex. The MHC sequencing consortium. Nature. 1999;401:921–923. doi: 10.1038/44853. [DOI] [PubMed] [Google Scholar]
- 37.Rock KL, Reits E, Neefjes J. Present yourself! By MHC Class I and MHC Class II Molecules. Trends Immunol. 2016;37:724–737. doi: 10.1016/j.it.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, et al. Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530:177–183. doi: 10.1038/nature16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Glanville KP, Coleman JR, Hanscombe KB, Euesden J, Choi SW, Purves KL, et al. Genetic variation in the Major Histocompatibility Complex and association with depression. bioRxiv: 2018;469023.
- 41.Genomes Project Consortium. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 43.Li Z, Zhang Z, He Z, Tang W, Li T, Zeng Z, et al. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio-x.cn) Cell Res. 2009;19:519–523. doi: 10.1038/cr.2009.33. [DOI] [PubMed] [Google Scholar]
- 44.Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97–98. doi: 10.1038/sj.cr.7290286. [DOI] [PubMed] [Google Scholar]
- 45.Nagel M, Jansen PR, Stringer S, Watanabe K, de Leeuw CA, Bryois J, et al. Meta-analysis of genome-wide association studies for neuroticism in 449,484 individuals identifies novel genetic loci and pathways. Nat Genet. 2018;50:920–927. doi: 10.1038/s41588-018-0151-7. [DOI] [PubMed] [Google Scholar]
- 46.Lee JJ, Wedow R, Okbay A, Kong E, Maghzian O, Zacher M, et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet. 2018;50:1112–1121. doi: 10.1038/s41588-018-0147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davies G, Lam M, Harris SE, Trampush JW, Luciano M, Hill WD, et al. Study of 300,486 individuals identifies 148 independent genetic loci influencing general cognitive function. Nat Commun. 2018;9:2098. doi: 10.1038/s41467-018-04362-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Savage JE, Jansen PR, Stringer S, Watanabe K, Bryois J, de Leeuw CA, et al. Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat Genet. 2018;50:912–919. doi: 10.1038/s41588-018-0152-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okbay A, Baselmans BM, De Neve JE, Turley P, Nivard MG, Fontana MA, et al. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat Genet. 2016;48:624–633. doi: 10.1038/ng.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jaffe AE, Straub RE, Shin JH, Tao R, Gao Y, Collado-Torres L, et al. Developmental and genetic regulation of the human cortex transcriptome illuminate schizophrenia pathogenesis. Nat Neurosci. 2018;21:1117–1125. doi: 10.1038/s41593-018-0197-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 53.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 54.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walters JT, Rujescu D, Franke B, Giegling I, Vasquez AA, Hargreaves A, et al. The role of the major histocompatibility complex region in cognition and brain structure: a schizophrenia GWAS follow-up. Am J Psychiatry. 2013;170:877–885. doi: 10.1176/appi.ajp.2013.12020226. [DOI] [PubMed] [Google Scholar]
- 56.Kendler KS, Gatz M, Gardner CO, Pedersen NL. Personality and major depression: a Swedish longitudinal, population-based twin study. Arch Gen Psychiatry. 2006;63:1113–1120. doi: 10.1001/archpsyc.63.10.1113. [DOI] [PubMed] [Google Scholar]
- 57.Kotov R, Gamez W, Schmidt F, Watson D. Linking “big” personality traits to anxiety, depressive, and substance use disorders: a meta-analysis. Psychol Bull. 2010;136:768–821. doi: 10.1037/a0020327. [DOI] [PubMed] [Google Scholar]
- 58.Genetics of Personality Consortium. de Moor MH, van den Berg SM, Verweij KJ, Krueger RF, Luciano M, et al. Meta-analysis of genome-wide association studies for neuroticism, and the polygenic association with major depressive disorder. JAMA Psychiatry. 2015;72:642–650. doi: 10.1001/jamapsychiatry.2015.0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luciano M, Hagenaars SP, Davies G, Hill WD, Clarke TK, Shirali M, et al. Association analysis in over 329,000 individuals identifies 116 independent variants influencing neuroticism. Nat Genet. 2018;50:6–11. doi: 10.1038/s41588-017-0013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nagel M, Watanabe K, Stringer S, Posthuma D, van der Sluis S. Item-level analyses reveal genetic heterogeneity in neuroticism. Nat Commun. 2018;9:905. doi: 10.1038/s41467-018-03242-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li M, Jaffe AE, Straub RE, Tao R, Shin JH, Wang Y, et al. A human-specific AS3MT isoform and BORCS7 are molecular risk factors in the 10q24.32 schizophrenia-associated locus. Nat Med. 2016;22:649–656. doi: 10.1038/nm.4096. [DOI] [PubMed] [Google Scholar]
- 62.Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, et al. Human demographic history impacts genetic risk prediction across diverse populations. Am J Hum Genet. 2017;100:635–649. doi: 10.1016/j.ajhg.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sillanpaa MJ, Auranen K. Replication in genetic studies of complex traits. Ann Hum Genet. 2004;68(Pt 6):646–657. doi: 10.1046/j.1529-8817.2004.00122.x. [DOI] [PubMed] [Google Scholar]
- 64.Traherne JA. Human MHC architecture and evolution: implications for disease association studies. Int J Immunogenet. 2008;35:179–192. doi: 10.1111/j.1744-313X.2008.00765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yasukochi Y, Satta Y. Current perspectives on the intensity of natural selection of MHC loci. Immunogenetics. 2013;65:479–483. doi: 10.1007/s00251-013-0693-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Luo XJ, Mattheisen M, Li M, Huang L, Rietschel M, Borglum AD, et al. Systematic integration of brain eQTL and GWAS identifies ZNF323 as a novel schizophrenia risk gene and suggests recent positive selection based on compensatory advantage on pulmonary function. Schizophr Bull. 2015;41:1294–1308. doi: 10.1093/schbul/sbv017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sullivan PF. The psychiatric GWAS consortium: big science comes to psychiatry. Neuron. 2010;68:182–186. doi: 10.1016/j.neuron.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bigdeli TB, Ripke S, Peterson RE, Trzaskowski M, Bacanu SA, Abdellaoui A, et al. Genetic effects influencing risk for major depressive disorder in China and Europe. Transl Psychiatry. 2017;7:e1074. doi: 10.1038/tp.2016.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li X, Luo Z, Gu C, Hall LS, McIntosh AM, Zeng Y, et al. Common variants on 6q16.2, 12q24.31 and 16p13.3 are associated with major depressive disorder. Neuropsychopharmacology. 2018;43:2146–2153. doi: 10.1038/s41386-018-0078-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Morris AP. Transethnic meta-analysis of genomewide association studies. Genet Epidemiol. 2011;35:809–822. doi: 10.1002/gepi.20630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ, Schaffner SF, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 72.Szpiech ZA, Hernandez RD. selscan: an efficient multithreaded program to perform EHH-based scans for positive selection. Mol Biol Evol. 2014;31:2824–2827. doi: 10.1093/molbev/msu211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19:1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fung TC, Olson CA, Hsiao EY. Interactions between the microbiota, immune and nervous systems in health and disease. Nat Neurosci. 2017;20:145–155. doi: 10.1038/nn.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wohleb ES, Franklin T, Iwata M, Duman RS. Integrating neuroimmune systems in the neurobiology of depression. Nat Rev Neurosci. 2016;17:497–511. doi: 10.1038/nrn.2016.69. [DOI] [PubMed] [Google Scholar]
- 76.Brandt DY, Aguiar VR, Bitarello BD, Nunes K, Goudet J, Meyer D. Mapping bias overestimates reference allele frequencies at the HLA genes in the 1000 Genomes Project Phase I Data. G3 (Bethesda) 2015;5:931–941. doi: 10.1534/g3.114.015784. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.