Abstract
The base excision repair DNA N-glycosylase MBD4 (also known as MED1), an interactor of the DNA mismatch repair protein MLH1, plays a central role in the maintenance of genomic stability of CpG sites by removing thymine and uracil from G:T and G:U mismatches, respectively. MBD4 is also involved in DNA damage response and transcriptional regulation. The interaction with other proteins is likely critical for understanding MBD4 functions. To identify novel proteins that interact with MBD4, we used tandem affinity purification (TAP) from HEK-293 cells. The MBD4-TAP fusion and its co-associated proteins were purified sequentially on IgG and calmodulin affinity columns; the final eluate was shown to contain MLH1 by western blotting, and MBD4-associated proteins were identified by mass spectrometry. Bands with molecular weight higher than that expected for MBD4 (~66kD) yielded peptides corresponding to MBD4 itself and the small ubiquitin-like molecule-1 (SUMO1), suggesting that MBD4 is sumoylated in vivo. MBD4 sumoylation was validated by co-immunoprecipitation in HEK-293 and MCF7 cells, and by an in vitro sumoylation assay. Sequence and mutation analysis identified three main sumoylation sites: MBD4 is sumoylated preferentially on K137, with additional sumoylation at K215 and K377. Patterns of MBD4 sumoylation were altered, in a DNA damage-specific way, by the anti-metabolite 5-fluorouracil, the alkylating agent N-Nitroso-N-methylurea and the crosslinking agent cisplatin. MCF7 extract expressing sumoylated MBD4 displays higher thymine glycosylase activity than the unmodified species. Of the 67 MBD4 missense mutations reported in The Cancer Genome Atlas, 14 (20.9%) map near sumoylation sites. These results indicate that MBD4 is sumoylated in vivo in a DNA damage-specific manner, and suggest that sumoylation serves to regulate its repair activity and could be compromised in cancer. This study expands the role played by sumoylation in fine-tuning DNA damage response and repair.
Keywords: MBD4/MED1, SUMO1, sumoylation, DNA damage
INTRODUCTION
Sumoylation is a post-translational modification involving the covalent attachment of small ubiquitin-like molecules (SUMO) to target proteins. It is a reversible process akin to ubiquitination, as they share overall structural similarities and activation cycle [1–3]. Sumoylation is highly conserved in evolution; invertebrates have just one SUMO gene, whereas mammalian genomes contain four genes, SUMO1–4 encoding SUMO proteins [4, 5].
All SUMO proteins are synthesized as inactive isoforms and then cleaved by isopeptidases SENPs to expose two Glycines in the C-terminus of the protein [6]. In an ATP-dependent reaction, SUMO is activated by activating enzyme E1, which is a heterodimer with two subunits, AOS1 and UBA2. SUMO is subsequently transferred onto a catalytic cysteine of the second cofactor of the sumoylation cycle, called E2, also known as UBC9, which moves SUMO to its target protein and for this reason it is called conjugating enzyme. The final step leads to the formation of a peptide bond between the glycine residue of SUMO and a lysine residue of the target protein. Sometimes, the sumoylation cycle requires the presence of another cofactor, the E3 ligase, whose main role is to enhance the identification of SUMO targets [7–9]. Most of SUMO target proteins show an acceptor lysine within the consensus site: ψKX(D/E), in which ψ is a large hydrophobic residue [10]. E3 can be omitted in in vitro experiments [7].
The molecular consequences of sumoylation are profoundly different from (poly)ubiquitination, which leads to protein degradation. In general, sumoylation has three main consequences: i) it may interfere with the interaction between a target and its partner, and in this case, the interaction is permissive in the absence of SUMO; ii) it may provide a non-covalent binding site for an interaction partner containing a SUMO-interacting motif (SIM); iii) it may promote a conformational change and modify the activity of the target. Sumoylation is involved in many cellular processes, including transcription, replication, chromosome segregation, cell cycle progression, DNA damage response and DNA repair [11–14].
The effects of sumoylation on base excision repair (BER) are substantial [15]. Sumoylation is particularly important for the regulation of the BER enzyme and epigenetic factor, Thymidine DNA Glycosylase (TDG). Sumoylation induces a conformational change in TDG, which leads to decreased binding to G:T, G:U mismatches and G:AP (abasic) product site; since many DNA Glycosylases are product-inhibited [16–21], it was proposed that sumoylation increases the turnover of TDG [22, 23]. Single-turnover experiments have shown that sumoylated TDG exhibits lower catalytic activity on G:T mismatches, as well as on the epigenetically relevant substrates G:formylcytosine (fC) and G:carboxylcytosine (caC) [24], involved in active DNA demethylation [25]. However, sumoylated TDG retains substantial affinity for fC and caC, raising the possibility that sumoylation converts TDG into a reader of these epigenetic modifications [24]. TDG is also known to interact with the transcriptional coactivators CREB-binding protein (CBP) and p300; sumoylation abrogates the ability of TDG to interact with these co-activators [26]. More recently, it has been shown that sumoylation affects the assembly of a BERosome in DNA demethylation [27].
MBD4 (also known as MED1) is a G:T and G:U mismatch glycosylase whose function is to maintain the integrity of CpG sites in the genome and prevent the mutagenic consequences of deamination of 5-methylcytosine and cytosine to thymine and uracil, respectively, i.e. prevent CpG to CpA/TpG transition mutations [17, 28]. MBD4 was identified independently as an interactor of the mismatch repair protein MLH1 [29] and as a protein containing a MethylCpG binding domain (MBD)[30]. Indeed, MBD4 has a tripartite structure with an N-terminal MBD and a C-terminal glycosylase domain, separated by a central region of low sequence complexity that appears to act as a spacer [31]. Recent studies have shown also its involvement in Immunoglobulin Class Switch Recombination [32, 33]. MBD4 has additional functions, including regulation of apoptosis of cells exposed to DNA-damaging agents [34, 35], modulation of the expression levels of core mismatch repair proteins [34], and transcriptional regulation [36]. MBD4 also shows a prominent activity in repairing halogenated pyrimidines and is essential for the cytotoxicity of the radiosensitizing agent, 5-iododeoxyuridine [37, 38]. Inactivation of MBD4 plays a role in tumorigenesis. In fact, secondary inactivating mutations of MBD4, causing frameshifts at A6 and A10 polyadenine tracks in its coding region, have been described in colorectal, endometrial, pancreatic and stomach cancers characterized by microsatellite instability [39–41]; and its promoter is silenced by hypermethylation in ovarian cancer silencing [42, 43]. More recently, biallelic inactivation of MBD4 has been described in hypermutable juvenile acute myelogenous leukemia [44] and was associated with outlier response of an uveal melanoma patient to immunotherapy [45].
We reasoned that identification of other interacting partners of MBD4 beside MLH1 may help clarify its multiple functions. For this reason, we performed tandem affinity purification, and identified several associated polypeptides, including prominently SUMO1, which led us to analyze in detail MBD4 sumoylation.
MATERIAL AND METHODS
Cell Culture –
HEK-293 and MCF7 cells were cultured and expanded in DMEM, supplemented with 10% fetal bovine serum, 2mM L-Glutamine, 0.1 mM non-essential amino acids, 1mM Sodium Pyruvate, 0.01mg/ml Bovine Insulin, 10 units/ml penicillin and 10 μg/ml streptomycin. Cells were grown at 37°C in 5% CO2.
Expression Plasmids –
For TAP experiments, human MBD4 cDNA was cloned in the EcoRI site of the vector pcDNA4 TO/TAP [46], to create an N-terminal tagged version of MBD4 (TAP-MBD4). Plasmids for HA-tagged MBD4 [29] and T7-tagged SUMO1 [47, 48] have been previously described. Site-directed mutants were generated using overlap extension PCR [49].
Transfections –
Cells were plated in 10 cm Petri dishes to obtain 80% of confluence (2.4 × 105 cells). Transfections were carried out using a total of 10 μg of plasmid DNA, 20 μL of Lipofectamine 2000 (Invitrogen) and Optimem (GIBCO).
Tandem affinity purification –
Tandem affinity purification was conducted as described in mammalian cells [46], with few modifications. Briefly, HEK-293 cells were transfected with TAP-MBD4 plasmid and, as negative control, empty pCDNA4/TO/TAP vector. After approximately 48 hours, the cells were lysed on ice in buffer containing 0.2% Nonidet P-40, 40 mM Tris, pH8, 150 mM NaCl, 10% glycerol, 1 mM ZnCl2, 10 mM NaF, and supplemented with Complete proteinase inhibitor (Roche), as previously described [29]. Lysates were loaded onto an IgG column and washed with lysis buffer without inhibitors. Subsequently, the protein A domain was cleaved off using TEV protease (New England Biolabs) during a 3-hour incubation with the enzyme in washing buffer supplemented with 0.5mM EDTA and 1mM DTT. Next, the TEV eluate was supplemented with 2 mM CaCl2 and loaded onto the calmodulin column, washed in calmodulin binding buffer (10 mM Tris, pH 8.0, 150 mM NaCl, 1 mM magnesium acetate, 1 mM imidazole, 2 mM CaCl2, 0.1% Nonidet P-40, 10 mM beta-mercaptoethanol) and eluted with calmodulin elution buffer (10 mM Tris, pH 8.0, 150 mM NaCl, 1 mM magnesium acetate, 1 mM imidazole, 10 mM EGTA, 0.1% Nonidet P-40, 10 mM beta-mercaptoethanol) in fractions. All volumes loaded onto the IgG and calmodulin columns and eluates were analyzed by Western blot to assess the purification process.
Mass spectrometry –
The affinity purified MBD4 complexes were resolved by SDS-PAGE and visualized by staining with colloidal Coomassie brilliant blue R250 (BioRad). Tryptic peptide mass mapping was used to identify the excised spots shown in Figure 1B. Bands excised from the gel were destained, reduced with dithiothreitol, alkylated with iodoacetamide and digested with trypsin [50, 51]. Aliquots of the digests were applied to Scout384 MALDI sample targets using alpha-cyano-4-hydroxy cinnamic acid as matrix. The Bruker Reflex IV mass spectrometer was programmed to collect mass spectra, perform internal mass calibration using trypsin autolysis products and write a peptide mass list for each sample. Peptide mass lists were used as input to the MASCOT sequence database search engine [52, 53].
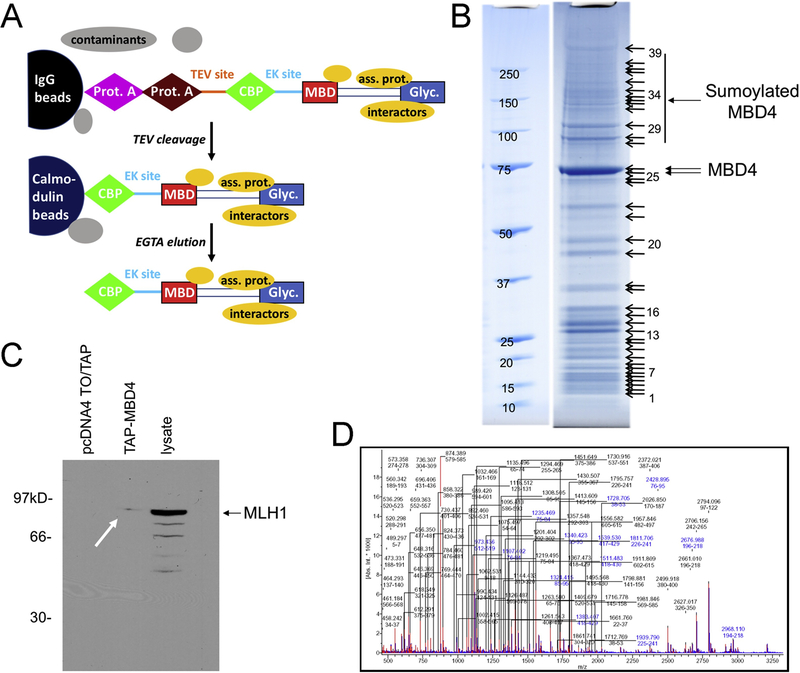
Figure 1 – Tandem affinity purification of MBD4-associated proteins.
(A) Schematic of TAP-tag strategy (modified from [56]). The TAP-tag-MBD4 fusion protein and the two sequential affinity columns are indicated; elution from the IgG column occurs by cleavage with TEV protease; elution from the calmodulin column occurs by calcium chelation by EGTA. Both covalently associated proteins and interactors (yellow) are co-purified with MBD4, whereas contaminants (grey) are eliminated. (B) SDS-PAGE, stained with Coomassie brilliant blue of acetone-precipitated TAP-tag eluate from HEK-293 cells, with bands submitted to mass spectrometry identification. Bands 25 and 26 correspond to unmodified MBD4; several of the higher molecular weight bands contain peptides identified as SUMO1 and MBD4 by mass spectrometry. (C) Detection of the MBD4 interactor MLH1 by western blotting of lysates from TAP-MBD4-transfected cells but not from empty pCDNA4/TO/TAP-transfected cells. Total lysate from HEK-293 cells is a positive control. (D) Tryptic peptide mass fingerprint showing well-defined peaks allowing reliable identification of peptides corresponding to MBD4.
Co-Immunoprecipitation and immunoblotting –
HEK-293 and MCF7 cells were washed three times with cold PBS (Phosphate buffered Saline) and lysed with cold RIPA buffer (50 mM Tris HCl pH 7.4, 150 mM NaCl, 1% sodium deoxycholate, 1% Triton X-100, 0.1% SDS, 10 mM NaF, 1 mM each of sodium pyrophosphate, sodium orthovanadate, dithiothreitol, and EDTA) plus protease inhibitors for 30 minutes on ice. Cells were scraped off the dish and sonicated for 30 seconds. After centrifugation for 30 minutes at max speed (14,000 rpm, in microcentrifuge), the supernatant was isolated and protein concentration was determined with Bradford assay (Bio-Rad) and BCA assay (Thermo Scientific Pierce), using bovine serum albumin as a standard. Immunoprecipitations were performed in 400 μL of RIPA buffer using 500 μg of protein lysate. For each protein lysate, 3 μL of anti-T7-tag antibody (Novagene) were added, and the lysates were incubated for 3 hours at 4°C on a rotating platform. Immune complexes were collected on protein A/G beads (Thermo Scientific) for 2 hours at 4°C. At the end of the incubation, the mix was gently spun, and the supernatant was discarded. The beads were washed three times with RIPA buffer, resuspended in 40 μL of Laemmli buffer (Novex) and boiled for 10 minutes. Immune complexes were resolved by 4–12% SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to PVDF membranes (Millipore). Membranes were blocked in 5% nonfat dry milk in PBS and incubated with primary antibodies (Anti-T7 tag, mouse, 1:2500 by Novagene; Anti-HA tag, rabbit, 1:5000 by Cell Signaling; anti-MBD4, rabbit, 1:500 in-house) overnight at 4°C, and then with secondary antibody (goat anti-mouse and goat anti-rabbit by Santa Cruz, 1:4000). Detection was performed using enhanced chemiluminescence (Pierce).
In vitro sumoylation –
We have previously described purification of recombinant activating E1 (Aos1-Uba2) and conjugating E2 (Ubc9) enzymes [47]; Yellow Fluorescent Protein-SUMO1 fusion was purified as described [47], using pET11d-YFP-SUMO (kind gift of Dr. Frauke Melchior). Purification of recombinant MBD4 [17] and TDG [54] has also been described. In vitro sumoylation reactions were carried out, as previously described [47], for 2hrs at 30°C in 50 mM Tris-Cl (pH 7.5) and 5 mM MgCl2, in the presence of an ATP regeneration system (2 mM ATP, 10 mM creatine phosphate disodium salt, 3.5 U/ml creatine kinase and 0.6 U/ml inorganic pyrophosphatase), with 500 ng Uba2/Aos1, 2 μg Ubc9, 2 μg YFP-SUMO1, 1 μg of recombinant MBD4 or recombinant TDG. Reactions were separated by SDS-PAGE and analyzed by immunoblotting with anti-SUMO1 antibody (Invitrogen).
Subcellular fractionation –
Subcellular fractionation was conducted with Nucbuster Protein Extraction Kit (Millipore), following the manufacturer’s recommendations.
Drug Treatments –
N-methyl-N-nitrosourea (MNU, Sigma-Aldrich) was dissolved in DMSO at the final concentration of 10 mg/mL; cisplatin (GensiaSicor Pharmaceuticals, 1 mg/ml) and 5-fluorouracil (American Pharmaceutical Partners, Inc., 50 mg/ml) were used as supplied by the manufacturer. Drug treatments were performed 24 hours after transfection by removing the medium from cultures of logarithmically growing cells and adding fresh medium containing drugs (2 μM). After 24 hours of drug exposure, cells were rinsed with PBS and harvested for analysis.
MBD4 repair activity –
MBD4/AP-Endonuclease activity of nuclear extracts was measured using a G:T mismatch-containing molecular beacon hairpin with quenching upon folding and fluorescence release upon base removal/cleavage at the lesion site [55]. The substrate 5’dT(FAM)-CCACTTGTGAATTGACAGCCCATGTGCATCAATTCACGAGTGG-T (Dabsyl)3’ (Biosearch Technologies, Novato, CA) was annealed by heating and slow-cooling in 4X BER buffer (1X: Hepes-KOH 25mM, KCl 150mM, Glycerol 1%, DTT 0.5mM)[55]. Substrate (0.56 μM) and nuclear extracts (50 μg) were mixed in 1X BER buffer supplemented with MgCl2 (1.2 mM) and DTT (10 mM). The reaction was run at 37°C in a real-time PCR machine (Applied Biosystems 7900 HT) and FAM fluorescence was recorded for 2 hrs at 0.5sec intervals.
Bioinformatics –
Prediction of sumoylation sites was conducted with SUMOplot (http://www.abgent.com/sumoplot).
RESULTS
Tandem affinity purification identifies sumoylation of MBD4
To purify the complex of MBD4-associated proteins, we conducted a tandem affinity purification tag (TAP-tag) strategy [56]. In this approach, cells are transfected with a plasmid expressing the protein of interest fused C-terminally to two IgG binding domains from S. aureus protein A, the cleavage site for the protease TEV, and the calmodulin binding peptide (Figure 1A). The presence of two high affinity tags in tandem separated by a protease site allows a two-step purification procedure of remarkable effectiveness not only in yeast but also in mammalian systems [46, 57]. We expressed in HEK-293 cells an N-terminal tagged MBD4 construct (MBD4-TAP), fused to protein A and calmodulin binding peptide (Suppl. Fig. 1). Total cellular lysate was first applied to an IgG beads affinity column; after washing, the MBD4 fusion protein and its interacting partners were released by cleavage with the TEV protease; the eluate was then applied onto calmodulin beads for a second affinity step; finally, the MBD4 complex was eluted with EGTA [46, 56], and fractions containing MBD4 were acetone-precipitated and separated by SDS-PAGE (Figure 1B). Western blotting demonstrated isolation of the MBD4 interactor, MLH1, thus further confirming that the interaction between these two proteins occurs in vivo in mammalian cells [29](Fig. 1C). Furthermore, mass spectrometry (MS) analysis of bands purified from the denaturing SDS-PAGE revealed that several bands with molecular weight higher than that predicted for MBD4 (~66kD) contained peptides corresponding to MBD4 itself and SUMO, suggesting that MBD4 is sumoylated in vivo (Fig. 1D). A list of additional interactors identified by MS and examples of protein ID determination are shown in Suppl. Table 1 and Suppl. Fig. 2, respectively.
MBD4 is sumoylated in vivo and in vitro
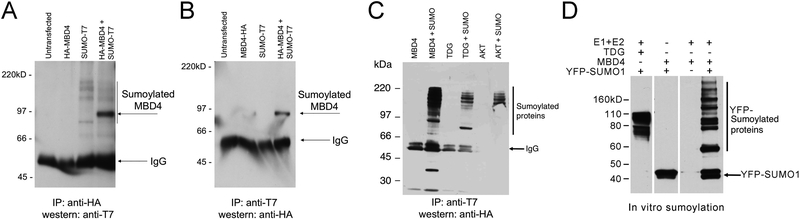
To verify the covalent interaction between MBD4 and SUMO1, stringent and reciprocal coimmunoprecipitation experiments, using RIPA lysates and SDS-PAGE, were performed in HEK-293 cells transfected with hemagglutinin-tagged (HA) human MBD4 [29] and T7-tagged SUMO1 [47, 48]. These experiments revealed detection of a band corresponding to T7-sumoylated HA-MBD4 (approximately 90kD) (Fig. 2A–B). This result was validated in a different cell line, MCF-7, in which coimmunoprecipitation experiments were conducted after transfecting cells with T7-tagged SUMO1 in combination with HA-tagged MBD4 and, as positive controls for sumoylation, TDG and the oncoprotein AKT1 [58, 59](Fig. 2C). To further confirm MBD4 sumoylation, we conducted an in vitro sumoylation assay, employing recombinant activating E1 (Aos1-Uba2) and conjugating E2 (Ubc9) enzymes, along with recombinant YFP-SUMO1 and MBD4 or, as positive control for sumoylation, TDG (Fig. 2D). These results indicate that MBD4 is sumoylated in vivo and in vitro.
Figure 2 – MBD4 is sumoylated in vivo and in vitro.
(A, B) HEK-293 cells were transfected with the indicated plasmids or left untransfected. Co-IP conducted in both directions (IP: antihemagglutinin / western: anti-T7; and IP: anti-T7 / western: anti-hemagglutinin), as indicated, revealed detection of an approximately 90kD band corresponding to sumoylated MBD4. (C) MCF-7 cells were transfected with hemagglutinin-tagged MBD4, TDG and AKT, in combination with T7-tagged SUMO1. Co-IP reveals sumoylation of MBD4 and positive controls TDG and AKT1 (upper panel). Western blotting shows approximately equal expression of hemagglutinin-tagged MBD4, TDG and AKT (lower panel). (D) In vitro sumoylation assay, in which the indicated activating E1 (Aos1-Uba2) and conjugating E2 (Ubc9) enzymes were incubated with recombinant MBD4, TDG and YFP-SUMO1; detection of reaction products was done by western blotting with anti-SUMO1 antibody.
Mapping of the main MBD4 sumoylation sites
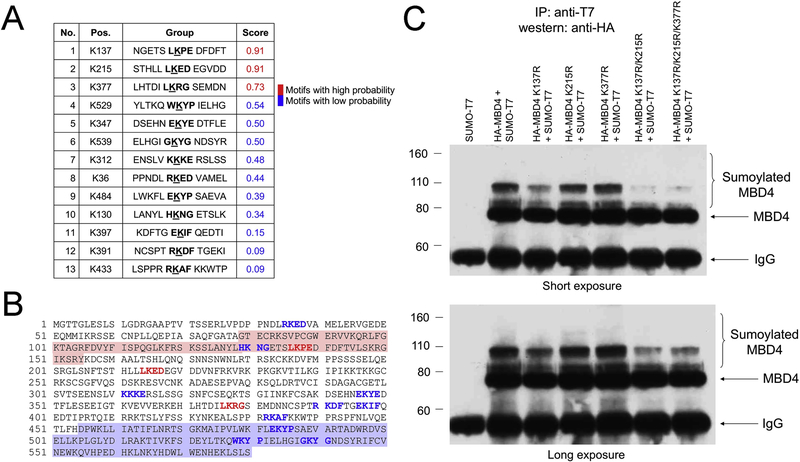
To map the MBD4 sumoylation sites, we first turned to sequence analysis. The sumoylation prediction software SUMOplot identified three high-probability and ten low-probability sites (Fig. 3A). The first of the three putative sumoylation sites, K137, maps to the MBD4 MBD, whereas the other two high-probability sites, K215 and K377, map to the linker region between the MBD and the glycosylase domain (Fig. 3B). These three sites were mutated by substituting lysine with arginine, alone or in combination, creating five different mutants (K137R, K215R, K377R, K137R/K215R and K137R/K215R/K377R). Co-immunoprecipitation and western blot analysis in transfected MCF-7 cells were then performed to determine if the mutated sites in MBD4 would prevent sumoylation. The results indicated that MBD4 is sumoylated preferentially at K137, as sumoylation was dramatically decreased in the K137R mutant; and additional sumoylation sites are located at K215R and K377R, as sumoylation was almost completely abrogated in the triple mutant (Figure 3C, top). However, it is likely that sumoylation occurs at least at one more lysine in MBD4, because a longer exposure revealed a band corresponding to residual sumoylation even in the triple mutant (Figure 3C, bottom).
Figure 3 – Mapping of the MBD4 sumoylation sites.
List (A) and location (B) of the high- (red) and low-probability (blue) sumoylation sites identified by the SUMOplot software. The MBD and glycosylase domain are shaded in light red and blue, respectively. (C) MCF-7 cells were transfected with hemagglutinin-tagged wild type MBD4 and indicated MBD4 mutants, in combination with T7-tagged SUMO1. Co-IP confirms that K137, K215 and K377 are the main sumoylation sites (upper panel, short exposure). However, long exposure (lower panel) shows residual sumoylation in the K137R/K215R/K377R, suggesting the existence of at least one more sumoylation site.
The sumoylation pattern of MBD4 is altered in response to DNA damage
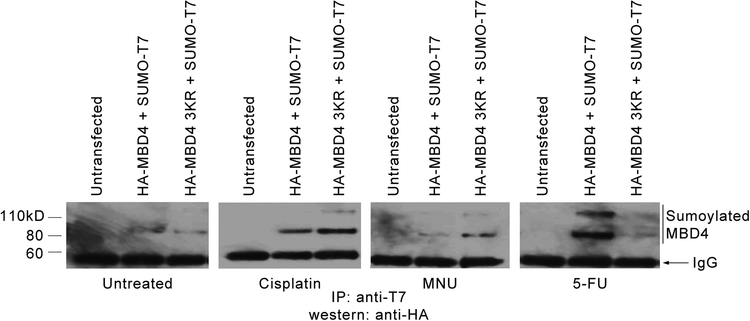
In addition to its repair function, MBD4 plays a role in the apoptotic response to DNA damage induced by alkylating agents, cisplatin and 5-fluorouracil (5-FU)[34, 35]. We then sought to determine if treatment with these agents affected sumoylation patterns. 24 hours after transfection with HA-MBD4 + SUMO1-T7 and HA-MBD4 K137R/K215R/K377R + SUMO1-T7, MCF7 cells were treated with cisplatin, N-Nitroso-N-methylurea (NMU) and 5-FU at a 2μM concentration for 24 hours, after which co-IPs were performed (IP anti-T7, WB anti-HA). The results revealed that 5FU increases the extent of sumoylation at the three main sites, whereas NMU and cisplatin increased sumoylation at additional site(s) when the three main sites are mutated, thus confirming the existence of at least one more site of sumoylation (Fig. 4).
Figure 4 – Sumoylation of MBD4 in response to DNA damage.
Co-immunoprecipitation analysis (IP: anti-T7 / western: anti-hemagglutinin) of MCF7 cells left untransfected and transfected with hemagglutinin-tagged wild type MBD4 or hemagglutinin-tagged K137R/K215R/K377R MBD4, as indicated, and treated with control DMSO or with cisplatin, NMU and 5-FU (2 μM for 24 hours).
Sumoylation increases the repair activity of MBD4
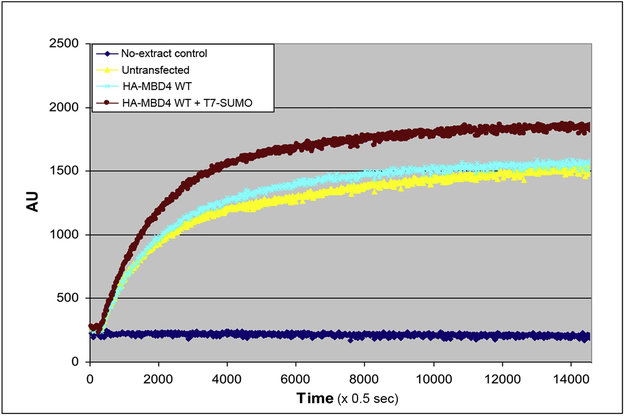
Since sumoylation may alter the biochemical activity of the modified protein, using a fluorescence-based, molecular beacon assay for G:T mismatch repair [60, 61], we assessed the repair activity of nuclear extracts from untransfected MCF7 cells, and from MCF7 cells transfected with HA-MBD4 or HA-MBD4 + SUMO1-T7. The results revealed robust G:T repair activity of nuclear extracts of untransfected MCF7 cells, which was slightly increased by overexpression of HA-MBD4, and further boosted by HA-MBD4 + SUMO1-T7 (Fig. 5). The results suggest that sumoylation of MBD4 increases its glycosylase activity on G:T mismatches.
Figure 5 – Sumoylation increases the repair activity of MBD4.
Molecular beacon assay for repair of G:T mismatches by extracts of untransfected MCF7 cells and MCF7 cells transfected with hemagglutinin-tagged MBD4 or hemagglutinin-tagged MBD4 plus SUMO-T7, showing time-dependent generation of fluorescence; fluorescence is in arbitrary units (AU); time is in 0.5-sec intervals.
MBD4 mutations near sumoylation sites in cancer
Inactivating mutations of the MBD4 gene in human cancer have been described by us and others [39–41]. The consequence of inactivating mutations of MBD4 is to increase the mutation rate at CpG sites [44, 62, 63] and provide an advantage to cancer cells, even in the context of inactivating MMR mutations [64]. Since sumoylation boosts MBD4 repair activity, we reasoned that it may be compromised in cancer. Examination of The Cancer Genome Atlas (TCGA), revealed a total of 67 MBD4 missense mutations reported to date (http://www.cbioportal.org/). Of these missense mutations, 14 (20.9%) map within 5 amino acids of sumoylation sites, and 6 are reported in endometrial cancer (Table 1). These results suggest that cancer-associated MBD4 mutations may affect sumoylation of its gene product.
Table 1.
Mutations near MBD4 sumoylation motifs
| Mutation. | Cancer | Group | Pos. |
|---|---|---|---|
| N131S | Colon adenocarcinoma | NGETS LKPE DFDFT | K137 |
| S135Y | Uterine endometrioid carcinoma | NGETS LKPE DFDFT | K137 |
| S209Y | Lung squamous cell carcinoma | STHLL LKED EGVDD | K215 |
| H211Y | Uterine mixed endometrial carcinoma | STHLL LKED EGVDD | K215 |
| D221H | Head and neck squamous cell carcinoma | STHLL LKED EGVDD | K215 |
| R378C | Uterine endometrioid carcinoma | LHTDI LKRG SEMDN | K377 |
| R546Q | Uterine endometrioid carcinoma | ELHGI GKYG NDSYR | K539 |
| R546Q | Breast invasive ductal carcinoma | ELHGI GKYG NDSYR | K539 |
| S308N | Rectal adenocarcinoma | ENSLV KKKE RSLSS | K312 |
| K311T | Cervical squamous cell carcinoma | ENSLV KKKE RSLSS | K312 |
| S487L | Cervical squamous cell carcinoma | LWKFL EKYP SAEVA | K484 |
| L124R | Papillary renal cell carcinoma | LANYL HKNG ETSLK | K130 |
| F399C | Uterine endometrioid carcinoma | KDFTG EKIF QEDTI | K397 |
| T439A | Uterine endometrioid carcinoma | LSPPR RKAF KKWTP | K433 |
█ Mutations near motifs with high probability
█ Mutations near motifs with low probability
DISCUSSION
Sumoylation plays an important role in cellular functions, and particularly in DNA repair, affecting activity, localization and stability of target proteins. Despite the fact that sumoylation was known to affect the function of base excision repair proteins, identification of MBD4 sumoylation was unexpected, as the MBD4 fusion protein expressed from the TAP-tag construct became sumoylated in transfected HEK-293 cells. Sumoylation of MBD4 was later confirmed by co-IP experiments in both HEK-293 and MCF7 cells and reconstituted in vitro, using recombinant proteins.
Sumoylation of MBD4 is complex. In addition to the three main sites predicted by software on the basis of the consensus sumoylation sequence ψKX(D/E), in which ψ is a large hydrophobic residue [10], additional sites with a lower probability score are present in MBD4. In fact, the triple mutant K137R/K215R/K377R (arginine substitutions at the three main acceptor lysine residues) shows evidence of being sumoylated in vivo. Several bands of increasing molecular weight were detected by mass spectrometry and by co-IP, suggesting the different, but not mutually exclusive possibilities, of addition of monomeric SUMO1 molecules at multiple sites or of poly-SUMO1 chains (polysumoylation)[65]. The possibility that (some of) these sites can be targeted by SUMO molecules other than SUMO1, with potentially different functional consequences, should be considered.
While K215 and K377 are located in the low-complexity linker region, whose function is unknown, K137 is part of the MBD; it is likely that sumoylation at this site will affect binding affinity of the MBD for 5-methylcytosine. Putative site(s) of sumoylation are located in the catalytic domain and may affect catalysis, as well as interaction with MLH1, which maps to this region [29]. In fact, only a small amount of MLH1 was identified in the TAP-MBD4 eluate (Fig. 1C), containing sumoylated MBD4.
Interestingly, sumoylation stimulated repair activity on a G:T mismatched substrate, a finding that is opposite to TDG sumoylation, which is known to decrease enzymatic activity [24]. This finding for MBD4 should be confirmed with recombinant proteins and with a precise measurement of the velocity of the repair reaction in the presence of unsumoylated or differentially sumoylated MBD4 molecules.
The pattern of MBD4 sumoylation changes in response to different kinds of DNA damage and, in particular, sumoylation at site(s) other than the three main sites is stimulated by cisplatin and MNU, when the three main sites are mutated, but not 5-FU. Since MBD4 affects the apoptotic response to DNA damage, but does not directly repair cisplatin or MNU damage, it is possible that sumoylation at different sites may serve to separate the apoptotic function of MBD4 from its repair function. To this end, it is interesting to note that 5-FU, which can be directly processed by MBD4 in the form of 5-FU:G mismatches [37], does not stimulate sumoylation at sites other than the three main sites. Again, it is possible that different SUMOs (i.e., SUMO1 vs. SUMO2/3 vs. SUMO4) may be involved in the apoptotic vs. repair role of MBD4).
Compromising both apoptotic and repair functions of MBD4 is likely to be important in tumorigenesis. We reported that a large fraction of MBD4 missense mutations (20.9%) occur near sumoylation sites, and therefore may impact apoptotic and/or repair activity of the enzyme. Future studies will clarify these possibilities. Mutations abrogating sumoylation do not strictly have to be at the modified lysine, but can be nearby as well. For instance, the E318K mutation in MITF, that impairs sumoylation and predisposes to melanoma and renal carcinoma, occurs at the glutamic acid at position +2 of the sumoylation consensus site, ψKX(D/E) [66, 67]. Also, in a recent pan-cancer analysis of lysine modifications, including sumoylation, a window similar to ours in Table 1 was used, i.e. from position −7 to +7 around the lysine [68].
In summary, to our knowledge, this is the first description of MBD4 sumoylation. Understanding the consequences of this modification for MBD4 functions may be important not only from the basic science standpoint, but also for translational research, as it may help devising novel diagnostic and/or therapeutic strategies for cancer. This study expands the role played by sumoylation in fine-tuning DNA damage response and repair, and constitutes the first step towards possible translational opportunities.
Supplementary Material
Highlights.
MBD4 is sumoylated at three main sites: K137, K215 and K377
MBD4 sumoylation is altered, in a DNA damage-specific way (5FU, NMU & cisplatin)
Sumoylation increases the G:T repair activity of MBD4 in cell extracts
Of the 67 MBD4 missense mutations in TCGA, 14 (20.9%) map near sumoylation sites
Acknowledgments —
We thank Dr. Rahul Prasad for critical reading of the manuscript; and L. Cathay for secretarial assistance. We thank the Genomic Facility and Cell Culture Facility at the Fox Chase Cancer Center. This study was supported by NIH grants CA78412, CA191956 and CA06927, DOD grant W81XWH-17–1-0136 and an appropriation from the Commonwealth of Pennsylvania to the Fox Chase Cancer Center. R. W. Sobol was supported by NIH grants CA148629 and ES025138, and is an Abraham A. Mitchell Distinguished Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD, Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1, Cell, 108 (2002) 345–356. [DOI] [PubMed] [Google Scholar]
- [2].Mossessova E, Lima CD, Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast, Mol. Cell, 5 (2000) 865–876. [DOI] [PubMed] [Google Scholar]
- [3].Bayer P, Arndt A, Metzger S, Mahajan R, Melchior F, Jaenicke R, Becker J, Structure determination of the small ubiquitin-related modifier SUMO-1, J. Mol. Biol, 280 (1998) 275–286. [DOI] [PubMed] [Google Scholar]
- [4].Guo D, Li M, Zhang Y, Yang P, Eckenrode S, Hopkins D, Zheng W, Purohit S, Podolsky RH, Muir A, Wang J, Dong Z, Brusko T, Atkinson M, Pozzilli P, Zeidler A, Raffel LJ, Jacob CO, Park Y, Serrano-Rios M, Larrad MT, Zhang Z, Garchon HJ, Bach JF, Rotter JI, She JX, Wang CY, A functional variant of SUMO4, a new I kappa B alpha modifier, is associated with type 1 diabetes, Nat Genet, 36 (2004) 837–841. [DOI] [PubMed] [Google Scholar]
- [5].Melchior F, SUMO--nonclassical ubiquitin, Annu. Rev. Cell Dev. Biol, 16 (2000) 591–626. [DOI] [PubMed] [Google Scholar]
- [6].Kunz K, Piller T, Muller S, SUMO-specific proteases and isopeptidases of the SENP family at a glance, J. Cell Sci, 131 (2018) 1–8. [DOI] [PubMed] [Google Scholar]
- [7].Gareau JR, Lima CD, The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition, Nat. Rev. Mol. Cell. Biol, 11 (2010) 861–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Pichler A, Fatouros C, Lee H, Eisenhardt N, SUMO conjugation - a mechanistic view, Biomolecular concepts, 8 (2017) 13–36. [DOI] [PubMed] [Google Scholar]
- [9].Cappadocia L, Lima CD, Ubiquitin-like Protein Conjugation: Structures, Chemistry, and Mechanism, Chem. Rev, 118 (2018) 889–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Rodriguez MS, Dargemont C, Hay RT, SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting, J. Biol. Chem, 276 (2001) 12654–12659. [DOI] [PubMed] [Google Scholar]
- [11].Mahajan R, Delphin C, Guan T, Gerace L, Melchior F, A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2, Cell, 88 (1997) 97–107. [DOI] [PubMed] [Google Scholar]
- [12].Matunis MJ, Coutavas E, Blobel G, A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex, J. Cell Biol, 135 (1996) 1457–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Boddy MN, Howe K, Etkin LD, Solomon E, Freemont PS, PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia, Oncogene, 13 (1996) 971–982. [PubMed] [Google Scholar]
- [14].Scurr LL, Haferkamp S, Rizos H, The Role of Sumoylation in Senescence, Adv. Exp. Med. Biol, 963 (2017) 215–226. [DOI] [PubMed] [Google Scholar]
- [15].Carter RJ, Parsons JL, Base Excision Repair, a Pathway Regulated by Posttranslational Modifications, Mol. Cell. Biol, 36 (2016) 1426–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Porello SL, Leyes AE, David SS, Single-turnover and pre-steady-state kinetics of the reaction of the adenine glycosylase MutY with mismatch-containing DNA substrates, Biochemistry (Mosc). 37 (1998) 14756–14764. [DOI] [PubMed] [Google Scholar]
- [17].Petronzelli F, Riccio A, Markham GD, Seeholzer SH, Stoerker J, Genuardi M, Yeung AT, Matsumoto Y, Bellacosa A, Biphasic kinetics of the human DNA repair protein MED1 (MBD4), a mismatch-specific DNA N-glycosylase, J. Biol. Chem, 275 (2000) 32422–32429. [DOI] [PubMed] [Google Scholar]
- [18].Petronzelli F, Riccio A, Markham GD, Seeholzer SH, Genuardi M, Karbowski M, Yeung AT, Matsumoto Y, Bellacosa A, Investigation of the substrate spectrum of the human mismatch-specific DNA N-glycosylase MED1 (MBD4): Fundamental role of the catalytic domain, J. Cell. Physiol, 185 (2000) 473–480. [DOI] [PubMed] [Google Scholar]
- [19].Fitzgerald ME, Drohat AC, Coordinating the initial steps of base excision repair. Apurinic/apyrimidinic endonuclease 1 actively stimulates thymine DNA glycosylase by disrupting the product complex, J. Biol. Chem, 283 (2008) 32680–32690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Waters TR, Swann PF, Kinetics of the action of thymine DNA glycosylase, J. Biol. Chem, 273 (1998) 20007–20014. [DOI] [PubMed] [Google Scholar]
- [21].Scharer OD, Nash HM, Jiricny J, Laval J, Verdine GL, Specific binding of a designed pyrrolidine abasic site analog to multiple DNA glycosylases, J. Biol. Chem, 273 (1998) 8592–8597. [DOI] [PubMed] [Google Scholar]
- [22].Steinacher R, Schar P, Functionality of human thymine DNA glycosylase requires SUMO-regulated changes in protein conformation, Current biology : CB, 15 (2005) 616–623. [DOI] [PubMed] [Google Scholar]
- [23].Hardeland U, Steinacher R, Jiricny J, Schar P, Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover, EMBO J, 21 (2002) 1456–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Coey CT, Drohat AC, Defining the impact of sumoylation on substrate binding and catalysis by thymine DNA glycosylase, Nucleic Acids Res, 46 (2018) 5159–5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bellacosa A, Drohat AC, Role of base excision repair in maintaining the genetic and epigenetic integrity of CpG sites, DNA Repair (Amst), 32 (2015) 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Mohan RD, Rao A, Gagliardi J, Tini M, SUMO-1-dependent allosteric regulation of thymine DNA glycosylase alters subnuclear localization and CBP/p300 recruitment, Mol. Cell. Biol, 27 (2007) 229–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Steinacher R, Barekati Z, Botev P, Kusnierczyk A, Slupphaug G, Schar P, SUMOylation coordinates BERosome assembly in active DNA demethylation during cell differentiation, EMBO J, 38 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A, The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites, Nature, 401 (1999) 301–304. [DOI] [PubMed] [Google Scholar]
- [29].Bellacosa A, Cicchillitti L, Schepis F, Riccio A, Yeung AT, Matsumoto Y, Golemis EA, Genuardi M, Neri G, MED1, a novel human methyl-CpG-binding endonuclease, interacts with DNA mismatch repair protein MLH1, Proc Natl Acad Sci U S A, 96 (1999) 3969–3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hendrich B, Abbott C, McQueen H, Chambers D, Cross S, Bird A, Genomic structure and chromosomal mapping of the murine and human Mbd1, Mbd2, Mbd3, and Mbd4 genes, Mamm. Genome, 10 (1999) 906–912. [DOI] [PubMed] [Google Scholar]
- [31].Bellacosa A, Role of MED1 (MBD4) Gene in DNA repair and human cancer, J. Cell. Physiol, 187 (2001) 137–144. [DOI] [PubMed] [Google Scholar]
- [32].Grigera F, Bellacosa A, Kenter AL, Complex relationship between mismatch repair proteins and MBD4 during immunoglobulin class switch recombination, PLoS One, 8 (2013) e78370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Grigera F, Wuerffel R, Kenter AL, MBD4 Facilitates Immunoglobulin Class Switch Recombination, Mol. Cell. Biol, 37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Cortellino S, Turner D, Masciullo V, Schepis F, Albino D, Daniel R, Skalka AM, Meropol NJ, Alberti C, Larue L, Bellacosa A, The base excision repair enzyme MED1 mediates DNA damage response to antitumor drugs and is associated with mismatch repair system integrity, Proc. Natl. Acad. Sci. USA, 100 (2003) 15071–15076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sansom OJ, Zabkiewicz J, Bishop SM, Guy J, Bird A, Clarke AR, MBD4 deficiency reduces the apoptotic response to DNA-damaging agents in the murine small intestine, Oncogene, 22 (2003) 7130–7136. [DOI] [PubMed] [Google Scholar]
- [36].Kondo E, Gu Z, Horii A, Fukushige S, The thymine DNA glycosylase MBD4 represses transcription and is associated with methylated p16(INK4a) and hMLH1 genes, Mol. Cell. Biol, 25 (2005) 4388–4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Turner DP, Cortellino S, Schupp JE, Caretti E, Loh T, Kinsella TJ, Bellacosa A, The DNA N-glycosylase MED1 exhibits preference for halogenated pyrimidines and is involved in the cytotoxicity of 5-iododeoxyuridine, Cancer Res, 66 (2006) 7686–7693. [DOI] [PubMed] [Google Scholar]
- [38].Valinluck V, Liu P, Kang JI Jr., Burdzy A, Sowers LC, 5-halogenated pyrimidine lesions within a CpG sequence context mimic 5-methylcytosine by enhancing the binding of the methyl-CpG-binding domain of methyl-CpG-binding protein 2 (MeCP2), Nucleic Acids Res, 33 (2005) 3057–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bader S, Walker M, Hendrich B, Bird A, Bird C, Hooper M, Wyllie A, Somatic frameshift mutations in the MBD4 gene of sporadic colon cancers with mismatch repair deficiency, Oncogene, 18 (1999) 8044–8047. [DOI] [PubMed] [Google Scholar]
- [40].Yamada T, Koyama T, Ohwada S, Tago K, Sakamoto I, Yoshimura S, Hamada K, Takeyoshi I, Morishita Y, Frameshift mutations in the MBD4/MED1 gene in primary gastric cancer with high-frequency microsatellite instability, Cancer Lett, 181 (2002) 115–120. [DOI] [PubMed] [Google Scholar]
- [41].Riccio A, Aaltonen LA, Godwin AK, Loukola A, Percesepe A, Salovaara R, Masciullo V, Genuardi M, Paravatou-Petsotas M, Bassi DE, Ruggeri BA, Klein-Szanto AJP, Testa JR, Neri G, Bellacosa A, The DNA repair gene MBD4 (MED1) is mutated in human carcinomas with microsatellite instability, Nat. Genet, 23 (1999) 266–268. [DOI] [PubMed] [Google Scholar]
- [42].Howard JH, Frolov A, Tzeng CD, Stewart A, Midzak A, Majmundar A, Godwin AK, Heslin MJ, Bellacosa A, Arnoletti JP, Epigenetic downregulation of the DNA repair gene MED1/MBD4 in colorectal and ovarian cancer, Cancer Biol. Ther, 8(1) (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lucci-Cordisco E, Neri G, Silent beginning: early silencing of the MED1/MBD4 gene in colorectal tumorigenesis, Cancer Biol Ther, 8 (2009) 192–193. [DOI] [PubMed] [Google Scholar]
- [44].Sanders MA, Chew E, Flensburg C, Zeilemaker A, Miller SE, Al Hinai AS, Bajel A, Luiken B, Rijken M, McLennan T, Hoogenboezem RM, Kavelaars FG, Frohling S, Blewitt ME, Bindels EM, Alexander WS, Lowenberg B, Roberts AW, Valk PJM, Majewski IJ, MBD4 guards against methylation damage and germline deficiency predisposes to clonal hematopoiesis and early-onset AML, Blood, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Rodrigues M, Mobuchon L, Houy A, Fievet A, Gardrat S, Barnhill RL, Popova T, Servois V, Rampanou A, Mouton A, Dayot S, Raynal V, Galut M, Putterman M, Tick S, Cassoux N, Roman-Roman S, Bidard FC, Lantz O, Mariani P, Piperno-Neumann S, Stern MH, Outlier response to anti-PD1 in uveal melanoma reveals germline MBD4 mutations in hypermutated tumors, Nat Commun, 9 (2018) 1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Cox DM, Du M, Guo X, Siu KW, McDermott JC, Tandem affinity purification of protein complexes from mammalian cells, Biotechniques, 33 (2002) 267–268, 270. [DOI] [PubMed] [Google Scholar]
- [47].Saha S, Chernoff J, Analysis of PTP1B sumoylation, Methods, 65 (2014) 201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Yip SC, Cotteret S, Chernoff J, Sumoylated protein tyrosine phosphatase 1B localizes to the inner nuclear membrane and regulates the tyrosine phosphorylation of emerin, J. Cell Sci, 125 (2012) 310–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Bellacosa A, Chan TO, Ahmed NN, Datta K, Malstrom S, Stokoe D, McCormick F, Feng J, Tsichlis P, Akt activation by growth factors is a multiple-step process: the role of the PH domain, Oncogene, 17 (1998) 313–325. [DOI] [PubMed] [Google Scholar]
- [50].Shevchenko A, Wilm M, Vorm O, Mann M, Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels, Anal. Chem, 68 (1996) 850–858. [DOI] [PubMed] [Google Scholar]
- [51].Spodik B, Seeholzer SH, Coleman TR, Using Xenopus Egg Extracts to Modify Recombinant Proteins, Cold Spring Harbor Laboratory Press, New York, 2002. [Google Scholar]
- [52].Hirosawa M, Hoshida M, Ishikawa M, Toya T, MASCOT: multiple alignment system for protein sequences based on three-way dynamic programming, Comput. Appl. Biosci, 9 (1993) 161–167. [DOI] [PubMed] [Google Scholar]
- [53].Perkins DN, Pappin DJ, Creasy DM, Cottrell JS, Probability-based protein identification by searching sequence databases using mass spectrometry data, Electrophoresis, 20 (1999) 3551–3567. [DOI] [PubMed] [Google Scholar]
- [54].Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, Abramowitz LK, Bartolomei MS, Rambow F, Bassi MR, Bruno T, Fanciulli M, Renner C, Klein-Szanto AJ, Matsumoto Y, Kobi D, Davidson I, Alberti C, Larue L, Bellacosa A, Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair, Cell, 146 (2011) 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Mancuso P, Tricarico R, Bhattacharjee V, Cosentino L, Kadariya Y, Jelinek J, Nicolas E, Einarson M, Beeharry N, Devarajan K, Katz RA, Dorjsuren DG, Sun H, Simeonov A, Giordano A, Testa JR, Davidson G, Davidson I, Larue L, Sobol RW, Yen TJ, Bellacosa A, Thymine DNA glycosylase as a novel target for melanoma, Oncogene, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B, A generic protein purification method for protein complex characterization and proteome exploration, Nat. Biotechnol, 17 (1999) 1030–1032. [DOI] [PubMed] [Google Scholar]
- [57].Seraphin B, Identification of transiently interacting proteins and of stable protein complexes, Adv. Protein Chem, 61 (2002) 99–117. [DOI] [PubMed] [Google Scholar]
- [58].Li R, Wei J, Jiang C, Liu D, Deng L, Zhang K, Wang P, Akt SUMOylation regulates cell proliferation and tumorigenesis, Cancer Res, 73 (2013) 5742–5753. [DOI] [PubMed] [Google Scholar]
- [59].de la Cruz-Herrera CF, Campagna M, Lang V, del Carmen Gonzalez-Santamaria J, Marcos-Villar L, Rodriguez MS, Vidal A, Collado M, Rivas C, SUMOylation regulates AKT1 activity, Oncogene, 34 (2015) 1442–1450. [DOI] [PubMed] [Google Scholar]
- [60].Mancuso P, Tricarico R, Bhattacharjee V, Cosentino L, Kadariya Y, Jelinek J, Nicolas E, Einarson M, Beeharry N, Devarajan K, Katz RA, Dorjsuren DG, Sun H, Simeonov A, Giordano A, Testa JR, Davidson G, Davidson I, Larue L, Sobol RW, Yen TJ, Bellacosa A, Thymine DNA glycosylase as a novel target for melanoma, Oncogene, 38 (2019) 3710–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Svilar D, Vens C, Sobol RW, Quantitative, real-time analysis of base excision repair activity in cell lysates utilizing lesion-specific molecular beacons, Journal of visualized experiments : JoVE, (2012) e4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Millar CB, Guy J, Sansom OJ, Selfridge J, MacDougall E, Hendrich B, Keightley PD, Bishop SM, Clarke AR, Bird A, Enhanced CpG mutability and tumorigenesis in MBD4-deficient mice, Science, 297 (2002) 403–405. [DOI] [PubMed] [Google Scholar]
- [63].Wong E, Yang K, Kuraguchi M, Werling U, Avdievich E, Fan K, Fazzari M, Jin B, Brown AM, Lipkin M, Edelmann W, Mbd4 inactivation increases C→T transition mutations and promotes gastrointestinal tumor formation, Proc. Natl. Acad. Sci. USA, 99 (2002) 14937–14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Tricarico R, Cortellino S, Riccio A, Jagmohan-Changur S, Van der Klift H, Wijnen J, Turner D, Ventura A, Rovella V, Percesepe A, Lucci-Cordisco E, Radice P, Bertario L, Pedroni M, Ponz de Leon M, Mancuso P, Devarajan K, Cai KQ, Klein-Szanto AJ, Neri G, Moller P, Viel A, Genuardi M, Fodde R, Bellacosa A, Involvement of MBD4 inactivation in mismatch repair-deficient tumorigenesis, Oncotarget, 6 (2015) 42892–42904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Ulrich HD, The fast-growing business of SUMO chains, Mol. Cell, 32 (2008) 301–305. [DOI] [PubMed] [Google Scholar]
- [66].Bertolotto C, Lesueur F, Giuliano S, Strub T, de Lichy M, Bille K, Dessen P, d’Hayer B, Mohamdi H, Remenieras A, Maubec E, de la Fouchardiere A, Molinie V, Vabres P, Dalle S, Poulalhon N, Martin-Denavit T, Thomas L, Andry-Benzaquen P, Dupin N, Boitier F, Rossi A, Perrot JL, Labeille B, Robert C, Escudier B, Caron O, Brugieres L, Saule S, Gardie B, Gad S, Richard S, Couturier J, Teh BT, Ghiorzo P, Pastorino L, Puig S, Badenas C, Olsson H, Ingvar C, Rouleau E, Lidereau R, Bahadoran P, Vielh P, Corda E, Blanche H, Zelenika D, Galan P, Aubin F, Bachollet B, Becuwe C, Berthet P, Bignon YJ, Bonadona V, Bonafe JL, Bonnet-Dupeyron MN, Cambazard F, Chevrant-Breton J, Coupier I, Dalac S, Demange L, d’Incan M, Dugast C, Faivre L, Vincent-Fetita L, Gauthier-Villars M, Gilbert B, Grange F, Grob JJ, Humbert P, Janin N, Joly P, Kerob D, Lasset C, Leroux D, Levang J, Limacher JM, Livideanu C, Longy M, Lortholary A, Stoppa-Lyonnet D, Mansard S, Mansuy L, Marrou K, Mateus C, Maugard C, Meyer N, Nogues C, Souteyrand P, Venat-Bouvet L, Zattara H, Chaudru V, Lenoir GM, Lathrop M, Davidson I, Avril MF, Demenais F, Ballotti R, Bressac-de Paillerets B, A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma, Nature, 480 (2011) 94–98. [DOI] [PubMed] [Google Scholar]
- [67].Yokoyama S, Woods SL, Boyle GM, Aoude LG, MacGregor S, Zismann V, Gartside M, Cust AE, Haq R, Harland M, Taylor JC, Duffy DL, Holohan K, Dutton-Regester K, Palmer JM, Bonazzi V, Stark MS, Symmons J, Law MH, Schmidt C, Lanagan C, O’Connor L, Holland EA, Schmid H, Maskiell JA, Jetann J, Ferguson M, Jenkins MA, Kefford RF, Giles GG, Armstrong BK, Aitken JF, Hopper JL, Whiteman DC, Pharoah PD, Easton DF, Dunning AM, Newton-Bishop JA, Montgomery GW, Martin NG, Mann GJ, Bishop DT, Tsao H, Trent JM, Fisher DE, Hayward NK, Brown KM, A novel recurrent mutation in MITF predisposes to familial and sporadic melanoma, Nature, 480 (2011) 99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Chen L, Miao Y, Liu M, Zeng Y, Gao Z, Peng D, Hu B, Li X, Zheng Y, Xue Y, Zuo Z, Xie Y, Ren J, Pan-Cancer Analysis Reveals the Functional Importance of Protein Lysine Modification in Cancer Development, Front Genet, 9 (2018) 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.