Abstract
Purpose of review
Pain researchers and clinicians increasingly encounter machine learning algorithms in both research methods and clinical practice. This review provides a summary of key machine learning principles, as well as applications to both structured and unstructured datasets.
Recent findings
Aside from increasing use in the analysis of electronic health record data, machine and deep learning algorithms are now key tools in the analyses of neuroimaging and facial expression recognition data used in pain research.
Summary
In the coming years, machine learning is likely to become a key component of evidence-based medicine, yet will require additional skills and perspectives for its successful and ethical use in research and clinical settings.
Keywords: deep learning, machine learning, pain, review
INTRODUCTION
In recent years, advances in machine learning have promulgated to many domains of biomedical informatics, including pain management [1■■]. In this article, we first review many core principles and definitions in the field of machine learning. Next, we examine the impact of machine learning approaches in the analyses of large electronic health record data sets. We end with a review of advanced machine and deep learning approaches to semi-structured and unstructured datasets, highlighting several exciting future directions in the use of machine learning to enhance our understanding and treatment of pain conditions.
OVERVIEW OF MACHINE LEARNING
In recent years, there has been an increasing interest in applying machine learning techniques in the healthcare domain. Today, a wide range of machine learning applications can be found, for example, selection of human blastocysts after in-vitro fertilization [2], predicting medical events based on electronic health records (EHR) [3,4■], and diabetic retinopathy detection [5]. In the pain management domain, many researchers have started to utilize such approaches, for rexample, recognizing pain facial expressions [6,7].
In the following primer, we provide a brief introduction of machine learning methods, followed by a discussion of pain management applications.
Definition
Machine learning is a sub-field of artificial intelligence, with the goal of learning a function from a set of data points by optimizing a certain performance metric, such as prediction accuracy. Each data point x, is composed of categorical or numerical features such as demographics, laboratory results, or preexisting conditions. Additionally, each data point might be associated with a label y. The label (category or outcome of interest) is either directly extracted from data, for example, postoperative outcome (0 = mortality, 1 = survival), or might be provided by an expert, for example, melanoma lesion diagnosis from photographic images (0 = not not present, 1 = present) [8].
Differentiation from statistics
Although machine learning and statistics share some key elements, they have been developed in different contexts to solve different problems. Classical statistical models were designed to handle data with a few dozen input variables and modest sample sizes, with an emphasis on experiment design [9].
On the other hand, machine learning models often concentrate on prediction from already available data by using general-purpose algorithms to find patterns in very rich and unwieldy data [9]. For example, modern deep learning models can detect and segment objects in high-dimensional image or video data, such as volumetric electron microscopy images [10], and can identify temporal patterns across many high-resolution physiological signals [4■].
TYPES OF MACHINE LEARNING TASKS
Machine learning approaches can be divided into three general categories: supervised learning, unsupervised learning, and reinforcement learning approaches. Beyond these, there are several other approaches, such as semi-supervised learning and active learning; we refer interested readers to references on these topics [11,12].
Supervised learning
Supervised learning models require a labeled dataset (the category or outcome is identified), where labels are used as supervision signals to guide the learning process [13]. The supervised task itself can be a classification task, where the target label is a nominal label (e.g. 0 = mortality, 1 = survival). Other supervised tasks include regression, where the target label (e.g. 0 = no stroke, 1 = stroke) is drawn from a continuous numerical range of values (e.g. blood pressure). Finally, the supervised task might be a ranking task, where the target is an ordinal value (e.g. ranking of pain management medications).
Many supervised machine learning techniques have been developed in the past few decades, including decision trees, random forests, support vector machines (SVM), neural networks, gradient boosting machines, among others [14■]. These methods have shown good performance on small/moderate sized and structured datasets in many domains related to traditional outcomes research including diagnosis management, prediction modeling, event detection, and risk evaluation, including the risk of severe pain following surgery [15–21].
Unsupervised learning
Unsupervised learning models, on the other hand, do not require labeled data, but rather will discover relationships in data in an unsupervised manner (e.g. by grouping similar data points together in a lower dimensional space (more homogenous subgroups)). Some examples include clustering, dimensionality reduction, and autoencoder techniques [14■,22]. Particularly in recent years, dimensionality reduction techniques, such as T-distributed Stochastic Neighbor Embedding (t-SNE)[23] and Uniform Manifold Approximation and Projection (UMAP) [24] have been used extensively in visualizing high-dimensional data (e.g. for visualization of genetic disease-phenotype similarity). The s-SNE and UMAP techniques ‘compress’ highly dimensional data into two dimensions that can be plotted on a more understandable x–y coordinate plane, all while preserving some of the mathematical relationships contained in the higher dimensional space. For instance, t-SNE could be used to compress the information contained within four-dimensional videos of ultrasound-guided nerve blocks (e.g. x, y, color depth, time) onto a two-dimensional plane to understand nonlinear relationships amongst the original four dimensions.
Reinforcement learning
Reinforcement learning models map a sequence of situations (i.e. states) into a sequence of actions by maximizing a reward signal [25]. Unlike supervised learning, in reinforcement learning problems, the correct actions are not labeled. Rather, the reinforcement learning model should infer the correct actions based on the reward signal, which might be provided in a delayed manner. Recent reinforcement learning models apply deep learning techniques to represent high-dimensional states (e.g. for diagnosing neural symptoms, or for optimizing medical dosing) [26,27]. Recent work in a simulated intensive care decision support model suggests these approaches may help improve clinical outcomes through improved, data-based decision support, although also highlighted the significant challenges in this approach in the healthcare setting [28].
MEASURING MACHINE LEARNING PERFORMANCE
To evaluate the performance of machine learning models, the original dataset is typically partitioned into three independent subsets: a training subset, a validation subset for fine-tuning parameters, and a test dataset solely used for reporting the performance. It is also possible to perform the data partitioning using cross-validation or stratified cross-validation methods. In stratified cross-validation, the folds contain proportional number of examples from each class based on considerations, such as prevalence. Several model performance metrics can be used, including classification accuracy, precision, recall, area under curve (AUC), F1 score, or mean absolute error (MAE) [29]. It is important to implement a rigorous testing approach to avoid overfitting problems, where data fits training data but fails to provide generalization in case of new data. Clinical machine learning models can greatly benefit from external validation and prospective clinical validation to avoid such issues [30].
DEEP LEARNING
Deep learning models are a relatively new class of machine learning models that aim to automatically learn data representations, and typically achieve higher performance compared with conventional machine learning models [31■■]. Most deep learning models learn a hierarchy of features, increasingly becoming more complex and building upon prior simpler features. For instance, recognizing a human in an image can involve finding representation of edges from pixels, contours and corners from edges, and facial features from corners and contours [32■■].
Most deep learning models are based on neural networks, where each layer is composed of several neurons. The input layer neurons represent input data (e.g. pixel values in an image) and the output neurons correspond to the target values, such as binary prediction of survival/mortality, or multi-pixel segmentation mask in a biomedical image. The middle layer neurons are called as hidden neurons. There are many different types of deep neural networks, including convolutional neural networks (CNN) for analyzing images, recurrent neural networks (RNN) for processing sequential and temporal data, and variational autoencoders for dimensionality reduction [31■■,32■■,33]. In most deep learning models, at least several thousand examples will be needed to develop a reasonable model [30].
STRUCTURED AND UNSTRUCTURED DATA
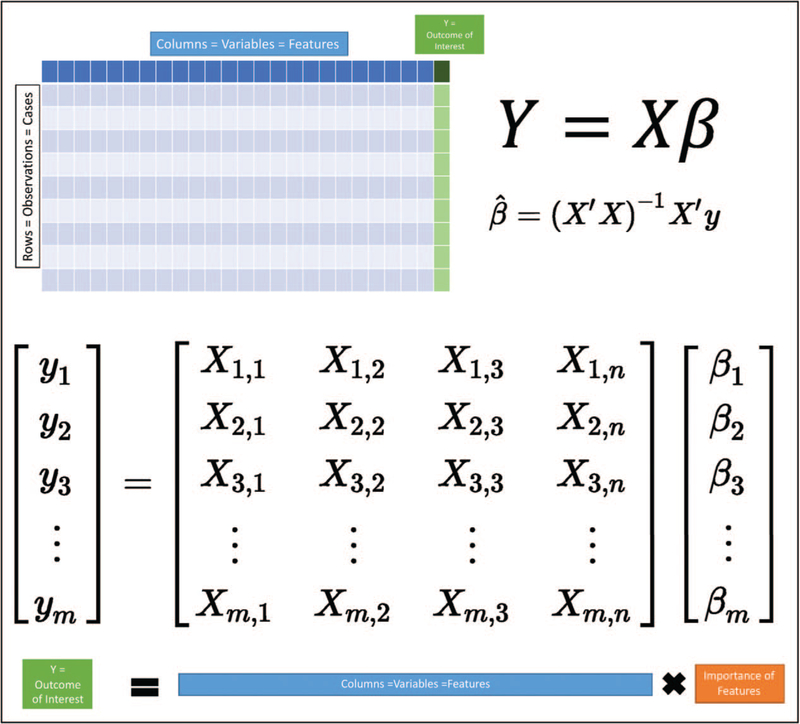
One of the key advances of machine learning over traditional statistical methods concerns the analyses of unstructured data. Structured data in this context refers to data organized in a traditional biomedical data model where columns commonly represent variables, and rows different patients, observations, or cases. Additionally, each element is generally curated and validated such that each variable is restricted to a single form and format. These organizational constructs lend themselves to traditional machine learning analyses given that their organization aligns well with the matrix-based solutions commonly used for solving regression-type problems (Fig. 2).
FIGURE 2.
Structured data. The organization of structured data lends itself well to solving traditional regression analyses.
Contrariwise, unstructured data is information that does not conform to such traditionally organized common data models. Examples of unstructured data in the perioperative environment include images, videos, waveforms, network structures, and clinical text documents. Without some type of decomposition or abstraction, unstructured data has no discretely labelled variables in which the information can be placed. Moreover, the information within unstructured data is typically accessible at several different scales. For instance, with imaging data, we may wish to consider information on a pixel-by-pixel level, but also to consider different shapes, their size, orientation, and relationship with other shapes.
Structured datasets, such as EHR records, readily provide most data points (features) in a numerical or categorical form. However, unstructured and semi-structured data, such as biomedical images, protein networks, clinical notes, and physiological time series data typically need to be converted into a suitable numerical format. The task of changing the form of representation for the purpose of learning is called representation learning or embedding, and is achieved using deep learning techniques [32■■,34].
EARLY EXPLORATIONS
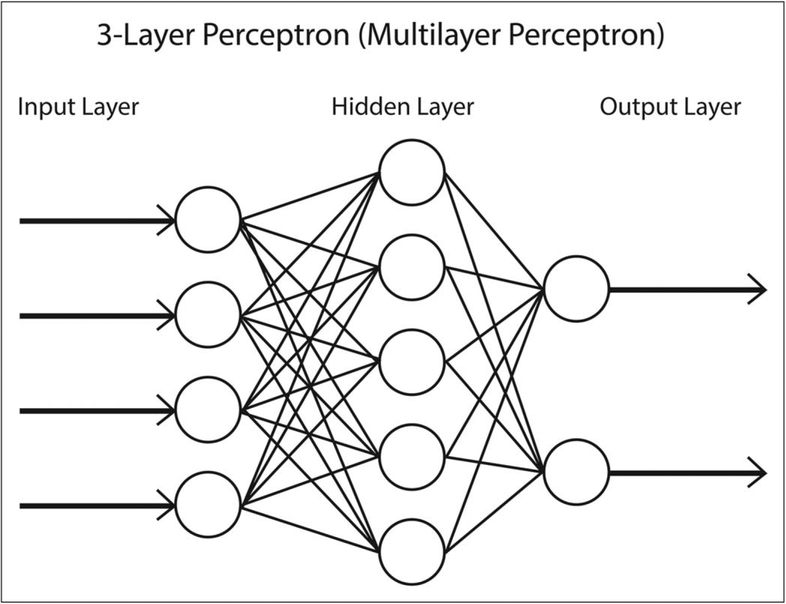
Early explorations with machine learning in health-care were developed by neurophysiologists and modeled after the complex function of the brain [35]. Neurons in the brain signal within networks by propagating suprathreshold signals across synapses by dispatching anterograde and retrograde messengers. Artificial neural network machine learning models were developed to explore this function of the brain. With only a few data features to use to predict an outcome, simple three-layer neural networks (Fig. 3) were developed that rivaled the performance of multiple regression mathematical models [36,37]. Since then, complex multilayer models (e.g. multilayer perceptron neural network) have been used in several clinical studies including to diagnose cancer [38,39].
FIGURE 3.
An example of a simple three-layer perceptron neural network. In this example, four input variables (input layer) have several interactions (hidden layer) and determine one of two possible outcomes (output layer).
Three basic reasons neural networks and other machine learning methods performed well on healthcare data are that they account for nonlinearity among parameters (e.g. nonlinear response of age), they account for important interactions among variables (e.g. smoking history and rates of surgical infection on hospital length of stay), and with increasing number of data features, predictive power can be retained despite a limited sample size (e.g. hundreds or even thousands of exposure variables can still result in a unbiased prediction models). Occasionally the complexity of the data precludes foreknowledge of which methods would be preferred, so multiple machine learning models can be developed as an initial screening strategy to identify the method that has the best performance.
Structured data studies
Postoperative pain is a difficult outcome to predict because of the number of possible contributing factors. Patient factors (e.g. age, sex, genetic profile, comorbid medical and psychological disease) and surgical factors (e.g. surgeon, operation, operative location, surgical and anesthetic techniques) can all contribute to the likelihood of severe acute or chronic postsurgical pain. Even though some patients are more likely than others to experience severe pain, resources and treatment are often applied to all patients because of an inability to identify before surgery those who are at risk.
Acute postsurgical knee pain
In a chart review of 349 patients who were to undergo anterior cruciate ligament (ACL) reconstruction, perioperative data (e.g. age, BMI, substance use, medications, open versus arthroscopic surgical approach, tourniquet time, anesthetic medications) were collected and multiple machine learning models [logistic regression, BayesNet, multilayer perceptron, support vector machine, alternating decision tree (ADTree)] created to predict which patients would experience severe acute postoperative pain, and therefore, require postoperative rescue pain treatment in the form of a femoral nerve block [21]. Machine learning models developed on structured data from the EHR outperformed logistic regression models identifying patients likely to experience severe postoperative pain after ACL repair.
Decision support tool for acute pain consultation
With structured data, machine learning methods can be useful decision support tools to replace less efficient human-driven operations. In the Tighe et al. [19] study, a cohort of machine learning models were developed and tested to predict the need for preoperative pain service consultation based solely on data elements from surgical posting schedules. Bayesian classifiers had the best performance on this data to predict which surgical cases should prompt a preoperative request for acute pain consultation with an accuracy AUC of 0.87 in a training time of 0.0018 s. This is noteworthy, given the training times required for other machine learning approaches, such as neural networks, which required over 30 s of training time per model, and highlight the need to consider outcomes, features, and algorithmic factors in designing machine learning solutions.
Unstructured data studies
Neuroimaging data
Within the domain of pain research, there have been several exciting forays into nontraditional, unstructured, and semistructured data using machine learning approaches. Machine learning analyses of neuroimaging data (e.g. images obtained through functional MRI – fMRI) can be based upon structural features, such as via volumetric pixel (voxel)-based morphometry indicating the size of specific brain regions, functional biomarkers, such as blood oxygenation (BOLD) or arterial spin labelling, and connectivity measures across brain regions [40,41]. Early work by Mackey and coworkers showed that the semi-structured data from fMRI studies, when analyzed using a support vector machine, could label the painful versus non-painful thermal stimuli with high degrees of accuracy (81–84%) [42]. Follow-up work by Wager et al.[43] extended these findings by identifying fMRI-based signatures of thermal pain that also discriminate from other aversive events and are sensitive to opioid analgesic effects. Notably, these machine learning approaches generally rely on human and/or algorithmic abstraction of neuroimaging features rather than deep learning approaches to feature processing.
Robinson, Hu, and others have raised important questions concerning disparities between machine learning-derived neuroimaging biomarkers of pain and patient self-report of pain [44■■,45]. Given the subjective nature of many pain experiences and ethical issues involved in under-treatment versus over-treatment of pain conditions, researchers and clinicians will need to seriously examine findings from this rapidly evolving field to ensure that appropriate relief of patient suffering is not lost to an overly simplified abstractions of quasi-validated models.
Facial recognition
One common approach to facial expression recognition concerns machine learning analyses on features coded as action units using the Facial Action Coding System, such that the action units serve as an interpretable abstraction layer for facial expressions [46]. Other approaches use deep neural networks to capture raw features direct from images of facial expressions, potentially accessing a richer set of features but at a cost of reduced interpretability [47]. Recent work by Chen et al. [48] has shown that mental representations of facial expressions of pain, as assessed via machine learning of facial action units, are consistent across cultures. This approach has also been extended to animal models of pain, such as with the Mouse Grimace Scale, which trained an Inception V3 convolutional neural net on a dataset of human-scored images of mice in various pain states [49,50].
These early investigations on facial recognition of pain are already translating towards the clinical environment, with special emphasis on pediatric populations. Sikka et al. [51] has used similar methods for pediatric postoperative pain, with detection of pain versus no pain model accuracy of AUC 0.84–0.94. Other teams have used similar approaches to neonatal pain, a particularly exciting advance given the range of painful procedures neonates experience during extended NICU stays [52,53].
ONGOING CHALLENGES AND OPPORTUNITIES
Despite the myriad advances offered by machine learning, these new analytical techniques have also forced a reckoning by physicians and researchers on fundamental challenges concerning the application of evidence-based medicine to individual patients.
Above all, physicians must remember that patients are more than just data. Accurate machine learning algorithms may enhance disease diagnosis, but they cannot deliver that diagnosis with compassion and understanding, with a recognition of the impact of that diagnosis on the patient and their future. Perhaps one of the greatest potential benefits of machine learning in medicine would be allowing physicians to attend more to the humanistic needs of our patients.
One final challenge concerns the ever-present potential for divergence of the observed from the expected. In prognosticating outcomes with grave consequences, physicians and patients will need to consider how to approach scenarios where the forecasted outcome is considered highly likely, but for reasons that may be murky. For instance, how should a physician counsel a patient who is 90% likely to succumb to an illness based upon a machine learning algorithm, but where the diagnosis is based upon 10 000 ‘-omic’ features that neither physician, nor patient, nor researcher can readily interpret? And what of the patient who is potentially concerned about more than one outcome, perhaps judging tradeoffs between life expectancy, function, and suffering? Although the future of machine learning in pain medicine is exciting, patients and physicians must first confront myriad ethical and operational questions before these tools can definitively improve patient health.
KEY POINTS.
In certain circumstances and contexts, machine learning may offer advantages over traditional techniques.
One of the key opportunities for machine learning in pain research and clinical practice pertains to the use of complex and unstructured data.
Patients, physicians, and researchers will need to address important ethical issues as machine learning methods and applications to pain continue to evolve.
Acknowledgements
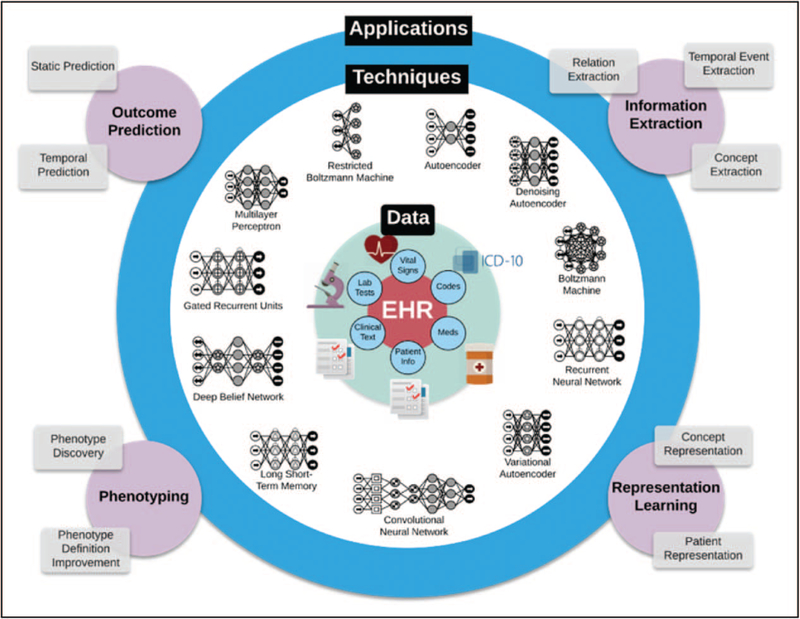
We gratefully acknowledge the assistance of Benjamin Shickel for his assistance in drafting Fig. 1. Portions of this work were supported by NIH/NIGMS R01 GM114290 (P.R., P.T.), NIH/NIBIB 1R21EB027344 (P.R., P.T.), and NSF CAREER 1750192 (P.R.). All authors have contributed and reviewed this work and report no commercial conflicts of interest to this submission.
FIGURE 1.
Different deep learning methods can be used to extract information from clinical data.
Financial support and sponsorship
None.
Footnotes
Conflicts of interest
There are no conflicts of interest.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
■ of special interest
■■ of outstanding interest
- 1. ■■.Lotsch J, Ultsch A. Machine learning in pain research. Pain 2018; 159:623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]; This extended review highlights numerous studies in the basic science and clinical pain literature.
- 2.Khosravi P, Kazemi E, Zhan Q, et al. Deep learning enables robust assessment and selection of human blastocysts after in vitro fertilization. NPJ Digit Med 2019; 2:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rajkomar A, Oren E, Chen K, et al. Scalable and accurate deep learning with electronic health records. NPJ Digit Med 2018; 1:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. ■.Shickel B, Loftus TJ, Adhikari L, et al. DeepSOFA: a continuous acuity score for critically ill patients using clinically interpretable deep learning. Sci Rep 2019; 9:1879. [DOI] [PMC free article] [PubMed] [Google Scholar]; A cutting-edge example of applying advanced deep learning algorithms on time series data to address challenging clinical problems.
- 5.Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 2016; 316:2402–2410. [DOI] [PubMed] [Google Scholar]
- 6.Davoudi A, Malhotra KR, Shickel B, et al. The intelligent ICU pilot study: using artificial intelligence technology for autonomous patient monitoring. arXiv preprint arXiv:180410201. 2018
- 7.Rodriguez P, Cucurull G, Gonalez J, et al. Deep pain: exploiting long short-term memory networks for facial expression classification. IEEE Trans Cybern 2017. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 8.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017; 542:115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bzdok D, Altman N, Krzywinski M. Statistics versus machine learning. Nat Methods 2018; 15:233–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beier T, Pape C, Rahaman N, et al. Multicut brings automated neurite segmentation closer to human performance. Nat Methods 2017; 14:101–102. [DOI] [PubMed] [Google Scholar]
- 11.Sammut C, Webb GI. Encyclopedia of machine learning and data mining. 2nd ed New York, NY: Springer; 2017. 1333 pp., 2 volumes (xvii). [Google Scholar]
- 12.Chapelle O, Schölkopf B, Zien A. Semi-supervised learning. Cambridge, MA: MIT Press; 2010; 508 pp. [Google Scholar]
- 13.Mohri M, Rostamizadeh A, Talwalkar A. Foundations of machine learning. Cambridge, MA: MIT press; 2018. [Google Scholar]
- 14. ■.Hastie T,Tibshirani R,Friedman JH.Theelementsofstatisticallearning:datamining, inference, and prediction. 2nd ed New York, NY: Springer; 2009;xxii, 745 pp. [Google Scholar]; This is a classic reference on the theoretical underpinnings of modern data science methods.
- 15.Bihorac A, Ozrazgat-Baslanti T, Ebadi A, et al. MySurgeryRisk: development and validation of a machine-learning risk algorithm for major complications and death after surgery. Ann Surg 2019; 269:652–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Golas SB, Shibahara T, Agboola S, et al. A machine learning model to predict the risk of 30-day readmissions in patients with heart failure: a retrospective analysis of electronic medical records data. BMC Med Inform Decis Mak 2018; 18:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torlay L, Perrone-Bertolotti M, Thomas E, Baciu M. Machine learning-XGBoost analysis of language networks to classify patients with epilepsy. Brain Inform 2017; 4:159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu J, Roy J, Stewart WF. Prediction modeling using EHR data: challenges, strategies, and a comparison of machine learning approaches. Med Care 2010; 48(6 Suppl):S106–S113. [DOI] [PubMed] [Google Scholar]
- 19.Tighe PJ, Lucas SD, Edwards DA, et al. Use of machine-learning classifiers to predict requests for preoperative acute pain service consultation. Pain Med 2012; 13:1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tighe PJ, Harle CA, Hurley RW, et al. Teaching a machine to feel postoperative pain: combining high-dimensional clinical data with machine learning algorithms to forecast acute postoperative pain. Pain Med 2015; 16:1386–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tighe P, Laduzenski S, Edwards D, et al. Use of machine learning theory to predict the need for femoral nerve block following ACL repair. Pain Med 2011; 12:1566–1575. [DOI] [PubMed] [Google Scholar]
- 22.Liou C-Y, Huang J-C, Yang W-C. Modeling word perception using the Elman network. Neurocomputing 2008; 71:3150–3157. [Google Scholar]
- 23.Lvd Maaten, Hinton G. Visualizing data using t-SNE. J Machine learning Res 2008; 9(Nov):2579–2605. [Google Scholar]
- 24.McInnes L, Healy J, Melville J. UMAP: uniform manifold approximation and projection for dimension reduction. arXiv e-prints [Internet]. 2018. 01 February 2018. Available at: https://ui.adsabs.harvard.edu/abs/2018ar-Xiv180203426M. [Accessed 30 July 2019]
- 25.Sutton RS, Barto AG. Reinforcement learning: an introduction. Cambridge, MA: MIT press; 2018. [Google Scholar]
- 26.Mnih V, Kavukcuoglu K, Silver D, et al. Human-level control through deep reinforcement learning. Nature 2015; 518:529–533. [DOI] [PubMed] [Google Scholar]
- 27.Nemati S, Ghassemi MM, Clifford GD, editors. Optimal medication dosing from suboptimal clinical examples: A deep reinforcement learning approach. 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC); 2016: IEEE. [DOI] [PubMed] [Google Scholar]
- 28.Komorowski M, Celi LA, Badawi O, et al. The Artificial Intelligence Clinician learns optimal treatment strategies for sepsis in intensive care. Nat Med 2018; 24:1716–1720. [DOI] [PubMed] [Google Scholar]
- 29.Powers DM. Evaluation: from precision, recall and F-measure to ROC, informedness, markedness and correlation. 2011.
- 30.Rajkomar A, Dean J, Kohane I. Machine learning in medicine. N Engl J Med 2019; 380:1347–1358. [DOI] [PubMed] [Google Scholar]
- 31. ■■.Shickel B, Tighe PJ, Bihorac A, Rashidi P. Deep EHR: a survey of recent advances in deep learning techniques for electronic health record (EHR) analysis. IEEE J Biomed Health Informat 2017; 22:1589–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is an extensive review of deep learning applications to electronic health record data.
- 32. ■■.Goodfellow I, Bengio Y, Courville A. Deep learning. Cambridge, Massachusetts: The MIT Press; 2016; xxii, 775 pp. [Google Scholar]; A classic text on the theory of deep learning algorithms, ranging from linear algebra to advanced research and practical applications of deep learning.
- 33.Pu Y, Gan Z, Henao R, et al. , editors. Variational autoencoder for deep learning of images, labels and captions. Advances in neural information processing systems; 2016. [Google Scholar]
- 34.Bengio Y, Courville A, Vincent P. Representation learning: a review and new perspectives. IEEE Trans Pattern Anal Mach Intell 2013; 35:1798–1828. [DOI] [PubMed] [Google Scholar]
- 35.Cleophas TJ, Zwinderman AH. Artificial intelligence, multilayer perceptron modeling BT Machine learning in medicine. New York: Springer; 2013; 145–156. [Google Scholar]
- 36.Cleophas TJ, Zwinderman AH. Machine learning in medicine-a complete overview. New York; 2015. [Google Scholar]
- 37.Minsky M. A framework for representing knowledge. 1974.
- 38.Finne P, Finne R, Auvinen A, et al. Predicting the outcome of prostate biopsy in screen-positive men by a multilayer perceptron network. Urology 2000; 56:418–422. [DOI] [PubMed] [Google Scholar]
- 39.Simpson H, McArdle C, Pauson A, et al. A noninvasive test for the pre-cancerous breast. Eur J Cancer 1995; 31:1768–1772. [DOI] [PubMed] [Google Scholar]
- 40.Boissoneault J, Sevel L, Letzen J, et al. Biomarkers for musculoskeletal pain conditions: use of brain imaging and machine learning. Curr Rheumatol Rep 2017; 19:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhong J, Chen DQ, Hung PS-P, et al. Multivariate pattern classification of brain white matter connectivity predicts classic trigeminal neuralgia. Pain 2018; 159:2076–2087. [DOI] [PubMed] [Google Scholar]
- 42.Brown JE, Chatterjee N, Younger J, Mackey S. Towards a physiology-based measure of pain: patterns of human brain activity distinguish painful from nonpainful thermal stimulation. PloS One 2011; 6:e24124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wager TD, Atlas LY, Lindquist MA, et al. An fMRI-based neurologic signature of physical pain. N Engl J Med 2013; 368:1388–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. ■■.Hu L, Iannetti GD. Painful issues in pain prediction. Trends Neurosci 2016; 39:212–220. [DOI] [PubMed] [Google Scholar]; Excellent summary of methodologicchallenges in forecasting pain-related outcomes.
- 45.Robinson ME, O’Shea AM, Craggs JG, et al. Comparison of machine classification algorithms for fibromyalgia: neuroimages versus self-report. J Pain 2015; 16:472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kunz M, Meixner D, Lautenbacher S. Facial muscle movements encoding pain—a systematic review. Pain 2019; 160:535–549. [DOI] [PubMed] [Google Scholar]
- 47.Kharghanian R, Peiravi A, Moradi F, editors. Pain detection from facial images using unsupervised feature learning approach. 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC); 2016: IEEE. [DOI] [PubMed] [Google Scholar]
- 48.Chen C, Crivelli C, Garrod OG, et al. Distinct facial expressions represent pain and pleasure across cultures. Proc Natl Acad Sci 2018; 115:E10013–E10021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szegedy C, Vanhoucke V, Ioffe S, et al. , editors. Rethinking the inception architecture for computer vision. Proceedings of the IEEE conference on computer vision and pattern recognition; 2016. [Google Scholar]
- 50.Tuttle AH, Molinaro MJ, Jethwa JF, et al. A deep neural network to assess spontaneous pain from mouse facial expressions. Mol Pain 2018; 14:1744806918763658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sikka K, Ahmed AA, Diaz D, et al. Automated assessment of children’s postoperative pain using computer vision. Pediatrics 2015; 136:e124–e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gholami B, Haddad WM, Tannenbaum AR. Relevance vector machine learning for neonate pain intensity assessment using digital imaging. IEEE Trans Biomed Eng 2010; 57:1457–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brahnam S, Chuang C-F, Sexton RS, Shih FY. Machine assessment of neonatal facial expressions of acute pain. Decis Support Syst 2007; 43:1242–1254. [Google Scholar]