Abstract
The de novo serine synthesis pathway is upregulated in many cancers. However, even cancer cells with increased serine synthesis take up large amounts of serine from the environment1 and we confirm that exogenous serine is needed for maximal proliferation of these cells. Here we show that even when enzymes in the serine synthesis pathway are genetically upregulated, the demand for oxidized NAD+ constrains serine synthesis, rendering serine-deprived cells sensitive to conditions that decrease the cellular NAD+/NADH ratio. Further, purine depletion is a major consequence of reduced intracellular serine availability, particularly when NAD+ regeneration is impaired. Thus, cells rely on exogenous serine consumption to maintain purine biosynthesis. In support of this explanation, providing exogenous purine nucleobases, or increasing NAD+ availability to facilitate de novo serine and purine synthesis, both rescue maximal proliferation even in the absence of extracellular serine. Together, these data indicate that NAD+ is an endogenous limitation for cancer cells to synthesize the serine needed for purine production to support rapid proliferation.
Many cancers exhibit altered serine metabolism2,3,4, including upregulated expression of de novo serine synthesis pathway enzymes5. The gene encoding the first enzyme in the serine synthesis pathway, phosphoglycerate dehydrogenase (PHGDH), exhibits copy number gain in some cancers5,6, most frequently in breast cancer and melanoma5,7. Serine synthesis enzymes can also be transcriptionally upregulated by the transcription factor ATF4, and many non-small cell lung cancers exhibit ATF4-mediated PHGDH upregulation downstream of deregulated NRF28. There is also evidence that environmental serine availability can be critical for proliferation of some tumor cells6,7,9,10,. In fact, environmental serine limitation can slow the growth of some tumors9, and even cells with upregulated serine synthesis appear to rely on extracellular serine for optimal proliferation10.
Serine is a precursor for many classes of biomolecules, and a major donor of one-carbon (1C) units, which can support methylation reactions and nucleotide synthesis. One 1C unit is needed to produce deoxythymidine monophosphate (dTMP), and two 1C units are needed to produce inosine monophosphate (IMP), the precursor for both AMP and GMP (Supplementary Fig. 1a). Indeed, purine levels are decreased in cells cultured without exogenous serine10, and blocking de novo serine synthesis can deplete nucleotides in some cells11.
Why cells differentially increase serine synthesis and whether the source of intracellular serine matters for cell proliferation remains unclear. Examining the metabolic constraints of serine synthesis could provide insight. A difference between serine consumption and synthesis is demand for redox cofactors: while extracellular serine uptake does not use NAD+, serine synthesis from glucose requires oxidation steps that consume NAD+ and produce NADH (Supplementary Fig. 1a).
Regeneration of NAD+ to support oxidation reactions and build biomass can be a metabolic constraint for cancer cell proliferation and tumor growth in some contexts12,13,14. This suggests that NAD+ availability could constrain flux through the serine synthesis pathway and necessitate exogenous serine consumption. In support of this notion, withdrawing extracellular serine can increase cancer cell sensitivity to the biguanides metformin and phenformin15, both of which inhibit NAD+ regeneration via Complex I of the mitochondrial electron transport chain (ETC)14. Furthermore, inhibiting NAD(H) production via the salvage pathway inhibits serine synthesis in breast cancer cells with PHGDH copy number gain16. NAD+ is also required to produce the 1C units needed in purine and dTMP synthesis (Supplementary Fig. 1a), raising the possibility that NAD+ insufficiency could exacerbate the effect of serine deprivation on nucleotide synthesis. Indeed, 1C unit production is impaired in cells with defective mitochondrial respiration17, a condition that causes decreased NAD+ availability. The cellular NAD+/NADH ratio can therefore dictate the metabolic processes available to cells18, and could influence dependence on extracellular serine consumption.
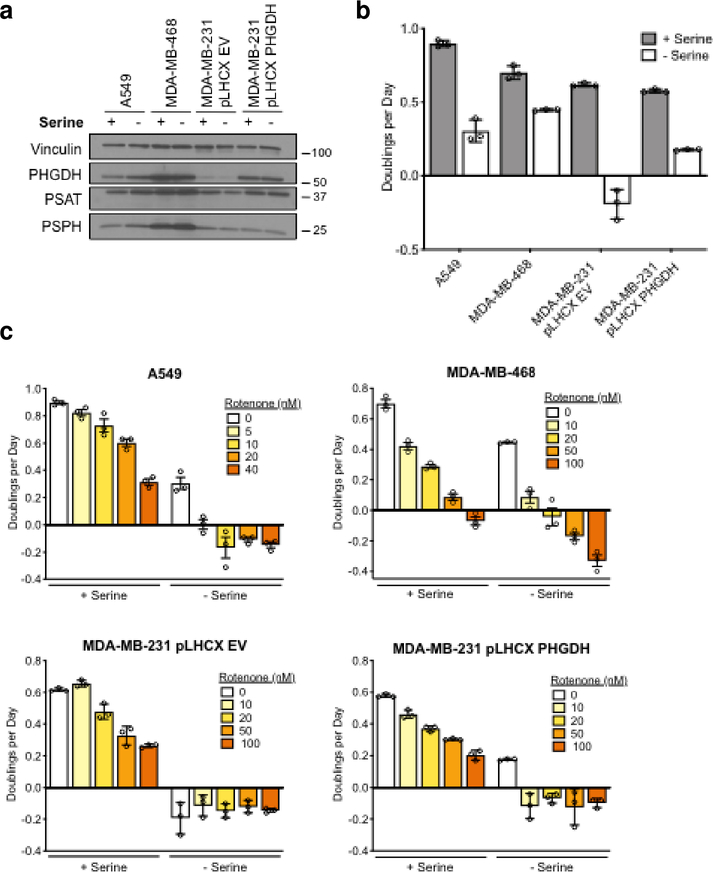
We evaluated the importance of extracellular serine consumption in cells in which serine synthesis is upregulated as a result of PHGDH gene copy number gain (MDA-MB-468)5 or increased via transcriptional regulation (A549)8. We also examined cells with low serine synthesis (MDA-MB-231)5,11 in which PHGDH was or was not exogenously expressed to increase serine synthesis (MDA-MB-231 pLHCX PHGDH and MDA-MB-231 pLHCX EV, respectively)11 (Fig. 1a). In each of these cell lines, as well as in other cells with high PHGDH expression, removing serine from the media substantially decreased proliferation (Fig. 1b and Supplementary Fig. 1b,c). PHGDH-expressing cells cultured in serine-free media maintain robust serine synthesis but cannot sustain the same levels of intracellular serine when extracellular serine is not available (Supplementary Fig. 1d, e). Together, these data indicate that even high expression of serine synthesis enzymes cannot sustain maximal proliferation.
Figure. 1: Withdrawal of exogenous serine limits proliferation and is exacerbated by inhibition of mitochondrial respiration.
a, Western blot analysis to assess expression of serine synthesis enzymes in A549 and MDA-MB-468 cells, and in MDA-MB-231 cells with (pLHCX PHGDH) and without (pLHCX EV) PHGDH expression as indicated. These experiments were repeated 3 times with similar results. b, Proliferation rates of the indicated cancer cells cultured in media with serine (+ Serine), or without serine (- Serine). c, Proliferation rates of A549 and MDA-MB-468 cells, and of MDA-MB-231 cells with (pLHCX PHGDH) and without (pLHCX EV) PHGDH expression at the indicated concentration of rotenone, in the presence or absence of exogenous serine as indicated. Data shown are mean (+/− standard deviation) of 3 biological replicates.
Serine deprivation has been shown to enhance the antiproliferative effects of the biguanides phenformin and metformin15, both of which inhibit Complex I of the mitochondrial ETC14. To examine whether increased sensitivity to serine withdrawal is generally observed with Complex I inhibitors, and whether this is affected by upregulated serine synthesis, we exposed cells to increasing concentrations of rotenone and metformin in the presence or absence of serine. Withdrawal of serine from the media increased sensitivity of cells to Complex I inhibitors (Fig. 1c and Supplementary Fig. 1f). We confirmed that expression of serine synthesis enzymes was not affected by mitochondrial inhibition (Supplementary Fig. 2a). These data suggest that extracellular serine can be limiting for cell proliferation even in cells with increased serine synthesis, and that this limitation can be exacerbated by inhibiting mitochondrial Complex I.
Production of nucleotides is an important fate of serine carbon to support proliferation10,11,19. To assess whether cells prefer to use either extracellular serine or newly synthesized serine to produce nucleotides, we traced the fate of 6-13C-glucose or 3-13C-serine. For cells cultured in 6-13C-glucose, newly synthesized serine has a mass value of M+1, and generates an M+1 1C unit. 6-13C-glucose can also label the ribose backbone of nucleotides. For cells cultured in 3-13C-serine, consumed serine has a mass of M+1, and generates an M+1 1C unit (Supplementary Fig. 1g). 1C units contribute to pyrimidine synthesis only when adding a methyl group to make dTMP. Thus, when cultured in the presence of 6-13C-glucose, whether dTMP is M+1 labeled relative to dCMP shows whether 1C units are derived from serine that was synthesized from glucose, while dTMP labeling from 3-13C-serine reflects dTMP production from exogenous serine. Consistent with only a small fraction of the serine in MDA-MB-468 cells being derived from glucose when extracellular serine is available (Supplementary Fig. 1e), the increased amount of dTMP labeled from 6-13C-glucose compared to dCMP labeled from 6-13C-glucose (resulting from 6-13C-glucose-derived 1C units) is small relative to the amount of dTMP labeled from extracellular serine (Supplementary Fig. 1g). This argues that cells do not preferentially use serine derived from one source for 1C unit production. It also raises the question of why extracellular serine is important for proliferation even when cells have genetically upregulated synthesis and unlimited access to glucose. Because glucose is abundant in cell culture media and these cells excrete most glucose carbon as lactate1, serine synthesis is unlikely to be limited by carbon substrate availability.
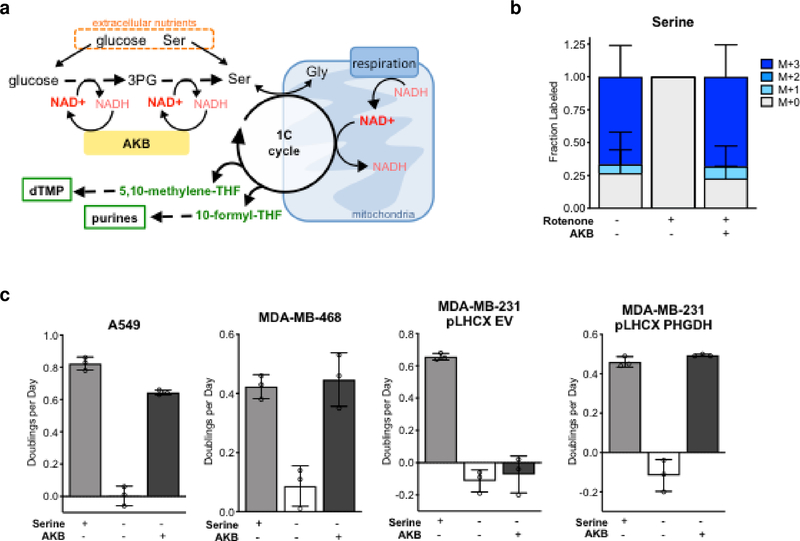
De novo serine synthesis and serine uptake have different redox cofactor requirements. Unlike the consumption of exogenous serine, serine production from glucose involves two oxidation reactions that consume NAD+ and produce NADH, and thus requires transfer of electrons to a waste product for every serine molecule produced (Fig. 2a). Thus, flux through serine synthesis should be sensitive to the cellular NAD+/NADH ratio. Of note, mitochondrial respiration allows transfer of electrons to oxygen and regenerates NAD+ from NADH. Thus, decreased NAD+/NADH ratio could explain why cells without access to exogenous serine are more sensitive to Complex I inhibition. To test this possibility, we compared serine production from U-13C-glucose in cells with and without Complex I inhibition (Supplementary Fig. 1d). While untreated serine-deprived cells displayed robust serine labeling from glucose, rotenone treatment completely eliminated U-13C-glucose incorporation into serine (Fig. 2b). As expected, rotenone treatment decreased the cellular NAD+/NADH ratio (Supplementary Fig. 4a). We next supplemented rotenone-treated cells with the exogenous electron acceptor α-ketobutyrate (AKB), which can be reduced to α-hydroxybutyrate (AHB) to regenerate NAD+ (Supplementary Fig. 2b)13. Indeed, providing AKB restored serine synthesis from glucose in rotenone-treated cells (Fig. 2b). We confirmed that cells took up AKB and used it to produce AHB (Supplementary Fig. 2c), and that AKB supplementation restored the NAD+/NADH ratio (Supplementary Fig. 4a), but did not cause cells to further upregulate expression of serine synthesis enzymes (Supplementary Fig. 2a). Addition of AKB also rescued proliferation of rotenone-treated and metformin-treated cells cultured in the absence of exogenous serine (Fig. 2c, Supplementary Fig. 2d,e, and Supplementary Fig. 5b). Together, these data indicate that NAD+ availability can limit serine synthesis and cell proliferation, particularly when the ability to transfer electrons to oxygen is impaired.
Figure. 2: Electron acceptor availability limits serine synthesis and cell proliferation.
a, Schematic showing the relationship between serine synthesis, the one-carbon (1C) cycle, and nucleotide synthesis (dTMP and purines), highlighting the NAD+-consuming steps involved in serine synthesis and 1C unit generation. Also shown is how NAD+ can be regenerated from NADH via mitochondrial respiration, or by the reduction of α-ketobutyrate (AKB) to α-hydroxybutyrate (AHB). Ser, serine; Gly, glycine; 3PG, 3-phosphoglycerate; THF, tetrahydrofolate. b, Fractional labeling of serine from U-13C-glucose in A549 cells cultured for 48 hours in serine-free media with or without 20 nM rotenone, and with or without AKB as indicated. Labeling was determined by LCMS analysis. c, Proliferation rates of the specified cells cultured in media containing 20nM rotenone, with or without serine and AKB, as indicated. Data shown are mean (+/− standard deviation) of 3 biological replicates.
Producing one-carbon (1C) units for nucleotide synthesis relies on functional mitochondrial respiration because mitochondrial NAD+ is needed to generate 1C units17 (Fig. 2a), as is the case in most cells20. Since serine is a major 1C unit donor, impaired serine synthesis following Complex I inhibition could exacerbate a decrease in 1C unit production. To test this possibility, we assessed how metabolite levels change in cells cultured in serine-replete compared to serine-free media, with or without rotenone treatment. Indeed, purine nucleotides were among the most depleted metabolites in serine-deprived cells, and rotenone treatment further exacerbated purine nucleotide depletion (Fig. 3a). To directly test whether de novo purine synthesis is impaired in serine-deprived cells, we measured the incorporation of 15N from amide-15N-glutamine into purines. Because the amide nitrogen of glutamine is incorporated in two steps of AMP synthesis and three steps of GMP synthesis, any newly synthesized adenylate or guanylate nucleotides have mass values of M+2 and M+3, respectively (Fig. 3b). Cells cultured in serine-replete media used de novo synthesis to turn over their purine pools by 48 hours (Fig. 3c and Supplementary Fig. 3a). In contrast, cells cultured in serine-free media displayed decreased de novo synthesis, leading to purine depletion over time. Consistent with previous reports, the purine synthesis intermediates GAR and AICAR also accumulated in serine-deprived cells10 (Fig. 3d). As these intermediates each precede 1C-requiring reactions in purine synthesis, this accumulation is consistent with insufficient 1C units limiting purine synthesis. Restoring 1C units by supplementing cells with formate alleviated this accumulation (Fig. 3d), as noted previously10,17.
Figure. 3: Purine nucleotide production downstream of serine metabolism constrains cell proliferation when NAD+ regeneration is impaired.
a, Hierarchical clustering and heat map indicating relative metabolite levels measured using LCMS from A549 cells cultured for 48 hours in media with or without serine and with or without 20 nM rotenone as indicated. b, Schematic showing how 15N label is incorporated from amide-15N-glutamine into the purines AMP and GMP. Two 15N labels are incorporated into newly synthesized AMP, while three 15N labels are incorporated into newly synthesized GMP. c, Total levels, and labeling of ATP and GTP from amide-15N-glutamine as determined by LCMS in A549 cells cultured in media with serine, or in media lacking serine for the indicated times. d, Levels of the purine synthesis intermediates GAR and AICAR measured using LCMS in A549 cells cultured with or without serine and with or without formate as indicated. GAR and AICAR each precede steps in purine synthesis that involve addition of a 1C unit derived from formate. GAR, glycineamide ribonucleotide; AICAR, 5-aminoimidazole-4-carboxamide ribonucleotide. e, Total levels and labeling of ATP and GTP from 5-13C-hypoxanthine in A549 cells cultured with or without serine, 20 nM rotenone, and 5-13C-hypoxanthine as indicated. Metabolites were analyzed by LCMS. f, Proliferation rates of A549 cells cultured with or without serine, formate, and/or hypoxanthine, and treated with DMSO or 20 nM rotenone as indicated. Data shown are mean (+/− standard deviation) of 3 biological replicates.
Salvage of purine nucleobases avoids the demand for serine-derived carbon to synthesize purines de novo. When 5-13C-hypoxanthine was provided, this was the primary carbon contributor to both adenylate and guanylate nucleotides regardless of serine availability (Fig. 3e and Supplementary Fig. 3b). Importantly, salvage of hypoxanthine rescued purine levels in serine-starved cells. Moreover, providing either formate to restore 1C unit availability or hypoxanthine to directly restore purine levels partially rescued cell proliferation in serine-free media (Fig. 3f). Formate or hypoxanthine supplementation also improved the proliferation of serine-deprived cells following rotenone or metformin treatment (Fig. 3f, Supplementary Fig. 3c,d, and Supplementary Fig. 5b). Importantly, formate or hypoxanthine supplementation did not restore the NAD+/NADH ratio (Supplementary Fig. 4a), suggesting that maintaining purine synthesis is a metabolic requirement downstream of NAD+ availability, and a reason why NAD+ is limiting for proliferation.
Serine synthesis requires NAD+ in the cytosol, so coupling this pathway to mitochondrial electron transport requires redox shuttles to transport electrons across the mitochondrial membranes. The malate-aspartate shuttle is one contributor to redox shuttling between the mitochondria and the cytosol, and disrupting this shuttle by knockdown of aspartate-glutamate carrier 1 (AGC1) expression21 did not affect proliferation of cells grown without exogenous serine, but did increase sensitivity of serine-deprived cells to Complex I inhibition (Supplementary Fig. 4b). These data are consistent with the malate-aspartate shuttle being important to maintain cytosolic NAD+ when mitochondrial NAD+ regeneration is impaired.
NAD+ availability can also be affected by nicotinamide salvage. Indeed, inhibiting nicotinamide salvage can inhibit serine synthesis in PHGDH-amplified breast cancer cells16. We therefore investigated whether nicotinamide salvage can increase oxidized NAD+ availability for serine synthesis. Addition of nicotinamide mononucleotide (NMN) did not restore proliferation of cells cultured without exogenous serine, and did not change the cellular NAD+/NADH ratio (Supplementary Fig. 4c). This may indicate that the NAD+/NADH ratio is a stronger constraint on serine synthesis than is the total amount of NAD(H) in cells.
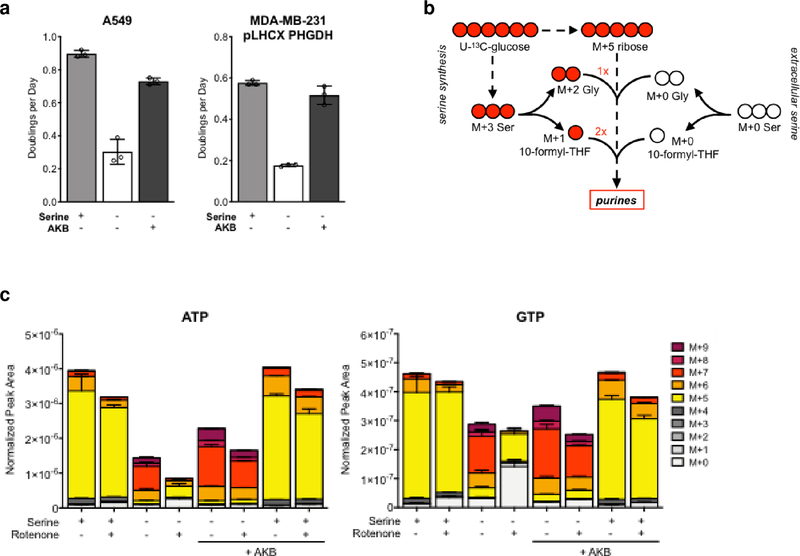
We next questioned whether basal NAD+ availability limits cell proliferation in the absence of exogenous serine. Indeed, increasing NAD+ availability by providing AKB increased proliferation of cells cultured in serine-free conditions (Fig. 4a and Supplementary Fig. 5a,b). To evaluate whether NAD+ availability constrains purine synthesis in serine-deprived cells, we measured U-13C-glucose incorporation into purines in cells grown in serine-free or serine-replete media with or without rotenone. In these experiments, newly synthesized nucleotides are labeled with a mass value of M+5 because the ribose sugar contains 5 13C carbons from U-13C-glucose. Moreover, this tracing can distinguish the source of serine used for purine production. When produced from glucose-derived serine, glycine is M+2 labeled, and each 1C unit is M+1 labeled. Thus, purines with mass values of M+6 or greater were produced using glucose-derived serine (Fig. 4b)11.
Figure. 4: Electron acceptors are an endogenous limitation for serine synthesis and proliferation in the absence of exogenous serine.
a, Proliferation rates of A549 and PHGDH-expressing MDA-MB-231 cells (pLHCX PHGDH) cultured in serine-replete or serine-free media with or without AKB as indicated. b, Schematic of how U-13C-glucose carbon can contribute label to serine and purines. Newly synthesized purines incorporate ribose containing 5 13C from labeled glucose. If serine that is synthesized from 13C-labeled glucose contributes to purine synthesis, up to 4 additional 13C-labeled carbons can be incorporated, because glycine produced from glucose-derived serine can contribute 2 13C-labeled carbons, and 10-formyl-THF from glucose-derived serine can contribute up to 2 13C labels. Thus, M+5 labeling suggests the purine is newly synthesized using exogenous serine, while species with >5 13C labels suggests the purine is synthesized using glucose-derived serine. c, Total levels and labeling of ATP and GTP from U-13C-glucose in A549 cells grown with or without serine, 20 nM rotenone, and AKB as indicated. Data shown are mean (+/− standard deviation) of 3 biological replicates.
By 48 hours of labeling, cells cultured in the presence of serine had purines that were mostly M+5 labeled (Fig. 4c and Supplementary Fig. 6a–d), suggesting that most of the serine used for purine synthesis in these cells is from the environment. Again, cells grown in the absence of serine had decreased purine pools. Furthermore, because serine-deprived cells must use glucose-derived serine for purine production, their purines had mass values of M+6 or greater. Consistent with NAD+ being limiting for serine synthesis and the production of 1C units for purines, rotenone treatment further decreased purine levels and decreased the levels of purines with mass values of M+6 or greater. For both untreated and rotenone-treated cells grown in serine-free media, addition of AKB increased intracellular purine levels, and increased contribution of glucose-derived serine to purines, resulting in increased labeling of purines with mass values of M+6 or greater. Notably, expression of serine synthesis enzymes was necessary for this rescue, as MDA-MB-231 cells required PHGDH expression to produce purines using synthesized serine (Supplementary Fig. 6b). These data confirm that increasing NAD+ availability can restore purine production using serine synthesized from glucose, and suggest that demand for NAD+ constrains the use of serine synthesis to support purine production and allow maximal cell proliferation.
The metabolic network available to cancer cells is defined by the enzymes and regulatory factors expressed, and thus is influenced by both the cell type which gave rise to the cancer, and the genetic and signaling changes which drive the cancer22,23. In addition, cancer cells in different tissues are exposed to different nutrient environments24. This suggests that different cancers experience different metabolic constraints, and may underlie why cancers show differential sensitivity to drugs targeting metabolic processes predicted to be important in all proliferating cells14,25.
We find that the extent to which cells can utilize the serine synthesis pathway is dictated by NAD+ availability. Mitochondrial respiration allows net regeneration of NAD+ from NADH, and this ability to regenerate NAD+ is an important way that respiration supports cell proliferation12,13,14. Previous work has focused on aspartate production as a precursor for proteins and nucleotides that could be limited by NAD+ regeneration12,13,26. The finding that serine metabolism for purine synthesis can also be limited by NAD+ regeneration demonstrates that aspartate production is not the only important use of NAD+ to support proliferation. Many NAD+-consuming reactions compete for NAD+, and whether production of aspartate, the generation of serine-derived 1C-units, or other processes requiring NAD+ as a cofactor are more limiting will depend on the cancer context. For example, if environmental serine is limited, then NAD+ consumption in serine synthesis may become required and select for adaptations to circumvent other NAD+-utilizing pathways.
The demand for extracellular serine is consistent with studies showing that dietary serine limitation can slow the growth of some, but not all, tumors. Indeed, dietary serine limitation appears to be more affective for slowing the growth of tumors arising in tissues with lower serine content6,9. Low-serine tissue environments appear to select for tumors that genetically upregulate serine synthesis6. While this increases intracellular serine and supports faster tumor growth6, the requirement for serine synthesis in these tumors could render them vulnerable to NAD+ depletion. Further, high rates of serine synthesis in these tumors might make NAD+ less available for other pathways needed to synthesize oxidized biomass, resulting in other metabolic limitations. Similarly, our data suggest that tumors growing in environments where NAD+ availability is more limited may be more susceptible to serine deprivation. In line with this idea, a serine-deficient diet enhances the efficacy of metformin treatment in an allograft cancer model15. Recognizing that forcing increased serine production can synergize with drugs such as metformin that inhibit NAD+ regeneration, not because they limit the carbon available for ATP production15 but rather because they cause a redox imbalance, suggests that other NAD+-depleting drugs could also enhance response to dietary serine limitation. By extension, a better understanding of how differential pathway use by cancer cells creates metabolic liabilities could suggest additional opportunities to target metabolism for therapeutic benefit.
Methods
Cell Culture
All cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (GIBCO) supplemented with 10% heat inactivated fetal bovine serum at 37°C with 5% CO2. Pyruvate or nucleotides were not included in the culture media except when indicated. Concentrations of metabolites added to treatment media are listed in Supplementary Table 1.
Proliferation Rates
Cells were trypsinized, counted and plated in 6-well plates (Denville) in 2 mL DMEM with 10% FBS and incubated overnight. Cells were plated at 20,000 cells per well (A549, 143B, H1299) or 40,000 (MDA-MB-468, MDA-MB-231, T.T, Carney, BT-20, H522, HS578T) cells per well in 6 well plates. The following day, one 6-well plate of each cell line was counted to determine the initial number of cells at the time of treatment. Cells were washed three times with 2 mL PBS and 4 mL of treatment media was added. Treatment media was made with 10% dialyzed FBS. Serine-free media was made by adding a mix of amino acids at DMEM concentrations lacking serine to DMEM without pyruvate or amino acids. The number of cells seeded for each cell line allowed for exponential growth over the course of the assay. After 4 days of treatment, final cell counts were measured using a Cellometer. The following formula was used to calculate proliferation rate:
Western Blots
106 cells were plated in 10 cm plates and incubated overnight to allow cells to adhere. The following day, cells were washed three times with PBS and 10 mL of treatment media containing 10% dialyzed FBS was added. After culturing cells for 24 hours in treatment media, protein lysates were prepared: cells were trypsinized, pelleted, and washed twice with PBS. Cells were then resuspended in ice-cold RIPA buffer with protease inhibitor. Lysed cells were centrifuged at maximum speed at 4°C and the protein lysate supernatant was removed and stored at −80°C. Proteins were separated using SDS-PAGE (12% acrylamide gels) and transferred to a nitrocellulose membrane using a wet transfer method. Membranes were blocked for 60 minutes using 5% bovine serum albumin (BSA) in Tris buffered saline with Tween (TBST). Membranes were incubated in primary antibody overnight at 4°C. The following primary antibodies were used: vinculin (Cell Signaling Technology), PHGDH (Sigma), PSAT (Abnova), and PSPH (Sigma Prestige). Antibodies were diluted in 5% BSA in TBST (vinculin and PSAT, 1:1000; PHGDH, 1:500; PSPH, 1:500). The following day, membranes were washed three times with TBST on a rocker for 10 minutes. Secondary antibodies were applied for 60 minutes. Anti-rabbit (Cell Signaling Technology) secondary antibody was used at a dilution of 1:5000 and anti-mouse (Cell Signaling Technology) secondary antibody was used at a dilution of 1:10000. Membranes were then washed again three times with TBST for 10 minutes on a rocker. Signal was detected using film. Uncropped Western blots are shown in Supplementary Figure 7.
LCMS Analysis
100,000 cells were plated in 6-well plates in 2 mL DMEM with 10% FBS and incubated overnight. The following day, cells were washed three times with PBS and 4 mL of treatment media was added. All treatment media was made with 10% dialyzed FBS. After the indicated time period, polar metabolites were extracted from cells: plates were placed on ice, cells were washed with ice-cold blood bank saline, and 500 μl of ice-cold 80% methanol in water with 250nM 13C/15N labeled amino acid standards (Cambridge Isotope Laboratories, Inc.) was added to each well. Cells were scraped, each sample was vortexed for 10 minutes at 4°C, and then centrifuged at maximum speed for 10 minutes at 4°C. Samples were dried under nitrogen gas and resuspended in 25 μl of a 50/50 acetonitrile/water mixture. Metabolites were measured using a Dionex UltiMate 3000 ultra-high performance liquid chromatography system connected to a Q Exactive benchtop Orbitrap mass spectrometer, equipped with an Ion Max source and a HESI II probe (Thermo Fisher Scientific). Samples were separated by chromatography by injecting 2–10 μl of sample on a SeQuant ZIC-pHILIC Polymeric column (2.1 × 150 mm 5 μM, EMD Millipore). Flow rate was set to 150 μl/min, temperatures were set to 25 °C for column compartment and 4 °C for autosampler sample tray. Mobile Phase A consisted of 20 mM ammonium carbonate, 0.1% ammonium hydroxide. Mobile Phase B was 100% acetonitrile. The mobile phase gradient (%B) was set in the following protocol: 0–20 min.: linear gradient from 80% to 20% B; 20–20.5 min.: linear gradient from 20% to 80% B; 20.5–28 min.: hold at 80% B. Mobile phase was introduced into the ionization source set to the following parameters: sheath gas = 40, auxiliary gas = 15, sweep gas = 1, spray voltage = −3.1kV, capillary temperature = 275 °C, S- lens RF level = 40, probe temperature = 350 °C. Metabolites were monitored in full-scan, polarity-switching, mode. An additional narrow range full-scan (220–700 m/z) in negative mode only was included to enhance nucleotide detection. The resolution was set at 70,000, the AGC target at 1,000,000, and the maximum injection time at 20 msec. Relative quantitation of metabolites was performed with XCalibur QuanBrowser 2.2 (Thermo Fisher Scientific) using a 5 ppm mass tolerance and referencing an in-house retention time library of chemical standards. Metabolite measurements were normalized to the internal 13C/15N labeled amino acid standard and to cell number.
Metabolomic Data Analysis
100,000 cells were plated in 6-well dishes and allowed to adhere overnight. The following day, cells were washed three times with PBS and 4mL of the specified media was added. After 48 hours, polar metabolites were extracted and analyzed using a Dionex UltiMate 3000 ultra-high performance liquid chromatography system connected to a Q Exactive benchtop Orbitrap mass spectrometer, as described in “LCMS Analysis.” Metabolite peak integration was performed as above using XCalibur QuanBrowser 2.2, and peak areas were normalized to an internal standard (13C/15N labeled amino acid standard) and to cell number. Differential and statistical analysis was performed using MetaboAnalystR. The following parameters were used: data normalization: normalization to sample median; data transformation: log normalization; data scaling: autoscaling. Hierarchical clustering in MetaboAnalystR was performed using the following parameters: similarity measure: Euclidean distance; clustering algorithm: Ward’s linkage.
NAD+/NADH Measurements
NAD+/NADH measurements were performed using the NAD/NADH Glo Assay (Promega) with a modified version of manufacturer instructions as reported previously13. Cells were plated as done for proliferation assays and incubated overnight. Cells were treated as indicated for 6 hours prior to preparation of cell extracts. For extraction, cells were washed 3 times in ice cold PBS, extracted in 100 μL ice cold lysis buffer (1% Dodecyltrimethylammonium bromide (DTAB) in 0.2 N NaOH diluted 1:1 with PBS). Each sample was flash-frozen in liquid nitrogen and immediately stored at −80°C. To measure NADH, 20 μL of sample was moved to PCR tubes and incubated at 75°C for 30 min where basic conditions selectively degrade NAD+. To measure NAD+, 20 μL of the samples was moved to PCR tubes containing 20 μL lysis buffer and 20 μL 0.4 N HCl and incubated at 60°C for 15 min, where acidic conditions selectively degrade NADH. Samples were then allowed to equilibrate to room temperature and quenched by neutralizing with 20 μL 0.25 M Tris in 0.2 N HCl (for NADH) or 20 μL 0.5 M Tris base (for NAD+). Manufacturer instructions were followed thereafter to measure NAD+/NADH.
Data Availability
The raw data that support the findings of this study are available from the corresponding author upon request. Raw data for Figure 1a, Supplementary Figure 1c, and Supplementary Figure 2a (uncropped Western blots) are shown in Supplementary Figure 7.
Reporting Summary
Further information about research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary Material
Acknowledgements
We thank members of the Vander Heiden Lab for helpful discussions. We thank H. Furkan Alkan for providing the A549 shAGC1 cells. F. F. D. acknowledges support from the NIH (F31CA236036). M. G. V. H. acknowledges support from a Faculty Scholar grant from the Howard Hughes Medical Institute, SU2C, the Lustgarten Foundation, the MIT Center for Precision Cancer Medicine, the Ludwig Center at MIT and the NIH (R01CA201276, R01CA168653 and P30CA14051).
Footnotes
Competing Interests
M. G. V. H. discloses that he is a consultant and SAB member for Agios Pharmaceuticals, Aeglea Biotherapeutics, and Auron Therapeutics.
References
- 1.Hosios AM et al. Amino Acids Rather than Glucose Account for the Majority of Cell Mass in Proliferating Mammalian Cells. Dev. Cell 36, 540–549 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang M & Vousden KH Serine and one-carbon metabolism in cancer. Nat. Rev. Cancer 16, 650–662 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Ducker GS & Rabinowitz JD One-Carbon Metabolism in Health and Disease. Cell Metab 25, 27–42 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Newman AC & Maddocks ODK One-carbon metabolism in cancer. Br. J. Cancer 116, 1499–1504 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Possemato R et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 476, 346–350 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sullivan MR et al. Increased Serine Synthesis Provides an Advantage for Tumors Arising in Tissues Where Serine Levels Are Limiting. Cell Metab (2019). doi: 10.1016/j.cmet.2019.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mattaini KR, Sullivan MR & Heiden MGV The importance of serine metabolism in cancer. J Cell Biol 214, 249–257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeNicola GM et al. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat. Genet 47, 1475–1481 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maddocks ODK et al. Modulating the therapeutic response of tumours to dietary serine and glycine starvation. Nature 544, 372–376 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Labuschagne CF, van den Broek NJF, Mackay GM, Vousden KH & Maddocks ODK Serine, but Not Glycine, Supports One-Carbon Metabolism and Proliferation of Cancer Cells. Cell Rep 7, 1248–1258 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Pacold ME et al. A PHGDH inhibitor reveals coordination of serine synthesis and 1-carbon unit fate. Nat. Chem. Biol 12, 452–458 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Birsoy K et al. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell 162, 540–551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sullivan LB et al. Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell 162, 552–563 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gui DY et al. Environment Dictates Dependence on Mitochondrial Complex I for NAD+ and Aspartate Production and Determines Cancer Cell Sensitivity to Metformin. Cell Metab 24, 716–727 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gravel S-P et al. Serine Deprivation Enhances Antineoplastic Activity of Biguanides. Cancer Res 74, 7521–7533 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Murphy JP et al. The NAD+ Salvage Pathway Supports PHGDH-Driven Serine Biosynthesis. Cell Rep 24, 2381–2391.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bao XR et al. Mitochondrial dysfunction remodels one-carbon metabolism in human cells. eLife 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hosios AM & Heiden MGV The redox requirements of proliferating mammalian cells. J. Biol. Chem jbc.TM117.000239 (2018). doi: 10.1074/jbc.TM117.000239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis CA et al. Tracing Compartmentalized NADPH Metabolism in the Cytosol and Mitochondria of Mammalian Cells. Mol. Cell 55, 253–263 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ducker GS et al. Reversal of Cytosolic One-Carbon Flux Compensates for Loss of the Mitochondrial Folate Pathway. Cell Metab 23, 1140–1153 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alkan HF et al. Cytosolic Aspartate Availability Determines Cell Survival When Glutamine Is Limiting. Cell Metab 28, 706–720.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayers JR et al. Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science 353, 1161–1165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vander Heiden MG & DeBerardinis RJ Understanding the Intersections between Metabolism and Cancer Biology. Cell 168, 657–669 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davidson SM et al. Environment Impacts the Metabolic Dependencies of Ras-Driven Non-Small Cell Lung Cancer. Cell Metab 23, 517–528 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muir A et al. Environmental cystine drives glutamine anaplerosis and sensitizes cancer cells to glutaminase inhibition. eLife 6, e27713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sullivan LB et al. Aspartate is an endogenous metabolic limitation for tumour growth. Nat. Cell Biol 20, 782–788 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data that support the findings of this study are available from the corresponding author upon request. Raw data for Figure 1a, Supplementary Figure 1c, and Supplementary Figure 2a (uncropped Western blots) are shown in Supplementary Figure 7.