Abstract
A new 8,8'-binaphthopyranone (mycopyranone, 1) was isolated from a solid fermentation of Phialemoniopsis sp. (fungal strain MSX61662), and the structure was elucidated via analysis of the NMR and HRESIMS data. The axial chirality of 1 was determined to be M by ECD. The central chirality at C-4/C-4' was assigned through a modified Mosher’s method, while the absolute configuration at C-3/C-3' was deduced based on analysis of the 3JH-3-H-4 values and NOESY correlations. Compound 1 was evaluated for its antimicrobial properties against Staphylococcus aureus SA1199 and a clinically relevant methicillin-resistant S. aureus strain (MRSA USA300 LAC strain AH1263). Compound 1 inhibited the growth of both strains in a concentration dependent manner with IC50 values in the low μM range. Molecular docking indicated that compound 1 binds to the FtsZ (tubulin-like) protein in the same pocket as viriditoxin (2), suggesting that 1 targets bacterial cell division.
Graphical Abstract
INTRODUCTION
Nosocomial infections caused by methicillin-resistant Staphylococcus aureus (MRSA) are a serious health problem.1–3 In the United States alone, over 80,000 severe MRSA infections were documented in 2013, 11,000 of which were fatal.4 Unfortunately, despite the imminent problem MRSA and similar drug resistant infections pose, most pharmaceutical companies do not focus on the discovery and development of new antimicrobial agents.5, 6 In this context, the World Health Organization has started a cooperative program with Academia to search for new molecules and strategies to combat antibiotic resistance.7, 8
One promising strategy to combat MRSA infections is targeting cell division via the inhibition of the proto ring protein FtsZ,2, 9 a vital and highly conserved protein involved in bacterial division and formation of new cells; it is a homolog of tubulin in eukaryotic cells.10, 11 However, despite the importance of this protein as a molecular target to combat infections against MRSA, just a few examples of molecules have been described as potential inhibitors, including natural products12–16 and synthetic probes.9, 17 Among natural products, viriditoxin (2), a 6,6'-binaphthopyranone, inhibited the polymerization of FtsZ, resulting in the inhibition of bacterial cell division, and this compound has been proposed as a promising lead for the development of anti-MRSA drugs.16, 18
As part of ongoing studies to identify structurally diverse and bioactive metabolites from fungal cultures, a new 8,8'-binaphthopyranone derivative (mycopyranone, 1) was isolated from Phialemoniopsis sp. (strain MSX61662), which was identified phylogenetically and morphologically using methods that were detailed previously.19, 20 The structure of 1 was established using a set of spectroscopic (1D and 2D NMR), spectrometric (HRESIMS), and chiroptic (ECD and OR) methods. The antimicrobial activity of 1 was evaluated against Staphylococcus aureus SA119921 and a clinically relevant methicillin-resistant S. aureus strain (MRSA USA300 LAC strain AH1263).22 Growth inhibition by 1 was noted in both strains in a concentration dependent manner with IC50 values in the low μM range. We suggest that 1 targets bacterial cell division, as molecular docking studies predicted that viriditoxin (2) and mycopyranone (1) bind to the protein FtsZ in the same pocket.
Compound 123 was isolated as an optically active (]) yellow amorphous powder, and its molecular formula was established as C40H46O12 via HRESIMS.24 Analysis of the 1H, 13C and HSQC NMR data indicated the presence of a chelated hydroxy group (δH 13.79), one phenolic proton (δH 9.67), two aromatic protons (δH 7.20, 6.79), two oxymethines (δH 4.48, 4.73), one aliphatic methine (δH 1.42), three methylene (δH 1.98, 1.29/1.65, 1.22/1.44), one methoxy (δH 3.86) and two methyl groups (δH 0.91, 0.94), as well as nine fully substituted carbons, including three oxygenated and a carbonyl (lactone) moiety (Table 1 and Figures S3, S4, and S6).
Table 1.
1H and 13C NMR data for compound 1 recorded in CDCl3 at 700 MHz
| Position | δC | type | δH, multiplicity (J = Hz) |
|---|---|---|---|
| 1 | 171.0 | C | |
| 3 | 83.1 | CH | 4.48, brt (7.0) |
| 4 | 66.9 | CH | 4.73, brs |
| 4a | 135.0 | C | |
| 5 | 117.3 | CH | 7.20, s |
| 5a | 140.1 | C | |
| 6 | 99.1 | CH | 6.79, s |
| 7 | 161.7 | C | |
| 8 | 109.4 | C | |
| 9 | 155.6 | C | |
| 9a | 109.2 | C | |
| 10 | 163.1 | C | |
| 10a | 97.8 | C | |
| 11 | 28.0 | CH2 | 1.98, m |
| 12a | 32.0 | CH2 | 1.29, m |
| 12b | 1.65, m | ||
| 13 | 34.5 | CH | 1.42, m |
| 14 | 29.5 | CH2 | 1.22, m |
| 1.44, m | |||
| 15 | 11.5 | CH3 | 0.91, t (7.3) |
| 16 | 19.2 | CH3 | 0.94, d (6.5) |
| 17 | 56.2 | CH3 | 3.86, s |
| 4-OH | 1.86, brs | ||
| 9-OH | 9.67, s | ||
| 10-OH | 13.79, s |
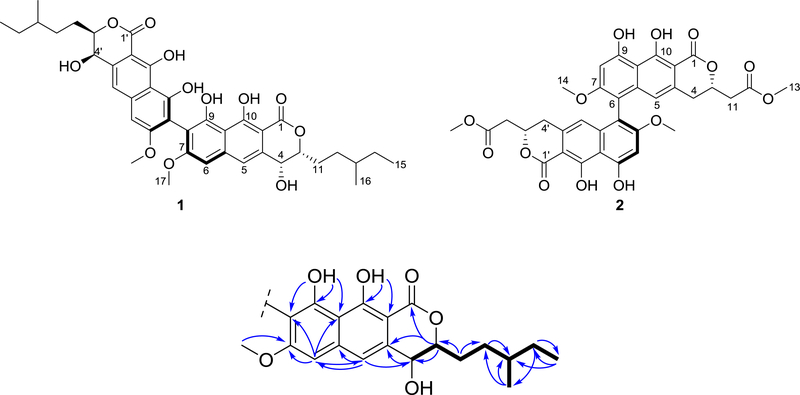
These data accounted for a molecular formula of C20H23O6, indicating that 1 was a symmetric dimer. In general, the 1H and 13C NMR data of 1 resembled those reported for viriditoxin (2),25 pigmentosin,26 talaroderxines A and B,27 vioxanthin,28 and cladiosporinone,29 with minor differences attributed to the aliphatic chain at C-3/C-3’. Analysis of the 2D NMR data, in particular COSY and HMBC experiments (Figures S5 and S7), allowed the identification of key fragments of the molecule (Figure 1).
Figure 1.
Structures of mycopyranone (1) and viriditoxin (2) and key COSY (bold bonds) and HMBC correlations (arrows) for compound 1.
For example, the COSY data permitted the assignment of the spin system shown in black (Figure 1), which was confirmed by the HMBC correlations observed between H3-15 to C-13 and C-14, H3-16 to C-12, C-13 and C-14, H2-14 to C-16, C-15 and C-13, H-13 to C-12, H2-12 to C-14, C-13 and C-11, and H2-11 to C-13, among others. The 9,10-dihydroxy-7-methoxy-naphthopyranone fragment was confirmed via HMBC correlations between H-3 to C-4a, C-4 and C-1, H-4 to C-4a, H-5 to C-6, C-5a and C-4, H-6 to, C-5, C-7, C-8 and C-9a, 9-OH to C-9a, C-9 and C-8, 10-OH to C-10a, and C-10, and H3-17 to C-7 (Figure 1). The connection between the 3-methylpentenyl moiety to the naphthopyranone fragment was established based on the HMBC correlations observed among H2-11 and C-3, and H-3 to C-11 and C-12 (Figure 1). Finally, the 8, 8' linkage between the homodimers was established based on the HMBC mutual correlation amongst H-6/H-6’ and C-8 and C-8'.
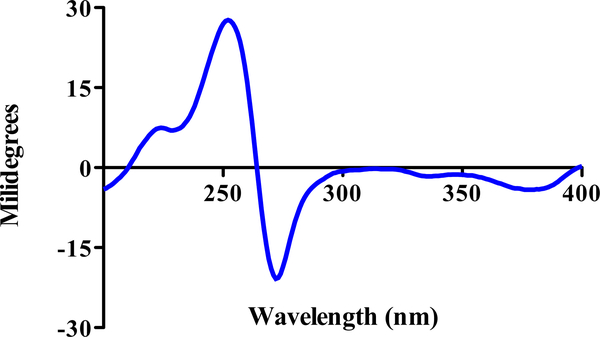
The axial chirality of the molecule was determined by ECD (Figure 2). Briefly, the spectrum of 1 in MeOH showed both positive and negative Cotton effects at 254 and 274 nm, respectively, similar to those of viriditoxin (2),25 talaroderxine B,27 and M-vioxanthin.28 The negative Cotton effect at 274 nm was attributed to the transitions of the naphthalene chromophores, indicating that the 8,8' axis in 1 was twisted in a counter-clockwise manner, characteristic for M axial chirality.25–27
Figure 2.
ECD spectrum of 1 recorded in MeOH (13 μM)
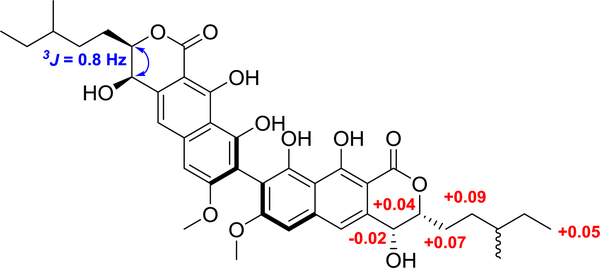
The absolute configuration at C-4/C-4' was determined as R based on the results of the modified Mosher´s ester method (Figure 3).30 The absolute configuration at C-3/C-3' was also determined as R by analysis of the NOESY experiment and analysis of the 3JH-3-H-4, value (0.8 Hz) (Figures 3 and S8). Unfortunately, the absolute configuration at C-13 was not determined due to the high flexibility of the chain. Thus, the structure of compound 1 was elucidated as depicted, and assigned the trivial name mycopyranone.
Figure 3.
ΔδH values [Δδ (in ppm) = δS − δR] (red) from the Mosher’s esters experiment and key NOESY correlation (blue arrow) and coupling constant, all observed for compound 1.
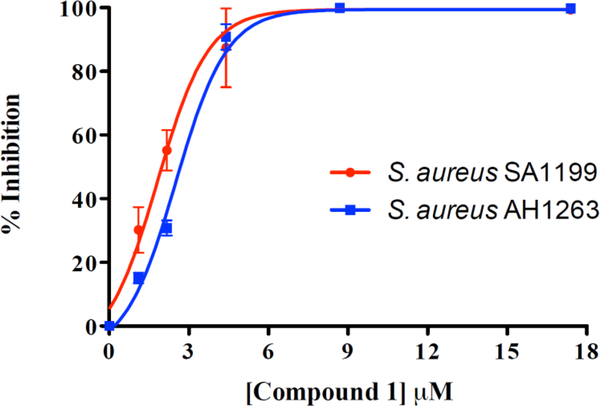
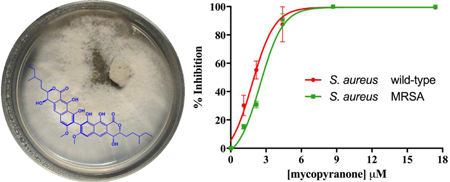
The activity of mycopyranone (1) was evaluated against Staphylococcus aureus (strain SA1199)21 and a clinically relevant methicillin-resistant strain (MRSA USA300 LAC, AH1263).22 Compound 1 displayed promising antibacterial activity against both strains. Due to structural similarity to 2, compound 1 likely functions as an FtsZ inhibitor.18 Since FtsZ is highly conserved across many Gram-positive organisms,18 the ability of pathogens to develop resistance to 6,6'- or 8,8' binaphthopyranone derivatives (including both compounds 1 and 2) may be limited. The minimum inhibitory concentration (MIC) of compound 1 was ≤ 8.7 μM against both strains, and IC50 values were in the lower μM range (2.0 μM and 2.7 μM against SA1199 and AH1263, respectively; Figure 4).
Figure 4.
Concentration-response curves of compound 1 against S. aureus SA1199 (red circles) and methicillin-resistant S. aureus AH1263 (blue squares). The minimum inhibitory concentration (MIC) of compound 1 was ≤ 8.7 μM against both S. aureus strains. The MIC of the positive control, berberine, was 223 μM against both S. aureus strains.
Berberine, which was previously shown to target FtsZ in Escherichia coli,13 was used as a positive control. Berberine’s antimicrobial activity (IC50 = 69.5 μM, MIC 223 μM; Supporting Information, Figure S10) was consistent with previous reports.13, 31, 32 In addition, the cytotoxic potential of compound 1 was evaluated against a panel of cancer cell lines, as described recently.33 It did not show potent activity (IC50 value > 50 μg/mL), suggesting that 1 lacks toxicity against mammalian cells.
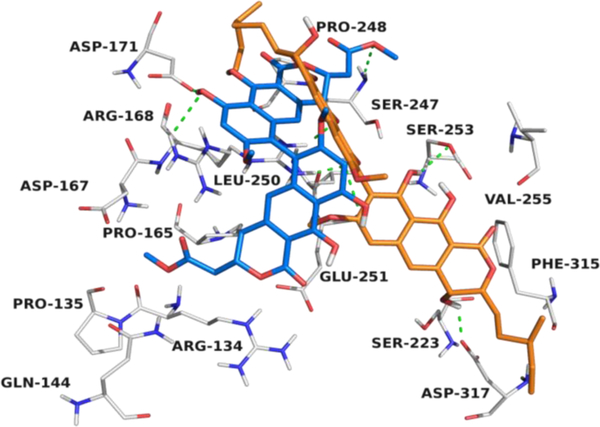
Based upon these results, and data from previous reports, we hypothesized that compound 1 binds to protein FtsZ, similar to viriditoxin (2), thereby inhibiting bacterial cell division.18 To gain additional information about the putative binding site of mycopyranone (1) and viriditoxin (2) in FtsZ, molecular docking studies were carried out. Briefly, the results indicated that 1 and 2 bind to FtsZ (−5.09. and −6.60 kcal/mol, respectively) in a hydrophobic pocket conformed by the amino acids Arg-134, Pro-135, Gln-144, Pro-165, Asp-167, Arg-168, Asp-171, Ser-223, Ser-247, Pro-248, Leu-250, Glu-251, Ser-253, Val-255, Phe-315 and Asp-317 (Figures 5 and S11).
Figure 5.
Structural model of the predicted binding pocket for viriditoxin (2) (blue) and mycopyranone 1 (orange) with FtsZ (PDB 4DXD).
The forces that govern the interactions are mainly hydrogen bonds and van der Waals interactions (Figure 5). This pocket differs from those reported previously for the synthetic compounds PC190723,9 TXA70934, 35 and TXA6101,17 revealing a potential new druggable site in the FtsZ protein of S. aureus.
Supplementary Material
HIGHLIGHTS.
A new 8,8'-binaphthopyranone (mycopyranone, 1) was discovered and elucidated.
Absolute configuration was elucidated via NOESY, Mosher’s esters, and ECD data.
Active in vitro vs a clinically-relevant methicillin-resistant S. aureus strain.
Molecular docking studies suggests binding to the FtsZ (tubulin-like) protein.
Acknowledgements
This research was supported in part by P01 CA125066 from the National Cancer Institute and T32 AT008938 (fellowship to LKC) from the National Center for Complementary and Integrative Health, both part of the National Institutes of Health, Bethesda, MD, USA. NMR studies were performed in part at the Joint School of Nanoscience and Nanoengineering (JSNN), a member of the Southeastern Nanotechnology Infrastructure Corridor (SENIC) and National Nanotechnology Coordinated Infrastructure (NNCI), which is supported by the National Science Foundation (Grant ECCS-1542174). We thank Dr. Joanna Burdette from the University of Illinois at Chicago for cytotoxicity testing.
Footnotes
Supplementary data
Supplementary data (Phylogenetic analysis of Phialemoniopsis sp., 1D and 2D NMR data of compound 1, HRESIMS of compound 1, concentration response curve of berberine, and structural model of compound 1-FtsZ) associated with this article can be found, in the online version.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Boucher HW; Talbot GH; Bradley JS; Edwards JE; Gilbert D; Rice LB; Scheld M; Spellberg B; Bartlett J Clin Infect Dis 2009, 48, 1. [DOI] [PubMed] [Google Scholar]
- 2.Liang S; He Y; Xia Y; Wang H; Wang L; Gao R; Zhang M Int J Infect Dis 2015, 30, 1. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization, http://www.who.int/mediacentre/factsheets/fs194/en/ 2016, Accesed September 2017.
- 4.Centers for Disease Control and Prevention: Antibiotic resistance threats in the United States, 2013, http://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf 2013, Accessed 04 September 2017.
- 5.Boucher HW; Talbot GH; Benjamin DK Jr.; Bradley J; Guidos RJ; Jones RN; Murray BE; Bonomo RA; Gilbert D Clin Infect Dis 2013, 56, 1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole ST Philos Trans R Soc Lond B Biol Sci 2014, 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chellat MF; Raguž L; Riedl R Angew Chem, Int Ed 2016, 55, 6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.World Health Organization, http://www.who.int/dg/speeches/2011/WHD_20110407/en/ 2011, Accesed September 2017.
- 9.Haydon DJ; Stokes NR; Ure R; Galbraith G; Bennett JM; Brown DR; Baker PJ; Barynin VV; Rice DW; Sedelnikova SE; Heal JR; Sheridan JM; Aiwale ST; Chauhan PK; Srivastava A; Taneja A; Collins I; Errington J; Czaplewski LG Science 2008, 321, 1673. [DOI] [PubMed] [Google Scholar]
- 10.Erickson HP; Anderson DE; Osawa M Microbiol Mol Biol Rev 2010, 74, 504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Margolin W Nat Rev Mol Cell Biol 2005, 6, 862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaiswal R; Beuria TK; Mohan R; Mahajan SK; Panda D Biochemistry 2007, 46, 4211. [DOI] [PubMed] [Google Scholar]
- 13.Domadia PN; Bhunia A; Sivaraman J; Swarup S; Dasgupta D Biochemistry 2008, 47, 3225. [DOI] [PubMed] [Google Scholar]
- 14.Urgaonkar S; La Pierre HS; Meir I; Lund H; RayChaudhuri D; Shaw JT Org Lett 2005, 7, 5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanoh K; Adachi K; Matsuda S; Shizuri Y; Yasumoto K; Kusumi T; Okumura K; Kirikae T J Antibiot 2008, 61, 192. [DOI] [PubMed] [Google Scholar]
- 16.Vollmer W Appl Microbiol Biotechnol 2006, 73, 37. [DOI] [PubMed] [Google Scholar]
- 17.Fujita J; Maeda Y; Mizohata E; Inoue T; Kaul M; Parhi AK; LaVoie EJ; Pilch DS; Matsumura H ACS Chem Biol 2017, 12, 1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J; Galgoci A; Kodali S; Herath KB; Jayasuriya H; Dorso K; Vicente F; González A; Cully D; Bramhill D; Singh S J Biol Chem 2003, 278, 44424. [DOI] [PubMed] [Google Scholar]
- 19.Details about the strain isolation and identification are given in the Supporting Information, and the sequence data for MSX61662 were deposited in GenBank (accession nos. MH700254, MH700255).
- 20.Raja HA; Miller AN; Pearce CJ; Oberlies NH J Nat Prod 2017, 80, 756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaatz GW; Seo SM Antimicrob Agents Chemother 1995, 39, 2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Junio HA; Todd DA; Ettefagh KA; Ehrmann BM; Kavanaugh JS; Horswill AR; Cech NB J Chromatogr B 2013, 930, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mycopyranone (1): yellow amorphous powder: 1H and 13C-NMR see Table 1, and Figures S3 and S4 (Supporting Information); HRESIMS, [M+H]+= 719.3052 (calcd. for C40H47O12, 719.3062)(Figure S9).
- 24.For details about the extraction and isolation of compound 1 see the Supporting Information file.
- 25.Suzuki K; Nozawa K; Nakajima S; Kawai K.-i. Chem Pharm Bull 1990, 38, 3180. [DOI] [PubMed] [Google Scholar]
- 26.Elix JA; Wardlaw JH Aust J Chem 2004, 57, 681. [Google Scholar]
- 27.Suzuki K; Nozawa K; Nakajima S; Udagawa S.-i.; Kawai K.-i. Chem Pharm Bull 1992, 40, 1116. [DOI] [PubMed] [Google Scholar]
- 28.Bode SE; Drochner D; Müller M Angew Chem, Int Ed 2007, 46, 5916. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y; Kurtan T; Yun Wang C; Han Lin W; Orfali R; Muller WEG; Daletos G; Proksch P J Antibiot 2016, 69, 702. [DOI] [PubMed] [Google Scholar]
- 30.Hoye TR; Jeffrey CS; Shao F Nat. Protocols 2007, 2, 2451. [DOI] [PubMed] [Google Scholar]
- 31.Yu HH; Kim KJ; Cha JD; Kim HK; Lee YE; Choi NY; You YO J Med Food 2005, 8, 454. [DOI] [PubMed] [Google Scholar]
- 32.Kellogg JJ; Todd DA; Egan JM; Raja HA; Oberlies NH; Kvalheim OM; Cech NB J Nat Prod 2016, 79, 376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rivera-Chávez J; Raja HA; Graf TN; Burdette JE; Pearce CJ; Oberlies NH J Nat Prod 2017, 80, 1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaul M; Mark L; Parhi AK; LaVoie EJ; Pilch DS Antimicrob Agents Chemother 2016, 60, 4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaul M; Mark L; Zhang Y; Parhi AK; Lyu YL; Pawlak J; Saravolatz S; Saravolatz LD; Weinstein MP; LaVoie EJ; Pilch DS Antimicrob Agents Chemother 2015, 59, 4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.