Abstract
Hematopoiesis is a complex process with a variety of different signaling pathways influencing every step of blood cell formation from the earliest precursors to final differentiated blood cell types. Formation of blood cells is crucial for survival. Blood cells carry oxygen, promote organ development and protect organs in different pathological conditions. Hematopoietic Stem and Progenitor Cells (HSPCs) are responsible for generating all adult differentiated blood cells. Defects in HSPCs or their downstream lineages can lead to anemia and other hematological disorders including leukemia. The zebrafish has recently emerged as a powerful vertebrate model system to study hematopoiesis. The developmental processes and molecular mechanisms involved in zebrafish hematopoiesis are conserved with higher vertebrates, and the genetic and experimental accessibility of the fish and the optical transparency of its embryos and larvae make it ideal for in vivo analysis of hematopoietic development. Defects in zebrafish hematopoiesis reliably phenocopy human blood disorders, making it a highly attractive model system to screen small molecules to design therapeutic strategies. In this review, we summarize the key developmental processes and molecular mechanisms of zebrafish hematopoiesis. We also discuss recent findings highlighting the strengths of zebrafish as a model system for drug discovery against hematopoietic disorders.
INTRODUCTION
Zebrafish offer several key advantages as a model system for studying hematopoiesis. These include external fertilization, optical transparency, genome editing, and easy high-resolution optical imaging in live animals. These features have led to many novel insights into hematopoietic development and differentiation. Large scale forward-genetic screens in zebrafish have identified several key genes essential for proper hematopoiesis. These same screens have also yielded mutants phenocopying human hematological disorders that have contributed significantly to our understanding of the molecular mechanisms behind these diseases.
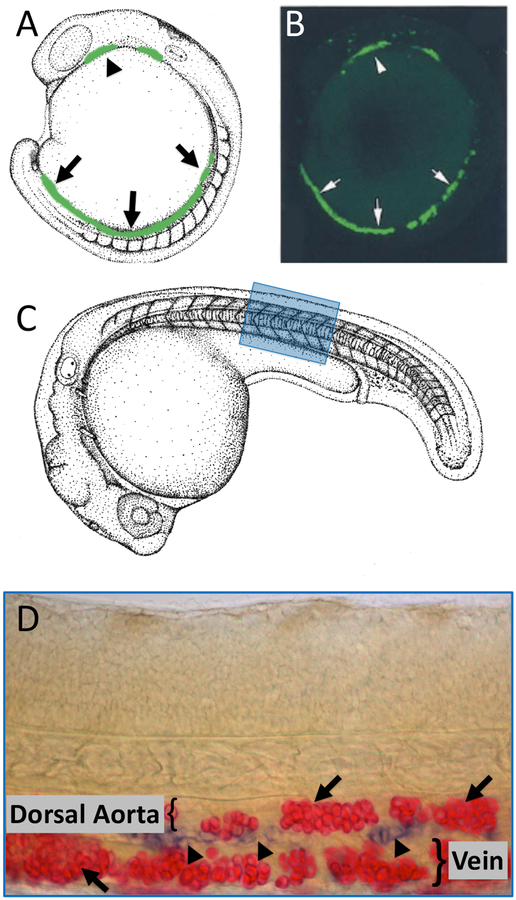
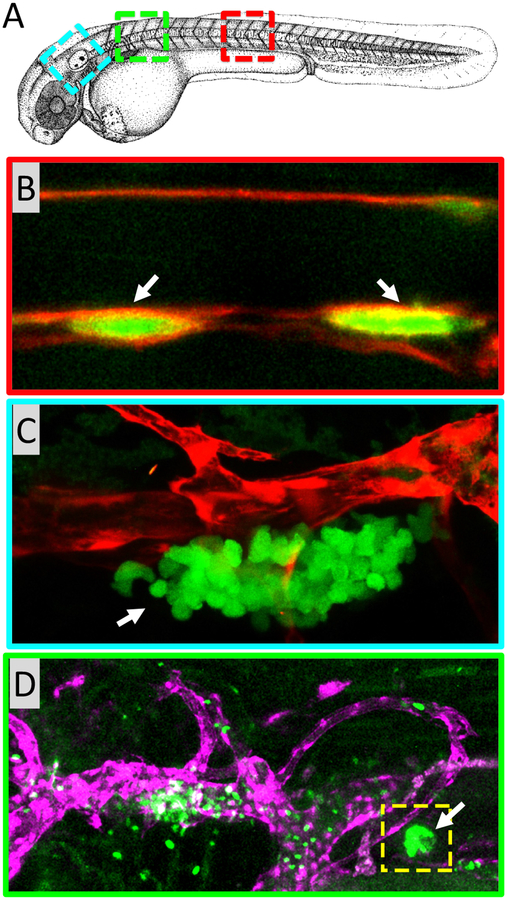
Hematopoiesis in zebrafish takes place primarily in two major waves (1). Primitive hematopoiesis takes place during early embryonic development and is primarily responsible for generating primitive erythroid and myeloid cell populations. Later during development definitive hematopoiesis generates hematopoietic stem and progenitor cells (HSPCs), which are responsible for generating all adult blood cells (Figure 1). The anatomical sites of hematopoiesis are different in zebrafish compared to mammals, although the molecular mechanisms behind hematopoiesis are highly conserved (1, 2) (Figures 2 and 3).
Figure 1:
Primitive hematopoiesis in zebrafish is responsible for generating the first populations of erythroid and myeloid cells. (A) Camera lucida drawing with superimposed green coloring depicting expression of an ETS transcription factor in the anterior (arrowhead) and posterior (arrows) lateral plate mesoderm. Primitive blood lineages are specified in anterior and posterior lateral plate mesoderm during early somitogenenesis. (B) Confocal image of a 16 hour-old Tg(fli1a:egfp)y1 embryo showing transgenic expression of EGFP in the anterior (arrowhead) and posterior (arrows) lateral plate mesoderm. (C) Camera lucida drawing of a 31 hpf zebrafish embryo with a blue box noting the approximate region of the trunk shown in panel D. (D) Double in situ hybridization staining for hbae1.1 primitive erythrocytes (red, arrows) and cmyb developing HSPCs (blue, arrowheads) in the ventral floor of the dorsal aorta of 32 hpf embryo.
Figure 2:
Definitive hematopoiesis in zebrafish is responsible for generating all types of blood cells. (A) Camera lucida drawing of a 48 hpf zebrafish embryo with red, blue, and green colored boxes noting the approximate regions of the embryo shown in panels B, C, and D, respectively. (B) HSPCs develop in the ventral wall of the dorsal aorta. Confocal image of the mid trunk of a 32 hpf Tg(runx1+23:EGFP); Tg(kdrl:mCherry-caax) double-transgenic embryo showing EGFP-positive developing HSPCs (green; arrows) in the ventral floor of the mCherry-positive dorsal aorta (red). (C) The developing zebrafish thymus. Lateral view of a 5 dpf Tg(lck:GFP); Tg(kdrl:mCherry) double-transgenic embryo, just ventral to the otolith, showing GFP-positive thymocytes (green; arrow) developing adjacent to an mCherry-positive blood vessel (red). (D) The zebrafish head kidney is a major organ of definitive hematopoiesis equivalent to the bone marrow of mammals. Confocal image of the from the anterior trunk of a 5 dpf Tg(runx1+23:EGFP); Tg(lyve1:dsRed) double-transgenic embryo, showing GFP-positive hematopoietic cells (green; yellow box, white arrow) developing adjacent to an mCherry-positive vein (purple).
Figure 3:
Schematic diagram illustrating the different sites of zebrafish hematopoiesis used throughout development. Stage of development is indicated in hours post fertilization (hpf) or days post fertilization (dpf). RBI, rostral blood islands; PLM, posterior lateral-plate mesoderm; ICM, intermediate cell mass; DA, dorsal aorta; CHT, caudal hematopoietic tissue.
In recent years, use of genome editing technologies such as TALENs and CRISPR/Cas9 systems have contributed significantly to the analysis of additional genes involved in this complex process. It is also relatively easy to make zebrafish transgenic lines labeling different types of blood cells with GFP or similar fluorescent proteins. The optical transparency and rapid external development of fish embryos and larvae permits visualization of hematopoiesis in living animals. Thanks in large part to these unique advantages, the zebrafish has contributed significantly to our understanding of how HSPCs are specified during definitive hematopoiesis, and how they enter circulation and migrate to their future destinations and settle in their niche (3). The availability of several transgenic lines labeling different types of immature and differentiated blood cells has allowed unique blood cell populations to be studied by enriching them using florescence activated cell sorting (FACS). FACS enrichment has facilitated genome wide gene expression analysis and transplantation studies in zebrafish, among other things.
In addition to genetic and molecular studies, zebrafish are also ideal for large-scale chemical screens to identify small molecules that influence different aspects of hematopoiesis. Zebrafish embryos develop externally and it is easy to obtain hundreds or thousands of fertilized eggs from wild-type or transgenic lines. Zebrafish embryos can be soaked in different concentrations of small molecules and screened for hematological phenotypes at a later time point. It is possible to easily screen though hundreds or even thousands of compounds in a few days. Whole embryo drug screens also have the added advantages of identifying toxicities and off-target effects that might not be evident in cell-based assays. These types of screens have identified compounds which affect hematopoietic cell numbers in zebrafish and that have been subsequently validated using mouse or cultured cell models (4, 5).
In this review, we discuss important developmental steps and signaling molecules involved in zebrafish hematopoiesis. We also summarize the key tools and technologies available for studying blood cell formation in the fish.
PRIMITIVE HEMATOPOIESIS
Primitive hematopoiesis, the initial wave of blood cell formation, produces erythrocytes that facilitate tissue oxygenation during rapid embryonic growth (6) and also macrophages that phagocytose pathogens and apoptotic cells arising as a natural byproduct of development (7). Primitive macrophages may also influence the morphology of the developing circulatory system in some contexts, shaping the developing vasculature by mediating blood vessel regression (8) or acting as endothelial cell chaperones to facilitate tip cell fusion (9). Microglia differentiating from primitive macrophages (10–12) are essential regulators of central nervous system neural development and function (reviewed in (13)). Consequently, although non-pluripotent, transient cell types arise from the initial hematopoietic wave, primitive hematopoietic defects may have far-reaching developmental consequences.
Common origin of endothelial and primitive hematopoietic cells
Based on their intimate association during early development, it was long ago hypothesized that angioblasts (endothelial cell precursors) and primitive hematopoietic cells arise during embryogenesis from a common ‘hemangioblast’ progenitor (2, 14, 15). In zebrafish, the existence of at least a transient bipotential progenitor cell during very early development is suggested by the identification of cells with common hematopoietic and endothelial gene expression (16), and shared regulatory factors that are required for the formation of both lineages, such as the PAS-domain-containing bHLH transcription factor npas4l (the mutated gene in cloche embryos) (17–19) lysocardiolipin acyltransferase lycat (20), and angiogenic factor with G-patch and FHA domains 1 (aggf1) (21). More conclusive fate-mapping experiments show that cells that give rise to both blood and endothelial cells are present within the ventral mesoderm of shield (early gastrula)-stage zebrafish embryos (22, 23). However, these same fate-mapping studies indicate that most zebrafish endothelial and primitive hematopoietic cells likely arise directly from ventral mesoderm, rather than from a transient common hemangioblast progenitor (22, 23). The Retinoic Acid (24, 25), BMP (26), JAK/STAT (27), and VEGF (28) signaling pathways have all been implicated in regulating zebrafish hemangioblast identity.
Primitive hematopoietic cell specification
Zebrafish primitive hematopoiesis occurs primarily in the so-called intermediate cell mass (ICM) blood islands, which form at the trunk midline from bilateral stripes of posterior lateral-plate mesoderm (PLM) (29). The ICM is situated below the notochord, in between the somites and above the yolk sac. It can be thought of as at least conceptually analogous to the extra-embryonic yolk sac blood islands of mammals and birds, and is responsible for generating most embryonic hematopoietic cells. Zebrafish primitive hematopoietic cell specification is subject to precise molecular regulation by a hierarchical cascade of transcription factors that are conserved throughout the vertebrates with clear mammalian orthologues. The earliest molecular marker of primitive hematopoiesis, the bHLH transcription factor stem cell leukemia (scl/tal1) is expressed as early as 10.5 hpf (2-somites) in the PLM (30, 31). Domains of gene expression become more refined at 11 hpf (3-somites), as mesoderm cell fate is progressively restricted to endothelial and hematopoietic lineages. At this stage, cells of the PLM demonstrate overlapping expression patterns of scl, other hematopoietic transcription factors such as LIM domain only 2 (lmo2), and vasculogenic genes including GATA binding protein 2a (gata2a) and the ETS family members fli1a, fli1b, and ets-related protein (etsrp/etv2) (24, 32, 33) (Figure 1A, B and Figure 3). Scl and its binding partner Lmo2 act downstream of the bHLH-PAS transcription factor cloche, which is required for both hematopoietic and endothelial development (17). Both are necessary for the formation of primitive hematopoietic cells (34–38) and sufficient to generate hemangioblasts from non-axial mesoderm (31, 39, 40). Etsrp also acts downstream of cloche in zebrafish, and upstream of scl and fli1a (41). Its depletion leads to reduced scl, fli1a, and vascular endothelial growth factor receptor kdrl angioblast gene expression and corresponding defects in vascular endothelial cell specification (17, 41–45). Conversely, overexpressing the mammalian etsrp homologue (Etv2) in zebrafish leads to ectopic scl and kdrl angioblast gene expression (46). Although morpholino-based knockdown studies indicate that fli1a and fli1b act in parallel to cloche to help regulate endothelial-specific cell fate (45, 47), fli1a and fli1b double homozygous zebrafish mutants do not exhibit angiogenic defects (33), indicating possible compensatory action by other Ets-related factors. Indeed, the angiogenic defects of etsrp zebrafish mutants recover as development proceeds, but only in the presence of intact Fli1b (33). gata2a-mutant zebrafish embryos exhibit reduced kdrl expression, and trunk circulatory defects resulting from impaired dorsal aorta morphogenesis (48, 49).
By 12 hpf, (5-somites) cell fate is irreversibly determined as ICM precursors adopt one of three fates: (i) erythroid cell fate, characterized by gata1a expression (29, 50), (ii) myeloid cell fate, characterized by pu.1 (spi1b) expression (7, 51–55), or (iii) angioblast (presumptive vascular) fate, which is characterized by kdrl expression (19, 30, 56). By 19 hpf (20-somites), overlapping expression of the granulocyte marker myeloid peroxidase (mpx) and gata1a in cells within the PLM, and later within the ICM, may indicate a primitive myeloerythroid cell population (54, 55, 57), although this remains to be tested. In zebrafish, primitive erythrocytes begin to enter circulation beginning at approximately 24 hpf (58). Unlike mammalian erythrocytes, they are oval-shaped and remain nucleated (59). Zebrafish primitive erythrocytes express embryonic hemoglobin genes (hbae1.1, hbae3, hbae5, hbbe1.1, hbbe1.2, hbbe1.3, hbbe2, hbbe3) (60–63), which allows them to be distinguished from their definitive hematopoietic counterparts.
In addition to the PLM, zebrafish also possess a second, more anterior hematopoietic site (Figure 1A,B). Known as the rostral blood islands (RBI), this site originates from anterior lateral-plate mesoderm (ALPM), and generates primitive macrophages, embryonic microglia, and neutrophilic granulocytes (7, 51, 64). At 10 hpf (tail bud), RBI cells express etsrp in an overlapping domain with scl, and the myeloid marker and ETS family transcription factor pu.1 (40, 51, 64). By 16–18 hpf (17 to 18-somites), these cells express the pan-leukocyte marker l-plastin (lcp1) (65–68), and macrophage progenitors spread out across the embryonic mesenchyme (68). Around this time, RBI-derived cells adopt more specific myeloid fates, expressing either interferon regulatory factor 8 (irf8) (69, 70), which is required to specify macrophage cell fate in zebrafish (69), or the granulocyte progenitor marker CCAAT/enhancer binding protein 1 (cebp1) (71, 72). Beginning at 21.5 hpf (25-somites) primitive macrophages express colony-stimulating factor receptor 1a (csf1ra/fms) (68), macrophage-expressed 1 (mpeg1) (66, 73), microfibrillar-associated protein 4 (mfap4) (73, 74), chemokine receptor cxcr3.2 (73), and ptpn6 (73). At 24 hpf, ALPM-derived granulocyte progenitors begin to differentiate, expressing lysozyme C (lyz) (7, 51, 64, 75), and mpx (72). By 36 hpf, (prim-25) lyz/mpx double-positive neutrophils stain strongly with Sudan Black, indicating that they are fully mature (72). Neutrophilic granulocytes are apparent within the trunk and tail by 48 hpf (long-pec) (53, 76). Beginning at 2.5 days post fertilization (dpf), microglia derived from the RBI colonize the developing zebrafish brain in response to neural cell death (77). Unlike in mice, where adult and embryonic microglia both arise from yolk sac blood islands (10, 78–80), fate mapping analyses in zebrafish indicate that RBI-derived embryonic microglia are eventually replaced by those originating from dorsal aorta hemogenic endothelium (12).
Different regulatory mechanisms are responsible for specifying ICM and RBI-derived granulocytes; Although etsrp does not regulate mpx myeloid gene expression within the ICM, loss of Etsrp leads to reduced mpx expression within the ALPM, and a concomitant reduction in neutrophil progenitors (57). Furthermore, fate mapping studies indicate that ALPM mpx-expressing cells are derived from cells that once expressed etsrp (52, 57).
Antagonism between gata1 and pu.1 regulates erythroid versus myeloid fate
The relationship between gata1 and pu.1 in specifying erythroid versus myeloid cell fate is well established (reviewed by (81). Gata1 represses myeloid differentiation, and is an essential regulator of erythroid cell fate (50, 82). In support of this, Gata1-depleted zebrafish embryos display expanded populations of granulocytic neutrophils and macrophages that occur at the expense of erythrocytes (50, 82). Biochemical data suggests that GATA1 and PU.1 proteins physically associate, transcriptionally inhibiting each other’s target genes (83–87). Evidence for this interaction also occurs in vivo, as Pu.1-depleted zebrafish embryos demonstrate ectopic ALPM gata1 expression (82), while Gata1-depleted zebrafish embryos demonstrate increased numbers of pu.1-positive cells, both within the ICM and ALPM (50, 82, 88). Notably, loss of function experiments show that Gata1 feedback maintains scl expression in the zebrafish ICM (88). Beyond Pu.1, the function of Gata1 in regulating erythroid cell fate is also modulated through its interactions with multiple other factors (reviewed by (89–92). These include Scl (93, 94), Friend of GATA (Fog1) (93, 95, 96), and multiple Kruppel-like transcription factors (Klfs) (81, 90, 91, 97–99). Genetic evidence suggests that Pu.1 dosage also influences macrophage versus neutrophilic granulocyte cell fate, as ALPM cells with high Pu.1 levels preferentially give rise to macrophages (72). Few identified factors have been shown to act upstream of gata1 or pu.1 to regulate the balance between erythroid and myeloid cell fate. However, the RNA-binding protein Elav1a has been shown to promote primitive erythroid cell fate in zebrafish by stabilizing gata1 mRNA (21). Furthermore, loss and gain of function analyses in zebrafish have implicated transcription intermediate factor 1γ (tif1γ) (100, 101), stat1b (102), as well as hox genes, and their cofactors Pbx2/4 and Meis1 (30, 103, 104) in regulating primitive erythropoiesis upstream of gata1.
DEFINITIVE HEMATOPOIESIS
Definitive hematopoiesis is the last wave of blood producing cells and although it starts as early as 26 hpf, this wave is responsible for generating self-renewable hematopoietic stem cells (HSPCs) that will maintain myeloid, erythroid and lymphoid lineages throughout the zebrafish life. As it has for primitive hematopoiesis, the zebrafish has also served as a solid model for uncovering genetic players and signaling pathways driving definitive hematopoiesis (105, 106). HSPCs in mammals and other vertebrates reside in the bone marrow, whereas zebrafish HSPCs seed the pronephros/kidney (107, 108). Despite this anatomical discrepancy, the signaling molecules, transcription factors and genetic programs controlling the definitive hematopoietic machinery are highly conserved between zebrafish and mammals. Zebrafish embryos bring a huge advantage when used to unveil gene regulators important for hematopoiesis, as they can survive without erythrocyte-dependent oxygen exchange up to 7 days, relying solely on passive diffusion. This feature has benefited the hematopoietic field, as a large variety of blood-related mutations were isolated in the classic 1996 zebrafish forward genetic screens and subsequent genetic screens while the function of other blood genes has been identified by reverse genetics. Functional work-up of many of these genes is difficult using mammalian models since most murine hematopoietic mutants are embryonic lethal (100, 105, 109–111). The advantages of the fish have gained more impact as several mutations discovered in zebrafish have later been shown to be present in human blood related diseases, ranging from anemic disorders to diverse types of leukemias and lymphomas (106, 112–123). Another factor propelling the zebrafish forward as a powerful animal model for hematopoiesis is the generation and use of transgenic reporter lines labeling specific HSPC-derived cell lineages, amplifying the advantages of the fish for in vivo imaging and lineage tracing (100, 108, 111, 124–133); See Table 1).
Table 1:
Transgenic lines available to study hematopoiesis in zebrafish
| Transgenic line | Cell type labeled | References |
|---|---|---|
| Tg(cmyb:EGFP) | HSPCs, myeloid | (5) |
|
Tg(−6.0itga2b:EGFP) aka Tg(cd41:EGFP) |
HSPCs, thrombocytes | (179) |
| Tg(runx1:EGFP) | HSPCs, anterior and posterior lateral plate mesoderm | (108) |
| Tg(runx1+23:EGFP) | HSPCs | (3, 180) |
|
TgBAC(gata2b:KalTA4)sd32 aka Tg(gata2b:GFP) |
HSPCs | (32) |
| Tg(scl-α:d2EGFP) | HSPCs, intermediate cell mass | (149) |
| Tg(scl-β:d2EGFP) | HSPCs, intermediate cell mass | (149) |
| Tg(lck:GFP) | T cells, lymphocytes | (126) |
| TgBAC(ikaros:EGFP) | T Cells, lymphocytes | (181) |
| Tg(rag1:GFP) | T Cells | (182) |
| Tg(rag2:GFP) | T Cells | (183) |
| Tg(mhc2dab:GFP)sd6 | B Cells, dendritic cells | (184) |
| Tg(mpx:GFP) | Neutrophils | (185) |
| Tg(lyz:EGFP) | Neutrophils | (133) |
| Tg(mpeg1:EGFP) | Macrophages | (186) |
| Tg(gata1:GFP) | Erythrocytes | (58) |
|
Tg(−20.7gata2:EGFP)la3 aka Tg(gata2a:GFP) |
Erythrocytes | (179) |
Sites and timing of definitive hematopoiesis
As development proceeds and circulation initiates at around 24 hpf, generation of hematopoietic cells derived from primitive hematopoiesis begins to subside, setting the stage for the definitive hematopoietic wave. The transition to definitive hematopoiesis occurs in two sequential events. First, a transient intermediate wave takes place in the posterior blood island (PBI), located ventro-caudally at the end of the yolk extension (55). This brief intermediate event generates gata1 and lmo2-expressing cells known as erythro-myeloid hematopoietic progenitors (EMPs) that will differentiate into erythroid and myeloid but not lymphoid lineages and populate the larval zebrafish (55, 134). Second, at around 28–30 hpf true multipotent self-renewable HSPCs become specified from endothelial cells in the ventral wall of the dorsal aorta (VDA), also known as the aorta-gonad-mesonephric (AGM) due to its mammalian anatomical counterpart. Specified HSPCs detach from the hemogenic endothelium by a dynamic process known as the endothelial-hematopoietic transition (EHT) (135, 136). Once detached, HSPCs enter circulation through the axial vein and will persist through adulthood as multipotent progenitors of all adult blood cell types (107, 124, 134, 136, 137). HSPCs seed three main hematopoietic organs, the caudal hematopoietic tissue (CHT) by 2 dpf, the thymus by 3 dpf, and the kidney marrow at ~4 dpf (Figures 2 and 3). HSPCs embedded in the CHT will serve as a source of embryonic macrophages, neutrophils and monocytes. The kidney, which corresponds to the mammalian bone marrow, will produce myeloid, erythroid, thromboid and lymphoid lineages, leaving the thymus to produce mature lymphoid T cells throughout adulthood.
Genetic signaling molecules in specification and lineage differentiation
Specific markers expressed by zebrafish EMPs and HSPCs facilitates their isolation and analysis. Transgenic lines based on gata1 and lmo2 promoters have been used to isolate EMPs from zebrafish embryos, while the distinctive expression of runx1, itga2a and c-myb on nascent HSPCs identifies these cells (124, 136, 137). Runx1 is considered one of the earliest markers of HSPCs, with cmyb acting downstream. Analysis of zebrafish mutants has demonstrated that cmyb expression is runx1-dependent. Runx1 mutants lose their HSPCs due to their incapacity to undergo EHT (136, 137). In the most severe cmyb loss of function animals, HSPCs are correctly specified, but fail to exit the dorsal aorta (138). Other cmyb mutants progressively lose adult hematopoietic lineages starting from 20 dpf but are viable up to at least 3 months old (137, 139). The induction of Runx1 relies on Notch1a/b receptor-mediated signaling in the hemogenic endothelium (140), and the requirement for Notch-Runx1 signaling is a key feature distinguishing HSPCs from EMPs. HSPCs are unable to form in the absence of Notch-Runx1 signaling, as demonstrated by analyzing mindbomb (mib) mutants and embryos treated with the γ-secretase inhibitor DAPT (141–143). Conversely, EMPs do not express the Notch receptor and their generation is not affected under these conditions (141, 142). As mib mutants and DAPT-treated embryos exhibit reduced arterial gene expression and ectopic venous gene expression within the dorsal aorta (143, 144), it is perhaps not surprising that they fail to produce HSPCs. However, transient activation of Notch signaling by induction of the Notch intracellular domain (NICD) in wild type embryos leads to the upregulation of HSPC sprouting from the AGM, without inducing ectopic arterial gene expression (142, 145), and jagged1-mutant zebrafish fail to produce HSPCs, but do not exhibit defects in specifying arterial cell fate (146). Combined, these data support the idea that there are arterial-venous patterning-independent functions for Notch signaling in definitive hematopoiesis.
Notch and downstream HSPC specification are also regulated by non-canonical Wnt signaling. The ligand Wnt16 induces the ligands deltaC and deltaD in the somites surrounding the dorsal aorta, leading to induction of runx1- and cmyb-expressing HSPCs. Knocking down wnt16 does not affect vasculature formation but leads to the loss of itga2b(+)/cmyb(+) HSPCs and lymphocytic lineages (147). Transforming growth factor β (tgfβ) is another signal coming not only from the dorsal aorta endothelium, but from the environment surrounding the AGM. Knocking down tgfβR2 leads to significant loss of runx1 and other HSPC markers and results in impairment of HSPC budding without affecting the arterial endothelium (148).
Additional factors act with or without Notch-Runx1 during specification of AGM-derived HSPCs. Mutations in cbfβ, the non-DNA binding beta-subunit of the Core Binding factor, showed that this Runx1 partner is also essential for HSPC’s sprouting (135). Using double specific scl promoter driven transgenic lines, it was shown that the transcription factor isoforms sclα/sclβ act consecutively during HSPC formation. sclβ expression, co-expressed with the endothelial kdrl marker, is required in the AGM for HSPC specification and EHT initiation, while expression of sclα only appears once the HSPCs detach from the AGM, and is important for their maintenance as evidenced by increased HSPC cell death noted in sclα morphants (149). The transgenic reporter lines available in the zebrafish have been very useful in advancing our knowledge of specific HSPC-derived cell lineages. For instance, thrombocytes populating the thymus were first imaged live using a line where GFP expression is driven by the itga2b promoter (a.k.a. CD41). This analysis revealed that nascent HSPCs express itga2b, showed by flow cytometry isolation that thrombocytes express high levels of CD41-GFP(+) and that thrombocyte formation is scl-dependent (150). Tissues surrounding the hemogenic endothelium also play important roles in HSPC specification. Using fli1a and sox10-driven transgenic lines, researchers observed that migratory neural crest cells (NCCs) come in touch with the dorsal aorta endothelium at the onset of and during HSPC specification. When NCC contact with the dorsal aorta was impeded, HSPC specification was lost. Pdfgra-dependent signaling from the migratory neural crest is required for this process. Pdgfra-deficient embryos display a lack of runx1 and cmyb positive HSPCs, as well as mature rag2 positive T-lymphocytes in the thymus (151).
Epigenetic signaling molecules in specification and lineage differentiation
Epigenetic control of gene regulation has gained significant interest in recent years, and recent evidence suggests that it plays important roles in hematopoiesis. Epigenetic adaptations include histone modifications, DNA methylation, small RNAs, and chromatin remodeling complexes (152). DNA methylation plays critical, evolutionarily conserved roles during vertebrate development (153, 154). Methyl marks are added to cytosines in CpG dinucleotides within DNA by specific enzymes called DNA methyltransferases (DNMTs). De novo DNMTs add methyl marks to previously unmarked DNA sequences, while maintenance DNMTs copy preexisting methyl marks onto newly synthesized strands during DNA replication. The zebrafish de novo DNA methyltransferase family consists of dnmt3aa, dnmt3ab, dnmt3ba, dnmt3bb.1, dnmt3bb.2, and dnmt3bb.3. Recent work has shown that dnmt3bb.1 is essential for maintenance of HSPC specification (124). Dnmt3bb.1 is the closest homologue of mammalian DNMT3b, and it is similarly expressed in developing HSPCs. Zebrafish dnmt3bb.1 mutants lose expression of cmyb by 72 hpf, followed by HSPC apoptosis and loss of downstream hematopoietic lineages as evidenced by a reduction of myeloid and lymphoid markers such as l-plastin and rag1, respectively (124). Bisulfite sequencing of dnmt3bb.1 mutants revealed that the intron1 CpG island of cmyb is a target of dnmt3bb.1 DNA methylation during definitive hematopoiesis. Dnmt3bb.1 expression is dependent on runx1 and ectopic expression of dnmt3bb.1 is sufficient to induce not only cmyb but downstream blood lineage specific markers in either wild type or runx1-deficient endothelium. Ectopic expression of dnmt3bb.1 during blastula stages demonstrated that dnmt3bb.1 is also capable of inducing cmyb in these naïve pre-gastrula cells, in addition to a battery of erythroid, myeloid, and lymphoid blood lineage markers, all without induction of non-hematopoietic mesodermal markers such as cadherin5 or etv2. Together, the findings from this zebrafish study indicate that once HSPCs detach from the AGM and begin to downregulate runx1 expression, continued maintenance of cmyb and downstream blood lineage marker expression depends on dnmt3bb.1 (124, 152). Interestingly, conditions defective epigenetic regulation has been shown to lead to hematopoietic malignancies like leukemias and lymphomas, and drugs targeting DNA methylation and other epigenetic modifications are currently being used as therapies for these conditions, as recently reviewed in detail elsewhere (152).
Later hematopoietic development in zebrafish
After being specified and detaching from the AGM, HSPCs seed a number of zebrafish hematopoietic organs and tissues, including the thymus and anterior kidney. Zebrafish possesses the same blood types found in mammals, and mostly they share similar anatomical locations and morphological and functional features. In teleost fish like the zebrafish the anterior portion of the kidney is the main source of blood cells, where HSPCs reside through adulthood, and it is comparable to the mammalian bone marrow (107, 108, 155). Zebrafish studies have been useful for studying juvenile and adult stages of hematopoiesis. The runx1W84X mutant described earlier loses all HSPC-derived lineages, but still has some cd41(+) cells in the AGM at larval stages. Although most homozygous mutants die by 20 dpf, a small percentage of runx1W84X/W84X homozygotes recover and survive to adulthood, forming all blood lineages as adults (137). ~10–12% of dnmt3bb.1 homozygous mutants also survive to adulthood, and, like the cmyb mutants described above, these show a strong reduction in hematopoietic cell number and malformation of the remaining erythrocytes, providing an interesting model for studying adult hematopoietic deficiency (124). Examples such as these open the door to investigate potential alternative hematopoietic sources in fish and may permit later juvenile and adult experiments not possible in mammalian models.
A variety of hematopoiesis-derived cells found away from the hematopoietic organs also have important roles in later-stage and adult zebrafish. Microglia, a brain resident macrophage, have an important role in protecting the CNS from toxic elements and cell debris and are also important effectors of neural development and function (156–158). In mice, they are known to arise during primitive hematopoiesis from yolk sac-derived macrophages (159). This is also the case in the larval zebrafish, where microglia arise from macrophages born in the rostral blood island (7, 68, 160). A recent study using a “switch line” to laser-induce a heat shock driving Cre recombination specifically in coro1-expressing leukocytes shed light on the origins of the adult population of zebrafish microglia. Temporally- and spatially-restricted activation of the switch line in wild type and pu1G242D, cmybhkz3 or runx1W84X mutant animals demonstrated that embryonic and adult microglia in zebrafish have different origins. While embryonic microglia formation occurs during primitive hematopoiesis and is pu.1-dependent and runx1-independent, adult microglia depend on runx1 and arise from the ventral wall of the dorsal aorta (12).
ZEBRAFISH AS A MODEL FOR HEMATOPOIETIC DISORDERS
Zebrafish has become an important model system for studying hematopoietic disorders and developing small molecule based therapeutic approaches. Conventional large-scale genetic screens have identified several zebrafish mutants that phenocopy human hematological disorders. In the last decade, this list has grown due to the relative ease of generating zebrafish mutants in genes linked to human diseases using CRISPR/Cas9 and other genome editing technologies. Hematopoiesis in zebrafish is very similar to that in higher vertebrates, and many of the mutated zebrafish orthologs of human disease genes have successfully phenocopied the human disease phenotypes. Since as noted above the small, externally developing zebrafish embryos and larvae can passively absorb oxygen and continue organ development for up to a week without red blood cells, the fish provides an ideal model system to study severe anemia and other disorders which might be difficult to study in higher vertebrates (161). The relative ease with which transgenic animals can be generated has also contributed significantly to the development of different leukemia models in zebrafish. Several research groups have used cell type-specific promoters to drive human oncogenes and fusion gene cDNAs to model different types of human blood cancers in zebrafish. These models have contributed significantly to our understanding of different aspects of these human diseases. In addition to studying disease biology, zebrafish leukemia models are being successfully used to develop epigenetic therapies (162). Table 2 lists a compilation of current zebrafish models of hematopoietic disorders.
Table 2:
Zebrafish models of hematological disorders
| Human Disease condition | Zebrafish transgenic line or mutant | Affected gene | Reference | ||
|---|---|---|---|---|---|
| Hereditary elliptocytosis | chablis and merlot | epb41b | (164) | ||
| Hereditary spherocytosis type 2 | riesling | b-spectrin | (167) | ||
| Hereditary spherocytosis type 4 | retsina | slc4a1 | (168) | ||
| Erythropoietic protoporphyria and X-linked sideroblastic anemia | sauternes | alas2 | (187) | ||
| hypochromic microcytic anemia | zinfandel | hbbe1.1 | (62) | ||
| hypochromic microcytic anemia | chianti | tfr1a | (188) | ||
| Hemochromatosis type 4 | weissherbst | slc40a1 | (189) | ||
| Erythropoietic protoporphyria | dracula | fech | (172) | ||
| Diamond-Blackfan anemia | rpl11 | rpl11 | (190) | ||
| Acute Myeloid leukemia | Tg(hsp:AML1-ETO) | runx1 | (175) | ||
| Acute Myeloid leukemia | Tg(spi1:lox-EGFP-lox-NUP98-HOXA9) | hoxa9 | (176) | ||
| Myelodysplastic Syndrome | Tet2 mutants | Tet2 | (191) | ||
| Myelodysplastic Syndrome | crimsonless | hspa9b | (192) | ||
| T- cell Acute lymphoblastic leukemia | Tg(rag2:EGFP-myc) | Myc-oncogene overexpression | (125) | ||
| T- cell Acute lymphoblastic leukemia | Tg(rag2:NOTCH1ICD-EGFP) | Notch1-intracellular domain overexpression | (178) | ||
| Pre-B cell Acute Lymphoblastic leukemia | Tg(ef1alpha:EGFP-TEL-AML1) | runx1 | (193) |
Zebrafish also offer unique advantages for large scale high throughput drug screening, including optical transparency, external fertilization and development, and the availability of a wide variety of transgenic reporter lines (Table 1). These advantages have made the use of zebrafish embryos in initial small molecule screens a standard practice around the world. A variety of different compounds that influence HSPCs and hematopoiesis in general have been identified through such screens (163). More recently, screens to isolate epigenetic modifiers have shown promise for developing new therapeutic targets.
Non-malignant models of hematopoietic disorders
Large scale forward genetic screens and reverse genetics using genome editing technologies have generated an array of zebrafish mutants that model different types of human anemia conditions. Zebrafish mutants chablis and merlot develop red blood cells (RBCs) with elliptical morphology that often contain two nuclei. Positional cloning revealed both mutants carry mutations in RBC membrane protein Epb41b (164). Erythrocytic membrane proteins play crucial roles in regulating RBC cytoskeleton and other membrane proteins (165). Human mutations in EPB42 are linked to hereditary elliptocytosis (166). Mutations in five human genes lead to Hereditary spherocytosis, a type of anemia with low number of RBCs typically caused due to hemolysis or structural defects in RBCs. Zebrafish mutations in beta-spectrin and slc4a phenocopy human hereditary spherocytosis. Zebrafish mutants in these genes show defective RBC morphology and abnormal cytokinesis (167, 168). Identification of these genes not only shed light on the human disease conditions but also revealed roles of these genes in RBC differentiation.
A number of zebrafish mutants model Diamond-Blackfan anemia, a bone marrow disorder caused due to mutations in ribosomal genes (169). People affected by Diamond-Blackfan syndrome carry increased risk of complications related to the bone marrow and have higher chances of developing secondary hematological conditions such as myelodysplastic syndrome and acute myeloid leukemia (170). Zebrafish models of Diamond-Blackfan syndrome have not only shed light on the molecular mechanisms behind this disease, but have proven useful in developing potential pharmacological therapies for this disease.
In humans, mutations in ferrochelatase (FCH) cause erythropoietic protoporphyria, a disorder in which RBCs accumulate excess phototoxic heme synthesis intermediates and undergo hemolysis after exposure to light (171). The toxic compounds released after lysis accumulate and damage the liver. Zebrafish ferrochelatase (dracula) mutants have strongly fluorescent RBCs that undergo lysis upon illumination (172). The mutant embryos also show abnormal liver development as in the human disease condition. Dracula mutant embryos provide a model system to screen small molecules that can block hemolysis (172).
Malignant models of hematopoietic disorders
Abnormal differentiation and uncontrolled growth of blood cells leads to blood disorders such as leukemia. Blood cancers are generally classified based on the type of blood cells involved in the disease. Many of these conditions arise through chromosomal translocations and insertions that lead to abnormal activation of oncogenes in HSPCs or in a specific downstream blood cell lineage (173). Many of these human fusion oncogenes cause blood disorders similar to human cancers when they are overexpressed in zebrafish (174). Inducible overexpression of AML1-ETO fusion protein in fish leads to defective erythropoiesis and granulopoiesis, a phenotype also seen in human acute myeloid leukemia (175). Overexpression of NUP98-HOXA9 fusion protein in zebrafish using a myeloid cell type-specific promoter causes overproliferation of HSPCs and myeloid cell expansion (176). Both the AML models were successfully used to identify small molecules that can reverse these conditions. Cyclooxygenase (COX) inhibitors and benzodiazepine Ro5–3335 treatment of NUP98-HOXA9 overexpressing embryos reverses myeloid cell over proliferation (177). Interestingly, epigenetic inhibitors targeting histone deacetylase or DNA methyltransferase activity reduces myeloid cell proliferation seen in AML1-ETO and NUP98-HOXA9 transgenic embryos respectively (115, 162). These results also highlight the potential role of epigenetics in these diseases.
Similar to the AML models described above, myc oncogene and Notch intracellular domain overexpression in zebrafish using lympohoid cell promoters leads to T-cell acute lymphoblastic leukemia (T-ALL) (125, 178). In these T-ALL zebrafish models, co-expression of GFP in leukemic cells allows high-resolution imaging of the behavior of these cancer cells in living animals, a difficult feat in murine models. GFP labelled cells are also easy to isolate from T-ALL zebrafish models using FACS techniques. Transplantation of the isolated cells in conditioned recipient zebrafish initiates full symptoms of leukemia, recapitulating human cancer cell-like behaviors (125, 178). Together, these zebrafish leukemia models provide powerful tools to identify molecular mechanisms behind these cancers.
CONCLUSIONS
Zebrafish has become an important model to study hematopoiesis. Over the last decade in particular many crucial new discoveries regarding different aspects of normal and pathological hematopoiesis have been made using zebrafish as a model system. Zebrafish is highly amenable to genetic manipulations and with recent advances in genome editing technologies it has become relatively easy to probe the functional roles of the different signaling molecules involved in hematopoiesis. An array of new transgenic zebrafish lines unique to specific hematopoietic cell populations have also been developed, and in addition to providing valuable research tools for studying basic molecular mechanisms of normal and pathologic hematopoiesis, they have already been used successfully in chemical screens to identify potential new therapeutic agents for human disease. With the availability of these powerful genome editing, transgenic, and chemical screening tools the fish seems likely to yield many more important new insights into the molecular mechanisms underlying malignant and non-malignant hematopoietic disorders.
There are still a few small limitations that need to be overcome to use zebrafish to study hematopoiesis. Culturing HSPCs and differentiating them into downstream lineages in vitro (similar to colony formation assays done in higher vertebrates) is still limited. Although there have been some successful attempts to culture zebrafish hematopoietic stem cells in vitro, more needs to be done to improve the culture conditions and make them straightforward and simple. Lack of reliable cell surface markers and antibodies against these markers is another problem for the zebrafish hematopoiesis field. FACS sorting of hematopoietic subpopulations using a broad array of antibodies targeting specific cell-surface proteins is widely used in mammalian studies. In fish most of the current hematopoietic cell isolation protocols are based on transgenic labeling, greatly limiting isolation of subpopulations of particular lineages. Developing many more reliable cell surface marker-specific antibodies would be very helpful in isolating specific hematopoietic cell populations. Despite these limitations, the zebrafish provides a superb model system to study hematopoiesis and hematopoietic disorders. In the future, the zebrafish will continue to greatly facilitate the discovery of new drugs and new molecular processes involved in hematopoiesis.
ACKNOWLEDGMENTS
We thank all the members of the Weinstein lab for their help and support. The Weinstein lab is supported by the intramural program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development. We apologize to authors whose work we could not cite due to space limitations.
REFERENCES
- 1.Paik EJ, Zon LI. Hematopoietic development in the zebrafish. Int J Dev Biol. 2010;54(6–7):1127–37. [DOI] [PubMed] [Google Scholar]
- 2.Ciau-Uitz A, Monteiro R, Kirmizitas A, Patient R. Developmental hematopoiesis: ontogeny, genetic programming and conservation. Exp Hematol. 2014;42(8):669–83. [DOI] [PubMed] [Google Scholar]
- 3.Tamplin OJ, Durand EM, Carr LA, Childs SJ, Hagedorn EJ, Li P, et al. Hematopoietic stem cell arrival triggers dynamic remodeling of the perivascular niche. Cell. 2015;160(1–2):241–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoggatt J, Singh P, Sampath J, Pelus LM. Prostaglandin E2 enhances hematopoietic stem cell homing, survival, and proliferation. Blood. 2009;113(22):5444–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.North TE, Goessling W, Walkley CR, Lengerke C, Kopani KR, Lord AM, et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447(7147):1007–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132(4):631–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herbomel P, Thisse B, Thisse C. Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development. 1999;126(17):3735–45. [DOI] [PubMed] [Google Scholar]
- 8.Lobov IB, Rao S, Carroll TJ, Vallance JE, Ito M, Ondr JK, et al. WNT7b mediates macrophage-induced programmed cell death in patterning of the vasculature. Nature. 2005;437(7057):417–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fantin A, Vieira JM, Gestri G, Denti L, Schwarz Q, Prykhozhij S, et al. Tissue macrophages act as cellular chaperones for vascular anastomosis downstream of VEGF-mediated endothelial tip cell induction. Blood. 2010;116(5):829–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330(6005):841–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Svahn AJ, Graeber MB, Ellett F, Lieschke GJ, Rinkwitz S, Bennett MR, et al. Development of ramified microglia from early macrophages in the zebrafish optic tectum. Dev Neurobiol. 2013;73(1):60–71. [DOI] [PubMed] [Google Scholar]
- 12.Xu J, Zhu L, He S, Wu Y, Jin W, Yu T, et al. Temporal-Spatial Resolution Fate Mapping Reveals Distinct Origins for Embryonic and Adult Microglia in Zebrafish. Dev Cell. 2015;34(6):632–41. [DOI] [PubMed] [Google Scholar]
- 13.Lyons DA, Talbot WS. Glial cell development and function in zebrafish. Cold Spring Harb Perspect Biol. 2014;7(2):a020586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lacaud G, Kouskoff V. Hemangioblast, hemogenic endothelium, and primitive versus definitive hematopoiesis. Exp Hematol. 2017;49:19–24. [DOI] [PubMed] [Google Scholar]
- 15.Nishikawa S. Hemangioblast: an in vitro phantom. Wiley Interdiscip Rev Dev Biol. 2012;1(4):603–8. [DOI] [PubMed] [Google Scholar]
- 16.Covassin L, Amigo JD, Suzuki K, Teplyuk V, Straubhaar J, Lawson ND. Global analysis of hematopoietic and vascular endothelial gene expression by tissue specific microarray profiling in zebrafish. Dev Biol. 2006;299(2):551–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reischauer S, Stone OA, Villasenor A, Chi N, Jin SW, Martin M, et al. Cloche is a bHLH-PAS transcription factor that drives haemato-vascular specification. Nature. 2016;535(7611):294–8. [DOI] [PubMed] [Google Scholar]
- 18.Stainier DY, Weinstein BM, Detrich HW 3rd, Zon LI, Fishman MC. Cloche, an early acting zebrafish gene, is required by both the endothelial and hematopoietic lineages. Development. 1995;121(10):3141–50. [DOI] [PubMed] [Google Scholar]
- 19.Liao W, Bisgrove BW, Sawyer H, Hug B, Bell B, Peters K, et al. The zebrafish gene cloche acts upstream of a flk-1 homologue to regulate endothelial cell differentiation. Development. 1997;124(2):381–9. [DOI] [PubMed] [Google Scholar]
- 20.Xiong JW, Yu Q, Zhang J, Mably JD. An acyltransferase controls the generation of hematopoietic and endothelial lineages in zebrafish. Circ Res. 2008;102(9):1057–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X, Lu YC, Dai K, Torregroza I, Hla T, Evans T. Elavl1a regulates zebrafish erythropoiesis via posttranscriptional control of gata1. Blood. 2014;123(9):1384–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vogeli KM, Jin SW, Martin GR, Stainier DY. A common progenitor for haematopoietic and endothelial lineages in the zebrafish gastrula. Nature. 2006;443(7109):337–9. [DOI] [PubMed] [Google Scholar]
- 23.Lee RK, Stainier DY, Weinstein BM, Fishman MC. Cardiovascular development in the zebrafish. II. Endocardial progenitors are sequestered within the heart field. Development. 1994;120(12):3361–6. [DOI] [PubMed] [Google Scholar]
- 24.de Jong JL, Zon LI. Use of the zebrafish system to study primitive and definitive hematopoiesis. Annu Rev Genet. 2005;39:481–501. [DOI] [PubMed] [Google Scholar]
- 25.Ma AC, Chung MI, Liang R, Leung AY. A DEAB-sensitive aldehyde dehydrogenase regulates hematopoietic stem and progenitor cells development during primitive hematopoiesis in zebrafish embryos. Leukemia. 2010;24(12):2090–9. [DOI] [PubMed] [Google Scholar]
- 26.McReynolds LJ, Gupta S, Figueroa ME, Mullins MC, Evans T. Smad1 and Smad5 differentially regulate embryonic hematopoiesis. Blood. 2007;110(12):3881–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma AC, Ward AC, Liang R, Leung AY. The role of jak2a in zebrafish hematopoiesis. Blood. 2007;110(6):1824–30. [DOI] [PubMed] [Google Scholar]
- 28.Liang D, Chang JR, Chin AJ, Smith A, Kelly C, Weinberg ES, et al. The role of vascular endothelial growth factor (VEGF) in vasculogenesis, angiogenesis, and hematopoiesis in zebrafish development. Mech Dev. 2001;108(1–2):29–43. [DOI] [PubMed] [Google Scholar]
- 29.Detrich HW 3rd, Kieran MW, Chan FY, Barone LM, Yee K, Rundstadler JA, et al. Intraembryonic hematopoietic cell migration during vertebrate development. Proc Natl Acad Sci U S A. 1995;92(23):10713–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davidson AJ, Ernst P, Wang Y, Dekens MP, Kingsley PD, Palis J, et al. cdx4 mutants fail to specify blood progenitors and can be rescued by multiple hox genes. Nature. 2003;425(6955):300–6. [DOI] [PubMed] [Google Scholar]
- 31.Gering M, Rodaway AR, Gottgens B, Patient RK, Green AR. The SCL gene specifies haemangioblast development from early mesoderm. EMBO J. 1998;17(14):4029–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Butko E, Distel M, Pouget C, Weijts B, Kobayashi I, Ng K, et al. Gata2b is a restricted early regulator of hemogenic endothelium in the zebrafish embryo. Development. 2015;142(6):1050–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Craig MP, Grajevskaja V, Liao HK, Balciuniene J, Ekker SC, Park JS, et al. Etv2 and fli1b function together as key regulators of vasculogenesis and angiogenesis. Arterioscler Thromb Vasc Biol. 2015;35(4):865–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dooley KA, Davidson AJ, Zon LI. Zebrafish scl functions independently in hematopoietic and endothelial development. Dev Biol. 2005;277(2):522–36. [DOI] [PubMed] [Google Scholar]
- 35.Patterson LJ, Gering M, Patient R. Scl is required for dorsal aorta as well as blood formation in zebrafish embryos. Blood. 2005;105(9):3502–11. [DOI] [PubMed] [Google Scholar]
- 36.Patterson LJ, Gering M, Eckfeldt CE, Green AR, Verfaillie CM, Ekker SC, et al. The transcription factors Scl and Lmo2 act together during development of the hemangioblast in zebrafish. Blood. 2007;109(6):2389–98. [DOI] [PubMed] [Google Scholar]
- 37.Qian F, Zhen F, Xu J, Huang M, Li W, Wen Z. Distinct functions for different scl isoforms in zebrafish primitive and definitive hematopoiesis. PLoS Biol. 2007;5(5):e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Juarez MA, Su F, Chun S, Kiel MJ, Lyons SE. Distinct roles for SCL in erythroid specification and maturation in zebrafish. J Biol Chem. 2005;280(50):41636–44. [DOI] [PubMed] [Google Scholar]
- 39.Gering M, Yamada Y, Rabbitts TH, Patient RK. Lmo2 and Scl/Tal1 convert non-axial mesoderm into haemangioblasts which differentiate into endothelial cells in the absence of Gata1. Development. 2003;130(25):6187–99. [DOI] [PubMed] [Google Scholar]
- 40.Liao EC, Paw BH, Oates AC, Pratt SJ, Postlethwait JH, Zon LI. SCL/Tal-1 transcription factor acts downstream of cloche to specify hematopoietic and vascular progenitors in zebrafish. Genes Dev. 1998;12(5):621–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sumanas S, Jorniak T, Lin S. Identification of novel vascular endothelial-specific genes by the microarray analysis of the zebrafish cloche mutants. Blood. 2005;106(2):534–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sumanas S, Lin S. Ets1-related protein is a key regulator of vasculogenesis in zebrafish. PLoS Biol. 2006;4(1):e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ren X, Gomez GA, Zhang B, Lin S. Scl isoforms act downstream of etsrp to specify angioblasts and definitive hematopoietic stem cells. Blood. 2010;115(26):5338–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gomez G, Lee JH, Veldman MB, Lu J, Xiao X, Lin S. Identification of vascular and hematopoietic genes downstream of etsrp by deep sequencing in zebrafish. PLoS One. 2012;7(3):e31658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pham VN, Lawson ND, Mugford JW, Dye L, Castranova D, Lo B, et al. Combinatorial function of ETS transcription factors in the developing vasculature. Dev Biol. 2007;303(2):772–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sumanas S, Gomez G, Zhao Y, Park C, Choi K, Lin S. Interplay among Etsrp/ER71, Scl, and Alk8 signaling controls endothelial and myeloid cell formation. Blood. 2008;111(9):4500–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu H, Traver D, Davidson AJ, Dibiase A, Thisse C, Thisse B, et al. Regulation of the lmo2 promoter during hematopoietic and vascular development in zebrafish. Dev Biol. 2005;281(2):256–69. [DOI] [PubMed] [Google Scholar]
- 48.Zhu C, Smith T, McNulty J, Rayla AL, Lakshmanan A, Siekmann AF, et al. Evaluation and application of modularly assembled zinc-finger nucleases in zebrafish. Development. 2011;138(20):4555–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kok FO, Shin M, Ni CW, Gupta A, Grosse AS, van Impel A, et al. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev Cell. 2015;32(1):97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Galloway JL, Wingert RA, Thisse C, Thisse B, Zon LI. Loss of gata1 but not gata2 converts erythropoiesis to myelopoiesis in zebrafish embryos. Dev Cell. 2005;8(1):109–16. [DOI] [PubMed] [Google Scholar]
- 51.Bennett CM, Kanki JP, Rhodes J, Liu TX, Paw BH, Kieran MW, et al. Myelopoiesis in the zebrafish, Danio rerio. Blood. 2001;98(3):643–51. [DOI] [PubMed] [Google Scholar]
- 52.Le Guyader D, Redd MJ, Colucci-Guyon E, Murayama E, Kissa K, Briolat V, et al. Origins and unconventional behavior of neutrophils in developing zebrafish. Blood. 2008;111(1):132–41. [DOI] [PubMed] [Google Scholar]
- 53.Lieschke GJ, Oates AC, Crowhurst MO, Ward AC, Layton JE. Morphologic and functional characterization of granulocytes and macrophages in embryonic and adult zebrafish. Blood. 2001;98(10):3087–96. [DOI] [PubMed] [Google Scholar]
- 54.Warga RM, Kane DA, Ho RK. Fate mapping embryonic blood in zebrafish: multi- and unipotential lineages are segregated at gastrulation. Dev Cell. 2009;16(5):744–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bertrand JY, Kim AD, Violette EP, Stachura DL, Cisson JL, Traver D. Definitive hematopoiesis initiates through a committed erythromyeloid progenitor in the zebrafish embryo. Development. 2007;134(23):4147–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sumoy L, Keasey JB, Dittman TD, Kimelman D. A role for notochord in axial vascular development revealed by analysis of phenotype and the expression of VEGR-2 in zebrafish flh and ntl mutant embryos. Mech Dev. 1997;63(1):15–27. [DOI] [PubMed] [Google Scholar]
- 57.Glenn NO, Schumacher JA, Kim HJ, Zhao EJ, Skerniskyte J, Sumanas S. Distinct regulation of the anterior and posterior myeloperoxidase expression by Etv2 and Gata1 during primitive Granulopoiesis in zebrafish. Dev Biol. 2014;393(1):149–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Long Q, Meng A, Wang H, Jessen JR, Farrell MJ, Lin S. GATA-1 expression pattern can be recapitulated in living transgenic zebrafish using GFP reporter gene. Development. 1997;124(20):4105–11. [DOI] [PubMed] [Google Scholar]
- 59.Kulkeaw K, Sugiyama D. Zebrafish erythropoiesis and the utility of fish as models of anemia. Stem Cell Res Ther. 2012;3(6):55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tiedke J, Gerlach F, Mitz SA, Hankeln T, Burmester T. Ontogeny of globin expression in zebrafish (Danio rerio). J Comp Physiol B. 2011;181(8):1011–21. [DOI] [PubMed] [Google Scholar]
- 61.Nefedochkina AV, Petrova NV, Ioudinkova ES, Kovina AP, Iarovaia OV, Razin SV. Characterization of the enhancer element of the Danio rerio minor globin gene locus. Histochem Cell Biol. 2016;145(4):463–73. [DOI] [PubMed] [Google Scholar]
- 62.Brownlie A, Hersey C, Oates AC, Paw BH, Falick AM, Witkowska HE, et al. Characterization of embryonic globin genes of the zebrafish. Dev Biol. 2003;255(1):48–61. [DOI] [PubMed] [Google Scholar]
- 63.Ganis JJ, Hsia N, Trompouki E, de Jong JL, DiBiase A, Lambert JS, et al. Zebrafish globin switching occurs in two developmental stages and is controlled by the LCR. Dev Biol. 2012;366(2):185–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lieschke GJ, Oates AC, Paw BH, Thompson MA, Hall NE, Ward AC, et al. Zebrafish SPI-1 (PU.1) marks a site of myeloid development independent of primitive erythropoiesis: implications for axial patterning. Dev Biol. 2002;246(2):274–95. [DOI] [PubMed] [Google Scholar]
- 65.Shi X, He BL, Ma AC, Guo Y, Chi Y, Man CH, et al. Functions of idh1 and its mutation in the regulation of developmental hematopoiesis in zebrafish. Blood. 2015;125(19):2974–84. [DOI] [PubMed] [Google Scholar]
- 66.He BL, Shi X, Man CH, Ma AC, Ekker SC, Chow HC, et al. Functions of flt3 in zebrafish hematopoiesis and its relevance to human acute myeloid leukemia. Blood. 2014;123(16):2518–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meijer AH, van der Sar AM, Cunha C, Lamers GE, Laplante MA, Kikuta H, et al. Identification and real-time imaging of a myc-expressing neutrophil population involved in inflammation and mycobacterial granuloma formation in zebrafish. Dev Comp Immunol. 2008;32(1):36–49. [DOI] [PubMed] [Google Scholar]
- 68.Herbomel P, Thisse B, Thisse C. Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M-CSF receptor-dependent invasive process. Dev Biol. 2001;238(2):274–88. [DOI] [PubMed] [Google Scholar]
- 69.Li L, Jin H, Xu J, Shi Y, Wen Z. Irf8 regulates macrophage versus neutrophil fate during zebrafish primitive myelopoiesis. Blood. 2011;117(4):1359–69. [DOI] [PubMed] [Google Scholar]
- 70.Mommaerts H, Esguerra CV, Hartmann U, Luyten FP, Tylzanowski P. Smoc2 modulates embryonic myelopoiesis during zebrafish development. Dev Dyn. 2014;243(11):1375–90. [DOI] [PubMed] [Google Scholar]
- 71.Jin H, Huang Z, Chi Y, Wu M, Zhou R, Zhao L, et al. c-Myb acts in parallel and cooperatively with Cebp1 to regulate neutrophil maturation in zebrafish. Blood. 2016;128(3):415–26. [DOI] [PubMed] [Google Scholar]
- 72.Jin H, Li L, Xu J, Zhen F, Zhu L, Liu PP, et al. Runx1 regulates embryonic myeloid fate choice in zebrafish through a negative feedback loop inhibiting Pu.1 expression. Blood. 2012;119(22):5239–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zakrzewska A, Cui C, Stockhammer OW, Benard EL, Spaink HP, Meijer AH. Macrophage-specific gene functions in Spi1-directed innate immunity. Blood. 2010;116(3):e1–11. [DOI] [PubMed] [Google Scholar]
- 74.Walton EM, Cronan MR, Beerman RW, Tobin DM. The Macrophage-Specific Promoter mfap4 Allows Live, Long-Term Analysis of Macrophage Behavior during Mycobacterial Infection in Zebrafish. PLoS One. 2015;10(10):e0138949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu F, Wen Z. Cloning and expression pattern of the lysozyme C gene in zebrafish. Mech Dev. 2002;113(1):69–72. [DOI] [PubMed] [Google Scholar]
- 76.Willett CE, Cortes A, Zuasti A, Zapata AG. Early hematopoiesis and developing lymphoid organs in the zebrafish. Dev Dyn. 1999;214(4):323–36. [DOI] [PubMed] [Google Scholar]
- 77.Xu J, Wang T, Wu Y, Jin W, Wen Z. Microglia Colonization of Developing Zebrafish Midbrain Is Promoted by Apoptotic Neuron and Lysophosphatidylcholine. Dev Cell. 2016;38(2):214–22. [DOI] [PubMed] [Google Scholar]
- 78.Kierdorf K, Erny D, Goldmann T, Sander V, Schulz C, Perdiguero EG, et al. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat Neurosci. 2013;16(3):273–80. [DOI] [PubMed] [Google Scholar]
- 79.Perdiguero EG, Klapproth K, Schulz C, Busch K, de Bruijn M, Rodewald HR, et al. The Origin of Tissue-Resident Macrophages: When an Erythro-myeloid Progenitor Is an Erythro-myeloid Progenitor. Immunity. 2015;43(6):1023–4. [DOI] [PubMed] [Google Scholar]
- 80.Hoeffel G, Ginhoux F. Ontogeny of Tissue-Resident Macrophages. Front Immunol. 2015;6:486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cantor AB, Orkin SH. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 2002;21(21):3368–76. [DOI] [PubMed] [Google Scholar]
- 82.Rhodes J, Hagen A, Hsu K, Deng M, Liu TX, Look AT, et al. Interplay of pu.1 and gata1 determines myelo-erythroid progenitor cell fate in zebrafish. Dev Cell. 2005;8(1):97–108. [DOI] [PubMed] [Google Scholar]
- 83.Nerlov C, Querfurth E, Kulessa H, Graf T. GATA-1 interacts with the myeloid PU.1 transcription factor and represses PU.1-dependent transcription. Blood. 2000;95(8):2543–51. [PubMed] [Google Scholar]
- 84.Rekhtman N, Radparvar F, Evans T, Skoultchi AI. Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev. 1999;13(11):1398–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stopka T, Amanatullah DF, Papetti M, Skoultchi AI. PU.1 inhibits the erythroid program by binding to GATA-1 on DNA and creating a repressive chromatin structure. EMBO J. 2005;24(21):3712–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang P, Behre G, Pan J, Iwama A, Wara-Aswapati N, Radomska HS, et al. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proc Natl Acad Sci U S A. 1999;96(15):8705–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang P, Zhang X, Iwama A, Yu C, Smith KA, Mueller BU, et al. PU.1 inhibits GATA-1 function and erythroid differentiation by blocking GATA-1 DNA binding. Blood. 2000;96(8):2641–8. [PubMed] [Google Scholar]
- 88.Lyons SE, Lawson ND, Lei L, Bennett PE, Weinstein BM, Liu PP. A nonsense mutation in zebrafish gata1 causes the bloodless phenotype in vlad tepes. Proc Natl Acad Sci U S A. 2002;99(8):5454–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tsiftsoglou AS, Vizirianakis IS, Strouboulis J. Erythropoiesis: model systems, molecular regulators, and developmental programs. IUBMB Life. 2009;61(8):800–30. [DOI] [PubMed] [Google Scholar]
- 90.Ferreira R, Ohneda K, Yamamoto M, Philipsen S. GATA1 function, a paradigm for transcription factors in hematopoiesis. Mol Cell Biol. 2005;25(4):1215–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lowry JA, Mackay JP. GATA-1: one protein, many partners. Int J Biochem Cell Biol. 2006;38(1):6–11. [DOI] [PubMed] [Google Scholar]
- 92.Cantor AB, Iwasaki H, Arinobu Y, Moran TB, Shigematsu H, Sullivan MR, et al. Antagonism of FOG-1 and GATA factors in fate choice for the mast cell lineage. J Exp Med. 2008;205(3):611–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kim SI, Bresnick EH. Transcriptional control of erythropoiesis: emerging mechanisms and principles. Oncogene. 2007;26(47):6777–94. [DOI] [PubMed] [Google Scholar]
- 94.Hoang T, Lambert JA, Martin R. SCL/TAL1 in Hematopoiesis and Cellular Reprogramming. Curr Top Dev Biol. 2016;118:163–204. [DOI] [PubMed] [Google Scholar]
- 95.Amigo JD, Ackermann GE, Cope JJ, Yu M, Cooney JD, Ma D, et al. The role and regulation of friend of GATA-1 (FOG-1) during blood development in the zebrafish. Blood. 2009;114(21):4654–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cantor AB, Orkin SH. Coregulation of GATA factors by the Friend of GATA (FOG) family of multitype zinc finger proteins. Semin Cell Dev Biol. 2005;16(1):117–28. [DOI] [PubMed] [Google Scholar]
- 97.Xue Y, Gao S, Liu F. Genome-wide analysis of the zebrafish Klf family identifies two genes important for erythroid maturation. Dev Biol. 2015;403(2):115–27. [DOI] [PubMed] [Google Scholar]
- 98.Oates AC, Pratt SJ, Vail B, Yan Y, Ho RK, Johnson SL, et al. The zebrafish klf gene family. Blood. 2001;98(6):1792–801. [DOI] [PubMed] [Google Scholar]
- 99.Woo AJ, Moran TB, Schindler YL, Choe SK, Langer NB, Sullivan MR, et al. Identification of ZBP-89 as a novel GATA-1-associated transcription factor involved in megakaryocytic and erythroid development. Mol Cell Biol. 2008;28(8):2675–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ransom DG, Bahary N, Niss K, Traver D, Burns C, Trede NS, et al. The zebrafish moonshine gene encodes transcriptional intermediary factor 1gamma, an essential regulator of hematopoiesis. PLoS Biol. 2004;2(8):E237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Monteiro R, Pouget C, Patient R. The gata1/pu.1 lineage fate paradigm varies between blood populations and is modulated by tif1gamma. EMBO J. 2011;30(6):1093–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Song H, Yan YL, Titus T, He X, Postlethwait JH. The role of stat1b in zebrafish hematopoiesis. Mech Dev. 2011;128(7–10):442–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Davidson AJ, Zon LI. The ‘definitive’ (and ‘primitive’) guide to zebrafish hematopoiesis. Oncogene. 2004;23(43):7233–46. [DOI] [PubMed] [Google Scholar]
- 104.Pillay LM, Forrester AM, Erickson T, Berman JN, Waskiewicz AJ. The Hox cofactors Meis1 and Pbx act upstream of gata1 to regulate primitive hematopoiesis. Dev Biol. 2010;340(2):306–17. [DOI] [PubMed] [Google Scholar]
- 105.Jing L, Zon LI. Zebrafish as a model for normal and malignant hematopoiesis. Dis Model Mech. 2011;4(4):433–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rasighaemi P, Basheer F, Liongue C, Ward AC. Zebrafish as a model for leukemia and other hematopoietic disorders. J Hematol Oncol. 2015;8:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Murayama E, Kissa K, Zapata A, Mordelet E, Briolat V, Lin HF, et al. Tracing hematopoietic precursor migration to successive hematopoietic organs during zebrafish development. Immunity. 2006;25(6):963–75. [DOI] [PubMed] [Google Scholar]
- 108.Lam EY, Hall CJ, Crosier PS, Crosier KE, Flores MV. Live imaging of Runx1 expression in the dorsal aorta tracks the emergence of blood progenitors from endothelial cells. Blood. 2010;116(6):909–14. [DOI] [PubMed] [Google Scholar]
- 109.Weinstein BM, Schier AF, Abdelilah S, Malicki J, Solnica-Krezel L, Stemple DL, et al. Hematopoietic mutations in the zebrafish. Development. 1996;123:303–9. [DOI] [PubMed] [Google Scholar]
- 110.Ransom DG, Haffter P, Odenthal J, Brownlie A, Vogelsang E, Kelsh RN, et al. Characterization of zebrafish mutants with defects in embryonic hematopoiesis. Development. 1996;123:311–9. [DOI] [PubMed] [Google Scholar]
- 111.Trede NS, Ota T, Kawasaki H, Paw BH, Katz T, Demarest B, et al. Zebrafish mutants with disrupted early T-cell and thymus development identified in early pressure screen. Dev Dyn. 2008;237(9):2575–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Teittinen KJ, Gronroos T, Parikka M, Ramet M, Lohi O. The zebrafish as a tool in leukemia research. Leuk Res. 2012;36(9):1082–8. [DOI] [PubMed] [Google Scholar]
- 113.He S, Jing CB, Look AT. Zebrafish models of leukemia. Methods Cell Biol. 2017;138:563–92. [DOI] [PubMed] [Google Scholar]
- 114.Harrison NR, Laroche FJ, Gutierrez A, Feng H. Zebrafish Models of Human Leukemia: Technological Advances and Mechanistic Insights. Adv Exp Med Biol. 2016;916:335–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Deveau AP, Bentley VL, Berman JN. Using zebrafish models of leukemia to streamline drug screening and discovery. Exp Hematol. 2017;45:1–9. [DOI] [PubMed] [Google Scholar]
- 116.Blake T, Adya N, Kim CH, Oates AC, Zon L, Chitnis A, et al. Zebrafish homolog of the leukemia gene CBFB: its expression during embryogenesis and its relationship to scl and gata-1 in hematopoiesis. Blood. 2000;96(13):4178–84. [PubMed] [Google Scholar]
- 117.Zhang Z, Jia H, Zhang Q, Wan Y, Zhou Y, Jia Q, et al. Assessment of hematopoietic failure due to Rpl11 deficiency in a zebrafish model of Diamond-Blackfan anemia by deep sequencing. BMC Genomics. 2013;14:896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang Z, Jia H, Zhang Q, Wan Y, Song B, Jia Q, et al. Transcriptome analysis of Rpl11-deficient zebrafish model of Diamond-Blackfan Anemia. Genom Data. 2014;2:173–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang Y, Ear J, Yang Z, Morimoto K, Zhang B, Lin S. Defects of protein production in erythroid cells revealed in a zebrafish Diamond-Blackfan anemia model for mutation in RPS19. Cell Death Dis. 2014;5:e1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wan Y, Zhang Q, Zhang Z, Song B, Wang X, Zhang Y, et al. Transcriptome analysis reveals a ribosome constituents disorder involved in the RPL5 downregulated zebrafish model of Diamond-Blackfan anemia. BMC Med Genomics. 2016;9:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Titus TA, Yan YL, Wilson C, Starks AM, Frohnmayer JD, Bremiller RA, et al. The Fanconi anemia/BRCA gene network in zebrafish: embryonic expression and comparative genomics. Mutat Res. 2009;668(1–2):117–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Titus TA, Selvig DR, Qin B, Wilson C, Starks AM, Roe BA, et al. The Fanconi anemia gene network is conserved from zebrafish to human. Gene. 2006;371(2):211–23. [DOI] [PubMed] [Google Scholar]
- 123.Taylor AM, Zon LI. Modeling Diamond Blackfan anemia in the zebrafish. Semin Hematol. 2011;48(2):81–8. [DOI] [PubMed] [Google Scholar]
- 124.Gore AV, Athans B, Iben JR, Johnson K, Russanova V, Castranova D, et al. Epigenetic regulation of hematopoiesis by DNA methylation. Elife. 2016;5:e11813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Langenau DM, Traver D, Ferrando AA, Kutok JL, Aster JC, Kanki JP, et al. Myc-induced T cell leukemia in transgenic zebrafish. Science. 2003;299(5608):887–90. [DOI] [PubMed] [Google Scholar]
- 126.Langenau DM, Ferrando AA, Traver D, Kutok JL, Hezel JP, Kanki JP, et al. In vivo tracking of T cell development, ablation, and engraftment in transgenic zebrafish. Proc Natl Acad Sci U S A. 2004;101(19):7369–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mathias JR, Dodd ME, Walters KB, Rhodes J, Kanki JP, Look AT, et al. Live imaging of chronic inflammation caused by mutation of zebrafish Hai1. J Cell Sci. 2007;120(Pt 19):3372–83. [DOI] [PubMed] [Google Scholar]
- 128.Mathias JR, Dodd ME, Walters KB, Yoo SK, Ranheim EA, Huttenlocher A. Characterization of zebrafish larval inflammatory macrophages. Dev Comp Immunol. 2009;33(11):1212–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mathias JR, Walters KB, Huttenlocher A. Neutrophil motility in vivo using zebrafish. Methods Mol Biol. 2009;571:151–66. [DOI] [PubMed] [Google Scholar]
- 130.Walters KB, Green JM, Surfus JC, Yoo SK, Huttenlocher A. Live imaging of neutrophil motility in a zebrafish model of WHIM syndrome. Blood. 2010;116(15):2803–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yoo SK, Huttenlocher A. Spatiotemporal photolabeling of neutrophil trafficking during inflammation in live zebrafish. J Leukoc Biol. 2011;89(5):661–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Renshaw SA, Loynes CA, Trushell DM, Elworthy S, Ingham PW, Whyte MK. A transgenic zebrafish model of neutrophilic inflammation. Blood. 2006;108(13):3976–8. [DOI] [PubMed] [Google Scholar]
- 133.Hall C, Flores MV, Storm T, Crosier K, Crosier P. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Dev Biol. 2007;7:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bertrand JY, Chi NC, Santoso B, Teng S, Stainier DY, Traver D. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature. 2010;464(7285):108–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bresciani E, Carrington B, Wincovitch S, Jones M, Gore AV, Weinstein BM, et al. CBFbeta and RUNX1 are required at 2 different steps during the development of hematopoietic stem cells in zebrafish. Blood. 2014;124(1):70–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kissa K, Herbomel P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature. 2010;464(7285):112–5. [DOI] [PubMed] [Google Scholar]
- 137.Sood R, English MA, Belele CL, Jin H, Bishop K, Haskins R, et al. Development of multilineage adult hematopoiesis in the zebrafish with a runx1 truncation mutation. Blood. 2010;115(14):2806–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhang Y, Jin H, Li L, Qin FX, Wen Z. cMyb regulates hematopoietic stem/progenitor cell mobilization during zebrafish hematopoiesis. Blood. 2011;118(15):4093–101. [DOI] [PubMed] [Google Scholar]
- 139.Soza-Ried C, Hess I, Netuschil N, Schorpp M, Boehm T. Essential role of c-myb in definitive hematopoiesis is evolutionarily conserved. Proc Natl Acad Sci U S A. 2010;107(40):17304–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kim AD, Melick CH, Clements WK, Stachura DL, Distel M, Panakova D, et al. Discrete Notch signaling requirements in the specification of hematopoietic stem cells. EMBO J. 2014;33(20):2363–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bertrand JY, Cisson JL, Stachura DL, Traver D. Notch signaling distinguishes 2 waves of definitive hematopoiesis in the zebrafish embryo. Blood. 2010;115(14):2777–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Burns CE, Traver D, Mayhall E, Shepard JL, Zon LI. Hematopoietic stem cell fate is established by the Notch-Runx pathway. Genes Dev. 2005;19(19):2331–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gering M, Patient R. Hedgehog signaling is required for adult blood stem cell formation in zebrafish embryos. Dev Cell. 2005;8(3):389–400. [DOI] [PubMed] [Google Scholar]
- 144.Lawson ND, Scheer N, Pham VN, Kim CH, Chitnis AB, Campos-Ortega JA, et al. Notch signaling is required for arterial-venous differentiation during embryonic vascular development. Development. 2001;128(19):3675–83. [DOI] [PubMed] [Google Scholar]
- 145.Lin MI, Price EN, Boatman S, Hagedorn EJ, Trompouki E, Satishchandran S, et al. Angiopoietin-like proteins stimulate HSPC development through interaction with notch receptor signaling. Elife. 2015;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Robert-Moreno A, Guiu J, Ruiz-Herguido C, Lopez ME, Ingles-Esteve J, Riera L, et al. Impaired embryonic haematopoiesis yet normal arterial development in the absence of the Notch ligand Jagged1. EMBO J. 2008;27(13):1886–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Clements WK, Kim AD, Ong KG, Moore JC, Lawson ND, Traver D. A somitic Wnt16/Notch pathway specifies haematopoietic stem cells. Nature. 2011;474(7350):220–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Monteiro R, Pinheiro P, Joseph N, Peterkin T, Koth J, Repapi E, et al. Transforming Growth Factor beta Drives Hemogenic Endothelium Programming and the Transition to Hematopoietic Stem Cells. Dev Cell. 2016;38(4):358–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhen F, Lan Y, Yan B, Zhang W, Wen Z. Hemogenic endothelium specification and hematopoietic stem cell maintenance employ distinct Scl isoforms. Development. 2013;140(19):3977–85. [DOI] [PubMed] [Google Scholar]
- 150.Lin HF, Traver D, Zhu H, Dooley K, Paw BH, Zon LI, et al. Analysis of thrombocyte development in CD41-GFP transgenic zebrafish. Blood. 2005;106(12):3803–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Damm EW, Clements WK. Pdgf signalling guides neural crest contribution to the haematopoietic stem cell specification niche. Nat Cell Biol. 2017;19(5):457–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gore AV, Weinstein BM. DNA methylation in hematopoietic development and disease. Exp Hematol. 2016;44(9):783–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Suzuki H, Tokino T, Shinomura Y, Imai K, Toyota M. DNA methylation and cancer pathways in gastrointestinal tumors. Pharmacogenomics. 2008;9(12):1917–28. [DOI] [PubMed] [Google Scholar]
- 154.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9(6):465–76. [DOI] [PubMed] [Google Scholar]
- 155.Zapata A. Ultrastructural study of the teleost fish kidney. Dev Comp Immunol. 1979;3(1):55–65. [DOI] [PubMed] [Google Scholar]
- 156.Prinz M Microglia and monocytes: molecularly defined. Acta Neuropathol. 2014;128(3):317–8. [DOI] [PubMed] [Google Scholar]
- 157.Prinz M, Priller J. Microglia and brain macrophages in the molecular age: from origin to neuropsychiatric disease. Nat Rev Neurosci. 2014;15(5):300–12. [DOI] [PubMed] [Google Scholar]
- 158.Prinz M, Tay TL, Wolf Y, Jung S. Microglia: unique and common features with other tissue macrophages. Acta Neuropathol. 2014;128(3):319–31. [DOI] [PubMed] [Google Scholar]
- 159.Gomez Perdiguero E, Schulz C, Geissmann F. Development and homeostasis of “resident” myeloid cells: the case of the microglia. Glia. 2013;61(1):112–20. [DOI] [PubMed] [Google Scholar]
- 160.Stachura DL, Traver D. Cellular dissection of zebrafish hematopoiesis. Methods Cell Biol. 2011;101:75–110. [DOI] [PubMed] [Google Scholar]
- 161.Rombough P, Drader H. Hemoglobin enhances oxygen uptake in larval zebrafish (Danio rerio) but only under conditions of extreme hypoxia. J Exp Biol. 2009;212(Pt 6):778–84. [DOI] [PubMed] [Google Scholar]
- 162.Deveau AP, Forrester AM, Coombs AJ, Wagner GS, Grabher C, Chute IC, et al. Epigenetic therapy restores normal hematopoiesis in a zebrafish model of NUP98-HOXA9-induced myeloid disease. Leukemia. 2015;29(10):2086–97. [DOI] [PubMed] [Google Scholar]
- 163.Robertson AL, Avagyan S, Gansner JM, Zon LI. Understanding the regulation of vertebrate hematopoiesis and blood disorders - big lessons from a small fish. FEBS Lett. 2016;590(22):4016–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Shafizadeh E, Paw BH, Foott H, Liao EC, Barut BA, Cope JJ, et al. Characterization of zebrafish merlot/chablis as non-mammalian vertebrate models for severe congenital anemia due to protein 4.1 deficiency. Development. 2002;129(18):4359–70. [DOI] [PubMed] [Google Scholar]
- 165.Bruce LJ, Ghosh S, King MJ, Layton DM, Mawby WJ, Stewart GW, et al. Absence of CD47 in protein 4.2-deficient hereditary spherocytosis in man: an interaction between the Rh complex and the band 3 complex. Blood. 2002;100(5):1878–85. [DOI] [PubMed] [Google Scholar]
- 166.Bouhassira EE, Schwartz RS, Yawata Y, Ata K, Kanzaki A, Qiu JJ, et al. An alanine-to-threonine substitution in protein 4.2 cDNA is associated with a Japanese form of hereditary hemolytic anemia (protein 4.2NIPPON). Blood. 1992;79(7):1846–54. [PubMed] [Google Scholar]
- 167.Liao EC, Paw BH, Peters LL, Zapata A, Pratt SJ, Do CP, et al. Hereditary spherocytosis in zebrafish riesling illustrates evolution of erythroid beta-spectrin structure, and function in red cell morphogenesis and membrane stability. Development. 2000;127(23):5123–32. [DOI] [PubMed] [Google Scholar]
- 168.Paw BH, Davidson AJ, Zhou Y, Li R, Pratt SJ, Lee C, et al. Cell-specific mitotic defect and dyserythropoiesis associated with erythroid band 3 deficiency. Nat Genet. 2003;34(1):59–64. [DOI] [PubMed] [Google Scholar]
- 169.O’Brien KA, Farrar JE, Vlachos A, Anderson SM, Tsujiura CA, Lichtenberg J, et al. Molecular convergence in ex vivo models of Diamond-Blackfan anemia. Blood. 2017;129(23):3111–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Danilova N, Bibikova E, Covey TM, Nathanson D, Dimitrova E, Konto Y, et al. The role of the DNA damage response in zebrafish and cellular models of Diamond Blackfan anemia. Dis Model Mech. 2014;7(7):895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lamoril J, Boulechfar S, de Verneuil H, Grandchamp B, Nordmann Y, Deybach JC. Human erythropoietic protoporphyria: two point mutations in the ferrochelatase gene. Biochem Biophys Res Commun. 1991;181(2):594–9. [DOI] [PubMed] [Google Scholar]
- 172.Childs S, Weinstein BM, Mohideen MA, Donohue S, Bonkovsky H, Fishman MC. Zebrafish dracula encodes ferrochelatase and its mutation provides a model for erythropoietic protoporphyria. Curr Biol. 2000;10(16):1001–4. [DOI] [PubMed] [Google Scholar]
- 173.Hasle H. Myelodysplastic and myeloproliferative disorders of childhood. Hematology Am Soc Hematol Educ Program. 2016;2016(1):598–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kwan W, North TE. Netting Novel Regulators of Hematopoiesis and Hematologic Malignancies in Zebrafish. Curr Top Dev Biol. 2017;124:125–60. [DOI] [PubMed] [Google Scholar]
- 175.Yeh JR, Munson KM, Chao YL, Peterson QP, Macrae CA, Peterson RT. AML1-ETO reprograms hematopoietic cell fate by downregulating scl expression. Development. 2008;135(2):401–10. [DOI] [PubMed] [Google Scholar]
- 176.Forrester AM, Grabher C, McBride ER, Boyd ER, Vigerstad MH, Edgar A, et al. NUP98-HOXA9-transgenic zebrafish develop a myeloproliferative neoplasm and provide new insight into mechanisms of myeloid leukaemogenesis. Br J Haematol. 2011;155(2):167–81. [DOI] [PubMed] [Google Scholar]
- 177.Cunningham L, Finckbeiner S, Hyde RK, Southall N, Marugan J, Yedavalli VR, et al. Identification of benzodiazepine Ro5–3335 as an inhibitor of CBF leukemia through quantitative high throughput screen against RUNX1-CBFbeta interaction. Proc Natl Acad Sci U S A. 2012;109(36):14592–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Chen J, Jette C, Kanki JP, Aster JC, Look AT, Griffin JD. NOTCH1-induced T-cell leukemia in transgenic zebrafish. Leukemia. 2007;21(3):462–71. [DOI] [PubMed] [Google Scholar]
- 179.Traver D, Herbomel P, Patton EE, Murphey RD, Yoder JA, Litman GW, et al. The zebrafish as a model organism to study development of the immune system. Adv Immunol. 2003;81:253–330. [PubMed] [Google Scholar]
- 180.Ng CE, Yokomizo T, Yamashita N, Cirovic B, Jin H, Wen Z, et al. A Runx1 intronic enhancer marks hemogenic endothelial cells and hematopoietic stem cells. Stem Cells. 2010;28(10):1869–81. [DOI] [PubMed] [Google Scholar]
- 181.Bajoghli B, Aghaallaei N, Hess I, Rode I, Netuschil N, Tay BH, et al. Evolution of genetic networks underlying the emergence of thymopoiesis in vertebrates. Cell. 2009;138(1):186–97. [DOI] [PubMed] [Google Scholar]
- 182.Jessen JR, Willett CE, Lin S. Artificial chromosome transgenesis reveals long-distance negative regulation of rag1 in zebrafish. Nat Genet. 1999;23(1):15–6. [DOI] [PubMed] [Google Scholar]
- 183.Jessen JR, Jessen TN, Vogel SS, Lin S. Concurrent expression of recombination activating genes 1 and 2 in zebrafish olfactory sensory neurons. Genesis. 2001;29(4):156–62. [DOI] [PubMed] [Google Scholar]
- 184.Wittamer V, Bertrand JY, Gutschow PW, Traver D. Characterization of the mononuclear phagocyte system in zebrafish. Blood. 2011;117(26):7126–35. [DOI] [PubMed] [Google Scholar]
- 185.Mathias JR, Perrin BJ, Liu TX, Kanki J, Look AT, Huttenlocher A. Resolution of inflammation by retrograde chemotaxis of neutrophils in transgenic zebrafish. J Leukoc Biol. 2006;80(6):1281–8. [DOI] [PubMed] [Google Scholar]
- 186.Ellett F, Pase L, Hayman JW, Andrianopoulos A, Lieschke GJ. mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood. 2011;117(4):e49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Brownlie A, Donovan A, Pratt SJ, Paw BH, Oates AC, Brugnara C, et al. Positional cloning of the zebrafish sauternes gene: a model for congenital sideroblastic anaemia. Nat Genet. 1998;20(3):244–50. [DOI] [PubMed] [Google Scholar]
- 188.Wingert RA, Brownlie A, Galloway JL, Dooley K, Fraenkel P, Axe JL, et al. The chianti zebrafish mutant provides a model for erythroid-specific disruption of transferrin receptor 1. Development. 2004;131(24):6225–35. [DOI] [PubMed] [Google Scholar]
- 189.Donovan A, Brownlie A, Zhou Y, Shepard J, Pratt SJ, Moynihan J, et al. Positional cloning of zebrafish ferroportin1 identifies a conserved vertebrate iron exporter. Nature. 2000;403(6771):776–81. [DOI] [PubMed] [Google Scholar]
- 190.Danilova N, Sakamoto KM, Lin S. Ribosomal protein L11 mutation in zebrafish leads to haematopoietic and metabolic defects. Br J Haematol. 2011;152(2):217–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Gjini E, Mansour MR, Sander JD, Moritz N, Nguyen AT, Kesarsing M, et al. A zebrafish model of myelodysplastic syndrome produced through tet2 genomic editing. Mol Cell Biol. 2015;35(5):789–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Craven SE, French D, Ye W, de Sauvage F, Rosenthal A. Loss of Hspa9b in zebrafish recapitulates the ineffective hematopoiesis of the myelodysplastic syndrome. Blood. 2005;105(9):3528–34. [DOI] [PubMed] [Google Scholar]
- 193.Sabaawy HE, Azuma M, Embree LJ, Tsai HJ, Starost MF, Hickstein DD. TEL-AML1 transgenic zebrafish model of precursor B cell acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2006;103(41):15166–71. [DOI] [PMC free article] [PubMed] [Google Scholar]