Abstract
Background:
Most chemicals in commerce have not been evaluated for their carcinogenic potential. The de facto gold-standard approach to carcinogen testing adopts the 2-y rodent bioassay, a time-consuming and costly procedure. High-throughput in vitro assays are a promising alternative for addressing the limitations in carcinogen screening.
Objectives:
We developed a screening process for predicting chemical carcinogenicity and genotoxicity and characterizing modes of actions (MoAs) using in vitro gene expression assays.
Methods:
We generated a large toxicogenomics resource comprising expression profiles corresponding to 330 chemicals profiled in HepG2 (human hepatocellular carcinoma cell line) at multiple doses and replicates. Predictive models of carcinogenicity and genotoxicity were built using a random forest classifier. Differential pathway enrichment analysis was performed to identify pathways associated with carcinogen exposure. Signatures of carcinogenicity and genotoxicity were compared with external sources, including Drugmatrix and the Connectivity Map.
Results:
Among profiles with sufficient bioactivity, our classifiers achieved 72.2% Area Under the ROC Curve (AUC) for predicting carcinogenicity and 82.3% AUC for predicting genotoxicity. Chemical bioactivity, as measured by the strength and reproducibility of the transcriptional response, was not significantly associated with long-term carcinogenicity in doses up to . However, sufficient bioactivity was necessary for a chemical to be used for prediction of carcinogenicity. Pathway enrichment analysis revealed pathways consistent with known pathways that drive cancer, including DNA damage and repair. The data is available at https://clue.io/CRCGN_ABC, and a portal for query and visualization of the results is accessible at https://carcinogenome.org.
Discussion:
We demonstrated an in vitro screening approach using gene expression profiling to predict carcinogenicity and infer MoAs of chemical perturbations. https://doi.org/10.1289/EHP3986
Introduction
Despite significant investments into cancer research over the last decades, million new cancer cases and 600,000 cancers deaths were estimated in the United States in 2017 alone (American Cancer Society 2017). Of these, 90–95% are not attributable to known heritable genetic factors, thus making environmental exposures a major suspect in driving cancer (Anand et al. 2008), notwithstanding recent studies pointing to the rate of cell replications as an important determinant of cancer development among different tissue types (Tomasetti and Vogelstein 2015; Tomasetti et al. 2017). Most research aimed at assessing cancer hazard from chemical exposure has primarily relied on epidemiological studies of past human exposures to suspected carcinogens in cancer clusters and on carcinogen screening based on the 2-y rodent-based bioassay. Epidemiological studies rely on observational data, and as such, it is often difficult to rule out the possibility of spurious associations due to confounding effects. They also require that exposure to a suspected carcinogen is documentable. Even when the nature of the chemical exposure and the exposure dose is known, epidemiological studies require long follow-up periods; hence, they are not appropriate for the evaluation of new chemicals on the market. Similarly, the 2-y rodent bioassay, the gold standard for carcinogen testing, is time-consuming and requires up to and animals per compound (Hodgson 2004; Meister 2005; Waters et al. 2010). As a result, of the chemicals registered in the Toxic Substances Control Act Chemical Substance Inventory have been tested by this approach (Bucher and Portier 2004; Gold et al. 2005; Huff et al. 2008).
High-throughput transcriptional profiles from short-term chemical exposures have proven useful for predicting long-term carcinogenicity and for capturing multiple biological modes of actions (MoAs) of long-term carcinogenicity. Many studies have explored the use of high-throughput transcriptional profiling in rodent models (Eichner et al. 2013; Ellinger-Ziegelbauer et al. 2008; Fielden et al. 2007; Gusenleitner et al. 2014; Kossler et al. 2015; Nie et al. 2006; Uehara et al. 2011). However, questions remain about the relevance of rodent models for characterizing human carcinogenicity, and most importantly, they are still excessively time-consuming and expensive for large-scale testing. In vitro–based screens would help address the time and cost constraints of carcinogen testing through automated high-throughput plating, exposure treatment, and assaying. The U.S. Environmental Protection Agency’s Toxcast (https://www.epa.gov/chemical-research/exploring-toxcast-data-downloadable-data) (Judson et al. 2010; Richard et al. 2016) and Tox21 (https://tox21.gov/) initiatives (Schmidt 2009; Tice et al. 2013) have used various reporter assays to characterize adverse effects across thousands of in vitro chemical exposures. However, while these efforts use high-throughput techniques with carefully selected gene, pathway, and adverse response–centric end points, the number of assays and the diversity of end points are limited. For instance, ToxCast used 624 in vitro end points mapped to 315 genes in Phase I (Judson et al. 2010) and an additional new end points in Phase II (Richard et al. 2016). Studies utilizing this data for the assessment of chemical carcinogenicity have emphasized the need to expand the assay set to better characterize diverse MoAs of certain carcinogens (Kleinstreuer et al. 2013). mRNA profiling, by assaying the entire transcriptome, or a large portion of it, represents a promising solution to this need by providing an agnostic view of which genes and pathways are relevant to chemical-induced carcinogenesis.
Given the technological advances in gene expression profiling and the development of cost-effective sequencing platforms, opportunities arise for their use in large-scale toxicological screenings (Reed et al. 2019; Zhang et al. 2017). One such solution is the Luminex 1,000 (L1000) platform (https://www.luminexcorp.com/) (Peck et al. 2006), a low-cost, high-throughput bead-based platform that measures the expression of landmark genes and infers the remaining genes in the transcriptome by imputation. This platform was used in the creation of the Connectivity Map (CMap) (Subramanian et al. 2017), which now includes 1.3 million perturbation profiles of drugs and small molecules and has been instrumental in the discovery of small-molecule MoAs. Due to its cost-effectiveness and appropriateness for large-scale perturbation screening, we adopted it for the profiling of chemical carcinogens.
We applied the L1000 platform to study the effects of chemical perturbations of previously validated rat liver carcinogens and noncarcinogens in HepG2 (human hepatocellular carcinoma) cell lines. The central hypothesis underlying our study design was that the long-term carcinogenicity of chemicals can be accurately predicted from gene expression profiles of short-term in vitro models. Our approach used machine-learning techniques to build predictive models of the long-term carcinogenicity of chemicals based on L1000-derived gene expression profiles of human cell lines exposed to the studied chemicals. Furthermore, we annotated the in vitro–derived gene signatures by performing pathway enrichment of carcinogens vs. noncarcinogens to identify MoAs associated with chemical-induced carcinogenesis. Signatures derived from this study were also compared to external gene signatures and chemical annotations from knowledge bases such as Drugmatrix (Ganter et al. 2006, 2005), CMap, and Tox21, to verify the consistency of results and expand the interpretation of findings. An overview of our experimental design and analysis aims is presented in Figure S1.
Methods
Chemical Selection and Annotation
In the chemical selection process, we prioritized chemicals with long-term rodent liver carcinogenicity annotation for inclusion in this experiment. Long-term carcinogenicity annotations were derived from the Carcinogenic Potency Database (CPDB) (Fitzpatrick 2008). Additional chemicals without carcinogenicity annotation were included on the basis of interest to the Superfund Research Program (environmental toxicants), presence in controversial commercial products (included for predictive purposes), and evidence of binding to the aryl hydrocarbon receptor (AhR), as the AhR is an important mediator of xenobiotics, including carcinogens. A complete list of chemicals and their annotations is provided in Excel Table S1. For CPDB annotations, the final carcinogenicity labels denote “” if carcinogenic in rat liver (female or male) or “” if noncarcinogenic in both rat and mouse (in female and male) across all tested organs in the CPDB. Genotoxicity labels denote “” if mutagenic or weakly mutagenic in the Salmonella assay, and “” otherwise. In total, 330 unique chemicals were used in the analysis, including 128 carcinogens, 168 noncarcinogens, 100 genotoxicants, and 161 nongenotoxicants.
Chemical Procurement and Data Generation
Chemicals were procured from the Tox21 library of the National Toxicology Program (NTP) when available, or from Sigma-Aldrich otherwise. Compound purity and identity were confirmed by UPLC-MS, an Acquity Classic UPLC coupled with an SQ mass spectrometer (Waters). Purity was measured by UV absorbance at or by evaporative light scattering. Identity was determined on a single quadrupole mass spectrometer by positive and/or negative electrospray ionization. Mobile phase A consisted of either 0.1% ammonium hydroxide or 0.05% trifluoroacetic acid in water, while mobile phase B consisted of either 0.1% ammonium hydroxide or 0.06% trifluoroacetic acid in acetonitrile. The gradient ran from 5% to 95% mobile phase B over 2.65 min at . An Acquity BEH C18 (Waters), , column was used with column temperature maintained at 65ºC. Compounds were dissolved in dimethylsulfoxide (DMSO) at a nominal concentration of , and of this solution was injected.
Detailed cell culture, plating, treatment and lysis protocols are described in https://assets.clue.io/resources/sop-cell.pdf (Subramanian et al. 2017). Briefly, HepG2 (ATCC® HB-8,065™) was used with medium RPMI1640 (10040CV; Mediatech) supplemented with 10% v/v fetal bovine serum (F4135; Sigma-Aldrich), penicillin-streptomycin-glutamine (Invitrogen 10378016), and incubated at humidified 5% atmosphere at 37°C. Cell cultures were plated with 4,000 cells ( of growth medium) per well on 384-well plates (3707; Corning) and incubated for 24 h before treatment. Cells were treated with of 1:100 diluted stock compound plates to final volume of and incubated for 24 h before lysis.
Each chemical perturbation was administered at six doses in triplicate wells per dose and chemical combination, starting from maximum dose ( stock diluted 1:1,000) for NTP chemicals and for chemicals procured from Sigma-Aldrich, in series of twofold dilutions. The sole exception to the standard dosage was 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), which had a starting dose of due to its extreme potency. The vehicle control used was DMSO with final dose of . Four positive controls were used (vorinostat, geldanamycin, mitoxantrone, withaferin-a) each in final doses of 10, 3.33, and , respectively. Four wells on each plate were reserved for L1000 pipeline assay controls. These include A01: bead-only control (negative control), B01: POSAMP control (hybridization/staining positive control), A02, and B02 (reference RNA control).
For cell lysis, of medium was aspirated, and TCL Lysis Buffer (1031576; Qiagen) was added. Plates were sealed and maintained at room temperature for 30 min and frozen in a freezer. Following treatment and lysis, the gene expression of the HepG2 cells was profiled using the L1000 platform, a high-throughput assay that measures the expression of landmark genes and computationally infers the expression of nonmeasured transcripts.
Following cell lysis, the exact L1000 protocol was followed as described in Subramanian et al. 2017. Briefly, each transcript of interest was targeted with upstream and downstream probe pairs with a 20-nucleotide gene-specific region, a unique identifying barcode, and a universal primer site. Gene-specific sequences were detected by coupling barcodes to Luminex (https://www.luminexcorp.com/) beads followed by ligation-mediated amplification and hybridization of amplicon to bead. The L1000 assay measured transcripts using 500 unique bead colors in a process called tag duo detection and peak deconvolution. Invariant genes were used as controls for data quality control and normalization.
For each perturbation and landmark gene, we computed the change in gene expression following the perturbation using a moderated z-score procedure as described in the CMap–L1000 workflow. Differential expression values were calculated as moderated z-scores for each landmark gene and each unique perturbation (chemical and dose combination) perturbation, collapsed to a single value across replicates (Subramanian et al. 2017).
Assessing the Transcriptional Strength of a Perturbation
We used the transcriptional activity score (TAS) as a summary measure of the impact of a chemical perturbation on landmark gene expression. TAS integrates signature strength, defined as the number of genes up-regulated or down-regulated by a particular perturbation above a given moderated z-score threshold, and replicate correlation, a measurement of similarity among triplicate profiles corresponding to the same perturbation (unique combination of chemical, dose, cell line, time). Formally, TAS is quantified as the geometric mean of the signature strength () and the replicate correlation (CCq75) in Equation 1. is defined as the number of landmark genes (referred to as card) with in Equation 2. ModZ is defined as the 978-element vector of replicate collapsed z-scores of landmark genes, and nrep is the number of replicates in Equation 3. CCq75 is the 75th percentile of the Spearman’s correlation between replicates in landmark space.
| (1) |
| (2) |
| (3) |
TAS was calculated for each aggregated profile (one unique score per chemical and dose combination). This metric takes value in the [0,1] range, with higher values of TAS taken to represent a higher level of chemical bioactivity.
Statistical Tests for Comparison of Transcriptional Activity Score across Profiles
We tested for the difference in TAS values among adjacent dose groups using a one-tailed Wilcoxon Signed-rank test (paired difference test), with the pairing determined by the unique chemical IDs to determine the statistical significance of strictly increased TAS levels between adjacent and increasing dose groups.
We next tested for difference in TAS between chemicals. In particular, for each dose rank, two-group comparisons of TAS scores between carcinogens and noncarcinogens, and between genotoxicants and nongenotoxicants, were conducted using one-tailed unpaired two-sample Wilcoxon test to determine the presence and significance of increased TAS for the carcinogenic compared to noncarcinogenic group, or for the genotoxic compared to nongenotoxic group.
Equivalent in Vitro Dose (Cmax) Estimation and Association with Transcriptional Activity Score
Finding the relationship between in vitro gene expression responses and adverse phenotypes in vivo is an important goal of this study. To this end, we assessed the relationship between in vitro transcriptional bioactivity (TAS) and corresponding in vivo dose used in the rodent bioassay from which carcinogenicity labels were derived. Using a toxicokinetic model (Pearce et al. 2017), we estimated the equivalent in vitro dose (Cmax) corresponding to the in vivo dose tested in the rat bioassay. Cmax values, maximum plasma concentrations, were estimated using a three-compartment model in the R package httk (version 1.8; R Project) (Pearce et al. 2017). For carcinogenic compounds, these values were derived from the CPDB-reported median toxic dose (TD50) administered in rats. For noncarcinogenicity compounds, Cmax values were derived from the CPDB-reported maximum dose administered in rats. Chemicals with missing TD50 (if carcinogenic) or maximum dose (if noncarcinogenic) were omitted from this analysis. It was assumed that dosing was once per day for 365 d. While these Cmax values were not used in the in vitro dosing scheme, they can be used in the interpretation of the aberrant behavior of some of our in vitro profiles.
To determine the association between TAS, in vivo carcinogenicity, and Cmax, we used the following linear regression model:
| (4) |
where TAS denotes the mean TAS for each chemical (across six doses), and CARC denotes the carcinogenicity status of the chemical in the rodent bioassay. We tested for significance of the coefficients , , and under the null hypotheses of zero-valued coefficients (no effect).
Supervised Learning for Prediction of Carcinogenicity and Genotoxicity
To build classifiers for the prediction of carcinogenicity and genotoxicity, we used the moderated z-scores of landmark genes as predictive features. The random forest classifier was used, as implemented in the R package caret (Kuhn 2008). The performance of the classifier was evaluated using a resampling scheme consisting of 25 random repeats of training on 70% of the samples and testing on the remaining 30%. The training and test set split was performed at the chemical level, so that all replicates of each chemical were only included either in the train or the test set, to avoid information leakage (overfitting). To assess the effect of chemicals’ bioactivity on the performance of the classifier, the evaluation was repeated on different subsets of profiles corresponding to different TAS thresholds (all profiles, , , ). Area under the receiver operating characteristic curve (AUC) was used for the assessment of a classifier performance, as it is a well-established metric that captures the trade-off between sensitivity and specificity across multiple thresholds.
We derived the top predictive features of the classifiers in the space of landmark genes using the variable importance metric. Variable importance was measured by the mean decrease in Gini index (“MeanDecreaseGini”) as defined in the function “importance” in the R package randomForest (Liaw and Wiener 2002).
Final predictions of carcinogenicity and genotoxicity were made using leave-one-(chemical)-out cross-validation (CV); that is, at each CV iteration, a single chemical’s profiles across multiple doses are left out, and a classifier is trained based on all remaining chemicals, then applied to the prediction of the left-out chemical’s profiles. This procedure was repeated with each of the TAS subsets.
Deriving Pathway Signatures of Carcinogenicity
We derived pathway activity scores using the R Bioconductor GSVA (Gene Set Variation Analysis) package (Hänzelmann et al. 2013). GSVA is a competitive test of gene set enrichment that takes as input a gene-by-sample expression matrix and generates a gene set–by-sample enrichment score matrix, with its entries representing the pathway enrichment of each sample with respect to each of a user-specified list of gene sets. Pathway enrichment scores were calculated for pathways in the MsigDB C2 Reactome pathway compendium (Croft et al. 2014; Fabregat et al. 2016; Liberzon et al. 2011). The gene set–projected matrix was then used as input for differential analysis with respect to sample phenotype labels (carcinogenicity or genotoxicity) using the R Bioconductor package limma (Ritchie et al. 2015; Smyth 2005) to identify pathways with differences in activity levels between chemical groups. This differential analysis was repeated from data inputs with various TAS thresholding (, 0.2, 0.3, 0.4). One-sided p-values consistent with the direction of change in pathway activity scores were estimated. The p-values across analyses from multiple TAS subsets were combined using the Fisher's method and adjusted for multiple hypothesis testing using false discovery rate (FDR) procedure (Benjamini and Hochberg 1995).
Comparison to Drugmatrix Signatures
Using gene set enrichment analysis (GSEA) (Subramanian et al. 2005), we compared how well our profiles recapitulated external signatures of carcinogenicity and genotoxicity extracted from the NTP Drugmatrix database (Ganter et al. 2006). The Drugmatrix is a compendium of microarray profiles of short-term chemical exposures in intact rat organs (liver samples used only) and in cell cultures (primary rat hepatocytes). The Drugmatrix-derived signatures were defined as the lists of genes in the Drugmatrix significantly associated with long-term carcinogenicity and genotoxicity. Data processing of the Drugmatrix data was consistent with methods described in Gusenleitner et al. (2014). Gene features were mapped from rat Ensembl (Zerbino et al. 2018) gene identifiers to human gene symbols using Biomart (Durinck et al. 2005). Differential expression analysis was conducted using limma (Ritchie et al. 2015; Smyth 2005) to identify markers of carcinogenicity and genotoxicity after correcting for the effect of dose and duration of exposure. For each comparison, a list of significant genes was derived using an FDR cutoff of 0.01 and absolute value of log fold change of 0.2, up to a maximum of 300 genes as ranked by FDR. Signatures of carcinogenicity and genotoxicity (direction sensitive: up-regulated/down-regulated) were derived for three Drugmatrix subsets: liver profiles, cell culture profiles, and low-dose cell culture profiles (), the latter consistent with the range of doses used in our experiment. The detailed gene lists included in the Drugmatrix signatures are documented in Excel Table S2. These gene signatures were tested for enrichment against our L1000 profiles in various subsets (, 0.2, 0.3, 0.4), using the binary phenotypes of carcinogenicity and genotoxicity and the GSEA method, with empirical p-values estimated based on 10,000 gene set permutations.
Comparison with Connectivity Map Signatures
We performed a systematic comparison of our signatures to those in the CMap database. To this end, we computed the connectivity score, a measure of similarity, between pairs of signatures, in this case, between each of our signatures and each of the perturbation signatures in the CMap, which comprises profiles corresponding to 19,811 drugs and small molecules, and 5,075 molecular (gene-specific knockdown and overexpression) perturbations across 3 to 77 cell lines (Subramanian et al. 2017). The connectivity scores were expressed as percentile values in the [, 100] range, wherein a score of 100 represented maximum signature overlap, represented maximum signature reversal, and 0 represented lack of concordance between signatures in either direction. Connectivity scores were computed with respect to both individual CMap perturbagens, and perturbagen classes (PCLs), defined as sets of perturbagens with similar MoAs or gene target annotations. Next, we performed differential connectivity analysis with respect to our chemical groups (carcinogens vs. noncarcinogens, genotoxicants vs. nongenotoxicants) using a one-tailed Wilcoxon rank-sum test to test for presence of increased connectivity in the positive class (carcinogenic or genotoxic). These tests were repeated for each TAS-based subset of our data, and FDR values were calculated. A minimum mean connectivity score of 60 for the positive class was used to filter out differential connectivity hits with low base connectivity scores.
Investigation Of Aryl Hydrocarbon Receptor Activation in L1000 Profiles
To examine the behavior of AhR-related chemicals included in the study, we tested whether these chemicals exhibit enriched activity of AhR-related gene sets compiled from independent sources. Lists of chemicals with known AhR activity were identified using multiple AhR-related Tox 21 reporter assays extracted from the tool Tox21 Enricher (Hur et al. 2018), or using custom chemical annotation with expert knowledge (referenced as “Sherr_AHR_agonist”). Lists of AhR target genes were compiled from literature, as annotated in Excel Table S3.
A one-directional weighted Kolmogorov-Smirnov (KS) test was performed to test for the enrichment of “AhR-positive” samples (profiles corresponding to AhR-related chemicals) among the top-ranked profiles sorted by descending AhR gene set activity scores. The activity scores represent the median scores across four individual AhR gene set scores calculated using GSVA.
Profiles corresponding to AhR-related chemicals in the list “Sherr_AHR_agonist” were clustered using the similarity matrix derived from the connectivity scores of the selected profiles (see previous section for the calculation of connectivity scores).
Statistical Reporting
All statements indicating significance are based on threshold of multiple hypothesis corrected , unless otherwise specified.
Results
Transcriptional Activity Score Analysis and Chemical Bioactivity
We used the TAS as a proxy for chemical bioactivity. Subsequent analyses were based on subsets of profiles at different TAS thresholds (, 0.2, 0.3, 0.4). is the standard cutoff for sufficient bioactivity adopted by the CMap–L1000 workflow (Subramanian et al. 2017), while and represent more stringent thresholds we used to assess the effect of increasing bioactivity on downstream analysis, such as classification and gene set enrichment. While the majority of our profiles showed low transcriptional bioactivity, a substantial percent of profiles achieved sufficient TAS. Among 330 chemicals represented across 1,972 replicated collapsed profiles, 133 chemicals (40.3%) achieved in at least one dose, 89 chemicals (26.97%) achieved , and 63 chemicals (19.09%) achieved .
The Effect of Chemical Dose on Transcriptional Bioactivity
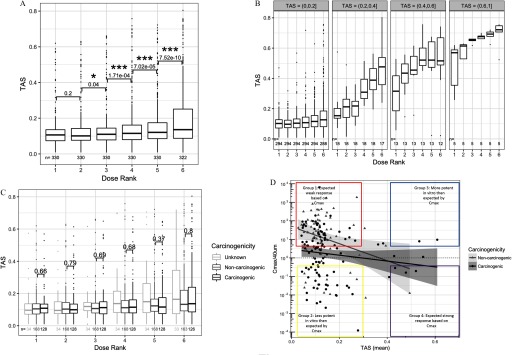
We performed statistical tests to compare TAS of adjacent dose groups and to evaluate how bioactivity is affected by dose. Statistically significantly higher TAS were found when comparing dose rank 3 with rank 2 (), rank 4 with 3, rank 5 with 4, and rank 6 with 5 () (Figure 1A). The consistent significance of TAS differences between adjacent dose groups implies that increasing the dose is effective at increasing the transcriptional bioactivity of profiles, with the maximum dose used in this experiment yielding the highest range of TAS scores. When binned by TAS range (Figure 1B), the monotonically increasing dose response of TAS was apparent across all bins and stronger for higher TAS ranges.
Figure 1.
Box plot of transcriptional activity scores (TAS) by sample subsets. (A) Box plot of TAS distributions for each dose level (; ). Numeric labels indicate the significance of paired one-sided two-group TAS comparison between adjacent dose groups, adjusted for multiple comparisons across doses using the false discovery rate method (FDR) (* ; *** ) (see “Methods” section). (B) Box plot of TAS distribution for each dose level, binned by TAS subsets. (C) Distribution of TAS grouped by chemical carcinogenicity within each dose level. p-Values indicate the significance of unpaired one-sided two-group TAS comparison between TAS of carcinogenic chemicals and TAS of noncarcinogenic chemicals within each dose group. (D) Scatterplot of mean TAS per chemical and the ratio of equivalent in vitro dose (Cmax) over maximum in vitro dose () (see “Methods” section for Cmax calculation). Box plots in Panels A, B, and C have the following specifications: the lower, middle, upper hinges corresponding to the 25th, 50th (median), and 75th percentiles, respectively; the upper and lower whiskers extend to the smaller and largest value at most 1.5 × IQR (interquartile range) from the hinge, and data points beyond the whiskers are represented as dots.
The Effect of Carcinogenicity and Genotoxicity on Transcriptional Bioactivity
Next, we evaluated whether the level of a chemical bioactivity as captured by TAS had any association with that chemical’s long-term carcinogenicity or genotoxicity. Remarkably, carcinogenicity showed no effect on TAS in all dose groups (Figure 1C). On the other hand, genotoxicity showed a marginally significant effect on TAS among profiles with dose rank 1 (lowest dose group) and dose rank 6 (highest dose group), where genotoxic chemicals had nominally significantly higher TAS compared to nongenotoxic chemicals (), although following multiple hypothesis testing (FDR method), no groups showed significance at (Figure S2). To discern possible modeling bias due to differing maximum dose depending on the source of chemical procurement, we repeated the test for the effect of carcinogenicity and genotoxicity on TAS in all dose groups in each of the two chemical groups separated by the procurement sources: Group 1: Sigma-Aldrich chemicals with maximum dose of (Figure S3A) and Group 2: NTP chemicals with maximum dose at (Figure S3B). Confirming the results of the bulk analysis in Figure 1C, carcinogenicity had no association with TAS in all dose groups for all dose ranks in the NTP group, as well as in the Sigma-Aldrich group. In the latter group, carcinogens had nominally significantly lower TAS compared to noncarcinogens in dose ranks 2 and 3 (), but these values became nonsignificant at the (multiple hypothesis corrected) FDR ( for dose rank 2, for dose rank 3).
Comparison of in Vivo Rat Bioassay Dosage with in Vitro Bioactivity
The lack of association between TAS and carcinogenicity motivated us to further investigate the relationship between the L1000 doses and the in vivo doses used in the rodent bioassay. To this end, we tested the association between in vitro bioactivity (TAS) and the estimated equivalent in vitro dose, Cmax (see “Methods” section), where Cmax represents the estimated in vitro dose corresponding to the in vivo dose tested in the rat bioassay. Cmax estimates could be calculated for 183 of the 330 chemicals included in our screen. The mean TAS of profiles for each chemical were plotted against the same chemical's (the ratio of estimated equivalent dose to the max in vitro dose) (Figure 1D).
We tested for the association between TAS, in vivo carcinogenicity, and Cmax, based on a linear model (Equation 4), and we found significant effects on Cmax of TAS (; ), and CARC (; ), and nonsignificant effect of the interaction of TAS and CARC (; ) (see “Methods” section).
As expected, TAS was negatively associated with Cmax. In other words, chemicals that required a low equivalent dose to elicit a carcinogenic response in the rodent bioassay tended to be more transcriptionally active in the in vitro assay. On the other hand, there were exceptions, as carcinogenic chemicals with low TAS and noncarcinogenic chemicals with high TAS were observed and can be explained by several unique pharmacokinetic properties.
We annotated the carcinogenic chemicals with low TAS based on their structural group membership, in vivo dose requirement for carcinogenicity labeling, and requirements for metabolic activation in HepG2. Carcinogenic chemicals with low TAS tended to fall in one or more of the following categories: a) small nitrosamines and other alkylating agents that form DNA adducts but are not adequately recognized by the DNA repair machinery (enriched in Group 2, Figure 1D), b) require bioactivation by CYP2E1 and other p450s that are not present at high levels in HepG2 cell culture (also enriched in Group2, Figure 1D), or c) require high equivalent in vitro dose (Cmax) to be carcinogenic, thus likely underdosed in our in vitro assay (enriched in Group 1 in Figure 1D).
Among noncarcinogenic chemicals with high TAS, we generally noted lower overall doses used in the rodent bioassays due to dose-limiting toxicity or early deaths at higher doses in the cancer bioassay, e.g., cyclosporin A (immune suppression and kidney toxicity) (Ryffel 1992), pyrimethamine, rhodamine 6G and rotenone (bone marrow suppression) (Abdo et al. 1988; National Toxicology Program 1978, 1989), and hexachlorocyclopentadiene (point of contact, pulmonary toxicity) (National Toxicology Program 1994). Thus, if higher doses were tolerated in rodent bioassays, it is possible that some of these chemicals would elicit a carcinogenic response in liver.
The Effect of Transcriptional Bioactivity on Prediction of Carcinogenicity and Genotoxicity
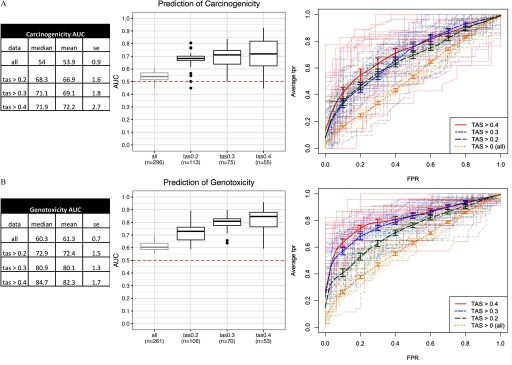
While chemical bioactivity level was not associated with long-term carcinogenicity, the most relevant question was whether a chemical’s bioactivity affected the ability of its expression profile to be predictive of carcinogenicity (and genotoxicity). To answer this question, we built multiple classifiers based on profiles with TAS values within various ranges, and used a random resampling scheme to assess their prediction performance. Datasets corresponding to different TAS ranges were randomly split into train (70%) and test (30%) sets multiple times (), classifiers were built on the train sets, and predictions were made on the test sets. The average area under the ROC curve (AUC), sensitivity, and specificity were then estimated over the 25 random resamples. The prediction AUC improved with higher stringencies of TAS (Figure 2). We achieved the highest predictive accuracy within the most stringent TAS subset (), with [] AUC for prediction of carcinogenicity (Figure 2A), and AUC for prediction of genotoxicity (Figure 2B). In addition to the AUC, we report the sensitivity (true positive rate) and specificity (true negative rate) at the cutoff of 0.5 in Figure S4. Higher specificities were observed at the expense of lower sensitivities in most TAS groups for both classifiers. This outcome is desirable for a preliminary screening strategy in which a higher false-positive rate can be tolerated.
Figure 2.
Performance of classifiers in predictive models of (A) carcinogenicity, and (B) genotoxicity. From left to right: a) Summary statistics tables of area under the ROC curve (AUC) for each transcriptional activity score (TAS) subsets; data represented are the median, mean, and SE (standard error) of the AUC scores; and b) box plots of AUC across resamples () for each TAS subset with the lower, middle, and upper hinges corresponding to the 25th, 50th (median), and 75th percentiles, respectively, the upper and lower whiskers extending to the smaller and largest value at most 1.5 × IQR (interquartile range) from the hinge, and data points beyond the whiskers represented as dots. Dotted line at 0.5 represents the expected AUC of a random classifier. Labels in each TAS group (“”) represent the number of unique chemicals in the model training and validation step. c) Receiver operating characteristic (ROC) curves [false positive rate (FPR) vs. average true positive rate (TPR)]. Thick lines represent vertical averaging of ROC curves across resamples in each TAS group shown with bars denoting the standard errors. Thin, semitransparent lines represent ROC curves of individual resamples in each TAS group.
Gene Markers for Prediction of Carcinogenicity and Genotoxicity
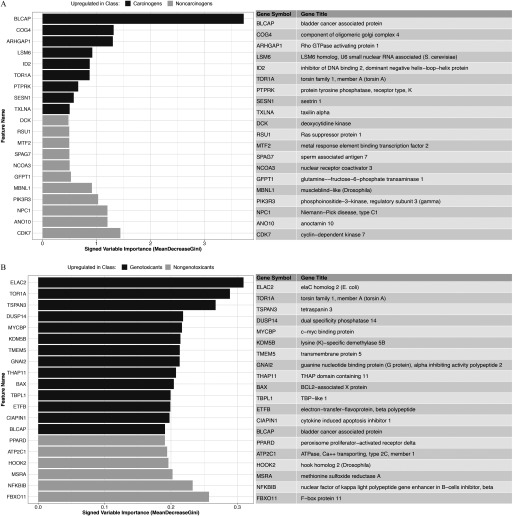
Final predictive models of carcinogenicity, genotoxicity, and genotoxicity within carcinogens were built using the entire set of profiles with . Landmark genes were ranked by variable importance (Excel Table S4), and the top 20 genes for each model were reported in Figure 3. In the carcinogenicity prediction model, top genes included BLCAP, an apoptosis-inducing gene, and SESN1, a target of p53 in response to DNA damage and oxidative stress (Figure 3A). Among the top 20 landmark genes for prediction of genotoxicity were pro-apoptotic regulators such as BLCAP and BAX (Figure 3B). BAX is regulated by p53 and has been shown to be involved in p53-mediated apoptosis, a hallmark of DNA damage response to genotoxic chemical exposure. Of note, the absolute magnitude of the variable importance of top markers is model dependent—thus not comparable between models—and is not informative about the comparative performance of different classifiers.
Figure 3.
Top 20 landmark gene features for prediction of (A) carcinogenicity, and (B) genotoxicity as ranked by variable importance (mean decrease in Gini index) in the predictive models of transcriptional activity scores (TAS) subset.
The markers in Figure 3 were the most predictive features of carcinogenicity and genotoxicity in the restricted space of the L1000 landmark genes, and as such, are not necessarily the most relevant to define chemicals’ MoAs. For a more thorough MoA analysis, see the “Pathway Enrichment Analysis to Characterize Modes of Actions of Carcinogenicity and Genotoxicity” section, where gene set scores were derived from the expression of all genes, including the L1000-inferred ones.
Final Predictions of Carcinogenicity and Genotoxicity in Bioactive Profiles
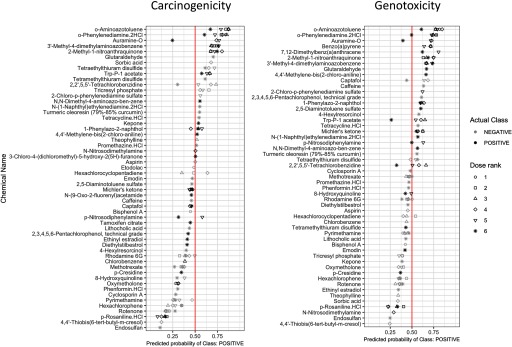
Final predictions of carcinogenicity and genotoxicity were made using a leave-one-chemical-out CV scheme, in which predictive models were trained based on all but one chemical, and predictions were made on the profiles of the left-out chemical (see “Methods” section). This procedure was repeated for all unique chemicals in profiles with to derive probability measurements of the profile being “Positive” for either carcinogenicity or genotoxicity (see “Methods” section) using a probability threshold of 0.5. Prediction probabilities for carcinogenicity and genotoxicity were reported along with the true class labels denoted by the dot colors (Figure 4). From this representation, we observed that predictions tended to be consistent across profiles of varying doses of the same chemical. Several exceptions existed in chemicals whose prediction probabilities were close to 0.5. In addition, prediction probabilities monotonically increasing as a function of dose were observed for some compounds, e.g., 3'-methyl-4-dimethylaminoazobenzene showed increased probability of genotoxicity prediction with increasing dose. However, this pattern did not generalize to all chemicals. Detailed predictions on profiles with are summarized in Excel Table S5.
Figure 4.
Dot plot of probabilities of predicted classes for hold-out chemicals in the transcriptional activity score (TAS) subset. Point outline colors represent actual class labels (carcinogenic vs. noncarcinogenic, genotoxic vs. nongenotoxic). Point shapes represent dose ranks (dose rank 6 represents the highest dose level for each chemical). x-Axis positions of points represent predicted probability of class “Positive” (carcinogenic in left column or genotoxic in right column), e.g., at the cutoff of 0.5 (vertical line), instances with values are predicted “Positive,” and those with are predicted “Negative."
Predictions of Unlabeled Chemicals
Using the final predictive models trained on all profiles with , predictions of carcinogenicity and genotoxicity were made for the chemicals without known CPDB annotation (Figure S5, with detailed summary in Excel Table S6 and Excel Table S7). The majority of unlabeled profiles were predicted “Positive” for both carcinogenicity and genotoxicity using a probability threshold of 0.5. This is likely due to bias in chemical selection. Sources of unknown chemicals include chemicals of interest to the Superfund Research Program (likely environmental toxicants), chemicals that were tested for either carcinogenicity or genotoxicity in the CPDB but whose labels cannot be determined, and controversial chemicals in commercial use (triclosan, Glycel). Many profiles have predicted probabilities between 0.5–0.65, indicating low confidence in prediction, potentially attributable to low bioactivity of profiles. When restricting predictions to unlabeled profiles with to be consistent with the subset used for model training, the separation of ranges of prediction probabilities becomes more evident. The top two ranked predicted carcinogens, benzo(a)pyrene and 7,12-dimethylbenz(a)anthracene (DMBA), are two polyaromatic hydrocarbons that have been shown to manifest carcinogenic and genotoxic properties.
Pathway Enrichment Analysis to Characterize Modes of Actions of Carcinogenicity and Genotoxicity
To identify pathway-level differences between carcinogens and noncarcinogens, and similarly, between genotoxicants and nongenotoxicants, we performed differential pathway enrichment analysis and ranked pathways according to the significance of their differential enrichment between chemical groups. In accordance with the breakdown of TAS subsets used in classification analysis, and based on the observation that increasing thresholds of TAS yield better classification performance, the differential pathway enrichment analysis was repeated for each of the TAS subsets previously considered (Excel Tables S8 and S9). With no TAS threshold (e.g., inclusion of all profiles), only a few pathways were differentially scored between carcinogens and noncarcinogens and between genotoxicants and nongenotoxicants. With increasing thresholds of TAS, the number of significantly expressed pathways increased. At TAS 0.2 and above, the identity of significant pathways became more stable, particularly for genotoxicity-related pathways, with many significant pathways shared across , 0.3, and 0.4. To quantify the similarity of significant pathways across TAS subsets, we measured by Jaccard index the overlapping proportion of significant () pathways among all possible TAS subset pairs, and then computed the mean Jaccard index of each TAS subset with respect to all other TAS subsets. The mean Jaccard index for carcinogenicity was 0.14, 0.36, 0.42, and 0.41 for , 0.2, 0.3, and 0.4, respectively. For genotoxicity, it was 0.23, 0.54, 0.54, and 0.52.
We derived an aggregated ranking score of differential pathway enrichment by combining p-values across all the TAS subsets (see “Methods” section), and the lists of differentially enriched pathways (combined ) with respect to carcinogenicity and genotoxicity are included in Excel Tables S6 and S7, respectively. When comparing carcinogens to noncarcinogens, we observed up-regulation of immune-related pathways (), cell death (apoptosis-induced DNA fragmentation), DNA repair (nucleotide excision repair), transcriptional regulation (RNA polymerase I-, II-, and III-related activity), and cell cycle checkpoints (p53-dependent G1 DNA damage checkpoint), and down-regulation of various metabolism-related pathways (phase II conjugation, phase I functionalization, peptide hormone biosynthesis), cell–cell organization and communication (cell–cell junction organization, integrin cell surface interactions, tight junction interactions), and G protein signaling. Among genotoxicants compared to nongenotoxicants, up-regulated pathways included DNA repair (nucleotide excision repair, formation of incision complex in GG-NER), protein kinase B signaling, programmed cell death, G1/S DNA damage checkpoints, innate immune response (interferon signaling, toll-like receptor signaling). Down-regulated pathways included xenobiotic metabolism (phase I and phase II metabolism), peptide hormone biosynthesis, cell–cell organization and cell–cell communication, innate immune response (complement cascade), and various hemostasis- and metabolism-related pathways.
From the differentially scored pathways of carcinogenicity (Excel Table S8) and genotoxicity (Excel Table S9), we identified a reduced set consisting of the top 40 up-regulated and top 40 down-regulated pathways with Reactome categories as ordered by the aggregated rankings, and visualized their enrichment scores across profiles with in Figure S6A (top pathways differentially enriched with respect to carcinogenicity) and Figure S6B (genotoxicity). Hierarchical clustering of samples revealed stratification by carcinogenicity status, with Cluster 1 significantly enriched for carcinogens compared to Cluster 2 (Fisher test ), and an even stronger stratification by genotoxicity status, with Cluster 1 significantly enriched for genotoxicants compared to Cluster 2 (Fisher test ).
Comparison of L1000 Signatures of Carcinogenicity and Genotoxicity with Signatures from Drugmatrix
We tested for enrichment of the Drugmatrix-derived signatures of carcinogenicity and genotoxicity against our L1000-based differential signatures of carcinogenicity and genotoxicity (see Excel Table S10). Both the directional concordance of signatures (column “direction_match,” e.g., are the genes up-regulated by carcinogens in Drugmatrix also up-regulated in L1000?) and the significance of signature enrichment (column “FDR.q.val”) were measured. Significant similarities were observed between signatures derived from Drugmatrix low-dose rat primary hepatocyte cell cultures and our L1000 profiles. For example, the signature of up-regulated genes in response to low-dose carcinogens in cell cultures (“UP_CARC_CELL_LOWDOSE”) was enriched in the L1000-profiled carcinogen subsets at , 0.3, and 0.2 (). Conversely, the signature of down-regulated genes in response to low-dose carcinogens in cell cultures (“DN_CARC_CELL_LOWDOSE”) was enriched in the L1000-profiled noncarcinogen subsets at and 0 (). Similarly, signature of genotoxicants in the Drugmatrix cell cultures (“UP_GTX_CELL_LOWDOSE”) was enriched in the L1000-profiled genotoxicant subsets at , 0.3, and 0. When repeating the analysis for signatures derived from all Drugmatrix cell culture profiles (including high doses), signatures of genotoxicity were directionally consistent with L1000 profiles (in all eight relevant tests), but signatures of carcinogenicity were inconsistent, and in fact sometimes behaved in the opposite direction (directions matched according to expectation in two out of eight relevant tests). For example, the Drugmatrix signature “UP_CARC_CELL” was enriched among noncarcinogens in the L1000 subset. This inconsistency is likely due to the use of extremely high doses for some of the chemicals in the Drugmatrix cell culture profiles. For reference, the mean dose in Drugmatrix cell culture profiles was , and the maximum dose was . In contrast, the maximum dose among L1000 profiles was .
Next, we compared signatures derived from the Drugmatrix in vivo rat liver profiles to the L1000 profiles. For carcinogenicity, the signature of down-regulated genes in response to carcinogens (“DN_CARC_LIVER”) was correctly enriched among noncarcinogens in L1000 , 0.3, 0.2, and 0 with . Similarly, the signature of up-regulated genes in response to carcinogens (“UP_CARC_LIVER”) was marginally enriched among L1000 (), and () carcinogens. On the other hand, the signatures of genotoxicity were largely not enriched in the right direction (e.g., “DN_GTX_LIVER” shows enrichment among genotoxicants of ).
To rule out the possibility that the observed signatures’ inconsistency was due to platform differences—since the Drugmatrix data were microarray-based, while our data were L1000-based—we compared Drugmatrix cell culture to Drugmatrix liver signatures of genotoxicity (both microarray based). We found that the down-regulated genotoxicant signature in liver was also behaving in the opposite direction compared to the cell culture signature (Excel Table S11A,B). This finding suggests that the signatures’ inconsistency between liver and cell line was likely due to differences between in vitro and in vivo responses to exposure rather than to differences in the profiling platform. Upon detailed inspection of the Drugmatrix liver signatures, we identified an enrichment of genes related to metabolism in both the up- and down-regulated gene signatures (lipid metabolism, cholesterol biosynthesis, Phase I metabolism in “UP_GTX_LIVER,” amino acid metabolism, fatty acid metabolism in “DN_GTX_LIVER”), supporting the conclusion that there may be substantial differences between metabolic activities in in vitro and in vivo models (Figure S7).
In summary, L1000-derived signatures of carcinogenicity and genotoxicity were concordant with Drugmatrix low-dose cell culture signatures but inconsistent with Drugmatrix liver signatures, with the differences largely driven by discrepancies in the expression of certain metabolism-related genes between in vitro and in vivo exposures.
Comparison of L1000 Signatures of Carcinogenicity and Genotoxicity with Drug Perturbation Signatures in the Connectivity Map
The availability of the CMap offered the opportunity to compare our profiles to a much larger database of pharmacologically annotated signatures and allowed us to predict MoAs or pharmacological properties based on signature similarity. To this end, we first computed the similarity of our signatures to each signature in the CMap using connectivity scores (see “Methods” section). We then identified the CMap signatures that showed significant difference in connectivity scores () between carcinogens and noncarcinogens and between genotoxicants and nongenotoxicants. The top CMap hits are summarized at the level of PCLs in Excel Table S12 (carcinogenicity) and Excel Table S13 (genotoxicity) and visualized in Figure 5, and at the level of individual chemical perturbations in Excel Table S14 (carcinogenicity) and Excel Table S15 (genotoxicity).
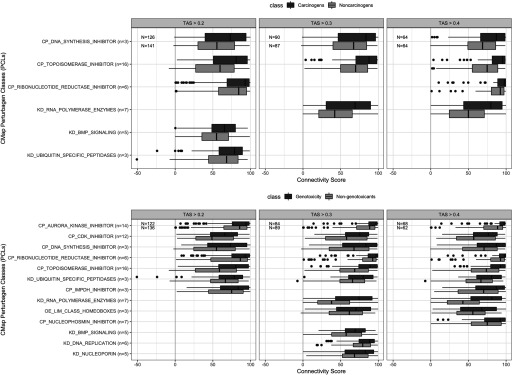
Figure 5.
Connectivity scores of top Connectivity Map (CMap) Perturbagen classes with differential connectivity [false discovery rate ] to carcinogens vs. noncarcinogens and genotoxicants vs. nongenotoxicants grouped by transcriptional activity scores (TAS) subsets. The lower, middle, and upper hinges of box plots correspond to the 25th, 50th (median), and 75th percentiles, respectively. The upper and lower whiskers extend to the smaller and largest value at most 1.5 × IQR (interquartile range) from the hinge, and data points beyond the whiskers are represented as dots.
Focusing on the significantly differential PCLs across all TAS subsets (, 0.3, 0.4), we found that carcinogens, compared to noncarcinogens, were significantly more connected to drug classes consisting of topoisomerase inhibitors, DNA synthesis inhibitors, and ribonucleotide reductase. Genotoxicants, compared to nongenotoxicants, were significantly more connected to the three aforementioned drug classes, as well as to cyclin-dependent kinase inhibitors, aurora kinase inhibitors, and ubiquitin specific peptidases (Figure 5).
Characterizing Aryl Hydrocarbon Receptor–Mediated Response in L1000 Gene Expression Profiles
Carcinogens and genotoxicants are sometimes recognized by cellular receptors such as the AhR. Given that the AhR is an important mediator of the toxicity of many chemicals represented in our dataset, we sought to investigate the effects of AhR-activated chemicals in terms of known AhR-regulated gene expression and the similarity of transcriptomic profiles among subgroups of AhR agonists.
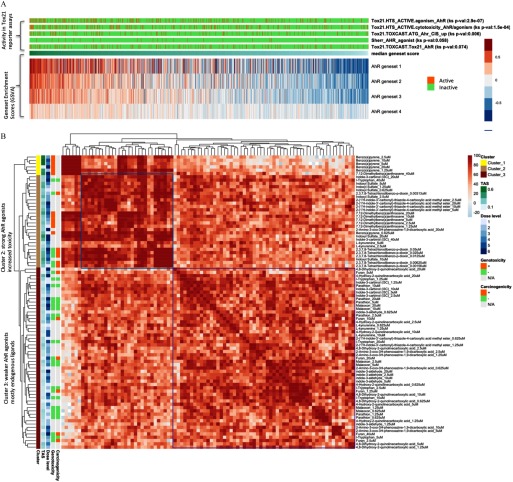
The L1000 profiles exhibited consistent enrichment of AhR-related gene set activity among chemicals labeled as AhR-active in several Tox21 reporter assays, namely, “HTS_ACTIVE.agonism_AhR” (), “HTS_ACTIVE.cytotoxicity_AhR/agonism” (), and “ATG_Ahr_CIS_up” () (Figure 6A).
Figure 6.
Investigation of profiles of aryl hydrocarbon receptor (AhR)–related chemical perturbations. (A) Profiles with AhR activity ranked by median gene set scores of AhR target gene lists. (B) AhR-related profiles clustered by connectivity scores.
Next, we examined an expert-curated set of AhR-related chemicals (Group: “Sherr_AHR_agonist”). Based on the similarity of their gene expression profiles as measured by the connectivity scores, we found two functionally distinct classes of AhR-related chemicals (Figure 6B). Cluster 1 contained five profiles (out of six) of perturbation by benzo(a)pyrene, a strong AhR agonist and known genotoxicant. Cluster 2 was enriched with profiles of strong exogenous AhR ligands, most with potent toxic effects, including DMBA and TCDD. It is not surprising that many of these chemicals also had high in vitro transcriptional bioactivity (high TAS). Interestingly, profiles of indoxyl sulfate clustered with this group of strong AhR agonists (Cluster 2).
Carcinogenome Portal: A Framework for Data Query and Visualization
All data described in this manuscript are available for public access. Data processed under the standard CMap-L1000 pipeline are available under https://clue.io/data/CRCGN_ABC. To facilitate the interactive querying of the downstream analysis results produced by this study, we developed a web portal (https://carcinogenome.org/HEPG2). The query and visualization functionalities supported by the portal include differential expression, gene set enrichment, and connectivity analysis against CMap signatures. This interface supports both marker-centered (genes, pathways, CMap signatures) and chemical-centered queries. For instance, one can find the top gene markers and pathways regulated by a particular perturbation, identify the top chemicals that up-regulate a particular gene or pathway of interest, or find CMap chemicals or chemical groups that are most similar to the profiles of a particular perturbation. In addition, the portal supports bulk query and visualization of groups of perturbations in the form of heatmaps.
Discussion
Prediction of Carcinogenicity and Genotoxicity
The results from the prediction of carcinogenicity and genotoxicity experiments provide strong evidence that transcriptional bioactivity as captured by TAS had a high impact on the classifier performance. In fact, while absolute levels of bioactivity were not associated with carcinogenicity in our experiments, a sufficiently high bioactivity was necessary to elicit enough transcriptional signal to use a chemical's expression profile for carcinogenicity prediction. Thus, when limiting to profiles with high TAS, the performance of our predictive models drastically improved. Among highly bioactive profiles (), our classifiers yielded mean AUC of 72.2% for prediction of carcinogenicity (Figure 2A), and 82.3% for prediction of genotoxicity (Figure 2B). These results confirm that short-term in vitro gene expression profiles of chemical perturbations, given sufficient transcriptional bioactivity, can accurately predict long-term chemical carcinogenicity, and to a greater extent, genotoxicity.
Of notice, when we applied our genotoxicity classifier to the prediction of unlabeled chemicals, the top ranked predicted genotoxicant was indoxyl sulfate, which is an endogenous tryptophan metabolite. Indoxyl sulfate has been shown to activate p53 expression through reactive oxygen species production and is a source of endogenous oxidative DNA damage (Shimizu et al. 2013). While indoxyl sulfate may not necessarily be considered a genotoxicant as it is a uremic solvent found in low concentrations () in the human serum normally, it activates the AhR, inducing cytochrome P450 enzymes. which metabolize other substrates, including mutagenic intermediates (see “Characterizing Aryl Hydrocarbon Receptor–Mediated Response in L1000 Gene Expression Profiles” in the “Results” section) (Shashar et al. 2017; Schroeder et al. 2010; Shivanna et al. 2016). Thus, prediction of indoxyl sulfate as a genotoxicant may be due to transcriptional activation of shared pathways involved in metabolism of genotoxic chemicals.
In Vitro Dose Recommendation
To boost the effective sample size used in classification, we outline the following dose selection strategy for improving bioactivity of in vitro gene expression profiles.
The selection of doses in short-term acute exposures for prediction of long-term in vivo phenotypes is a challenging task. In this experiment, we chose to adopt a standard six-dose titration, starting from or , depending on source of chemicals. The sole exception to the standard dosing was TCDD, whose starting concentration was efficiency of standardized dosing using the L1000 platform.
One alternative dosing scheme is to determine unique doses for each chemical using the MTT assay. For instance, a previous study of genotoxicity prediction based on in vitro experiments selected doses based on a MTT assay resulting in 80% viability at 72 h incubation, or maximum dose of in the case of lack of cytotoxicity (Magkoufopoulou et al. 2012). Some chemicals used in that study were administered at doses that vastly exceeded the or dose limit adopted in our experimental setup. Furthermore, the lack of plateau effect in dose response as a function of TAS (proxy for bioactivity) suggests that doses exceeding the or threshold may indeed yield profiles with higher bioactivity and increase the power to detect gene and pathway markers for prediction of carcinogenicity and genotoxicity without experiencing saturation effects (response plateauing) or excessive cell death. Although standardizing dosage across chemicals was the logistically and cost-effective solution for this experiment, going forward, MTT assays are highly recommended for maximizing biological signal across transcriptional profiles. Estimation of the appropriate in vitro dose from toxicokinetic modeling of the in vivo doses tested in animal bioassays, when available, is another viable alternative, as shown in Figure 1D and associated discussion. We offer these dose recommendations in the context of accurate hazard prediction, for which this study has shown that sufficient signal (transcriptional bioactivity) is necessary. For effective risk assessment and translation, human relevant doses should be considered.
Acute vs. Chronic Response
Through analysis of TASs between carcinogens and noncarcinogens (Figure 1C), we observed that long-term carcinogenicity, as established from long-term in vivo rodent studies, had no effect on transcriptional bioactivity in our short-term assay (Figure 2A). This observation supports the conclusion that bioactivity as defined by TAS at less than is not associated with carcinogenicity, and consequently, a short-term chemical perturbation with minimal transcriptional response cannot be assumed safe.
While TAS alone was not predictive of carcinogenicity, it was instrumental to the selection of those compounds with sufficient bioactivity to allow us to build an accurate gene expression–based classifier of carcinogenicity (up to 72.2% AUC). It was also instrumental to capturing important MoAs of carcinogenicity, as shown by our pathway enrichment analysis, which highlighted the up-regulation of response, cell death, DNA repair, and transcriptional regulation (RNA polymerase I, II, III) pathways in response to carcinogen exposure, as well as down-regulation of phase I and phase II metabolism and cell–cell organization and communication pathways. Overall, we observed a stronger signal of genotoxicity compared to carcinogenicity, which is to be expected, as the latter is a more heterogeneous phenotype and thus harder to capture as a binary distinction; this is also evidenced by the higher accuracy of the genotoxicity classifier (82.3%) as well as by the higher TAS among genotoxicants compared to nongenotoxicants.
Higher TAS levels were also associated with the identification of more stably enriched pathways across TAS subsets when performing differential enrichment analysis between carcinogens and noncarcinogens, and between genotoxicants and nongenotoxicants (see “Pathway Enrichment Analysis to Characterize Modes of Actions of Carcinogenicity and Genotoxicity” section). The increase in the number and in the overlap of significant pathways at higher TAS is likely due to the associated stronger signal. At lower TAS, the larger number of false positives increased the noise level and heterogeneity of the transcriptional response, and likely led to the reduction in the number of pathways found to be significantly enriched.
High-Resolution Characterization of Chemical Classes’ Response
Given the presence of several AhR ligands in our panel, we used this set of chemicals to assess our platform’s ability to go beyond supervised classification of a binary phenotype and toward taxonomy discovery within classes of related compounds (Figure 6B). Interestingly, in this analysis indoxyl sulfate was clustered with the group of strong AhR agonists (Cluster 2). While this chemical is an endogenous AhR ligand, it can be considered a uremic toxin that is observed at elevated levels in patients with chronic renal failure (Niwa et al. 1999). Cluster 3 contained endogenous AhR ligands (l-kynurenine, indole-3-carbonyl, kynurenic acid, xanthurenic acid, and cinnabarinic acid). Since l-tryptophan is not an AHR ligand, its presence in this latter group suggests that it is metabolized to one of the kynurenine pathway metabolites that are AhR ligands (L-kynurenine, kynurenic acid, xanthurenic acid, and cinnabarinic acid).
These results show promise for our platform to be used not only as a general predictor of active transcriptional pathways such as the AhR signaling pathway, but also to distinguish, with finer granularity, classes of AhR agonists according to the transcriptomic profile they induce. Furthermore, the fact that the L1000 profiles exhibited consistent enrichment of AhR-related gene set activity among chemicals labeled as AhR-active in several Tox21 reporter assays validates the ability of unbiased gene expression profiling to accurately capture end points from more specific and targeted assays such as those in the Tox21 library.
Implication of Findings in Context of Tumor Initiation and Promotion
Chemical carcinogens can be classified into tumor initiators and promoters. Initiators cause changes to the DNA (mutagens) and promoters drive the proliferation of the cell, typically by interacting with receptors to affect pathways leading to cell proliferation. We derived labels of carcinogenicity based on long-term rodent studies, which includes both tumor initiators and promoters. However, it is important to understand that we used short-term human cell line gene expression patterns to predict long-term rodent carcinogenicity. Pathways relevant to tumor initiation were accurately captured by the short-term in vitro gene expression data (DNA repair, DNA damage, etc.). As for tumor promotion, promoters typically interact with receptors to mediate cell proliferation, and our cell culture model contained a subset of receptors that mediate these processes (Yueh et al. 2014). However, one limitation is that culture conditions were already a promotion environment (high growth) that might limit the detection of promoting agents. Another limitation is that tumor promoters mediated by receptors not expressed in a culture system may elicit reproducible but not biologically accurate patterns of gene expression in the short-term in vitro assay, although they may be correctly classified by our machine-learning approach. Mechanistic expert judgement will need to be applied to evaluate the relevance of these findings to human carcinogenicity.
Interfacing with the Connectivity Map
One of the important features of the perturbation experiment data we generated is the data’s support for guilt by association inference of chemical function by signature-based comparison to the CMap’s PCLs, as illustrated in Figure 5.
For example, we showed that carcinogens were significantly more connected than noncarcinogens to the PCL consisting of topoisomerase inhibitors. These represent a specific class of DNA synthesis inhibitors, which are mainly recognized as chemotherapeutic drugs that preferentially inhibit the topoisomerase enzymes (commonly topoisomerase I or II) in cancer cells to slow their rate of replication. Topoisomerase I or II introduce single- or double-strand DNA breaks in cells undergoing replication, and form topoisomerase-DNA complexes. Most topoisomerase inhibitors function by trapping these complexes, leading to increased strand breaks but incomplete DNA replication, subsequently provoking DNA damage response and DNA repair (Pommier et al. 2006; Pommier 2013; Wang and Eastmond 2002). Thus, DNA damage response induced by topoisomerase inhibitors is expected to mimic the response to genotoxic carcinogens.
Other relevant PCLs also exhibit shared MoAs with carcinogens and genotoxicants. Aurora kinase inhibitors play a major role in cell cycle regulation through the induction of G1 arrest and apoptosis (Bavetsias and Linardopoulos 2015). Ubiquitin-specific peptidases, specifically USP24, have been shown to play a role in DNA damage response (Zhang and Gong 2015).
Challenges and Future Developments
This experiment aimed to accelerate short-term in vitro testing approaches to predict long-term chemical carcinogenicity. We showed that short-term in vitro gene expression profiling is not only capable to accurately predict carcinogenicity and genotoxicity, but is also useful to characterize important mechanisms of carcinogenic response, particularly DNA damage and repair, and changes in cell cycle and cell–cell organization and communication. Other general biological processes that may be relevant for carcinogenic response, including inflammatory response, immune dysfunction, metabolic disruption, and endocrine disruption, require further investigation in other in vitro contexts.
The choice of HepG2 as our primary cell line model was driven by the abundance of chemical annotations for liver carcinogenicity and the appropriateness of HepG2 for the study of liver toxicity. However, there are limitations in its use. Firstly, the expression of genes involved in phase I and phase II metabolism vary between passages, and results relating to xenobiotic metabolism may be difficult to determine (Soldatow et al. 2013); this was also seen in the comparison of our genotoxicity-related signatures to Drugmatrix liver signatures. One potential contribution to this effect is the low bioactivation capacity in HepG2 compared to in vivo. As an alternative, the hepatoma cell line, HepaRG, which has a liver-like bioactivation, could be used as an in vitro liver model for studying carcinogens and genotoxicants (Guillouzo et al. 2007). One study has shown that while HepG2 performs better in discriminating signatures between genotoxic and nongenotoxic carcinogens, HepaRG is a more suitable in vitro liver model for biological interpretation of effects of chemical exposures (Jennen et al. 2010). Secondly, since HepG2 is a cancer cell line, the exposures of carcinogens in this line may show differences as compared to a nontransformed cell line. For the purpose of predictive modeling, we contend that these cell line–specific nuances may be overlooked as long as the performance of the classifier is adequate.
Other cell line candidates for follow up studies should include more realistic hepatocyte models, such as induced pluripotent stem cells–derived hepatocytes, or organoids (Davidson et al. 2015; Underhill and Khetani 2018). Alternatively, hepatic stem cells such as oval cells could be considered given that the stem cell theory of cancer initiation and maintenance is well supported (Fábián et al. 2013; Tan et al. 2006).
While liver carcinogenicity prediction was the adverse phenotype of choice for this study, this experiment provided us with many valuable insights to facilitate future experiments, including the logistics of large chemical panels procurement, and chemical and dose selection for tissue-specific carcinogenicity. It also set the stage for in vitro based exposure studies of additional adverse phenotypes. For instance, we initiated the in vitro screening of mammary gland carcinogenicity through the use of a nontumorigenic human mammary epithelial cell line, MCF10A and p53-deficient MCF10A. The experimental and computational pipeline we established, paired with the cost-effective technology we used for chemical exposure and gene expression profiling, paves the way for the screening of large chemical panels for exposure-based experiments in other organ, disease, and adverse outcome contexts.
Conclusions
Long-term tests for chemical carcinogens based on epidemiology and rat studies are expensive and time-consuming and not feasible for scaling to a large number of chemicals. In this study, we detailed a high-throughput gene expression profiling of liver carcinogens and noncarcinogens in a short-term in vitro exposure model. These gene expression profiles, given sufficient transcriptional bioactivity, were capable of accurate prediction of long-term carcinogenicity and even more accurate prediction of genotoxicity. Pathway enrichment analysis revealed similarities between pathway level response captured by the short term in vitro exposures and known MoAs of carcinogenesis, particularly genotoxic mechanisms such as DNA damage and repair.
Supplementary Material
Acknowledgments
The authors would like to thank the members of the CMap team, L. Litichevskiy, N. Lyons, and M. Macaluso, for their help in data generation and processing and participation in many fruitful discussions. This study was supported in part by the Find the Cause Breast Cancer Foundation (https://findthecausebcf.org/), the Evans Center for Interdisciplinary Biomedical Research ARC on Genomics Models of Environmental Chemical Carcinogenicity at Boston University (http://www.bumc.bu.edu/evanscenteribr/), the Superfund Research Program at Boston University (P42 ES007381-21), and a Library of Integrated Network-Based Cellular Signatures (LINCS)/National Institute of Environmental Health Sciences (NIEHS) Admin Supplement (P42 ES007381-19S2).
References
- Abdo KM, Eustis SL, Haseman J, Huff JE, Peters A, Persing R. 1988. Toxicity and carcinogenicity of rotenone given in the feed to F344/N rats and B6C3F1 mice for up to two years. Drug Chem Toxicol 11(3):225–235, PMID: 3181037, 10.3109/01480548809017879. [DOI] [PubMed] [Google Scholar]
- American Cancer Society. 2017. Cancer Facts & Figures. https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2017.html [accessed 3 December 2018].
- Anand P, Kunnumakkara AB, Sundaram C, Harikumar KB, Tharakan ST, Lai OS, et al. 2008. Cancer is a preventable disease that requires major lifestyle changes. Pharm Res 25(9):2097–2116, PMID: 18626751, 10.1007/s11095-008-9661-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavetsias V, Linardopoulos S. 2015. Aurora kinase inhibitors: current status and outlook. Front Oncol 5:278, PMID: 26734566, 10.3389/fonc.2015.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57(1):289–300, 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- Bucher JR, Portier C. 2004. Human carcinogenic risk evaluation, Part V: the national toxicology program vision for assessing the human carcinogenic hazard of chemicals. Toxicol Sci 82(2):363–366, PMID: 15456919, 10.1093/toxsci/kfh293. [DOI] [PubMed] [Google Scholar]
- Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, et al. 2014. The Reactome pathway knowledgebase. Nucleic Acids Res 42(Database issue):D472–D477, PMID: 24243840, 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson MD, Ware BR, Khetani SR. 2015. Stem cell-derived liver cells for drug testing and disease modeling. Discov Med 19(106):349–358, PMID: 26105698. [PMC free article] [PubMed] [Google Scholar]
- Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, et al. 2005. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21(16):3439–3440, PMID: 16082012, 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- Eichner J, Kossler N, Wrzodek C, Kalkuhl A, Bach Toft D, Ostenfeldt N, et al. 2013. A toxicogenomic approach for the prediction of murine hepatocarcinogenesis using ensemble feature selection. PLoS One 8(9):e73938, PMID: 24040119, 10.1371/journal.pone.0073938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinger-Ziegelbauer H, Gmuender H, Bandenburg A, Ahr HJ. 2008. Prediction of a carcinogenic potential of rat hepatocarcinogens using toxicogenomics analysis of short-term in vivo studies. Mutat Res 637(1-2):23–39, PMID: 17689568, 10.1016/j.mrfmmm.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Fábián Á, Vereb G, Szöllősi J. 2013. The hitchhikers guide to cancer stem cell theory: markers, pathways and therapy. Cytometry A 83(1):62–71, PMID: 22997049, 10.1002/cyto.a.22206. [DOI] [PubMed] [Google Scholar]
- Fabregat A, Sidiropoulos K, Garapati P, Gillespie M, Hausmann K, Haw R, et al. 2016. The Reactome pathway Knowledgebase. Nucleic Acids Res 44(D1):D481–D487, PMID: 26656494, 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielden MR, Brennan R, Gollub J. 2007. A gene expression biomarker provides early prediction and mechanistic assessment of hepatic tumor induction by nongenotoxic chemicals. Toxicol Sci 99(1):90–100, PMID: 17557906, 10.1093/toxsci/kfm156. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick RB. 2008. CPDB: Carcinogenic Potency Database. Med Ref Serv Q 27(3):303–311, PMID: 19042710, 10.1080/02763860802198895. [DOI] [PubMed] [Google Scholar]
- Ganter B, Snyder RD, Halbert DN, Lee MD. 2006. Toxicogenomics in drug discovery and development: mechanistic analysis of compound/class-dependent effects using the DrugMatrix database. Pharmacogenomics 7(7):1025–1044, PMID: 17054413, 10.2217/14622416.7.7.1025. [DOI] [PubMed] [Google Scholar]
- Ganter B, Tugendreich S, Pearson CI, Ayanoglu E, Baumhueter S, Bostian KA, et al. 2005. Development of a large-scale chemogenomics database to improve drug candidate selection and to understand mechanisms of chemical toxicity and action. J Biotechnol 119:219–244, 10.1016/j.jbiotec.2005.03.022. [DOI] [PubMed] [Google Scholar]
- Gold LS, Manley NB, Slone TH, Rohrbach L, Garfinkel GB. 2005. Supplement to the Carcinogenic Potency Database (CPDB): results of animal bioassays published in the general literature through 1997 and by the National Toxicology Program in 1997-1998. Toxicol Sci 85(2):747–808, PMID: 15800034, 10.1093/toxsci/kfi161. [DOI] [PubMed] [Google Scholar]
- Guillouzo A, Corlu A, Aninat C, Glaise D, Morel F, Guguen-Guillouzo C. 2007. The human hepatoma HepaRG cells: a highly differentiated model for studies of liver metabolism and toxicity of xenobiotics. Chem Biol Interact 168(1):66–73, PMID: 17241619, 10.1016/j.cbi.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Gusenleitner D, Auerbach SS, Melia T, Gómez HF, Sherr DH, Monti S. 2014. Genomic models of short-term exposure accurately predict long-term chemical carcinogenicity and identify putative mechanisms of action. PLoS One 9(7):e102579, PMID: 25058030, 10.1371/journal.pone.0102579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänzelmann S, Castelo R, Guinney J. 2013. GSVA: gene set variation analysis for microarray and RNA-sSeq data. BMC Bioinformatics 14:7, PMID: 23323831, 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson E. 2004. A Textbook of Modern Toxicology. Hoboken, NJ:John Wiley & Sons. [Google Scholar]
- Huff J, Jacobson MF, Davis DL. 2008. The limits of two-year bioassay exposure regimens for identifying chemical carcinogens. Environ Health Perspect 116(11):1439–1442, PMID: 19057693, 10.1289/ehp.10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur J, Danes L, Hsieh J-H, McGregor B, Krout D, Auerbach S. 2018. Tox21 Enricher: Web-based chemical/biological functional annotation analysis tool based on Tox21 toxicity screening platform. Mol Inform 37:e1700129, 10.1002/minf.201700129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennen DG, Magkoufopoulou C, Ketelslegers HB, van Herwijnen MH, Kleinjans JC, van Delft JH. 2010. Comparison of HepG2 and HepaRG by whole-genome gene expression analysis for the purpose of chemical hazard identification. Toxicol Sci 115(1):66–79, PMID: 20106945, 10.1093/toxsci/kfq026. [DOI] [PubMed] [Google Scholar]
- Judson RS, Houck KA, Kavlock RJ, Knudsen TB, Martin MT, Mortensen HM, et al. 2010. In vitro screening of environmental chemicals for targeted testing prioritization: the ToxCast project. Environ Health Perspect 118(4):485–492, PMID: 20368123, 10.1289/ehp.0901392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstreuer NC, Dix DJ, Houck KA, Kavlock RJ, Knudsen TB, Martin MT, et al. 2013. In vitro perturbations of targets in cancer hallmark processes predict rodent chemical carcinogenesis. Toxicol Sci 131(1):40–55, PMID: 23024176, 10.1093/toxsci/kfs285. [DOI] [PubMed] [Google Scholar]
- Kossler N, Matheis KA, Ostenfeldt N, Bach Toft D, Dhalluin S, Deschl U, et al. 2015. Identification of specific mRNA signatures as fingerprints for carcinogenesis in mice induced by genotoxic and nongenotoxic hepatocarcinogens. Toxicol Sci 143(2):277–295, PMID: 25410580, 10.1093/toxsci/kfu248. [DOI] [PubMed] [Google Scholar]
- Kuhn M. 2008. Building predictive models in R using the caret package. J Stat Softw 28(5):1–26, 10.18637/jss.v028.i05.27774042 [DOI] [Google Scholar]
- Liaw A, Wiener M. 2002. Classification and regression by randomForest. R News 2/3:18–22. [Google Scholar]
- Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. 2011. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27(12):1739–1740, PMID: 21546393, 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magkoufopoulou C, Claessen SM, Tsamou M, Jennen DG, Kleinjans JC, van Delft JH. 2012. A transcriptomics-based in vitro assay for predicting chemical genotoxicity in vivo. Carcinogenesis 33(7):1421–1429, PMID: 22623647, 10.1093/carcin/bgs182. [DOI] [PubMed] [Google Scholar]
- Meister KA. 2005. America’s War on “Carcinogens”: Reassessing the Use of Animal Tests to Predict Human Cancer Risk. New York, NY:American Council on Science and Health. [Google Scholar]
- National Toxicology Program. 1978. Bioassay of pyrimethamine for possible carcinogenicity. Natl Cancer Inst Carcinog Tech Rep Ser 77:1–107, PMID: 12830220. [PubMed] [Google Scholar]
- National Toxicology Program. 1989. NTP Toxicology and Carcinogenesis Studies of Rhodamine 6G (C.I. Basic Red 1) (CAS No. 989-38-8) in F344/N rats and B6C3F1 mice (feed studies). Natl Toxicol Program Tech Rep Ser 364:1–192, PMID: 12692640. [PubMed] [Google Scholar]
- National Toxicology Program. 1994. NTP Toxicology and Carcinogenesis Studies of Hexachlorocyclopentadiene (CAS No. 77-47-4) in F344/N rats and B6C3F1 mice (inhalation studies). Natl Toxicol Program Tech Rep Ser 437:1–308, PMID: 12595926. [PubMed] [Google Scholar]
- Nie AY, McMillian M, Parker JB, Leone A, Bryant S, Yieh L, et al. 2006. Predictive toxicogenomics approaches reveal underlying molecular mechanisms of nongenotoxic carcinogenicity. Mol Carcinog 45(12):914–933, PMID: 16921489, 10.1002/mc.20205. [DOI] [PubMed] [Google Scholar]
- Niwa T, Aoyama I, Takayama F, Tsukushi S, Miyazaki T, Owada A, et al. 1999. Urinary indoxyl sulfate is a clinical factor that affects the progression of renal failure. Miner Electrolyte Metab 25(1-2):118–122, PMID: 10207273, 10.1159/000057433. [DOI] [PubMed] [Google Scholar]
- Pearce RG, Setzer RW, Strope CL, Sipes NS, Wambaugh JF. 2017. httk: R package for high-throughput toxicokinetics. J Stat Softw 79(4):1–26, PMID: 30220889, 10.18637/jss.v079.i04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck D, Crawford ED, Ross KN, Stegmaier K, Golub TR, Lamb J. 2006. A method for high-throughput gene expression signature analysis. Genome Biol 7(7):R61, PMID: 16859521, 10.1186/gb-2006-7-7-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y. 2013. Drugging topoisomerases: lessons and challenges. ACS Chem Biol 8(1):82–95, PMID: 23259582, 10.1021/cb300648v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y, Barcelo J, Rao VA, Sordet O, Jobson AG, Thibaut L, et al. 2006. Repair of topoisomerase I-mediated DNA damage. Prog Nucleic Acid Res Mol Biol 81:179–229, PMID: 16891172, 10.1016/S0079-6603(06)81005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed ER, Moses E, Xiao X, Liu G, Campbell J, Perdomo C, et al. 2019. Assessment of a highly multiplexed RNA sequencing platform and comparison to existing high-throughput gene expression profiling techniques. Front Genet 10, 10.3389/fgene.2019.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard AM, Judson RS, Houck KA, Grulke CM, Volarath P, Thillainadarajah I, et al. 2016. ToxCast chemical landscape: paving the road to 21st century toxicology. Chem Res Toxicol 29(8):1225–1251, PMID: 27367298, 10.1021/acs.chemrestox.6b00135. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43(7):e47, PMID: 25605792, 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryffel B. 1992. The carcinogenicity of ciclosporin. Toxicology 73(1):1–22, PMID: 1589877, 10.1016/0300-483X(92)90166-C. [DOI] [PubMed] [Google Scholar]
- Schmidt CW. 2009. TOX 21: new dimensions of toxicity testing. Environ Health Perspect 117(8):A348–A353, PMID: 19672388, 10.1289/ehp.117-a348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder JC, Dinatale BC, Murray IA, Flaveny CA, Liu Q, Laurenzana EM, et al. 2010. The uremic toxin 3-indoxyl sulfate is a potent endogenous agonist for the human aryl hydrocarbon receptor. Biochemistry 49(2):393–400, PMID: 20000589, 10.1021/bi901786x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shashar M, Belghasem ME, Matsuura S, Walker J, Richards S, Alousi F, et al. 2017. Targeting STUB1-tissue factor axis normalizes hyperthrombotic uremic phenotype without increasing bleeding risk. Sci Transl Med 9(417):eaam8475, PMID: 29167396, 10.1126/scitranslmed.aam8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu H, Yisireyili M, Higashiyama Y, Nishijima F, Niwa T. 2013. Indoxyl sulfate upregulates renal expression of ICAM-1 via production of ROS and activation of NF-κB and p53 in proximal tubular cells. Life Sci 92(2):143–148, PMID: 23201429, 10.1016/j.lfs.2012.11.012. [DOI] [PubMed] [Google Scholar]
- Shivanna S, Kolandaivelu K, Shashar M, Belghasim M, Al-Rabadi L, Balcells M, et al. 2016. The aryl hydrocarbon receptor is a critical regulator of tissue factor stability and an antithrombotic target in uremia. J Am Soc Nephrol 27(1):189–201, PMID: 26019318, 10.1681/ASN.2014121241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. 2005. limma: linear models for microarray data. In: Statistics for Biology and Health: Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, eds. New York, NY:Springer, 397–420. [Google Scholar]
- Soldatow VY, LeCluyse EL, Griffith LG, Rusyn I. 2013. In vitro models for liver toxicity testing. Toxicol Res (Camb) 2(1):23–39, PMID: 23495363, 10.1039/C2TX20051A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, et al. 2017. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell 171(6):1437–1452.e17, PMID: 29195078, 10.1016/j.cell.2017.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102(43):15545–15550, PMID: 16199517, 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan BT, Park CY, Ailles LE, Weissman IL. 2006. The cancer stem cell hypothesis: a work in progress. Lab Invest 86(12):1203–1207, PMID: 17075578, 10.1038/labinvest.3700488. [DOI] [PubMed] [Google Scholar]
- Tice RR, Austin CP, Kavlock RJ, Bucher JR. 2013. Improving the human hazard characterization of chemicals: a Tox21 update. Environ Health Perspect 121(7):756–765, PMID: 23603828, 10.1289/ehp.1205784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasetti C, Li L, Vogelstein B. 2017. Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science 355(6331):1330–1334, PMID: 28336671, 10.1126/science.aaf9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasetti C, Vogelstein B. 2015. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 347(6217):78–81, PMID: 25554788, 10.1126/science.1260825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Minowa Y, Morikawa Y, Kondo C, Maruyama T, Kato I, et al. 2011. Prediction model of potential hepatocarcinogenicity of rat hepatocarcinogens using a large-scale toxicogenomics database. Toxicol Appl Pharmacol 255(3):297–306, PMID: 21784091, 10.1016/j.taap.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Underhill GH, Khetani SR. 2018. Bioengineered liver models for drug testing and cell differentiation studies. Cell Mol Gastroenterol Hepatol 5(3):426–439.e1, PMID: 29675458, 10.1016/j.jcmgh.2017.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Eastmond DA. 2002. Catalytic inhibitors of topoisomerase II are DNA-damaging agents: induction of chromosomal damage by merbarone and ICRF-187. Environ Mol Mutagen 39(4):348–356, PMID: 12112387, 10.1002/em.10072. [DOI] [PubMed] [Google Scholar]
- Waters MD, Jackson M, Lea I. 2010. Characterizing and predicting carcinogenicity and mode of action using conventional and toxicogenomics methods. Mutat Res 705(3):184–200, PMID: 20399889, 10.1016/j.mrrev.2010.04.005. [DOI] [PubMed] [Google Scholar]
- Yueh MF, Taniguchi K, Chen S, Evans RM, Hammock BD, Karin M, et al. 2014. The commonly used antimicrobial additive triclosan is a liver tumor promoter. Proc Natl Acad Sci U S A 111(48):17200–17205, PMID: 25404284, 10.1073/pnas.1419119111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, et al. 2018. Ensembl 2018. Nucleic Acids Res 46:D754–D761, 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Gong F. 2015. Involvement of USP24 in the DNA damage response. Mol Cell Oncol 3(1):e1011888, PMID: 27308530, 10.1080/23723556.2015.1011888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Huang K, Zhu L, Luo Y, Xu W. 2017. Precision toxicology based on single cell sequencing: an evolving trend in toxicological evaluations and mechanism exploration. Arch Toxicol 91(7):2539–2549, PMID: 28451740, 10.1007/s00204-017-1971-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.