Abstract
Background:
Available toxicity data can be optimally interpreted if they are integrated using computational approaches such as systems biology modeling. Such approaches are particularly warranted in cases where regulatory decisions have to be made rapidly.
Objectives:
The study aims at developing and applying a new integrative computational strategy to identify associations between bisphenol S (BPS), a substitute for bisphenol A (BPA), and components of adverse outcome pathways (AOPs).
Methods:
The proposed approach combines a text mining (TM) procedure and integrative systems biology to comprehensively analyze the scientific literature to enrich AOPs related to environmental stressors. First, to identify relevant associations between BPS and different AOP components, a list of abstracts was screened using the developed text-mining tool AOP-helpFinder, which calculates scores based on the graph theory to prioritize the findings. Then, to fill gaps between BPS, biological events, and adverse outcomes (AOs), a systems biology approach was used to integrate information from the AOP-Wiki and ToxCast databases, followed by manual curation of the relevant publications.
Results:
Links between BPS and 48 AOP key events (KEs) were identified and scored via 31 references. The main outcomes were related to reproductive health, endocrine disruption, impairments of metabolism, and obesity. We then explicitly analyzed co-mention of the terms BPS and obesity by data integration and manual curation of the full text of the publications. Several molecular and cellular pathways were identified, which allowed the proposal of a biological explanation for the association between BPS and obesity.
Conclusions:
By analyzing dispersed information from the literature and databases, our novel approach can identify links between stressors and AOP KEs. The findings associating BPS and obesity illustrate the use of computational tools in predictive toxicology and highlight the relevance of the approach to decision makers assessing substituents to toxic chemicals. https://doi.org/10.1289/EHP4200
Introduction
Integrative computational approaches that combine systems biology and toxicology can increase our understanding of the links between environmental chemical exposure and human health. Systems biology and advanced bioinformatics tools generate new hypotheses. They furnish new insights into and predictions of biological mechanisms induced by chemical substances, including drugs and environmental pollutants. Compelling evidence indicates that a number of chemical substances may play a causative role in diseases (Heindel and Blumberg 2018). Computational sciences, including systems toxicology, can speed up the identification of linkage between adverse outcome pathways (AOPs) and a chemical stressor as well as its effects on health (Ankley et al. 2010).
The concept of an AOP was originally proposed by Ankley et al. (2010). AOPs integrate various key events (KEs) to connect biological perturbations, at the molecular or cellular levels, to toxicity events [i.e., adverse outcomes (AOs)] at organismal and population levels. The use of clearly identified AOPs for decision-making is part of a global methodological initiative, which has, among its goals, the reduction of animal use in toxicity testing. AOPs are expected to be used more and more in regulatory frameworks since they provide evidence-based mechanistic insights (Bopp et al. 2018). The AOPs, which have been identified, are stored in the AOP-Wiki online database (SAAOP 2016). The database is part of a collaborative program that involves the Organisation for Economic Co-operation and Development (OECD) and the European Commission. The AOP knowledge database (AOP-KB) is another tool from the OECD program for AOP development, to support and share information to the scientific community and harmonize the format of generated novel AOP (OECD). All the terms defined in the AOPs are standardized according to structured ontologies (Ives et al. 2017).
Although the development of AOPs has a great potential to address existing knowledge gaps, AOP development and assembly is laborious and time-consuming, since extensive toxicity data need to be gathered. Much of the information that is accumulating derives from omics technologies, high-throughput testing with robots [ToxCast (U.S. EPAa) (Judson et al. 2010)], and novel databases derived from the compilation of heterogeneous information such as the Comparative Toxicogenomics database (CTD) (Davis et al. 2018). Therefore, the development of innovative computing methodologies that allow the prioritization of chemicals according to their inferred threats is highly relevant both for the research community and for health agencies (Richard et al. 2016; Thomas et al. 2013). Such in silico methods that use available data sources also can accelerate the description of new AOPs and provide integrated data to increase the information content of existing AOPs (Berggren et al. 2015; OECD 2014).
The breadth of the currently available scientific literature and diversity of synonyms for chemicals complicates meaningful integration of the information. Thus, it can be difficult to completely and accurately acquire the information on a selected topic, even if specific databases related to a given field have been compiled and information stored [e.g., the Developmental and Reproductive Toxicology database, DART (NIH and TOXNETb)]. In addition to enriching toxicological databases, there is a need for tools allowing better exploration of available databases (including available published literature), and to improve text mining (TM) is such a way to facilitate the establishment of links between a chemical and relevant AOP components. Such tools should be able to explore a wider range of data and have the potential to prioritize the chemical–health outcome connections. We describe here a strategy that integrates a new tool called AOP-helpFinder version 1.0, downloadable on github (https://github.com/jecarvaill/aop-helpFinder). Using the available literature, AOP-helpFinder can automatically find, extract, and score links between chemical substances (i.e., stressors) and diverse biological elements, i.e., molecular initiating events (MIEs), KEs, and AOs, which are the components of AOPs (Villeneuve et al. 2014). The novelty of AOP-helpFinder is that it consists of a hybrid approach that combines TM and graph theory to explore the contents of abstracts for the identification of reliable associations between a chemical and AOPs. The main objective of AOP-helpFinder is to assist toxicologists and biologists in the identification of relevant associations between AOP components and small molecules through the analysis of large-scale, existing (published in peer-review journals and databases), text-based knowledge (in vitro, in vivo, and in silico data). As a proof of concept, we applied AOP-helpFinder to bisphenol S (BPS), a structural analog of bisphenol A (BPA) that is suspected to have endocrine-disrupting properties (Karrer et al. 2018). The biological mode of action (MoA) and potential toxicity of BPS are still poorly characterized. Using our computational method, links between BPS and AOP components have been uncovered.
Methods
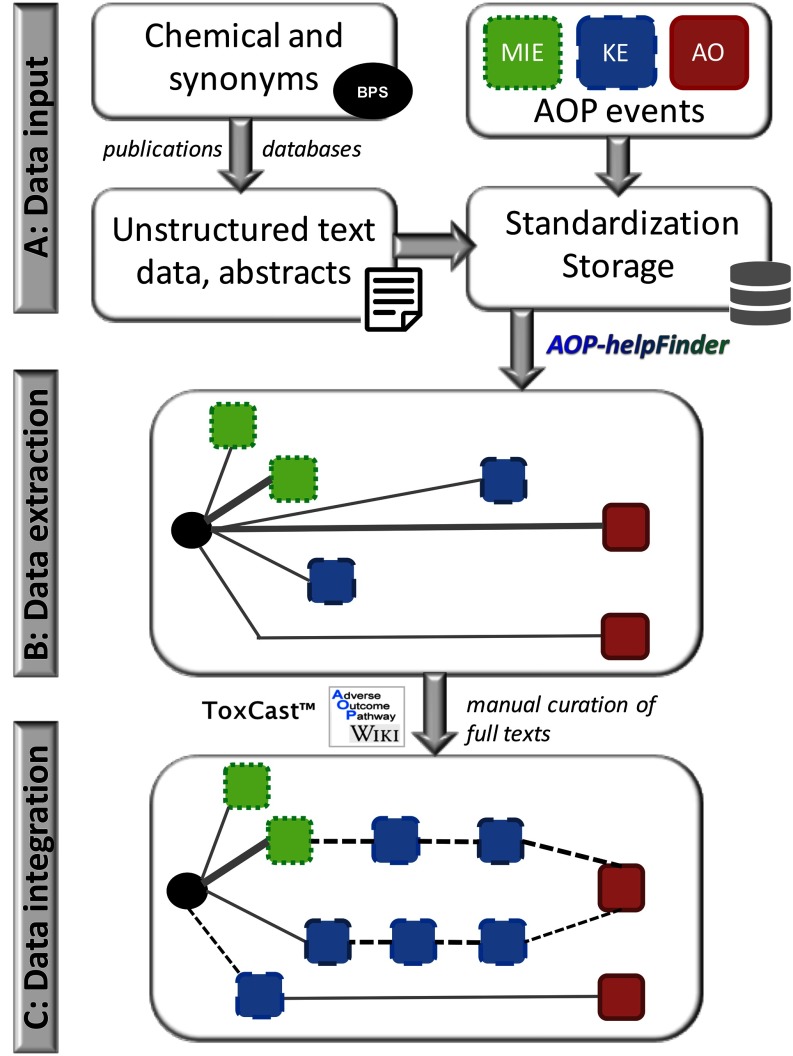
A workflow of the strategy is shown in Figure 1.
Figure 1.
Workflow of the strategy for linking bisphenol S (BPS) to health effects. (A) Data input: we compiled a) published data from multiple sources related to BPS and its associated toxic effects, and b) adverse outcome pathway (AOP) events to prepare a dictionary of AOP events (standardization and storage in an in-house database), which includes molecular initiating events [MIEs (squares with small dots)], key events [KEs (squares with large dots)], and adverse outcomes [AOs (square with solid lines)] as defined in the AOP-Wiki database. (B) Data extraction: we performed preprocessing followed by text mining of the data using the developed AOP-helpFinder tool in order to identify links between BPS (black circle) and different AOP events (MIE, KE, and AO). Scores, represented by the width of the edges, were calculated to indicate the confidence of the association. (C) Data integration: To fill potential gaps between two nodes (such as a KE and an AO), existing AOP information was integrated into the network as well as information from the ToxCast database. Full-text manual curation of the identified publications (by text mining) was performed by experts, thus increasing the availability of knowledge (dashed nodes/edges lines).
Data Input: Data Description and Preprocessing
Development of the adverse outcome pathway dictionary.
In order to explore associations between AOP-related terminologies (e.g., “obesity”) and a term of interest (here a chemical), an AOP dictionary that includes AOP events, i.e., MIEs, KEs, and AOs (Karrer et al. 2018), was generated. From the AOP-Wiki database, we downloaded the available .xml file that contains all AOP information (aop-wiki-xml-2017.gz, 3 July 2017). We extracted the AOP identifier (for example, “72”), the key event name (i.e., “obesity”), the key event identifier (“1447” for obesity), and the key event type (that is, whether the term is a MIE/KE or AO; for example, adipogenesis is defined as an AO). The AOP dictionary contained two files: one containing the AO names and one with the MIE/KE names. MIEs and KEs were combined into one unique table.
Development of the disease dictionary.
Controlled disease vocabulary from the U.S. National Library of Medical Subject Headings (MeSH, https://www.nlm.nih.gov/mesh/meshhome.html) was used, which represented 11,850 disease terms (downloaded from the CTD database as of September 2017) (Davis et al. 2017).
Development of the bisphenol S dictionary.
According to the data sources, BPS is identified by different terms for its name, synonyms, and Chemical Abstracts Service (CAS) registry number. In order to capture, as fully as possible, the existing biological and toxicological information related to BPS, chemical terms were retrieved using the PubChem database (NIH) (Table S1).
Text-based toxicological data.
To compile abstracts linked to BPS, we used an integrative approach that consisted of manual searches of five specific toxicological databases; some from the U.S. Toxnet platform [the Chemical Carcinogenesis Research Information System (CCRIS) (NIH and TOXNETa), DART, Toxicology Literature Online (TOXLINE) (NIH and TOXNETd), and Hazardous Substances Data Bank (HSDB) (NIH and TOXNETc) databases, which are collections of publicly available information] and the Registry of Toxic Effects of Chemical Substances (RTECS) database (Biovia). For each database, using the BPS dictionary, we extracted information concerning abstracts, authors, journal, year of publication, title, toxic effects, targets, and species. According to the databases, these data were mentioned under various fields. To avoid the duplication of information (since the same article can be present in several data sources) and subsequent overestimation of MIE/KE/AO, the parsing of extracted data was necessary since the toxicological data are not collected under the same field in the different data sources. For example, in the CCRIS format, the field “Target data” was found, and in the RTECS source, the field “Toxics Effects” was found. Therefore, to facilitate further analysis, three fields that contained toxicological information were retained to be screened against AOPs. These three fields were “Abstract,” “Toxic effects,” and “Target data,” since MIE, KE, and AO can be found in all these fields. The DART database provided more than journal references related to teratology and other aspects of developmental and reproductive toxicology. The CCRIS database contained data curated from published studies that possess chemical records that have carcinogenicity, mutagenicity, tumor promotion, and tumor inhibition test results. TOXLINE is a database based on bibliographic data, which includes specialized journals. It provided references from diverse fields, including toxicological effects of environmental chemicals. The HSDB is a database focusing on the toxicology of potentially hazardous chemicals. The RTECS database is a comprehensive collection of basic toxicity information that includes various types of chemicals such as drugs, food additives, and environmental pollutants. It covers six categories of toxicity data: acute toxicity, tumorigenicity, mutagenicity, skin and eye irritation, reproductive effects, and multiple dose effects.
Development of a relational database for storing data.
All selected information were stored in an in-house database to facilitate further analyses. Data were entered once in a relational SQLite, version 3.27.2, (https://www.sqlite.org/index.html) database to avoid any redundancy. The architecture of the database consisted of twelve tables, in which the previously compiled information [database source, first author, journal, year of publication, title, data text (abstract, target text, and toxic effects information), related animal and sex information, and MIE, KE, and AO data linked to AOP] were stored and connected together.
Data Extraction: The AOP-helpFinder Tool
To identify reliable associations between MIE/KE/AO and BPS, we developed a new method called AOP-helpFinder, which is based on Natural Language ToolKit (NLTK, version 3.2.5) (https://www.nltk.org/) and on Dijkstra graph theory (Dijkstra 1959). NLTK is a leading platform for building programs that employ human language data. It contains a suite of libraries and small programs for symbolic and statistical natural language processing under the Python programming language. NLTK initially was created for fields such as cognitive science, artificial intelligence, and machine learning, to mention a few areas. Dijkstra’s algorithm identifies the shortest path between two nodes in a graph, and in the AOP-helpFinder, it was used to determine the shortest path between words (by computing the distance between the terms, for example between a substance and an AOP event in an abstract). The AOP-helpFinder is a multistep TM procedure, consisting of the following:
-
1.
data preprocessing
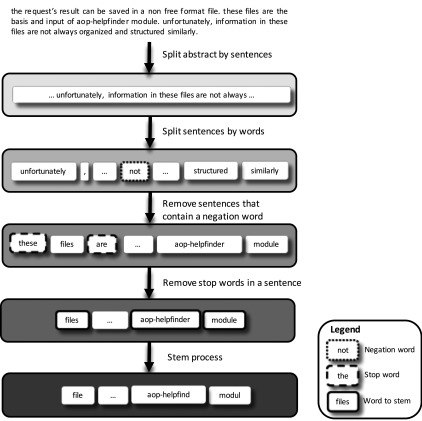
To maximize the chances of matching MIEs, KEs, and AOs in text data, it was necessary to clean and simplify them (Figure 2). This consisted of filtering out the noise and stemming the words remaining (the stem is the root or main part of a word to which inflections or formative elements are added), as well as considering spaces, conjunction, and punctuation. For example, for AOP 7, the text “Aromatase (Cyp19a1) reduction leading to impaired fertility in adult females” was simplified to “aromata cyp19a1 reduct lead impair fertil adult female.” This multistep preprocessing consisted of a) dividing an abstract into sentences, b) splitting all sentences into words, c) removing sentences that contain a negation word (never, neither, no, not, did not, hasn’t, should not, …) to reduce the risk of false positives, d) deleting stop words (coordinating conjunction, punctuation, most common words) from sentences, and e) stemming the remaining words. As a result, we obtained stemmed data for information related to AOPs and text data related to the chemical of interest ready to be screened against the AOPs’ data stored into the database.
-
2.
identification of association and scoring function
Two different approaches were developed to screen for AO and MIE/KE terms. As AO is more related to a few words and MIE/KE to complex sentences, the approaches were based on the calculation of scoring functions that took into consideration the complexity of the terms/sentences related to AO/KE/MIE.
Figure 2.
Data preprocessing in the AOP-helpFinder tool. Before identifying and scoring associations between a chemical and an adverse outcome pathway (AOP) event, text information collected from multiple sources (publications, databases) was preprocessed in order to obtain stemmed data, which were stored in a relational in-house database. Negation words (never, neither, no, not, did not, hasn’t, should not, …) were identified with squares surrounded with small dots, stop words (coordinating conjunctions, punctuations, most common words such as “the”) with squares surrounded with large dots, and “words to stem” with solid lines (e.g., files file or finder find, considering that a word has a single stem, namely the part of the word that is common to all its inflected variants).
Matching AO data.
The stemmed AO were screened in the “abstracts,” “targets,” and “toxic effects” fields. Associations between chemicals and AO are likely to have different meanings if they are found in the beginning or at the end of an abstract. Generally, in published studies, a working hypothesis is found at the beginning of an abstract. However, chemical–AO associations co-mentioned at the end of an abstract are more likely to be considered as true positives because results and findings are often cited at the end of an abstract. Therefore, in order to differentiate working hypothesis from findings, we calculated a score based on the position () using . Both the position of the AO term relative to the other words () and the total number of words in the text mined abstract () are taken into consideration. An optimal value is around 1.0, which corresponded to the last position in the abstract. The more the AO is placed toward the end of the abstract, the more it can be considered as a result and not a working hypothesis.
Matching molecular initiating event and key event data.
On the average, a MIE/KE consisted of four terms in the AOP-Wiki database. Fifty-four percent of the MIE/KE contained at least four terms (for example, “allergic contact dermatitis challenge”), and among these, 21% were composed of exactly four terms. Moreover, 40% of the MIEs/KEs had between four and six terms. The minimum number of terms was one (such as “obesity”), and the maximum was 21 terms [KE 1,119: failure gamma glutamyl carboxylation glutamine residues clotting factors ii vii ix x under carboxylation clotting factors (gamma carboxy prothrombins)]. Based on these data, when a MIE/KE was composed of terms, we assumed that 25% of the information could be missing (for example, for “allergic contact dermatitis challenge,” we retrieved “allergic contact dermatitis”), whereas for a MIE/KE having , we assumed that every term was important. Therefore, we applied a threshold of 75% of stemmed terms in order to identify relevant MIE/KE in the screened abstracts. The matching process continued under these conditions; otherwise, it was stopped.
The search for MIE/KE in an abstract was based on the creation of acyclic weighted graphs that use the positions of stemmed terms. The edges characterizing these graphs had a weight that was the distance between two nodes (positions of two different terms). Given the notion of weight on edges, we decided to use Dijkstra’s algorithm because it supports finding the shortest path in a weighted acyclic graph (Dijkstra 1959). The acyclic property was composed of a step-by-step process, starting from the term to the term , without returning to the nth term (unidirectional). Since the graphs that are formed are acyclics, the algorithmic complexity was solved by a maximum of calculations, where represents the number of nodes, and m the number of edges in a graph O. Dijkstra’s theory is represented by a quadratic complexity, i.e., . Considering all the existing MIE/KE, which have an average of four terms, the use of Dijkstra’s algorithm resulted in a faster calculation compared with the use of a table. Therefore, the weight of the edges was used to find the shortest paths of the stemmed terms. The nearer the stemmed terms are to one another, the lower the total weight is. In order to identify a MIE/KE in texts and to classify the MIE/KE scores, we calculated a weighted score () using . The is based on the lowest total weight calculated using graph theory () and the total number of words in a MIE or KE (). The ideal score is 1.0. At this score, all the stemmed terms are present and close to one another; that is, the expression/sentence found in the abstract is very similar to the MIE/KE of the reference (AOP-Wiki). The more the score varies from 1.0 (higher or lower), the less the MIE/KE is likely. Scores indicate that there are missing MIE/KE stemmed terms in a sentence of an abstract. Scores indicate that MIE/KE stemmed terms are far from each other. Scores indicate that stemmed terms are missing (if a stemmed term is missing, a minus sign is added).
Performance of the AOP-helpFinder.
Manual curation of the abstracts was used to assess the ability of the AOP-helpFinder to find relevant associations.
Data Integration: Linking a Chemical to Health Effects
Integrating known adverse outcome pathways and ToxCast information.
A biological network of the key AOP event names for BPS was developed using the events (MIE, KE, and AO) identified with the AOP-helpFinder. To fill potential gaps between a MIE and a KE, or a KE and an AO, integration of an updated version of the AOP-Wiki (23 April 2018) was performed. We also integrated additional evidence of molecular events related to BPS using the ToxCast database (April 2018). In this 2018 version, ToxCast provided data for 9,076 chemicals, tested on 359 assays, and information for 1,192 high-throughput end-point components.
Integrating published data by manual curation.
AOP-helpFinder identified relevant publications. To estimate their toxicological relevance as well as the importance of the experiments carried out for BPS, scientific experts manually analyzed the abstracts of the publications. For example, articles that used BPS to introduce a study or as a reference molecule in a study that involved other compounds were not taken into consideration for further analysis. Following this first manual curation, articles were classified according to the AOs to which they belong, and the corresponding full texts were read. This step highlighted the molecular events that were most often described, and particular attention was paid to those that are confirmed repeatedly by different articles. Based on the extracted results, AOP networks were suggested, and experts examined additional publications (cited references in the selected publications) to identify potential indirect linkages between events (KEs).
Results
Data Preparation
We first had to retrieve the AOP terms on one hand and the BPS synonyms on the other hand. Annotations of AOPs were retrieved by downloading the full AOP-Wiki database. As of July 2017, there were 1,073 biological events (MIEs and KEs) and 61 defined AOs, some of which were under development, and others were included in the OECD work plan. These events were at different biological levels and ranged from the molecular (ID 18: Activation, AhR) to cellular (ID 52: decreased, calcium influx), tissue (ID 68: accumulation, collagen), organ (ID 161: increase, liver and splenic hemosiderosis), individual (ID 270: induction, sustained hepatotoxicity), and population (ID 417: skewed, sex ratio). To create the list of terms related to BPS (i.e., synonyms), we queried the PubChem database (Kim et al. 2019). We were able to retrieve 129 terms that include the MeSH vocabulary and depositors’ names (see Table S1). For example, BPS has several synonyms such as bis(4-hydroxyphenyl)sulfone and 4,4’-sulfonyldiphenol, and has the CAS number of 80-09-1. Raw text data that mention BPS or synonyms were acquired from five toxicological databases (CCRIS, DART, TOXLINE, HSDB, and RTECS), and processed in an SQLite3 database. A total of 109 publications involving BPS and toxicological effects were identified (Table S2). Then, in order to identify co-occurrence between the BPS compound and the AOP terms in the 109 abstracts extracted from five databases, the BPS dictionary and the AOP dictionary were used.
Process of the AOP-helpFinder Tool
The central feature of the AOP-helpFinder was its ability to find associations between compounds (e.g., BPS) and AOP terms using a NLTK TM approach. After the calculation of scoring functions using the Djikstra graph theory, only the most relevant information to support linkage of a compound to an AOP was kept. As a result, the relationships between a chemical (i.e., a stressor) and a MIE, which is the initial component of an AOP (e.g., chemical binding to a receptor or a protein), were identified. Other types of associations also were uncovered between BPS, AOP KEs, and AOs, which are the ultimate component of an AOP that impacts health at an individual or a population level.
Performance of the AOP-helpFinder
To evaluate the performance of the method, we manually curated the 109 abstracts. We obtained a success rate of 76% with 85 true positive (existing MIE/KEs that were found) and 27 false positive (MIE/KEs found by the AOP-helpFinder, but not present in the 109 publications). The sensitivity of the method, i.e., the ability to find the right AOP related term, was 67%, with 41 false negatives (not found by AOP-helpFinder, but existing in at least one of the abstracts; for example, “reproductive defects” was not found but could be linked to the KE “decreased reproductive success”).
The relevance of the position score was also evaluated. In other words, we evaluated the hypothesis that the position of the AO terms in the abstract relates to the context, that is, whether the statement was a working hypothesis or a finding. We manually checked the abstracts that co-mention BPS and at least one AO, and found a good correlation between the pS and the real position of the AO term. All the high pS scores (close to 1) were related to a result in an abstract, whereas low pS related to the introduction or a hypothesis in the abstract. For example, a pS of 0.95 was obtained for steatosis from the study of Héliès-Toussaint et al. (2014). Examination of the abstract revealed that BPS and steatosis were co-mentioned in the last sentence (“the findings suggest that both BPA and BPS could be involved in obesity and steatosis processes, but through two different metabolic pathways”).
Linking Bisphenol S to Health Effects
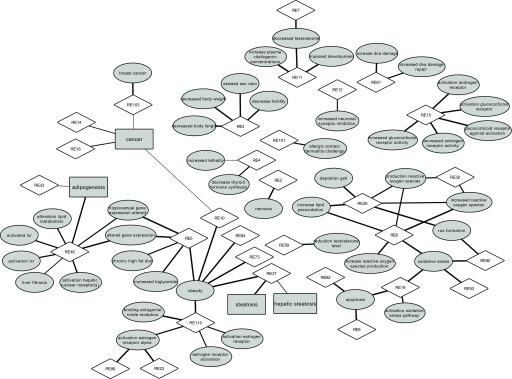
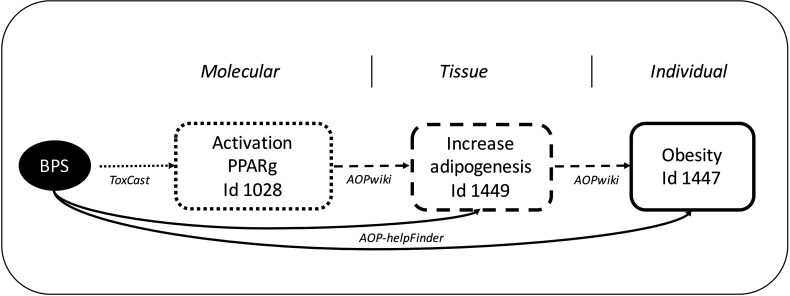
The 109 publications that were obtained were filtered using AOP-helpFinder and only the ones mentioning associations between AOP-related terms (MIEs, KEs, and AOs) and BPS were kept (see Table S2). Among the 109 publications, seven co-mention BPS and an AO term (e.g., “steatosis”), and 46 co-mention BPS and MIE/KE terms (for example, “activation estrogen receptor alpha” or “decrease body weight”). Very few AOP event duplicates were retrieved. Therefore, for further analysis, we kept a total of four unique AOs (for example, the AO “cancer” was retrieved in four of the seven publications that co-mention BPS and an AO term) and 45 unique MIE/KEs. We also removed the publications with the insignificant term “decrease” found among the MIE/KE, when it was alone, as it could not be mapped to a specific biological event. Thus, we ended up with four AOs and 44 MIEs/KEs (Table 1) present in 31 references (Table S2). To visualize these findings, we displayed bipartite networks using Cytoscape, version 3.5.1 (https://cytoscape.org/) (Figure 3). Among others, an association was found between BPS and “decrease thyroid hormone synthesis.” This corresponded to KE 277 in the AOP-Wiki database “Thryroid hormone synthesis, Decreased” with a score of 0.75. The most relevant health outcomes resulting from our study were reproductive effects (decreased testosterone, skewed sex ration, decreased fertility), endocrine disruption (estrogen and androgen receptor activation, as observed biologically with BPA), and metabolism impairment (adipogenesis, increased triglycerides, obesity). Among the latter, the most optimal score was associated with obesity (KE 1,447 in the AOP-Wiki database, belonging to the AOP 72) with a wS value of 1. Furthermore, obesity was mentioned in six of the 31 publications (Figure 3) (Table S2). Few published articles link directly BPS to obesity (Héliès-Toussaint et al. 2014); nevertheless, our approach allowed us to highlight this connection. Since BPS is a substitute for BPA, which is suspected to be an obesogen, the link between BPS and obesity that we found is of interest. The AOP-helpFinder tool allowed us to identify publications that mention a link between BPS and increased adipogenesis (ID 1,449 in the AOP-Wiki database). In order to have a more accurate assessment of the MoA leading to obesity, an integrative systems biology approach was used. We integrated information from current available sources such as the AOP-Wiki and ToxCast databases (as of April 2018). In the AOP-Wiki database, a MIE involving the peroxisome proliferator–activated receptor gamma (PPARγ), “PPARγ, activation” (ID 1028 in the AOP-Wiki database) was identified as being connected to other AOP terms such as “increase adipogenesis” and “obesity.” Indeed, PPARγ activation is a known initiating event for increased adipogenesis (Lefterova et al. 2014). PPARγ also is known to be involved in the regulation of adipocyte differentiation and has been implicated in several pathologies including obesity, diabetes, and cancer (Polvani et al. 2016). The investigation of the ToxCast database allowed us to identify positive associations between BPS and PPARγ through two assays, i.e., ATG-PPARγ_TRANS_up (mRNA activation in human HepG2 cell lines, ) and NVS_NR_hPPARγ (active binding, ) (Figure 4).
Table 1.
List of the adverse outcomes (AOs) and molecular initiative and key events (MIEs and KEs) associated with bisphenol S (BPS).
| Name | Score (pS or wS)a | AOP-Wiki ID |
|---|---|---|
| AO name | ||
| Adipogenesis | 0.96 | 16 |
| Cancer | 0.47 | 11 |
| Hepatic steatosis | 0.28 | 17 |
| Steatosis | 0.95 | 18 |
| MIE/KE name | ||
| Activated LXR | 3 | 1,421 |
| Activation androgen receptor | 1.6 | 785 |
| Activation estrogen receptor | 4 | 1,181 |
| Activation estrogen receptor alpha | 1,065 | |
| Activation glucocorticoid receptor | 2 | 122 |
| Activation hepatic nuclear receptor(s) | 1,157 | |
| Activation LXR | 3 | 167 |
| Activation oxidative stress pathway | 1,238 | |
| Allergic contact dermatitis challenge | 312 | |
| Alteration lipid metabolism | 2.66 | 1,060 |
| Altered gene expression | 1.66 | 1,239 |
| Apoptosis | 1 | 1,262 |
| Binding antagonist NMDA receptors | 201 | |
| Breast cancer | 1 | 1,193 |
| Chronic high-fat diet | 1,454 | |
| Decrease fertility | 2 | 330 |
| Decrease thyroid hormone synthesis | 277 | |
| Decreased androgen receptor activity | 742 | |
| Decreased body length | 1.66 | 315 |
| Decreased body weight | 1.66 | 864 |
| Decreased testosterone | 1 | 808 |
| Depletion of GSH | 1 | 130 |
| Estrogen receptor activation | 4 | 1,180 |
| Glucocorticoid receptor agonist activation | 494 | |
| Hippocampal gene expression altered | 756 | |
| Impaired development | 1 | 577 |
| Increase DNA damage | 1 | 1,194 |
| Increase lipid peroxidation | 2.3 | 1,445 |
| Increase plasma vitellogenin concentrations | 220 | |
| Increase reactive oxygen species production | 257 | |
| Increased DNA damage repair | 1,281 | |
| Increased lethality | 8.5 | 342 |
| Increased neuronal synaptic inhibition | 1,015 | |
| Increased reactive oxygen species | 1,115 | |
| Increased triglyceride | 2 | 881 |
| Liver fibrosis | 5.5 | 344 |
| Increased glucocorticoid receptor activity | 1,396 | |
| Necrosis | 1 | 1,263 |
| Obesity | 1 | 1,447 |
| Oxidative stress | 1 | 210 |
| Production reactive oxygen species | 249 | |
| Reduction testosterone level | 2.3 | 446 |
| ROS formation | 1 | 1,278 |
| Skewed sex ratio | 1 | 417 |
Note: GSH, glutathione; LXR, liver X receptor; NMDA, N-methyl-d-aspartate; ROS, reactive oxygen species.
The position score (pS) reflects the position of the AO term in the abstract. The weighted score (wS) allows the user to quantify the extent to which a MIE/KE expression is retrieved in an abstract.
Figure 3.
Bipartite networks of the relevant references and adverse outcome pathway (AOP) events for bisphenol S (BPS), identified by text mining. The bipartite networks consisting of 48 AOP terms: 4 AOs (rectangle), 44 molecular initiating events (MIEs)/key events (KEs) (ellipse), and 31 references (RE) (diamond) (see Table S2 for the corresponding references). The width of each AOP term–reference edge is proportional to the corresponding scores: position score (pS) for AO and weighted score (wS) for MIE/KE. Note: LXR, liver X receptor; NMDA, N-methyl-d-aspartate.
Figure 4.
Potential mode of action for bisphenol S (BPS) leading to obesity resulting from text mining and integrated systems toxicology (data integration from the AOP-Wiki and ToxCast databases). Small dots (arrows and square lines) indicate information from the ToxCast database, large dots from the AOP-Wiki database, and solid lines, information identified using AOP-helpFinder.
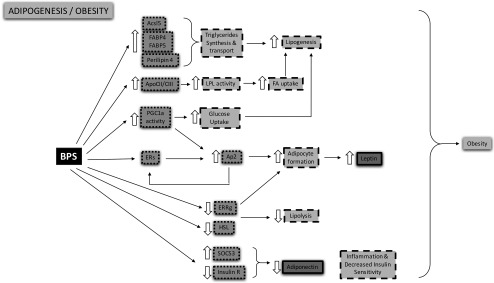
We attempted to identify other biological pathways that link BPS to obesity. We sought to build patterns with coherent MoA using the full text of the publications that were identified by AOP-helpFinder through the screening of abstracts (Figure 5). Several molecular and cellular pathways, whose disruption could be linked to obesity, were identified: a) formation of adipocytes, b) increased lipogenesis, or c) decreased lipolysis (Table 2). Regarding adipogenesis, the activation of estrogen receptor alpha () by BPS was shown to trigger the expression of several adipogenic markers in MCF-7 cells (Molina-Molina et al. 2013), HEK293T cells (Teng et al. 2013), and preadipocytes (Boucher et al. 2016a), such as Ap2 (adipocyte protein 2, a carrier protein for fatty acids). Since Ap2 was also shown to increase the expression of in ER-negative MDA-MB-231 cells (McPherson and Weigel 1999), the existence of a positive feedback loop contributing to adipogenesis could be suggested. Interestingly, Ap2 was also shown to be a transcriptional target of PPARγ in 3T3-L1 adipocytes (Rival et al. 2004), whose pathway was activated by BPS (Boucher et al. 2016a). Further, the expression of the PPARγ coactivator 1 α, which is recruited by PPARγ to transactivate its target genes, was shown to be specifically up-regulated by BPS in the 3T3-L1 cell line (Héliès-Toussaint et al. 2014).
Figure 5.
Identification of the mechanistic effects linking BPS to obesity. The AOP-helpFinder tool allowed us to select relevant publications that associated BPS, key events (KEs), and obesity. A manual curation of these studies by experts led to the identification of potential molecular targets of BPS (small dots surrounding squares), molecular processes (large dots surrounding squares), and hormone disruption (solid lines). Thick arrows: “increased” or “decreased.” Note: ApoC, apolipoprotein C-III; Ap2, adipocyte protein 2; Ascl5, Acyl-CoA synthetase long-chain family member 5; BPS, bisphenol S; ERRg, estrogen-related receptor gamma; ERs, estrogen receptors; FA, fatty acids; FABP, fatty acid–binding proteins; HSL, hormone-sensitive lipase; insulin R, insulin receptor; LPL, lipoprotein lipase; PGC1a: peroxisome proliferator–activated receptor gamma coactivator 1-alpha; SOCS3, suppressor of cytokine signaling 3.
Table 2.
List of the seven publications, identified with AOP-helpFinder, fully explored to identify biological linkage between bisphenol S and obesity.
| References | Detailed MIE–KE |
|---|---|
| Molina-Molina et al. 2013 | Activation of human and by BPS |
| Boucher et al. 2016a | Activation of the expression of the adipogenic marker, adipocyte protein 2 (Ap2) (blocked by antagonist) |
| Teng et al. 2013 | Activation of ( of 2.2 ) |
| Héliès-Toussaint et al. 2014 | BPS increased lipid content in the 3T3-L1 cell line and decreased the expression of |
| Boucher et al. 2016a | BPS transactivates the expression of FABPs and perilipin 4 (microarray data, differentially expressed genes). |
| Crump et al. 2016 | BPS transactivates the expression of ACSL5. |
| Ivry Del Moral et al. 2016 | BPS decreased the expression of HSL. |
Note: ACSL5, acyl-CoA synthetase long-chain family member 5; BPS, bisphenol S; , half-maximal effective concentration; ER, estrogen receptor; ; estrogen-related receptor gamma; FABPs, fatty acids binding proteins; HSL, hormone-sensitive lipase; KE, key event; MIE, molecular initiating event.
Additional observations collected from the retrieved articles could also contribute to the increased adipogenesis outcome, notably through metabolic disruption: BPS promotes the cellular uptake of glucose in two cell lines (3T3-L1 and HepG2), which could contribute to the production of glycerol-3-phosphate and then subsequently to the synthesis of triglycerides (Héliès-Toussaint et al. 2014). One study also has linked the activation of PPARγ to an increased uptake of glucose (Zhang et al. 2017). Several key factors that regulate lipogenesis and lipolysis (including fatty acids binding proteins (FABP4 and FABP5) (Boucher et al. 2016b), perilipin 4 (Crump et al. 2016; Boucher et al. 2016b), or acyl-CoA synthetase long-chain family member 5 (ACSL5) (Crump et al. 2016) also were found to be associated with BPS exposure by AOP-helpFinder. Together, these proteins could contribute to an increased fatty acid uptake in preadipocytes (Boucher et al. 2016a) and to the production of triglycerides and, subsequently, of lipid droplets. Furthermore, BPS negatively regulates the expression of the hormone-sensitive lipase (HSL) in adipose tissue of C57BL/6 mice (Ivry Del Moral et al. 2016) and estrogen-related receptor gamma () in the 3T3-L1 cell line (Héliès-Toussaint et al. 2014). regulates positively the expression of uncoupling protein 1 during the browning of 3T3-L1 adipocytes, which suggests that its down-regulation contributes to the increase in white adipose tissue, whereas down-regulation of HSL contributes to decreased lipolysis (Akter et al. 2008). In addition to metabolic disruption, AOP-helpFinder identified references that showed that BPS could be associated to the establishment of insulin resistance through both decreased expression of the insulin receptor and an increased inflammatory response in adipose tissue of C57BL/6 mice (Ivry Del Moral et al. 2016). Taken together, the studies retrieved through AOP-helpFinder suggested a link between BPS exposure and an AOP network related to adipogenesis and metabolic disruption.
In order to test additional applications of the AOP-helpFinder tool, we screened other data sources such as PubMed and included more disease terms than those in the AOP dictionary. Specifically, we used AOP-helpFinder to investigate BPS in the 109 selected publications, but instead of using AOP terms, the 11,850 MeSH disease terms were used (downloaded from the CTD database as of 28 September 2017). Obesity (MESH: D009765) was retrieved with a pS value of 0.93. Other disorders or phenotypes were also found, such as “diabetes mellitus” (pS of 0.84) or “body weight” (pS of 0.73) (see full list in Table S3).
Thus, our computational approach highlighted the need to integrate various data sources (publications, databases) in order to capture as much information as possible when studying links between a selected chemical and AOs.
Discussion
We have developed a novel computational approach using TM, graph theory, and systems biology to improve our capacity to link chemicals with AOPs. The AOP-helpFinder tool is particularly well suited to explore putative toxicity of chemicals to be used as replacements for toxic compounds, since studies exploring the human effects of the substitutes usually are scarce, whereas public decisions concerning the suitability of their use are urgently needed. The ability to identify chemical–biological associations is illustrated using BPS as an example. There is a large amount of published data for BPA. However, much less information is available for alternatives to BPA (BPS, for example), even though there has been a recent surge in the number of publications due to increased use of alternatives in consumer products (Rochester and Bolden 2015). We mined available information from multiple sources using the AOP-helpFinder tool. The ability to make novel observations or to confirm suspected effects using this tool, combined with integrative systems biology, is clearly illustrated by our observations on putative links between BPS and obesity. This method increases our knowledge, may give orientation for further experimental analysis, and supports the development of models that provide alternatives to animal testing [Replacement, Reduction and Refinement (3Rs)]. The advantages of our approach include the rapid exploration and the integration of existing information from multiple sources (databases and literature). It therefore has the potential to accelerate information gathering and is useful particularly when data are limited and present in diverse sources, which is the case for a large number of chemicals.
The strategy described here is complementary to existing systems toxicological models already developed for the identification and prediction of linkages between chemicals and human health. Some of these models are hybrid methods that combine multiple methods (Krysiak-Baltyn et al. 2014). Others are chemical structure based (Thomas et al. 2013; Ball et al. 2016), involving the physicochemical and reactivity properties of the chemicals [read-across and quantitative structure–activity relationships (QSARs)]. Both are important for the interactions with specific biological targets and pathways, and therefore allow the prediction of toxic effects (Dang et al. 2017; Zang et al. 2017). Following the OECD recommendations for the construction of robust QSARs (OECD 2014), several models are now available such as the OPERA version 1.5 (Mansouri et al. 2018) or the VEGA platform (www.vegahub.eu). Recently, a new tool that integrates both chemical similarity and biosimilarity has been developed, the Chemical In vitro-In vivo Profiling (CIIPro) (Russo et al. 2017). This tool profiles compounds of interest utilizing biological data from public resources. It uses the data for read-across assessment with the aim of predicting complex bioactivities. Several web-based tools, such as the REACHAcross™ tool (Hartung 2016) and the ChemProt database (using virtual screening) (Kim Kjærulff et al. 2013), are available.
Other methods are primarily based on the delineation of the MoA and on systems biology. Chemicals can disrupt a variety of pathways and lead to complex manifestation of toxicities. However, different chemicals can have similar MoA if they dysregulate the same target or targets that belong to the same signaling pathway (Bopp et al. 2018). Such pathway-based approach is recapitulated in AOPs and can be addressed by systems biology. Integrative systems biology models, which combine toxicogenomics, protein–chemical associations, protein–protein interactions (Audouze et al. 2010; Audouze and Grandjean 2011), disease phenotype information, genome-wide association, and genetic disease similarities (Audouze et al. 2013), have revealed molecular mechanisms of xenobiotics and have linked them to diseases such as obesity and endocrine disruption. Recently, data-driven approaches have been designed to generate and enrich AOP descriptions. For example, Nymark et al. (2017) proposed a multistep procedure based on an in silico pipeline to identify a network of functional elements for pulmonary fibrosis-associated genes. This generated novel AOP-linked molecular pathways (WP3624 in WikiPathways). Another recent study based on a systems toxicology profiling approach using information from the U.S. Environmental Protection Agency database (ToxRefDB) (U.S. EPAb), the ToxCast database, and a comprehensive literature analysis, identified links between environmental chemicals, molecular targets, and AOs for male reproduction (Leung et al. 2016). Another type of data-driven approach is the use of TM and frequent item-set mining (FIM) to extract fragmented information in texts (e.g., abstracts or the full text of publications) (Jensen et al. 2012) or large datasets (clinical data, biobank, electronic patient records) (Jiang et al. 2019) and establish relevant associations. Currently, such methods are used widely in the biomedical area to identify information regarding biological entities (e.g., genes and proteins, metabolites, phenotypes, pathways) (Jensen et al. 2006; Krallinger et al. 2008). These approaches have been used less frequently in the toxicological field. However, in the case of U.S. biomonitoring surveys and Danish clinical studies, a study based on FIM has led to the successful identification and prioritization of connections between environmental chemicals, biomarkers, and human disorders (Krysiak-Baltyn et al. 2014). FIM has been also used to create computationally predicted AOPs, using the chemicals as the common aggregators between data (Oki and Edwards 2016), employing information from the in vitro ToxCast HTS assays and disease information from the CTD database (Davis et al. 2017). As a case study, they predicted an uncharacterized connection between the aryl hydrocarbon receptor and glaucoma resulting from changes in CYP1B1. This connection is in agreement with experimental data that shows an association between rare CYP1B1-activating mutations and congenital glaucoma (Alsaif et al. 2018; Stoilov et al. 1997).
As compared to the abovementioned structural or systems biology–based methods, the added value of the presented approach is that it combines the exploration of several databases (including ToxCast) with an improved method of TM that is based on the scoring of connections between chemicals and relevant AOP components. Therefore, it explores a wider range of data and has the potential to prioritize the chemical–health outcome connections, thus allowing further targeted studies. It also provides hints as to potential MoAs and, therefore, could be of use in risk assessment. We believe that such a proposed approach could be improved by screening more available data (the PubMed database, for example) and by including other disease terms in the disease dictionary that was initially limited to the AOP terms. As an example, when we used MeSH disease terms, the method was also efficient and confirmed the links between BPS and obesity.
In addition to providing a new tool to explore the putative toxicity of chemicals and chemical mixtures, our study highlights the need for methods improvements and for additional tools. Novel computational systems toxicology models are needed to better characterize and predict the complex toxic effects of the chemicals to which humans are exposed. The current lack of high-quality data for some of the environmental chemicals and the current limitation of defined AOPs restricts approaches such as the one described here. An extension of AOP-helpFinder, which would implement a comprehensive dictionary of synonyms related to AOP terms, would increase the sensitivity of the method. Furthermore, the manual curation, used here, of the full texts of the selected publications could be automated by a TM approach. A recent comprehensive comparison of text mining of 15 million full-text articles vs. their corresponding abstracts from the period 1,823–2016 concluded that access to the full text improved findings (Westergaard et al. 2018).
We believe that the most relevant applications of the method described here are the delineation of chemical mixtures effects and a more rapid and efficient evaluation of the safety of substitutes to toxic chemicals. In both cases, it is critical to determine whether different chemicals exhibit similar MoA, and this can be accelerated by computational methods. Indeed, a critical issue in mixtures studies is to determine whether the compounds have similar modes of action (Kortenkamp and Faust 2018) because, in such a case, dose addition is the most appropriate method to assess the global effect of the mixture. Similarly, it is critical to determine whether substitutes to toxic chemicals exhibit an MoA that is similar to that of the parent compound. We have illustrated this aspect here by the study of BPS, which appears to be a putative obesogen, like BPA and other bisphenol compounds.
Conclusion
Exposure to chemical substances that can produce multiple health effects, such as endocrine disruption or metabolic disruption, represents one of the most critical public health threats at present. Novel, innovative computational methods, such as the AOP-helpFinder, are useful resources for exploring and predicting potential links among environmental chemicals and molecular targets, biological events, and AOs. We believe that the development and improvement of such in silico approaches has the potential to significantly impact relevant frameworks to assess chemical toxicity, such as the integrated approaches for testing and assessment, the reduction of animal testing, and the identification of safe chemical substituents.
Supplementary Material
Acknowledgments
The authors would like to acknowledge HBM4EU (https://www.hbm4eu.eu/), a project funded by the European Union’s Horizon 2020 research and innovation program under grant agreement no. 733032. This work was also supported by the University of Paris Descartes-USPC, INSERM, and Assistance Publique-Hôpitaux-de-Paris.
References
- Akter MH, Yamaguchi T, Hirose F, Osumi T. 2008. Perilipin, a critical regulator of fat storage and breakdown, is a target gene of estrogen receptor-related receptor alpha. Biochem Biophys Res Commun 368(3):563–568, PMID: 18243128, 10.1016/j.bbrc.2008.01.102. [DOI] [PubMed] [Google Scholar]
- Alsaif HS, Khan AO, Patel N, Alkuraya H, Hashem M, Abdulwahab F, et al. 2018. Congenital glaucoma and CYP1B1: an old story revisited. Hum Genet 1–7. [DOI] [PubMed] [Google Scholar]
- Ankley GT, Bennett RS, Erickson RJ, Hoff DJ, Hornung MW, Johnson RD, et al. 2010. Adverse outcome pathways: a conceptual framework to support ecotoxicology research and risk assessment. Environ Toxicol Chem 29(3):730–741, PMID: 20821501, 10.1002/etc.34. [DOI] [PubMed] [Google Scholar]
- Audouze K, Brunak S, Grandjean P. 2013. A computational approach to chemical etiologies of diabetes. Sci Rep 3:2712, PMID: 24048418, 10.1038/srep02712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audouze K, Grandjean P. 2011. Application of computational systems biology to explore environmental toxicity hazards. Environ Health Perspect 119(12):1754–1759, PMID: 21846611, 10.1289/ehp.1103533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audouze K, Juncker AS, Roque F, Krysiak-Baltyn K, Weinhold N, Taboureau O, et al. 2010. Deciphering diseases and biological targets for environmental chemicals using toxicogenomics networks. PLoS Comput Biol 6(5):e1000788, PMID: 20502671, 10.1371/journal.pcbi.1000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball N, Cronin MTD, Shen J, Blackburn K, Booth ED, Bouhifd M, et al. 2016. Toward Good Read-Across Practice (GRAP) guidance. ALTEX 33(2):149–166, PMID: 26863606, 10.14573/altex.1601251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berggren E, Amcoff P, Benigni R, Blackburn K, Carney E, Cronin M, et al. 2015. Chemical safety assessment using read-across: assessing the use of novel testing methods to strengthen the evidence base for decision making. Environ Health Perspect 123(12):1232–1240, PMID: 25956009, 10.1289/ehp.1409342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biovia. Registry of Toxic Effects of Chemical Substances (RTECS) database. http://www.3dsbiovia.com/products/collaborative-science/databases/bioactivity-databases/rtecs.html [accessed 25 August 2017].
- Bopp SK, Barouki R, Brack W, Dalla Costa S, Dorne JCM, Drakvik PE, et al. 2018. Current EU research activities on combined exposure to multiple chemicals. Environ Int 120:544–562, PMID: 30170309, 10.1016/j.envint.2018.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher JG, Ahmed S, Atlas E. 2016a. Bisphenol S induces adipogenesis in primary human preadipocytes from female donors. Endocrinology 157(4):1397–1407, PMID: 27003841, 10.1210/en.2015-1872. [DOI] [PubMed] [Google Scholar]
- Boucher JG, Gagné R, Rowan-Carroll A, Boudreau A, Yauk CL, Atlas E. 2016b. Bisphenol A and bisphenol S induce distinct transcriptional profiles in differentiating human primary preadipocytes. PLoS One 11(9):e0163318, PMID: 27685785, 10.1371/journal.pone.0163318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump D, Chiu S, Williams KL. 2016. Bisphenol S alters embryonic viability, development, gallbladder size, and messenger RNA expression in chicken embryos exposed via egg injection. Environ Toxicol Chem 35(6):1541–1549, PMID: 26606162, 10.1002/etc.3313. [DOI] [PubMed] [Google Scholar]
- Dang NL, Hughes TB, Miller GP, Swamidass SJ. 2017. Computational approach to structural alerts: furans, phenols, nitroaromatics, and thiophenes. Chem Res Toxicol 30(4):1046–1059, PMID: 28256829, 10.1021/acs.chemrestox.6b00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, Grondin CJ, Johnson RJ, Sciaky D, King BL, McMorran R, et al. 2017. The Comparative Toxicogenomics Database: update 2017. Nucleic Acids Res 45(D1):D972–D978, PMID: 27651457, 10.1093/nar/gkw838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, Wiegers TC, Wiegers J, Johnson RJ, Sciaky D, Grondin CJ, et al. 2018. Chemical-induced phenotypes at CTD help inform the pre-disease state and construct adverse outcome pathways. Toxicol Sci 165(1):145–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkstra EW. 1959. A note on two problems in connexion with graphs. Numer Math 1(1):269–271, 10.1007/BF01386390. [DOI] [Google Scholar]
- Hartung T. 2016. Making big sense from big data in toxicology by read-across. ALTEX 33(2):83–93, PMID: 27032088, 10.14573/altex.1603091. [DOI] [PubMed] [Google Scholar]
- Heindel JJ, Blumberg B. 2018. Environmental obesogens: mechanisms and controversies. Annu Rev Pharmacol Toxicol 59:89–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Héliès-Toussaint C, Peyre L, Costanzo C, Chagnon MC, Rahmani R. 2014. Is bisphenol S a safe substitute for bisphenol A in terms of metabolic function? An in vitro study. Toxicol Appl Pharmacol 280(2):224–235, PMID: 25111128, 10.1016/j.taap.2014.07.025. [DOI] [PubMed] [Google Scholar]
- Ives C, Campia I, Wang RL, Wittwehr C, Edwards S. 2017. Creating a structured adverse outcome pathway knowledgebase via ontology-based annotations. Appl In Vitro Toxicol 3(4):298–311, PMID: 30057931, 10.1089/aivt.2017.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivry Del Moral L, Le Corre L, Poirier H, Niot I, Truntzer T, Merlin JF, et al. 2016. Obesogen effects after perinatal exposure of 4,4′-sulfonyldiphenol (Bisphenol S) in C57BL/6 mice. Toxicology 357–358:11–20, PMID: 27241191, 10.1016/j.tox.2016.05.023. [DOI] [PubMed] [Google Scholar]
- Jensen PB, Jensen LJ, Brunak S. 2012. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet 13(6):395–405, PMID: 22549152, 10.1038/nrg3208. [DOI] [PubMed] [Google Scholar]
- Jensen LJ, Saric J, Bork P. 2006. Literature mining for the biologist: from information retrieval to biological discovery. Nat Rev Genet 7(2):119–129, PMID: 16418747, 10.1038/nrg1768. [DOI] [PubMed] [Google Scholar]
- Jiang L, Audouze K, Romero Herrera JA, Angquist LH, Kim-Kjaerulff S, Izarzugaza JMG, et al. 2019. Conflicting associations between dietary patterns and changes of anthropometric traits across subgroups of middle-aged women and men. Clin Nutr, 10.1016/j.clnu.2019.02.003. [DOI] [PubMed] [Google Scholar]
- Judson RS, Houck KA, Kavlock RJ, Knudsen TB, Martin MT, Mortensen HM, et al. 2010. In vitro screening of environmental chemicals for targeted testing prioritization: the ToxCast project. Environ. Environ Health Perspect 118(4):485–492, PMID: 20368123, 10.1289/ehp.0901392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrer C, Roiss T, von Goetz N, Gramec Skledar D, Peterlin Mašič L, Hungerbühler K. 2018. Physiologically based pharmacokinetic (PBPK) modeling of the bisphenols BPA, BPS, BPF, and BPAF with new experimental metabolic parameters: comparing the pharmacokinetic behavior of BPA with its substitutes. Environ Health Perspect 126(7):077002, PMID: 29995627, 10.1289/EHP2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Kjærulff S, Wich L, Kringelum J, Jacobsen UP, Kouskoumvekaki I, Audouze K, et al. 2013. ChemProt-2.0: visual navigation in a disease chemical biology database. Nucleic Acids Res 41(Database issue):D464–D469, PMID: 23185041, 10.1093/nar/gks1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, et al. 2019. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res 47(D1):D1102–D1109, PMID: 30371825, 10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortenkamp A, Faust M. 2018. Regulate to reduce chemical mixture risk. Science 361(6399):224–226, PMID: 30026211, 10.1126/science.aat9219. [DOI] [PubMed] [Google Scholar]
- Krallinger M, Valencia A, Hirschman L. 2008. Linking genes to literature: text mining, information extraction, and retrieval applications for biology. Genome Biol 9(Suppl 2):S8, PMID: 18834499, 10.1186/gb-2008-9-s2-s8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysiak-Baltyn K, Nordahl Petersen T, Audouze K, Jørgensen N, Angquist L, Brunak S. 2014. Compass: a hybrid method for clinical and biobank data mining. J Biomed Inform 47:160–170, PMID: 24513869, 10.1016/j.jbi.2013.10.007. [DOI] [PubMed] [Google Scholar]
- Lefterova MI, Haakonsson AK, Lazar MA, Mandrup S. 2014. PPARγ and the global map of adipogenesis and beyond. Trends Endocrinol Metab 25(6):293–302, Jun, PMID: 24793638, 10.1016/j.tem.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung MCK, Phuong J, Baker NC, Sipes NS, Klinefelter GR, Martin MT, et al. 2016. Systems toxicology of male reproductive development: profiling 774 chemicals for molecular targets and adverse outcomes. Environ Health Perspect 124(7):1050–1061, PMID: 26662846, 10.1289/ehp.1510385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri K, Grulke CM, Judson RS, Williams AJ. 2018. OPERA models for predicting physicochemical properties and environmental fate endpoints. J Cheminformatics 10(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson LA, Weigel RJ. 1999. AP2alpha and AP2gamma: a comparison of binding site specificity and trans-activation of the estrogen receptor promoter and single site promoter constructs. Nucleic Acids Res 27(20):4040–4049, PMID: 10497269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-Molina JM, Amaya E, Grimaldi M, Sáenz JM, Real M, Fernández MF, et al. 2013. In vitro study on the agonistic and antagonistic activities of bisphenol-S and other bisphenol-A congeners and derivatives via nuclear receptors. Toxicol Appl Pharmacol 272(1):127–136, PMID: 23714657, 10.1016/j.taap.2013.05.015. [DOI] [PubMed] [Google Scholar]
- NIH (National Institutes of Health). PubChem Database. https://pubchem.ncbi.nlm.nih.gov/ [accessed 28 July 2017].
- NIH, TOXNET (Toxicology Data Network)a. Chemical Carcinogenesis Research Information System (CCRIS). https://toxnet.nlm.nih.gov/newtoxnet/ccris.htm [accessed 28 July 2017].
- NIH, TOXNETb. Developmental and Reproductive Toxicology Database (DART). https://toxnet.nlm.nih.gov/newtoxnet/dart.htm [accessed 28 July 2017].
- NIH, TOXNETc. Hazardous Substances Data Bank (HSDB). https://toxnet.nlm.nih.gov/cgi-bin/sis/htmlgen?HSDB [accessed 28 July 2017].
- NIH, TOXNETd. Toxicology Literature Online (TOXLINE). https://toxnet.nlm.nih.gov/cgi-bin/sis/htmlgen?TOXLINE [accessed 28 July 2017].
- Nymark P, Rieswijk L, Ehrhart F, Jeliazkova N, Tsiliki G, Sarimveis H, et al. 2017. A data fusion pipeline for generating and enriching Adverse Outcome Pathway descriptions. Toxicol Sci 162(1):264–275, PMID: 29149350, 10.1093/toxsci/kfx252. [DOI] [PubMed] [Google Scholar]
- OECD (Organisation for Economic Co-operation and Development). 2014. The Guidance Document for Using the OECD (Q)SAR Application Toolbox to Develop Chemical Categories According to the OECD Guidance on Grouping Chemicals. Paris, France:OECD Publishing. [Google Scholar]
- OECD. AOP Knowledge Base. https://aopkb.oecd.org.
- Oki NO, Edwards SW. 2016. An integrative data mining approach to identifying adverse outcome pathway signatures. Toxicology 350–352:49–61, PMID: 27108252, 10.1016/j.tox.2016.04.004. [DOI] [PubMed] [Google Scholar]
- Polvani S, Tarocchi M, Tempesti S, Bencini L, Galli A. 2016. Peroxisome proliferator activated receptors at the crossroad of obesity, diabetes, and pancreatic cancer. World J Gastroenterol 22(8):2441–2459, PMID: 26937133, 10.3748/wjg.v22.i8.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard AM, Judson RS, Houck KA, Grulke CM, Volarath P, Thillainadarajah I, et al. 2016. ToxCast chemical landscape: paving the road to 21st century toxicology. Chem Res Toxicol 29(8):1225–1251, PMID: 27367298, 10.1021/acs.chemrestox.6b00135. [DOI] [PubMed] [Google Scholar]
- Rival Y, Stennevin A, Puech L, Rouquette A, Cathala C, Lestienne F, et al. 2004. Human adipocyte fatty acid-binding protein (aP2) gene promoter-driven reporter assay discriminates nonlipogenic peroxisome proliferator-activated receptor γ ligands. J Pharmacol Exp Ther 311(2):467–475, PMID: 15273253, 10.1124/jpet.104.068254. [DOI] [PubMed] [Google Scholar]
- Rochester JR, Bolden AL. 2015. Bisphenol S and F: a systematic review and comparison of the hormonal activity of bisphenol A substitutes. Environ Health Perspect 123(7):643–650, PMID: 25775505, 10.1289/ehp.1408989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo DP, Kim MT, Wang W, Pinolini D, Shende S, Strickland J, et al. 2017. CIIPro: a new read-across portal to fill data gaps using public large-scale chemical and biological data. Bioinformatics 33(3):464–466, PMID: 28172359, 10.1093/bioinformatics/btw640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAAOP (Society for the Advancement of Adverse Outcome Pathways). AOP-Wiki. Updated 4 December 2016. https://aopwiki.org/.
- Stoilov I, Akarsu AN, Sarfarazi M. 1997. Identification of three different truncating mutations in cytochrome P4501B1 (CYP1B1) as the principal cause of primary congenital glaucoma (Buphthalmos) in families linked to the GLC3A locus on chromosome 2p21. Hum Mol Genet 6(4):641–647, PMID: 9097971, 10.1093/hmg/6.4.641. [DOI] [PubMed] [Google Scholar]
- Teng C, Goodwin B, Shockley K, Xia M, Huang R, Norris J, et al. 2013. Bisphenol A affects androgen receptor function via multiple mechanisms. Chem Biol Interact 203(3):556–564, PMID: 23562765, 10.1016/j.cbi.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas RS, Philbert MA, Auerbach SS, Wetmore BA, Devito MJ, Cote I, et al. 2013. Incorporating new technologies into toxicity testing and risk assessment: moving from 21st century vision to a data-driven framework. Toxicol Sci 136(1):4–18, PMID: 23958734, 10.1093/toxsci/kft178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. EPA (Environmental Protection Agency)a. ToxCast Dashboard. https://www.epa.gov/chemical-research/toxcast-dashboard [accessed 26 April 2018].
- U.S. EPAb. ToxRefDB - Release user-friendly web-based tool for mining. Updated 8 May 2018. https://cfpub.epa.gov/si/si_public_record_report.cfm?Lab=NCCT&dirEntryId=227139 [accessed 28 July 2017].
- Villeneuve DL, Crump D, Garcia-Reyero N, Hecker M, Hutchinson TH, LaLone CA, et al. 2014. Adverse outcome pathway (AOP) development I: strategies and principles. Toxicol Sci 142(2):312–320, PMID: 25466378, 10.1093/toxsci/kfu199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergaard D, Stærfeldt H-H, Tønsberg C, Jensen LJ, Brunak S. 2018. A comprehensive and quantitative comparison of text-mining in 15 million full-text articles versus their corresponding abstracts. PLoS Comput Biol 14(2):e1005962, PMID: 29447159, 10.1371/journal.pcbi.1005962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang Q, Mansouri K, Williams AJ, Judson RS, Allen DG, Casey WM, et al. 2017. In silico prediction of physicochemical properties of environmental chemicals using molecular fingerprints and machine learning. J Chem Inf Model 57(1):36–49, PMID: 28006899, 10.1021/acs.jcim.6b00625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Xu Y, Xu Q, Shi H, Shi J, Hou Y. 2017. PPARδ promotes tumor progression via activation of Glut1 and SLC1-A5 transcription. Carcinogenesis 38(7):748–755, PMID: 28419191, 10.1093/carcin/bgx035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.