Abstract
It has been known for more than a century that, in adult vertebrates, the maintenance of taste buds depends on their afferent nerves. However, the initial formation of taste buds is proposed to be nerve-independent in amphibians, and evidence to the contrary in mammals has been endlessly debated, mostly due to indirect and incomplete means to impede innervation during the protracted perinatal period of taste bud differentiation. Here, by genetically ablating, in mice, all somatic (i.e. touch) or visceral (i.e. taste) neurons for the oral cavity, we show that the latter but not the former are absolutely required for the proper formation of their target organs, the taste buds.
Research organism: Mouse
Introduction
Taste buds are onion-shaped clusters of 60–100 taste receptors and support cells, embedded in epidermal papillae and distributed in a punctate pattern in the tongue and soft palate epithelia. They sense nutrients in the oral cavity and transmit taste information to the termini of sensory neurons, through conventional (Finger et al., 2005) and non-conventional (Ma et al., 2018; Romanov et al., 2018) synapses. Taste receptors and their support cells have a limited life span of 8 to 20 days depending on cell types (Perea-Martinez et al., 2013) and are constantly renewed from progenitors situated outside (Okubo et al., 2009; Ohmoto et al., 2017; Perea-Martinez et al., 2013) and, for a small minority, inside (Perea-Martinez et al., 2013) (and references therein) the taste bud. This process depends on the sensory afferents as demonstrated by the degeneration and regeneration of taste buds triggered, respectively, by the degeneration and regeneration of their surgically severed nerves (Oakley and Witt, 2004). While nerve-dependency has been uncontroversial for more than a century concerning the maintenance of taste buds in adult animals (Jacobson, 1991), it is still debated concerning their initial development in the embryo. Mature taste buds have been observed to form in the absence of nerves in amphibians (Barlow et al., 1996). In mammals, the case for nerve-dependency of taste bud development was mostly made through genetic invalidations of neurotrophins or their receptors, which hamper, to various extents, the outgrowth of sensory afferents towards the taste buds and simultaneously entail large losses of taste buds on the tongue at postnatal stages (Oakley and Witt, 2004). However, the protracted development of lingual taste buds, most of which mature after birth, left open the possibility that their postnatal loss reflected a failure of ‘maintenance’ or ‘maturation’ after a process of nerve-independent embryonic development (Barlow and Northcutt, 1998; Kapsimali and Barlow, 2013). To resolve this decades-old controversy, we completely blocked sensory innervation of the oral cavity in mouse by genetically preventing the birth of cranial sensory neurons, and examined the formation of taste buds — including palatal ones, whose development is almost entirely embryonic.
Results and discussion
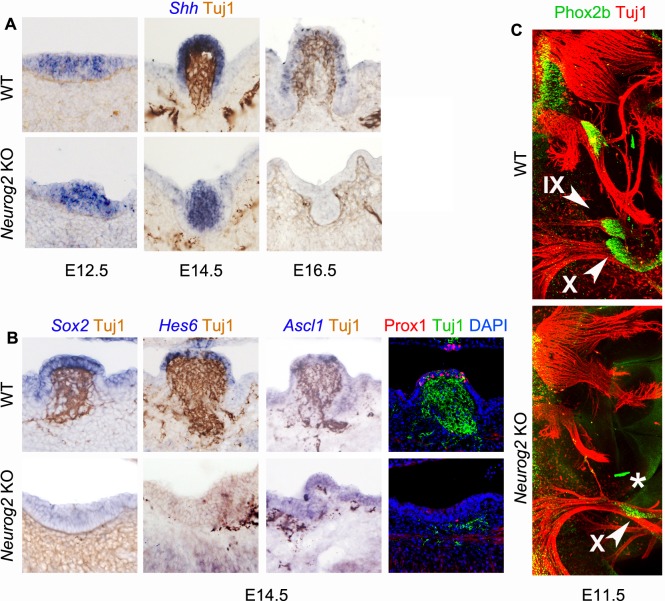
Taste organs (taste papillae and their resident taste buds) of the anterior tongue and soft palate are innervated by somatic sensory neurons (for touch and pain) located in the trigeminal ganglion and visceral sensory neurons (for taste) located in the geniculate ganglion (Watson et al., 2012). The circumvallate papilla receives innervation from visceral sensory neurons in the distal (petrosal) ganglion of the glossopharyngeal nerve, while its somatosensory ones, projecting in the same nerve (Frank, 1991) (and references therein) are probably located in the proximal (‘superior’) ganglion. To suppress innervation of taste organs, we exploited the developmental dependency of cranial sensory neurons on the proneural transcription factors Neurog1 and Neurog2. As early as day 9.5 of embryonic development (E9.5), the knockout of Neurog1 blocks neuronal differentiation in the trigeminal, superior and jugular ganglia (Ma et al., 1998) which harbor somatic sensory neurons. Inactivation of its paralogue Neurog2, expressed in epibranchial placodes, blocks the formation of the geniculate and petrosal ganglia, which harbor visceral sensory neurons (Fode et al., 1998). In each single Neurog1 and Neurog2 knockout at E16.5, the epithelium of the tongue and of the soft palate retained nerve fascicles at regularly spaced locations (corresponding to the taste organs) from, presumably, visceral or somatic sensory fibers, respectively (which thus navigate to their target independently of each other) (Figure 1—figure supplement 1). In double Neurog1/Neurog2 knockouts, which lose all sensory innervation of the head (Espinosa-Medina et al., 2014), all nerve fibers had disappeared from the epithelium and underlying lamina propria of the palate and tongue, as expected (Figure 1—figure supplement 1).
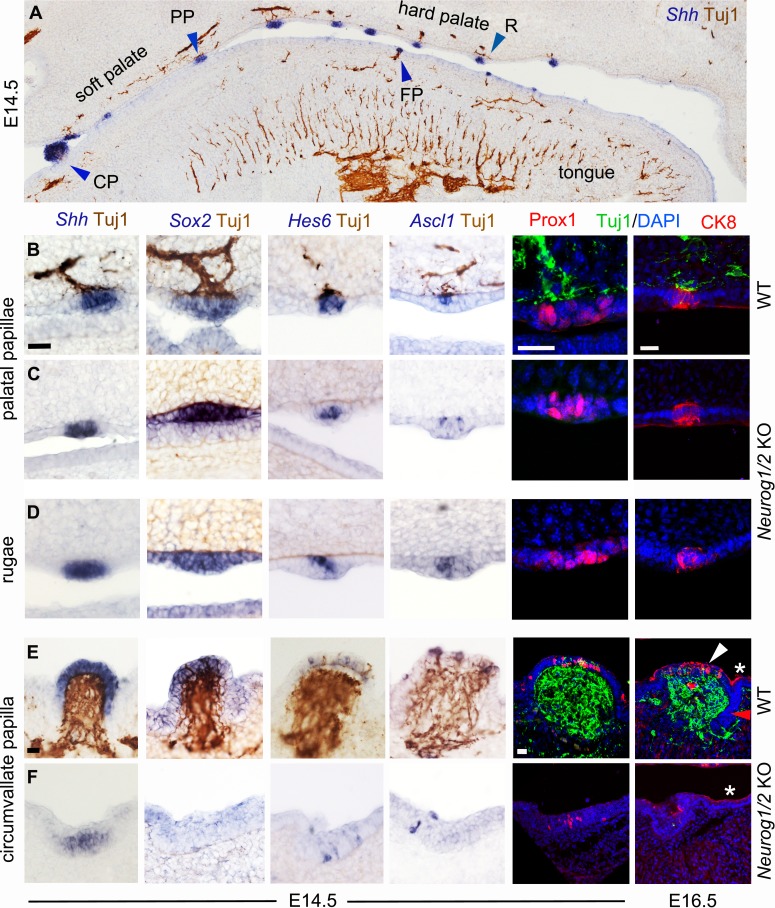
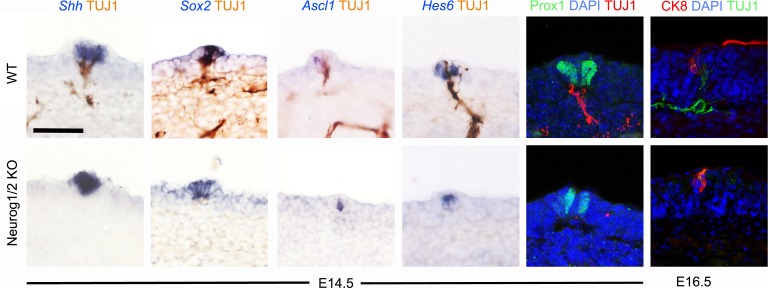
Taste organs differentiate from taste placodes, specialized patches of the oral epithelium, thickened by apico-basal elongation of the cells, which transform into dome-shaped papillae with a mesenchymal core — most dramatically at the site of the single circumvallate papilla, on a smaller scale for fungiform papillae of the tongue, and also in the palate albeit less conspicuously (Rashwan et al., 2016) — and express a number of markers. Expression of the signaling molecule sonic hedgehog (Shh), after a diffuse phase throughout the oral epithelium, resolves around E12.5 into a punctate pattern corresponding to taste placodes, which also start expressing Prox1 and high levels of Sox2 (Thirumangalathu et al., 2009; Nakayama et al., 2008; Liu et al., 2013; Okubo et al., 2006). Two days later, Shh, Sox2 and Prox1 were still expressed in the same pattern in the placodes — by then about to become papillae (hereafter placodes/papillae) — (Figure 1A,B, Figure 1—figure supplement 2), and the transcription factors Ascl1 and Hes6 (Seta et al., 2003) were switched on in just a few cells (Figure 1B, Figure 1—figure supplement 2). All these markers were expressed in a normal pattern in the soft palate and anterior tongue of double Neurog1/2 knockouts at E14.5, except for Sox2 whose expression was stronger and expanded in the palate (Figure 1C, Figure 1—figure supplement 2). Expression of these markers was not restricted to taste placodes/papillae but also occurred in the incipient ridges (rugae) of the hard palate (which never give rise to taste buds) and this expression was also essentially unchanged in Neurog1/2 knockouts (Figure 1A,D). Between E14.5 and E16.5, a cluster of cells in each taste papilla or ruga had switched on cytokeratine 8 (CK8) in both wild type and Neurog1/2 KO (Figure 1B,D, Figure 1—figure supplement 2). Thus, fungiform and palatal taste papillae (whose morphology and, as we show here, gene expression program are similar to that of palatal rugae) are epithelial specializations that form in the absence of any nerve, in agreement with prior observations of fungiform placodes/papillae development in cultured tongue explants (Farbman and Mbiene, 1991, Mbiene et al., 1997, Hall et al., 2003). The single circumvallate papilla of the posterior tongue stood in contrast. In the wild type, it displayed the same gene expression events as fungiform and palatal papillae on its dorsal surface (Figure 1A,E). However, in this case, as in rugae, expression of Shh, Sox2, Hes6, Ascl1, Prox1 and CK8 does not prefigure the later differentiation of taste bud cells, which takes place after birth, mostly in the semi-circular trenches, not at the dorsal surface. In the Neurog1/2 knockouts at E14.5 (a day after arrival of nerve, thickening of the placode and onset of Shh expression (as previously observed on tongue explants [Mistretta et al., 2003] and see Figure 1—figure supplement 3A), the sparse expression of Hes6, Ascl1 and Prox1 was preserved, but Shh and Sox2 was not upregulated and morphogenetic events leading to papilla formation were stalled (Figure 1F), corroborating and extending a previous observation of circumvallate papilla atrophy in Brain-Derived Neurotrophic Factor/Neurotrophin three double knockouts (Ito et al., 2010). A similar phenotype was obtained in single Neurog2 KO (Figure 1—figure supplement 3A,B), which lack petrosal ganglia (Figure 1—figure supplement 3C). Therefore, the circumvallate papilla, already known to differ from fungiform papillae by its ontogenetic requirement for an Fgf10 signal from the mesenchyme (Petersen et al., 2011), also differs from both fungiform and palatal papillae by requiring its afferent nerve for its formation.
Figure 1. Soft palate taste papillae as well as hard palate rugae, but not the tongue circumvallate papilla form without innervation.
(A–F) Combined immunohistochemistry for β-III tubulin (Tuj1, brown) and in situ hybridization (blue) for the indicated probes (top panel and left four columns), or immunofluorescence for Prox1 or CK8, combined with immunofluorescence for β-III tubulin and a counterstain with DAPI (two right columns) in wild type (A,B,E) and double Neurog1/2 KO (C,D,F) at E14.5 or E16.5 as indicated. In the circumvallate papilla, markers are expressed on the dorsal surface (white arrowhead for CK8) but not at the lower part of the trenches (red arrowhead in the right column), where taste buds will develop after birth. CK8 is also expressed in flattened cells of the periderm (asterisks). For every probe two animals were examined. CP : circumvallate papilla; FP : fungiform papilla ; PP : palatal papilla ; R : ruga. Scale bars: 20 μm.
Figure 1—figure supplement 1. Sensory innervation of the oral cavity in Neurog1, Neurog2 and double Neurog1/Neurog2 knockouts.
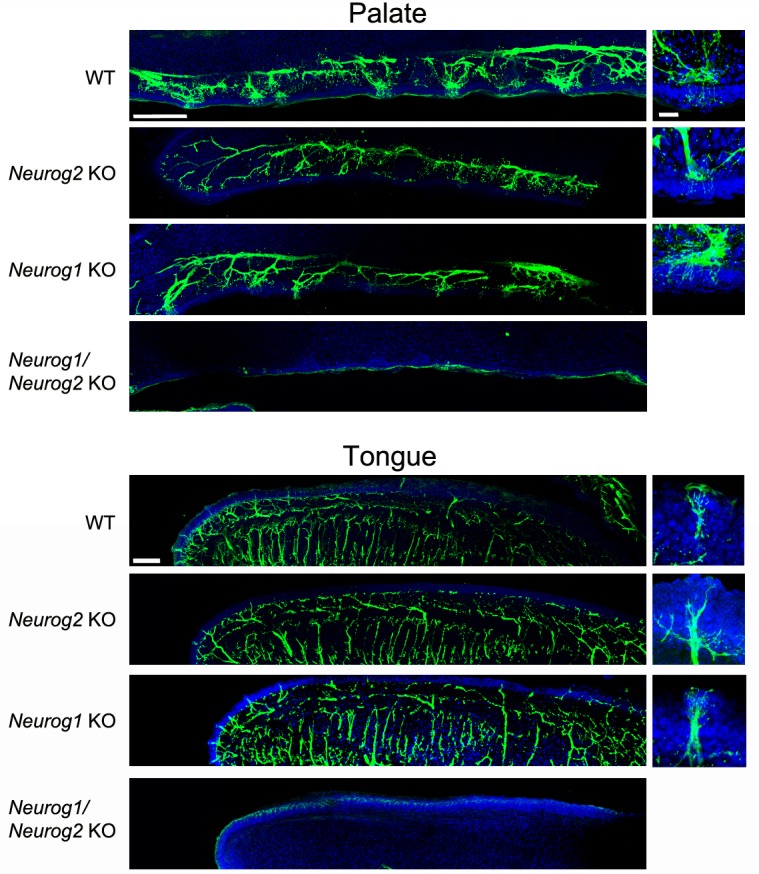
Figure 1—figure supplement 2. Placodal/papillary gene expression is nerve-independent in fungiform papillae.
Figure 1—figure supplement 3. The circumvallate papilla does not form in the absence of visceral innervation.
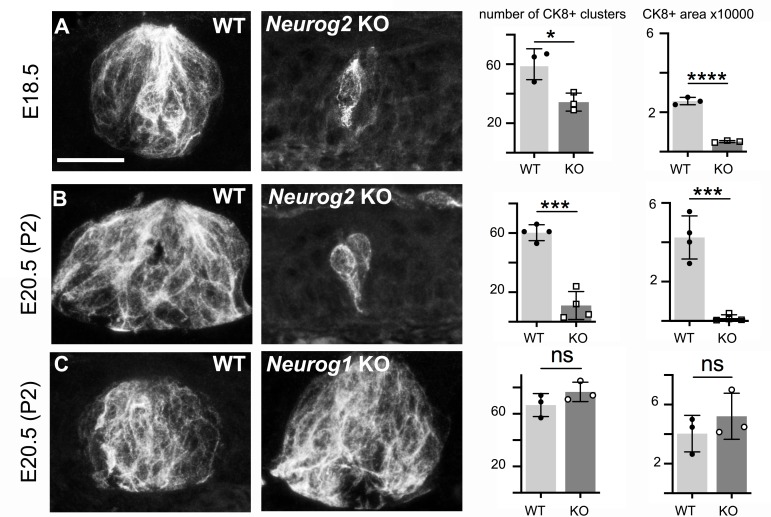
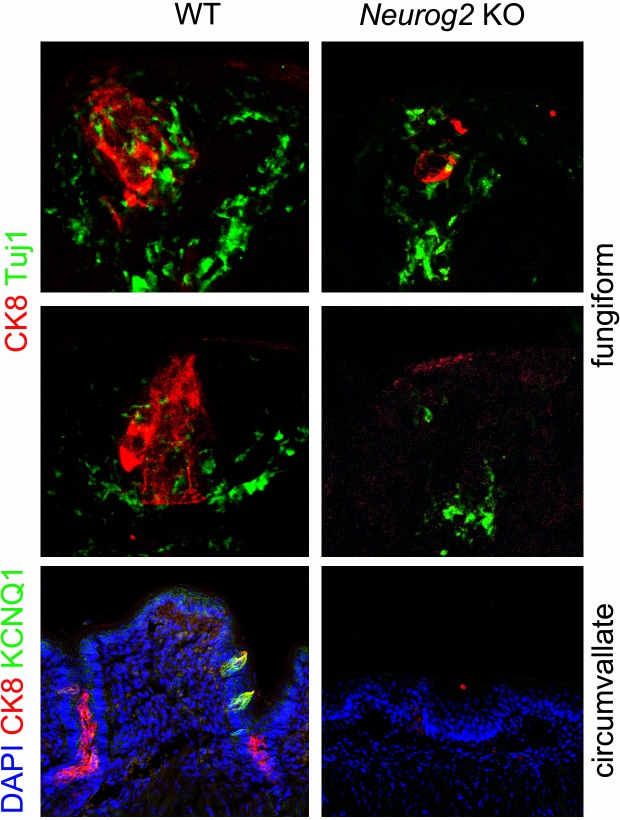
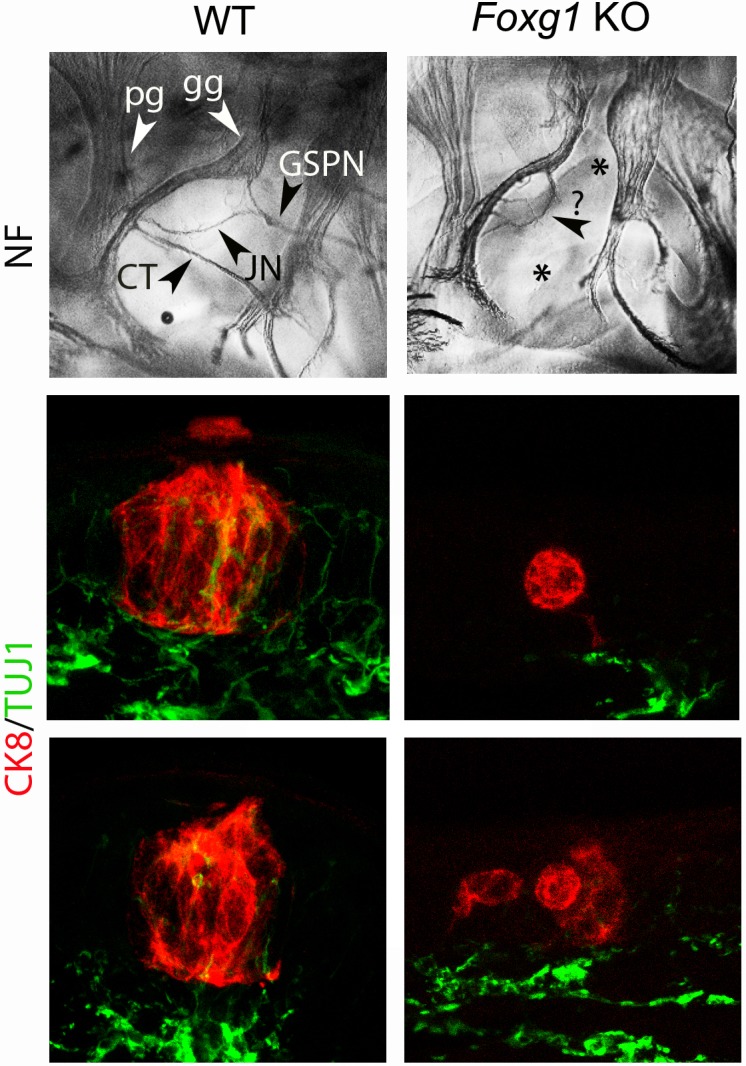
By E18.5, in wild type embryos, taste buds had begun to form, as onion shaped clusters of many cells expressing CK8, spanning the height of the oral epithelium, to different extents in different locations. From this developmental stage on, we will designate these taste bud anlagen, irrespective of their size and degree of maturation, as ‘CK8-positive (CK8+) cell clusters’. In the palate, they almost reach their mature size and structure by E18.5 (Rashwan et al., 2016) (Figure 2A). In Neurog2 knockouts, that is in the absence of visceral nerves, there was a dramatic deficit in both the number of CK8+ cell clusters detected in every other section (43% fewer) in the palate (this number can be taken as an approximation of the number of taste buds in the palate, see Materials and methods) and their cross-sectional area (66% smaller) so that the overall area occupied by CK8+ cells was 80% smaller than in wild types (Figure 2A). The latter measure, on two-dimensional projections of confocal stacks through 20 μm-thick sections, likely underestimates the extent of the deficit. To rule out that the massive shortfall in taste cells in Neurog2 KO at E18.5 reflected developmental delay rather than failure, and to circumvent the neonatal death of Neurog2 KO, we prolonged pregnancies by 2 days with Delvosteron delivered to the dams, and examined the embryos at E20.5 (equivalent to post-natal day 2 (P2) for the normal pregnancy of C57BL/6 mice). The degree of atrophy was even more pronounced at E20.5 than at E18.5, due to a decrease in the number of CK8+ cell clusters (now 89% fewer than in wild type) and a stagnation of their average size (while their wild type counterparts had enlarged), so that the total area occupied by CK8+ cells in the soft palate was smaller by 96% relative to wild type (Figure 2B). CK8+ cell clusters in the tongue, which are harbored in fungiform papillae and are still immature at this stage in wild type, were similarly atrophic in the mutants (Figure 2—figure supplement 1). Those of the circumvallate papilla were absent (Figure 2—figure supplement 1). To verify that the taste bud phenotype of Neurog2 knockouts does not stem from a cell-autonomous action of Neurog2 in taste placodes (through some undetected transient expression there), we devised a second genetic strategy to prevent visceral innervation of the soft palate. We found that ablating the transcription factor Foxg1, expressed in epibranchial placodes (Hatini et al., 1999), causes defects in epibranchial ganglia, and a fully penetrant (n = 5) lack of the greater superficial petrosal nerve as early as E11.5 — thus of the visceral innervation to the soft palate (Figure 2—figure supplement 2). In Foxg1 KO embryos at E18.5 the palatal taste buds were atrophic in a manner comparable to Neurog2 KO embryos (Figure 2—figure supplement 2). In contrast to visceral (taste) innervation, somatic (touch) innervation was dispensable for taste bud formation, as demonstrated by the normal number of palatal CK8+ cell clusters and total area occupied by CK8+ cells in E20.5 Neurog1 knockouts (Figure 2C).
Figure 2. Formation of palatal taste buds requires visceral but not somatic innervation.
Typical examples of CK8+ cell clusters detected by immunofluorescence in wild type and Neurog2 or Neurog1 KO, at E18.5 (A) or E20.5 (B,C), and quantification of the number of clusters throughout the soft palate on alternate sections, and overall surface (in μm2) occupied by CK8+ cells (in alternate sections) throughout the soft palate. *: p<0.05, ***: p<0.001, ****: p<0.0001 and ns: p>0.05; error bars presented as mean ± SD (n = 3 for A, C and n = 4 for B). The individual results for each animal are represented by dots. Scale bar: 20 μm.
Figure 2—figure supplement 1. CK8+ cell clusters are atrophic in fungiform papillae and absent in the circumvallate papilla of Neurog2 KO.
Figure 2—figure supplement 2. Taste buds do not form in the soft palate of Foxg1 knockouts.
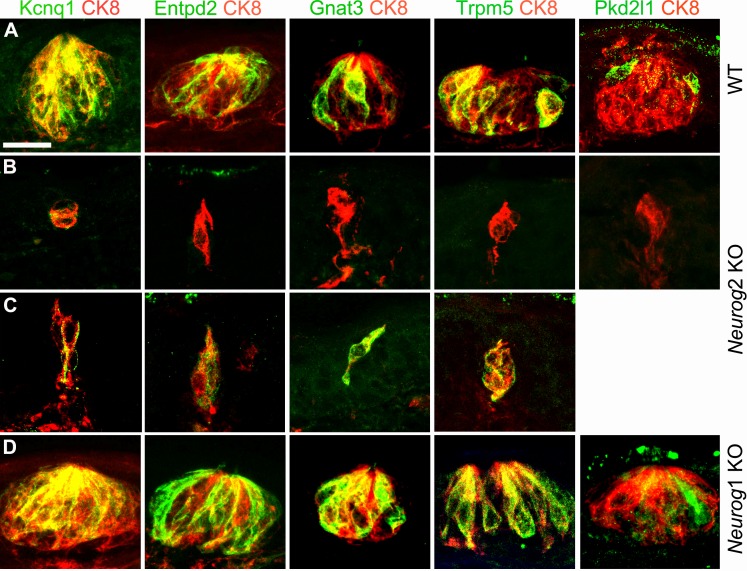
At E20.5 (P2), many palatal taste buds in wild types are fully mature and probably functional (Rashwan et al., 2016). In line with this, we could detect type I, II and III cells recognizable by the expression, respectively, of the ecto-ATPase Entpd2, the Gustducin α-chain Gnat3 or the cation channels Trpm5 and Pkd2l1, all cells sharing the expression of the potassium channel Kcnq1 (Matsumoto et al., 2011) (Figure 3A). In Neurog2 knockouts, expression of each marker was missing in many of the residual CK8+ clusters (Figure 3B), but present, except for Pkd2l1, in one or two cells of others (Figure 3C). Neurog1 KO pups had a normal complement of differentiated cells in their fully formed buds (Figure 3D). The cells in the Neurog2 mutants that display one marker or other of mature taste receptors argue that taste nerves are indispensable, not for the differentiation of CK8+ cells into taste bud cells (by the criterion of the markers tested), but for the constitution, maintenance or proliferation of the pool of progenitors required for bud formation, possibly reminiscent of the role of parasympathetic nerves in the organogenesis of salivary glands (Knox et al., 2010). In line with this, high Sox2 expression in perigemmal cells (i.e. closely apposed to the taste buds), a hallmark of taste cell progenitors (Okubo et al., 2006; Ohmoto et al., 2017), was massively reduced in Neurog2 KO (although a few residual CK8+ cells were themselves Sox2+) (Figure 4A), together with that of the proliferation marker Ki67 (Figure 4B). We could not detect any sign of cell death at E16.5 or E18.5 in or around the taste cell clusters of wild type or mutant pups by the TUNEL reaction or immunofluorescence against Caspase 3.
Figure 3. Taste bud cell differentiation is impeded, but not always, in the absence of visceral innervation.
Sections through taste buds in the soft palate of wild types (A), Neurog2 KO (B,C) and Neurog1 KO (D) at E20.5 (P2), immunostained with the indicated antibodies. Kcnq1, Entpd2, Gnat3 and Trpm5 are occasionally detected in residual CK8+ cells of Neurog2 KO embryos. In Neurog1 KOs, taste buds contained the normal complement of markers. Scale bar: 20 μm.
Figure 4. Taste nerves are required for Sox2 expression and cell proliferation around taste bud anlagen.
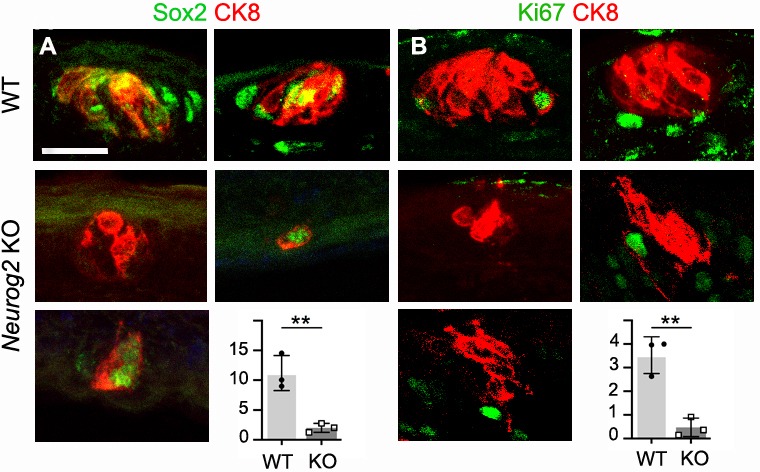
(A,B) Immunofluorescence against CK8 and Sox2, or CK8 and the proliferation marker Ki67 on representative CK8+ cell clusters in the soft palate from wild type, and Neurog2 KO, at E18.5. The graphs show the counts of Sox2High and Ki67+ cells inside or within two cell diameters of CK8+ cell clusters. **: p<0.01; error bars presented as mean ± SD (for Sox2High cells, a sample of 72 wild type and all (n = 100) mutant clusters were counted, on three animals; for Ki67+ cells, a sample of 71 wild type and all (n = 101) mutant clusters were counted on three animals). The individual results for each animal are represented by dots. Scale bar: 20 μm.
The scattered residual CK8+ cell clusters of aneural oral epithelia resembled, by their small size yet expression of terminal differentiation markers, those induced by ectopic expression of Shh (Castillo et al., 2014). Such clusters might thus represent the endpoint, in the absence of subsequent innervation, of taste bud organogenesis triggered by local Shh (endogenous to the taste placodes, or ectopically induced in the epithelium). Along the same lines (and barring a myriad of possible species differences), the most parsimonious reconciliation of our finding with the apparently opposite one in axolotl would be that the ectopic aneural taste buds obtained in that species are much smaller than normal ones — as appears from Figure 2B versus Figure 4D of Barlow et al. (1996); or that in axolotl, taste buds would be so much smaller than in mouse as to not require a pool of progenitors.
All in all, we show that formation of taste buds in the mammalian embryo is as dependent on innervation as their renewal and regeneration are in the adult. In line with this, the time between the arrival of nerves and taste bud formation (2 to 3 days in the mouse palate; Rashwan et al., 2016) is in the same range as that between re-innervation and taste bud regeneration in gerbils (Cheal and Oakley, 1977) — the only species, to our knowledge, where the lag between regrowth of nerves and reformation of taste buds was measured. In light of our results, ‘maintenance’, in the case of taste buds, can be interpreted as an ongoing ontogeny. The developmental role that we document for nerves is neuron-type specific: exerted by visceral sensory neurons, derived from epibranchial placodes, not by somatosensory neurons, derived from the neural crest. This specificity makes less likely the possibility, that we cannot exclude however, that the nerve associated cells (Schwann cell precursors and Schwann cells) would have the inducing role, rather than the nerve fibers themselves. At the molecular level, nerve-derived Shh has been implicated in the maintenance of adult taste buds (Castillo-Azofeifa et al., 2017; Lu et al., 2018), with a proposed action, however, too slow and too limited to underlie the dramatic effect of nerve cutting on the integrity of taste buds — or on expression of Shh in taste bud cells (Miura et al., 2004). The nerve-derived trophic ‘hormone-like’ substance(s) postulated a century ago (Olmsted, 1920) to induce taste buds — in the adult for their continuous renewal, and, as we show here, in the embryo for their ontogeny — are likely yet to be discovered.
Materials and methods
Key resources table.
| Reagent type (species) or resource |
Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Anti- Neurofilament (mouse monoclonal) | Hybridoma Bank | Hybridoma Bank (#2H3)RRID:AB_531793 | one in 500 |
| Antibody | Anti-Gustducin (Gnat3) (goat polycolonal) | MyBioSource | MyBioSource (#MBS421805) RRID:AB_10889192 | one in 500 |
| Antibody | Anti-Entpd2 (rabbit polycolonal) | http://ectonucleotidases-ab.com/(Bartel et al., 2006) | RRID:AB_2314986 | one in 500 |
| Antibody | Anti-Pkd2l1 (rabbit polycolonal) | Millipore | Millipore (#AB9084) RRID:AB_571091 |
one in 1000 |
| Antibody | Anti-Prox1 (rabbit polyclonal) | Millipore | Millipore (#AB5475) RRID:AB_177485 | one in 1000 |
| Antibody | Cytokeratin8 (Troma-I) (rat monoclonal) | Developmental Studies Hybridoma Bank (DSHB) | RRID:AB_531826 | one in 400 |
| Antibody | Anti-Kcnq1 (rabbit polyclonal) | Millipore | Millipore (#AB5932) RRID:AB_92147 | one in 1000 |
| Antibody | Anti-Ki67 (rabbite polycolonal) | abcam | abcam (#ab15580) RRID:AB_443209 | one in 200 |
| Antibody | Anti-Sox2 (goat polyclonal) | R and D Systems | R and D Systems (#AF2018) RRID:AB_355110 | one in 500 |
| Antibody | Anti-Trpm5 (guinea pig polyclonal) | Obtained from ER Liman’s lab, USC | one in 500 | |
| Antibody | Anti-βIII Tubulin (Tuj1) (mouse monoclonal) | Covance | Covance (#MMS-435P) RRID:AB_2313773 | one in 500 |
| Antibody | Anti-Phox2b (Rabbit polyclonal) | Pattyn et al., Development, 124,4065–4075 (1997) | one in 500 | |
| Antibody | Donkey anti-goat A488 | Thermo Fisher | Thermo Fisher (#A-11055) RRID:AB_2534102 | one in 500 |
| Antibody | Donkey anti- guinea pig A488 | Jackson Immunoresearch Laboratories | Jackson Immunoresearch Laboratories (#706-545-148) | one in 500 |
| Antibody | Donkey anti- mouse A488 | Invitrogen | Invitrogen (#A-21202) RRID:AB_141607 | one in 500 |
| Antibody | Donkey anti- rabbit A488 | Jackson Immunoresearch Laboratories | Jackson Immunoresearch Laboratories (#711-545-152) | one in 500 |
| Antibody | Donkey anti-rat Cy3 | Jackson Immunoresearch Laboratories | Jackson Immunoresearch Laboratories (#712-165-153) | one in 500 |
| Commercial kit | Vectastain Elite ABC Kits | Vector Laboratories | Vector Laboratories, PK-6101 and PK-6012 | |
| Chemical compound | DAB (3,3’-Diaminobenzidine) | Sigma | Sigma (#SLBP9645V) | |
| Commercial reagent | Proligesterone (Delvesterone) | MSD Animal Health | Delvosteron: NaCl (0.9%)=1:1 |
Histology
In situ hybridization and immunochemistry on sections or wholemounts have been described (Coppola et al., 2010). Immunofluorescence on cryostat or vibratome sections was performed as previously described (Espinosa-Medina et al., 2014). Wholemount immunofluorescent staining was made of cleared embryos following the 3DISCO protocol (Ertürk et al., 2012) and imaged with an Sp8 confocal microscope. 3D reconstructions were performed using the IMARIS imaging software.
Mouse embryos (E11.5, E12.5, E14.5 and E16.5) or 4% paraformaldehyde (PFA)-perfused pups (at P0 (E18.5) and P2 (E20.5)) were fixed in 4% PFA overnight at 4°C, and the tongue or palate was dissected out. For cryostat sections, tissues were embedded in 7% gelatine/15% sucrose and sectioned at 20 μm. For vibratome sections, tissues were embedded in 3% agarose (in PBS) and cut at 100 μm.
Immunohistochemical reactions were done with the Vectastain Elite ABC Kits (PK-6101 and PK-6012; Vector Laboratories) and color revealed by DAB (3,3’-Diaminobenzidine).
For anti-Pkd2l1 and anti-Sox2 immunoreactions, light fixation was required with 4% PFA at 4°C for 3 hr.
Antisense RNA Probes used were Hes6 (obtained from Ryoichiro Kageyama), Ascl1 (obtained from François Guillemot), Shh (obtained from Andrew McMahon) and Sox2 (obtained from Dr. Robin Lovell-Badge).
Transgenic mouse lines
Foxg1Cre (BF-1) KO (Hébert and McConnell, 2000): a knock-in of Cre in the Foxg1 locus. RRID:MGI:5806112
Neurog1 KO (Ma et al., 1998): a fragment of the Neurog1 ORF was replaced by GFP and a PGK-neo cassette. RRID:MGI:3639866
Neurog2 KO (Florio et al., 2012): knock in of CreERT2 into the Neurog2 locus, allowing the expression of a tamoxifen inducible Cre from the Neurog2 promoter and creating a null allele. RRID:MGI:5432590
Neurog1/2 KO were bred by intercrossing single Neurog1 and Neurog2 heterozygous knockouts, then Neurog1/2 double heterozygotes.
All lines were maintained by crossing with C57BL/6 x DBA/2 F1 mice.
Animal treatment
Proligestone (Delvosteron) was used to prolong the gestation of Neurog1 and Neurog2 KO mice. At E16.5, 100 μL of diluted Delvosteron (MSD Animal Health) solution (Delvosteron: NaCl (0.9%)=1:1) was injected subcutaneously in each pregnant mouse. Pups were surgically delivered on day 20.5 of embryonic development.
Statistical analyses
For the quantification of CK8+ cell clusters, one cryosection out of two through the entire soft palate was analyzed in Neurog1 and Neurog2 KO embryos, wild types serving as controls. This raw number was used as an estimate of the number of CK8+ cell clusters in the palate, without multiplying by two and performing the Abercrombie correction. Indeed, all the clusters we counted in the wild type spanned the whole height of the epithelium and/or had a visible pore, de facto excluding objects that would be tangentially sectioned. This amounts to a systematic loss of ‘caps’, an important source of errors in the Abercrombie correction (Hedreen, 1998). Our method makes it next to impossible that clusters (with an average width of 28 μm (E18.5) and 38 μm (E20.5), see Source data 1) would be counted more than once on alternate 20 μm sections. On the other hand, a few clusters were probably lost, if they were centered on one of the uncounted section. The accuracy of our method is verified by the fact that we found the same number of clusters at E20.5 and E18.5 despite the 36% increase in average diameter, and also that we found (not shown) the same number of clusters on alternate 30 μm sections (as opposed to 20 μm).
For each genotype, all CK8+ clusters (if there were less than 10), or up to ten (if there were more) were imaged, for the wild type in 2 to 3 sections around the midline of the soft palate (where taste buds were the densest), for the mutants, in one out of two sections throughout the palate. Confocal imaging was performed on a Leica SP5 microscope or a LSM 880 Airyscan Zeiss microscope. The surface occupied by each CK8+ cluster was automatically outlined and measured with the Fiji software, and the mean cross-sectional area was calculated for each genotype. To calculate the entire surface occupied by CK8+ cell clusters on alternate sections of one half of soft palate, the mean value of the calculated surfaces of CK8+ cell clusters was multiplied by the raw, uncorrected number of CK8+ clusters. Statistical analysis was performed using unpaired two-tailed t-test. Results are expressed as mean ± SD. All graphs were performed with GraphPad Prism software. For the reason that we did not count ‘caps’ of wild type CK8+ clusters (see above), the decrease in total area is likely underestimated.
Acknowledgements
We thank the Imaging Facility of IBENS, which is supported by grants from Fédération pour la Recherche sur le Cerveau, Région Ile-de-France DIM NeRF 2009 and 2011 and France-BioImaging. We wish to thank the animal facility of IBENS, C Goridis for helpful comments on the manuscript and all the members of the Brunet laboratory for discussions. This study was supported by the CNRS, the École Normale Supérieure, INSERM, ANR award 17-CE160006-01 (to J-FB), FRM award DEQ2000326472 (to J-FB), the ‘Investissements d’Avenir’ program of the French Government implemented by the ANR (referenced ANR-10-LABX-54 MEMO LIFE and ANR-11-IDEX-0001–02 PSL Research University). D-F received a fellowship from the China Scholarship Council. GGC's lab was funded by the Italian Telethon Foundation grant GGP13146.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Jean-François Brunet, Email: jfbrunet@biologie.ens.fr.
Jeremy Nathans, Johns Hopkins University School of Medicine, United States.
Marianne E Bronner, California Institute of Technology, United States.
Funding Information
This paper was supported by the following grants:
Agence Nationale de la Recherche ANR-12-BSV4-0007-01 to Jean-François Brunet.
Agence Nationale de la Recherche ANR-10-LABX-54 MEMOLIFE to Jean-François Brunet.
Agence Nationale de la Recherche ANR-11-IDEX-0001-02 PSL research University to Jean-François Brunet.
Fondation pour la Recherche Médicale DEQ 2000326472 to Jean-François Brunet.
Agence Nationale de la Recherche ANR-17-CE16-00006-01 to Jean-François Brunet.
China Scholarship Council to Di Fan.
Additional information
Competing interests
No competing interests declared.
Author contributions
Conceptualization, Investigation, Writing—original draft.
Validation, Investigation, Visualization, Methodology, Writing—review and editing, Involved in every step of the production, acquisition and analysis of the data.
Resources, Methodology, Writing—review and editing, Provided the main genetic tool for this study, the Neurog2 KO.
Conceptualization, Supervision, Funding acquisition, Visualization, Writing—original draft, Project administration.
Ethics
Animal experimentation: All animal studies were done in accordance with the guidelines issued by the French Ministry of Agriculture and have been approved by the Direction Départementale des Services Vétérinaires de Paris.
Additional files
Data availability
All data generated or analysed during this study are included in the manuscript, and in the source data files.
References
- Barlow LA, Chien CB, Northcutt RG. Embryonic taste buds develop in the absence of innervation. Development. 1996;122:1103–1111. doi: 10.1242/dev.122.4.1103. [DOI] [PubMed] [Google Scholar]
- Barlow LA, Northcutt RG. Reply. Trends in Neurosciences. 1998;21:338. doi: 10.1016/S0166-2236(98)01256-9. [DOI] [PubMed] [Google Scholar]
- Bartel DL, Sullivan SL, Lavoie EG, Sévigny J, Finger TE. Nucleoside triphosphate diphosphohydrolase-2 is the ecto-ATPase of type I cells in taste buds. The Journal of Comparative Neurology. 2006;497:1–12. doi: 10.1002/cne.20954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo D, Seidel K, Salcedo E, Ahn C, de Sauvage FJ, Klein OD, Barlow LA. Induction of ectopic taste buds by SHH reveals the competency and plasticity of adult lingual epithelium. Development. 2014;141:2993–3002. doi: 10.1242/dev.107631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Azofeifa D, Losacco JT, Salcedo E, Golden EJ, Finger TE, Barlow LA. Sonic hedgehog from both nerves and epithelium is a key trophic factor for taste bud maintenance. Development. 2017;144:3054–3065. doi: 10.1242/dev.150342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheal M, Oakley B. Regeneration of fungiform taste buds: temporal and spatial characteristics. The Journal of Comparative Neurology. 1977;172:609–625. doi: 10.1002/cne.901720405. [DOI] [PubMed] [Google Scholar]
- Coppola E, Rallu M, Richard J, Dufour S, Riethmacher D, Guillemot F, Goridis C, Brunet JF. Epibranchial ganglia orchestrate the development of the cranial neurogenic crest. PNAS. 2010;107:2066–2071. doi: 10.1073/pnas.0910213107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertürk A, Becker K, Jährling N, Mauch CP, Hojer CD, Egen JG, Hellal F, Bradke F, Sheng M, Dodt HU. Three-dimensional imaging of solvent-cleared organs using 3disco. Nature Protocols. 2012;7:1983–1995. doi: 10.1038/nprot.2012.119. [DOI] [PubMed] [Google Scholar]
- Espinosa-Medina I, Outin E, Picard CA, Chettouh Z, Dymecki S, Consalez GG, Coppola E, Brunet JF. Parasympathetic ganglia derive from schwann cell precursors. Science. 2014;345:87–90. doi: 10.1126/science.1253286. [DOI] [PubMed] [Google Scholar]
- Farbman AI, Mbiene JP. Early development and innervation of taste bud-bearing papillae on the rat tongue. The Journal of Comparative Neurology. 1991;304:172–186. doi: 10.1002/cne.903040203. [DOI] [PubMed] [Google Scholar]
- Finger TE, Danilova V, Barrows J, Bartel DL, Vigers AJ, Stone L, Hellekant G, Kinnamon SC. ATP signaling is crucial for communication from taste buds to gustatory nerves. Science. 2005;310:1495–1499. doi: 10.1126/science.1118435. [DOI] [PubMed] [Google Scholar]
- Florio M, Leto K, Muzio L, Tinterri A, Badaloni A, Croci L, Zordan P, Barili V, Albieri I, Guillemot F, Rossi F, Consalez GG. Neurogenin 2 regulates progenitor cell-cycle progression and purkinje cell dendritogenesis in cerebellar development. Development. 2012;139:2308–2320. doi: 10.1242/dev.075861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fode C, Gradwohl G, Morin X, Dierich A, LeMeur M, Goridis C, Guillemot F. The bHLH protein NEUROGENIN 2 is a determination factor for epibranchial placode-derived sensory neurons. Neuron. 1998;20:483–494. doi: 10.1016/S0896-6273(00)80989-7. [DOI] [PubMed] [Google Scholar]
- Frank ME. Taste-responsive neurons of the glossopharyngeal nerve of the rat. Journal of Neurophysiology. 1991;65:1452–1463. doi: 10.1152/jn.1991.65.6.1452. [DOI] [PubMed] [Google Scholar]
- Hall JMH, Bell ML, Finger TE. Disruption of sonic hedgehog signaling alters growth and patterning of lingual taste papillae. Developmental Biology. 2003;255:263–277. doi: 10.1016/S0012-1606(02)00048-9. [DOI] [PubMed] [Google Scholar]
- Hatini V, Ye X, Balas G, Lai E. Dynamics of placodal lineage development revealed by targeted transgene expression. Developmental Dynamics. 1999;215:332–343. doi: 10.1002/(SICI)1097-0177(199908)215:4<332::AID-AJA5>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Hébert JM, McConnell SK. Targeting of cre to the Foxg1 (BF-1) locus mediates loxP recombination in the telencephalon and other developing head structures. Developmental Biology. 2000;222:296–306. doi: 10.1006/dbio.2000.9732. [DOI] [PubMed] [Google Scholar]
- Hedreen JC. What was wrong with the abercrombie and empirical cell counting methods? A review. The Anatomical Record. 1998;250:373–380. doi: 10.1002/(SICI)1097-0185(199803)250:3<373::AID-AR12>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Ito A, Nosrat IV, Nosrat CA. Taste cell formation does not require gustatory and somatosensory innervation. Neuroscience Letters. 2010;471:189–194. doi: 10.1016/j.neulet.2010.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson M. Developmental Neurobiology. London: Springer; 1991. [DOI] [Google Scholar]
- Kapsimali M, Barlow LA. Developing a sense of taste. Seminars in Cell & Developmental Biology. 2013;24:200–209. doi: 10.1016/j.semcdb.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox SM, Lombaert IM, Reed X, Vitale-Cross L, Gutkind JS, Hoffman MP. Parasympathetic innervation maintains epithelial progenitor cells during salivary organogenesis. Science. 2010;329:1645–1647. doi: 10.1126/science.1192046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HX, Ermilov A, Grachtchouk M, Li L, Gumucio DL, Dlugosz AA, Mistretta CM. Multiple shh signaling centers participate in fungiform papilla and taste bud formation and maintenance. Developmental Biology. 2013;382:82–97. doi: 10.1016/j.ydbio.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu WJ, Mann RK, Nguyen A, Bi T, Silverstein M, Tang JY, Chen X, Beachy PA. Neuronal delivery of hedgehog directs spatial patterning of taste organ regeneration. PNAS. 2018;115:E200–E209. doi: 10.1073/pnas.1719109115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Chen Z, del Barco Barrantes I, de la Pompa JL, Anderson DJ. Neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron. 1998;20:469–482. doi: 10.1016/S0896-6273(00)80988-5. [DOI] [PubMed] [Google Scholar]
- Ma Z, Taruno A, Ohmoto M, Jyotaki M, Lim JC, Miyazaki H, Niisato N, Marunaka Y, Lee RJ, Hoff H, Payne R, Demuro A, Parker I, Mitchell CH, Henao-Mejia J, Tanis JE, Matsumoto I, Tordoff MG, Foskett JK. CALHM3 is essential for rapid ion Channel-Mediated purinergic neurotransmission of GPCR-Mediated tastes. Neuron. 2018;98:547–561. doi: 10.1016/j.neuron.2018.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto I, Ohmoto M, Narukawa M, Yoshihara Y, Abe K. Skn-1a (Pou2f3) specifies taste receptor cell lineage. Nature Neuroscience. 2011;14:685–687. doi: 10.1038/nn.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbiene JP, Maccallum DK, Mistretta CM. Organ cultures of embryonic rat tongue support tongue and gustatory papilla morphogenesis in vitro without intact sensory ganglia. The Journal of Comparative Neurology. 1997;377:324–340. doi: 10.1002/(SICI)1096-9861(19970120)377:3<324::AID-CNE2>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Mistretta CM, Liu HX, Gaffield W, MacCallum DK. Cyclopamine and jervine in embryonic rat tongue cultures demonstrate a role for shh signaling in taste papilla development and patterning: fungiform papillae double in number and form in novel locations in dorsal lingual epithelium. Developmental Biology. 2003;254:1–18. doi: 10.1016/S0012-1606(02)00014-3. [DOI] [PubMed] [Google Scholar]
- Miura H, Kato H, Kusakabe Y, Tagami M, Miura-Ohnuma J, Ninomiya Y, Hino A. A strong nerve dependence of sonic hedgehog expression in basal cells in mouse taste bud and an autonomous transcriptional control of genes in differentiated taste cells. Chemical Senses. 2004;29:823–831. doi: 10.1093/chemse/bjh248. [DOI] [PubMed] [Google Scholar]
- Nakayama A, Miura H, Shindo Y, Kusakabe Y, Tomonari H, Harada S. Expression of the basal cell markers of taste buds in the anterior tongue and soft palate of the mouse embryo. The Journal of Comparative Neurology. 2008;509:211–224. doi: 10.1002/cne.21738. [DOI] [PubMed] [Google Scholar]
- Oakley B, Witt M. Building sensory receptors on the tongue. Journal of Neurocytology. 2004;33:631–646. doi: 10.1007/s11068-005-3332-0. [DOI] [PubMed] [Google Scholar]
- Ohmoto M, Ren W, Nishiguchi Y, Hirota J, Jiang P, Matsumoto I. Genetic lineage tracing in taste tissues using Sox2-CreERT2 strain. Chemical Senses. 2017;42:547–552. doi: 10.1093/chemse/bjx032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okubo T, Pevny LH, Hogan BL. Sox2 is required for development of taste bud sensory cells. Genes & Development. 2006;20:2654–2659. doi: 10.1101/gad.1457106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okubo T, Clark C, Hogan BL. Cell lineage mapping of taste bud cells and keratinocytes in the mouse tongue and soft palate. Stem Cells. 2009;27:442–450. doi: 10.1634/stemcells.2008-0611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmsted JMD. The results of cutting the seventh cranial nerve in amiurus nebulosus (lesueur) Journal of Experimental Zoology. 1920;31:369–401. doi: 10.1002/jez.1400310403. [DOI] [Google Scholar]
- Perea-Martinez I, Nagai T, Chaudhari N. Functional cell types in taste buds have distinct longevities. PLOS ONE. 2013;8:e53399. doi: 10.1371/journal.pone.0053399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen CI, Jheon AH, Mostowfi P, Charles C, Ching S, Thirumangalathu S, Barlow LA, Klein OD. FGF signaling regulates the number of posterior taste papillae by controlling progenitor field size. PLOS Genetics. 2011;7:e1002098. doi: 10.1371/journal.pgen.1002098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashwan A, Konishi H, El-Sharaby A, Kiyama H. Ontogeny and innervation of taste buds in mouse palatal gustatory epithelium. Journal of Chemical Neuroanatomy. 2016;71:26–40. doi: 10.1016/j.jchemneu.2015.11.003. [DOI] [PubMed] [Google Scholar]
- Romanov RA, Lasher RS, High B, Savidge LE, Lawson A, Rogachevskaja OA, Zhao H, Rogachevsky VV, Bystrova MF, Churbanov GD, Adameyko I, Harkany T, Yang R, Kidd GJ, Marambaud P, Kinnamon JC, Kolesnikov SS, Finger TE. Chemical synapses without synaptic vesicles: purinergic neurotransmission through a CALHM1 channel-mitochondrial signaling complex. Science Signaling. 2018;11:eaao1815. doi: 10.1126/scisignal.aao1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seta Y, Seta C, Barlow LA. Notch-associated gene expression in embryonic and adult taste papillae and taste buds suggests a role in taste cell lineage decisions. The Journal of Comparative Neurology. 2003;464:49–61. doi: 10.1002/cne.10787. [DOI] [PubMed] [Google Scholar]
- Thirumangalathu S, Harlow DE, Driskell AL, Krimm RF, Barlow LA. Fate mapping of mammalian embryonic taste bud progenitors. Development. 2009;136:1519–1528. doi: 10.1242/dev.029090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson C, Paxinos G, Puelles L. The Mouse Nervous System. Elsevier; 2012. [DOI] [Google Scholar]