Abstract
SETD2, an epigenetic tumor suppressor, is frequently mutated in MLL-rearranged (MLLr) leukemia and relapsed acute leukemia (AL). To clarify the impact of SETD2 mutations on chemotherapy sensitivity in MLLr leukemia, two loss-of-function (LOF) Setd2-mutant alleles (Setd2F2478L/WT or Setd2Ex6-KO/WT) were generated and introduced, respectively, to the Mll-Af9 knock-in leukemia mouse model. Both alleles cooperated with Mll-Af9 to accelerate leukemia development that resulted in resistance to standard Cytarabine-based chemotherapy. Mechanistically, Setd2-mutant leukemic cells showed downregulated signaling related to cell cycle progression, S, and G2/M checkpoint regulation. Thus, after Cytarabine treatment, Setd2-mutant leukemic cells exit from the S phase and progress to the G2/M phase. Importantly, S and G2/M cell cycle checkpoint inhibition could resensitize the Mll-Af9/Setd2 double-mutant cells to standard chemotherapy by causing DNA replication collapse, mitotic catastrophe, and increased cell death. These findings demonstrate that LOF SETD2 mutations confer chemoresistance on AL to DNA-damaging treatment by S and G2/M checkpoint defects. The combination of S and G2/M checkpoint inhibition with chemotherapy can be explored as a promising therapeutic strategy by exploiting their unique vulnerability and resensitizing chemoresistant AL with SETD2 or SETD2-like epigenetic mutations.
Introduction
Cytarabine (Ara-C)-based chemotherapy has remained as the first-line treatment for acute myeloid leukemia (AML) for over four decades. Even with great success in disease remission using this strategy, the 10-year overall survival (OS) and event-free survival (EFS) for children/adolescents after induction therapy is about 55–65%. Treated with more conventional chemotherapy alone, only 20–40% of the patients gain disease-free survival of >5 years in adulthood AML [1, 2].
Although new targeted therapies and immune therapies are promising, the cost, the technical availability, and the presence of appropriate cell surface targets for immunotherapy significantly limit their clinical utility, as compared with traditional chemotherapy [3–6]. Therefore, research on less toxic treatments or chemotherapy in combination with targeted therapies is urgently needed [7]. Previously, we had identified a frequency of 6% SETD2 mutations in AML and acute lymphoid leukemia (ALL), and 22% in MLL leukemia [8]. Another study reported that SETD2 mutations were enriched at relapse in pediatric B-cell ALL [9]. A recent follow-up study indicated that SETD2 loss results in an impaired DNA damage response (DDR) after exposure to cytotoxic chemotherapy, which leads to reduced apoptosis [10]. These findings could partially explain how SETD2 mutations contribute to chemotherapy resistance and relapse [11]. However, the mechanisms of how SETD2 mutations confer chemoresistance are still not fully understood. More importantly, a better understanding of how sensitivity to chemotherapy can be restored in SETD2-mutant leukemia will help to design a better therapeutic strategy for refractory/relapsed acute leukemia (AL).
In this study, we generated two novel loss-of-function (LOF) Setd2 mutation alleles (Setd2F2478L and Setd2Ex6-KO) in mice to model LOF conditions of SETD2 in vivo. We found that both mutant alleles showed similar epigenetic, cellular, and growth retardation phenotypes. They also cooperated with Mll-Af9 to accelerate the development of leukemia that resulted in resistance to standard cytarabine-based chemotherapy by altering S and G2/M cell cycle checkpoints. Importantly, S and G2/M cell cycle checkpoint inhibition, by either WEE1 or CHK1 inhibitors, resensitized Mll-Af9/Setd2 double-mutant cells to standard chemotherapy by causing the DNA replication collapse, mitotic catastrophe, and increasing cell death. Thus, the combination of cell cycle checkpoint inhibitors with conventional chemotherapeutic agents may provide a promising therapeutic strategy for the treatment of refractory or relapsed leukemia patients with mutations in SETD2 or SETD2-like epigenetic mutations.
Materials and methods
Animals
Setd2F2478L/WT and Setd2Ex6-KO/WT mice were generated by the Cincinnati Children’s Hospital Transgenic Core. Mll-Af9 knock-in mice, B6-SJL (CD45.1+), and NSGS (NOD/SCID IL2Rγ−/− SGM3) mice were purchased from the Jackson Laboratory. All mice were housed in the rodent barrier facility at Cincinnati Children’s Hospital Medical Center (CCHMC). All animal studies were conducted according to an approved Institutional Animal Care and Use Committee protocol in accordance with federal regulations. Bone marrow cell transplantations were performed as previously described [12].
Chemotherapy reagents
Chemotherapy drugs (Doxorubicin, Ara-C, and Daunorubicin) were obtained from the clinical pharmacy at Cincinnati Children’s Hospital. WEE1 inhibitor MK-1775 and CHK1 inhibitor MK-8776 were obtained from Selleckchem.
Detailed methods are described in the Supplementary information.
Results
Two novel mouse models with Setd2 loss-of-function mutations present similar phenotypes
We previously corroborated the results of others demonstrating that similar frequencies of missense mutations and nonsense/frameshift mutations of SETD2 are observed in acute leukemia patients [8]. To model the function of SETD2 mutations in leukemia development and chemotherapy resistance, two Setd2-mutant alleles were generated using CRISPR/Cas9-mediated genome editing in fertilized embryos: (1) the Setd2F2478L allele, which is equivalent to the SETD2F2505L mutation found in an AML patient [8] (Fig. 1a and Materials and Methods in Supplementary information). The Setd2F2478L mutation, located in the SRI domain, loses the interaction with the C-terminal domain (CTD) of RNA polymerase II [13]. (2) The Setd2Ex6-KO allele, resulting in a frameshift and nonsense-mediated decay of Setd2 mRNA (Fig. 1b and Materials and Methods in Supplementary information).
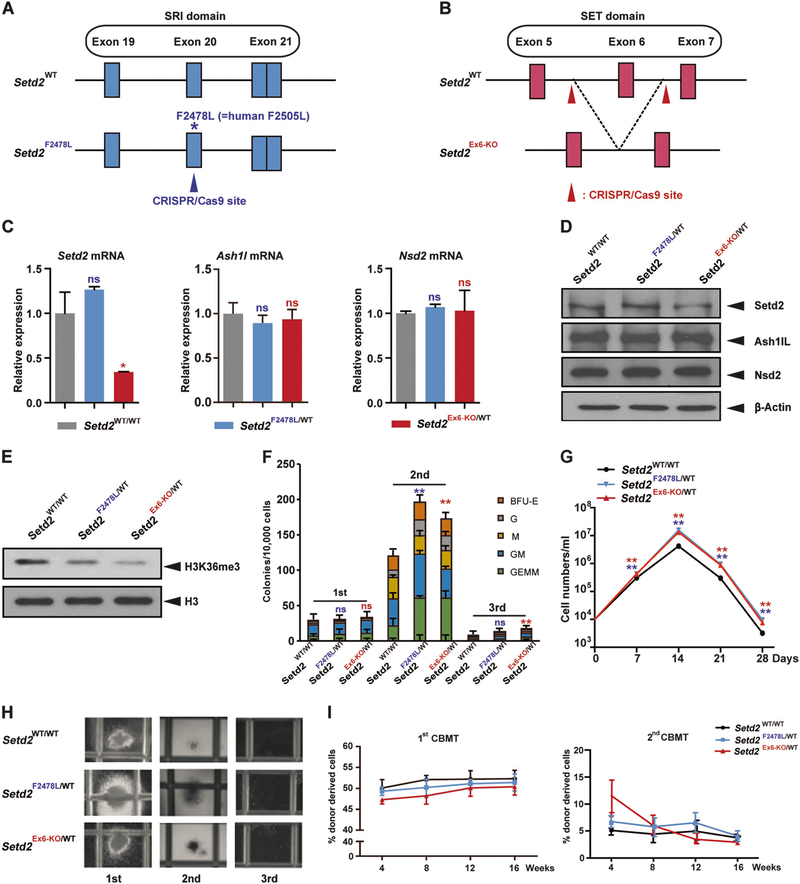
Fig. 1.
Mouse models with two distinct loss-of-function Setd2 mutations show similar phenotypes. a, b Schemes of the wild-type Setd2 locus (top) and Setd2 mutation locus (bottom) after Cas9–CRISPR- mediated modification. The Setd2F2478L point mutation locates within exon 20 in the SRI domain (a). Setd2 exon 6 locates in the SET domain (b). c Relative Setd2, Ashl1, and Nsd2 mRNA levels in c-Kit positive bone marrow cells measured by quantitative real-time PCR (Q-PCR) and normalized to β-actin levels using the Δ–CT method. d, e Endogenous Setd2, Ash1L, and Nsd2 protein expression (d), and H3K36me3 levels (e), in c-Kit positive bone marrow cells from wild type, Setd2F2478L/WT, and Setd2Ex6-KO/WT mice determined by western blot. f Bone marrow cells from wild type, Setd2F2478L/WT, and Setd2Ex6-KO/WT mice were analyzed with M3434 methylcellulose-based medium. A total of 10,000 bone marrow cells were plated in triplicate in M3434. Colony count scoring and replating was repeated every 7 days. g Cell number counts of CFUs in serial replating in (f). h Representative image of colonies in (f). i First and second competitive bone marrow transplantation. WT recipient mice were intravenously injected with 1.5 × 106 BM-MNCs from WT, Setd2F2478L/WT, or Setd2Ex6-KO/WT mice (CD45.2+) together with an equal number of CD45.1+ competitor cells. Peripheral blood cells were collected from recipients monthly and analyzed by FACS for the presence of CD45.2+ donor-derived cells. Three biological replicates of each genotype are performed in triplicate and the data are presented as the mean ± SD values. **P < 0.01
Both Setd2F2478L/F2478L and Setd2Ex6-KO/Ex6-KO homozygous mutations showed early embryonic lethality, which is consistent with a previous report stating that homozygous deletion of exon 4 and exon 5 of Setd2 results in embryonic lethality at E10.5–E11.5 [14] (Supplementary Table 1A, B, and data not shown). Interestingly, mice that were heterozygous for either mutation showed the same growth retardation phenotypes. The body weights of both types of heterozygous mice were consistently lower, as compared with wild-type littermates (Supplementary Fig. 1A). At 12 weeks, both Setd2 heterozygous mutant mouse models were significantly smaller than their wild-type littermates in both genders (Supplementary Fig. 1B, and data not shown). We also measured the weights of different organs in males (Supplementary Fig. 1C). The livers were smaller (Supplementary Fig. 1D) and bowels were shorter, as compared with their wild-type littermates (Supplementary Fig. 1E). The major weight differences were from the bones and muscles (data not shown).
Although the Setd2F2478L mutation loses the interaction with the C-terminal domain (CTD) of RNA polymerase II [13], the expression of Setd2 in heterozygous mice was similar to that in wild-type mice, at both the mRNA and protein levels. However, Setd2 expression in exon 6-KO mice decreased to 50% at both the mRNA and protein levels, indicating nonsense-mediated mRNA decay (NMD) (Fig. 1c, d). Other major SETD2 family H3K36 methyltransferases, such as ASH1L and NSD subfamily members, were not affected significantly in mice heterozygous for either Setd2 mutation (Fig. 1c, d; Supplementary Fig. 2A, B).
In mice heterozygous for either Setd2 mutation, we found a similar decrease in H3K36me3 modification in purified c-Kit+ bone marrow cells (Fig. 1e). H3K36me3 is involved in active transcription [15]. We thus performed the CFU assay to test whether the decrease of this epigenetic mark affects hematopoiesis. We found that bone marrow cells from mice with either Setd2 mutation had increased CFU and cell numbers in the second passage, but all underwent terminal differentiation in the third passage. This indicated that the Setd2 mutations may increase the number, or proliferation, of progenitors, but cannot transform them alone (Fig. 1f, g, h). To further determine whether Setd2F2478L/WT and Setd2Ex6-KO/WT HSPCs exhibit enhanced self-renewal, standard competitive bone marrow transplantation (CBMT) assays were performed to examine the engraftment potential of HSPCs in vivo (Fig. 1i). The frequency of donor-derived reconstitution with mononuclear cells (MNCs), showed no significant difference, as compared with MNCs from wild-type mice at all time points detected in both the first and second CBMT (Fig. 1i). This finding is consistent with data showing that the percentage of LK and LSK populations in the total bone marrow cells was not obviously changed in mice heterozygous for either Setd2 mutation (Supplementary Fig. 3A–C).
Setd2 mutants cooperate with an Mll-Af9 knock-in allele to accelerate leukemia
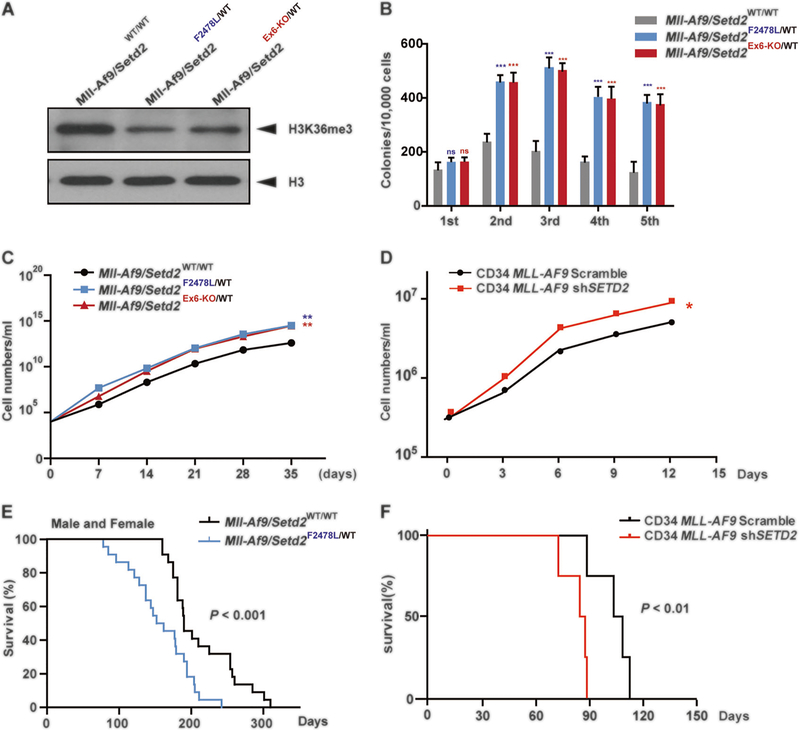
A higher frequency of Setd2 mutations in MLL-rearranged patients with leukemia (22.2%, 6 out of 27) was observed compared with patients in the cohort with leukemia that did not have MLL rearrangements (4.6%, 8 out of 173) [8, 9, 16]. Thus, we used our new genetic alleles to investigate whether these Setd2-mutant alleles cooperate with Mll-Af9 to accelerate leukemia development. Mice with Setd2 mutations were bred with Mll-Af9 knock-in mice [17]. When the mice were harvested at the end stage of leukemia, both Mll-Af9/Setd2F2478L/WT and Mll-Af9/Setd2Ex6-KO/WT mutant mice developed similar pathological features. Both had >25% of leukemic blasts present in the bone marrow (Supplementary Fig. 4A–D) and decreased levels of H3K36me3 modification in bone marrow c-Kit+ cells compared with Setd2 wild-type controls (Fig. 2a). CFU assays were performed to measure the cooperative effects of Setd2 mutations with Mll-Af9 on cell differentiation and proliferation. As shown in Fig. 2b, c, bone marrow cells from mice heterozygous for either Setd2 mutation, along with Mll-Af9 knock-in, exhibited a significantly higher yield of total colonies, dense clones, and a growth advantage in multiple rounds of plating assays, as compared with cells with only Mll-Af9 knock-in. Similar results were found in CD34 MLL-AF9 cells with SETD2 knockdown using shRNA (Fig. 2d, Supplementary Fig. 6F). The results indicate that the growth advantage of double-mutant cells may contribute to increased numbers of leukemic stem cells and accelerated development of leukemia. Consistent with this in vitro result, mice with both Setd2 mutation and Mll-Af9 knock-in developed AML and died faster, as compared with mice with the Mll-Af9 knock-in alone, in separate male and female cohorts (Supplementary Fig. 5A, B). The data for males and females combined showed that Mll-Af9/ Setd2F2478L/WT mice died much faster than mice with the Mll-Af9 knock-in. Both showed similar white blood cell counts in peripheral blood at the end point (Fig. 2e; Supplementary Fig. 6D). To further confirm the impact of SETD2 mutations in human CD34 MLL-AF9 leukemic progression, CD34 MLL-AF9 cells with SETD2 knockdown (SETD2-KD) were transplanted into NSGS (NOD/SCID IL2Rγ−/− SGM3) mice under a myeloablative condition [18]. The results were consistent with murine AML model data and confirmed that Setd2 mutations accelerate Mll-Af9 leukemia (Fig. 2f) without changing the obvious monocytic AML phenotype. No significant difference in differentiation was observed, both in vivo and in vitro, in murine or in human leukemic cells (Supplementary Fig. 4A–D, and data not shown).
Fig. 2.
Setd2 mutants cooperate with the Mll-Af9 knock-in allele to accelerate leukemia. a H3K36me3 levels in c-Kit positive bone marrow cells from Mll-Af9/Setd2WT/WT, Mll-Af9/Setd2F2478L/WT, and Mll-Af9/Setd2Ex6-KO/WT mice at 6 months, the end stage of leukemia. b CFU replating assays of bone marrow cells from Mll-Af9 and two Mll-Af9/ Setd2-mutant mice. c Cell number counts of CFUs in the serial replating in (b). d Cell number counts of serial replating of CD34 MLL-AF9/Scrambled and CD34 MLL-AF9/shSETD2 cells. Three biological replicates of each genotype are performed in triplicate and the data are presented as the mean ± SD values. *P < 0.05; **P < 0.01; ***P < 0.001. e Survival curves for Mll-Af9 (n = 16) and Mll-Af9/ Setd2Ex6-KO/WT (n = 20) transgenic mutant mice, regardless of gender. f Survival curves for NSGS mice receiving bone marrow transplantation with CD34 MLL-AF9/Scrambled (n = 4) or CD34 MLL-AF9/ shSETD2 (n = 4) cells
Setd2 mutations confer resistance to chemotherapy in Mll-Af9 AML
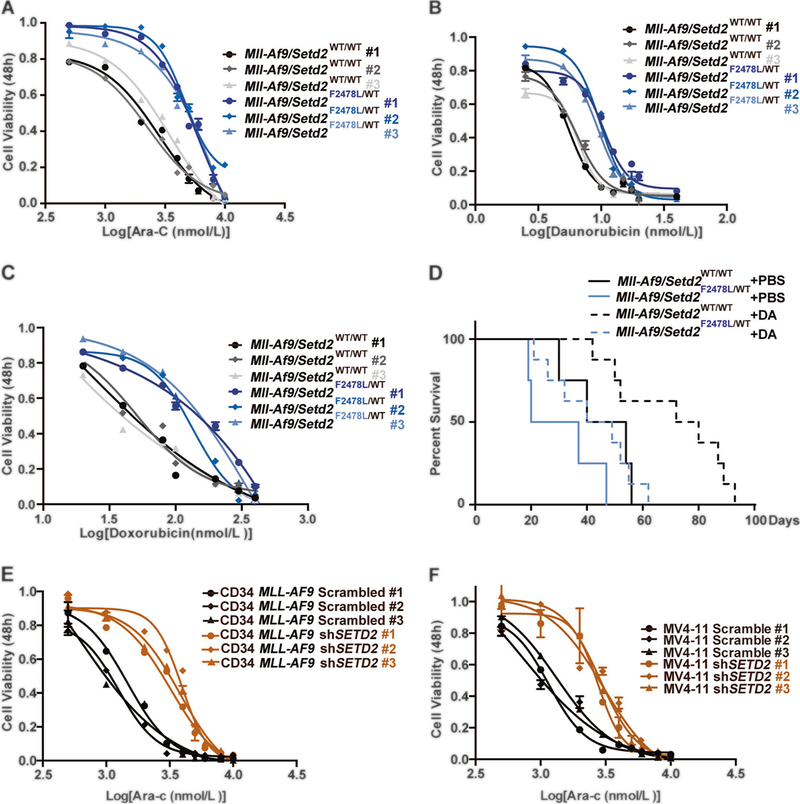
Setd2 mutations are frequently found in relapsed ALL and AML patients [8, 9], indicating that Setd2 mutations might play a role in refractory/relapsed acute leukemia and resistance to chemotherapy. Thus, in vitro experiments were performed to test the hypothesis that Setd2 mutations may lead to chemoresistance of Mll-Af9 AML. We used the standard clinical chemotherapy drugs, Ara-C and anthracyclines (Daunorubicin or Doxorubicin), to treat AML cells isolated from Mll-Af9 and Mll-Af9/Setd2F2478L/WT mouse bone marrow. Strikingly, Mll-Af9/Setd2F2478L/WT cells exhibited a higher tolerance to three major chemotherapeutic agents than Mll-Af9 cells (Fig. 3a–c, Supplementary Fig. 6A–C, Supplementary Tables 3, 4, and data not shown). Transplantations of Mll-Af9 and Mll-Af9/ Setd2F2478L/WT bone marrow cells, at similar leukemic stages, were performed to develop AML in vivo (Supplementary Fig. 6D). Importantly, 3 weeks after BMT, Doxorubicin and Ara-C (DA) were administered to these BMT mice along with controls. Treatment with DA significantly improved survival in mice transplanted with Mll-Af9 leukemia; however, mice transplanted with Mll-Af9 leukemic cells bearing Setd2 mutations had a survival length two-thirds that of the Mll-Af9 controls with the same dose of DA treatment, suggesting resistance to chemotherapy in vivo (Fig. 3d, Supplementary Fig. 6E). Furthermore, human CD34-MLL-AF9 and MV4–11 cell lines with SETD2 knockdown (KD) by shRNA were used to recapitulate chemotherapy resistance to in vitro treatments (Supplementary Fig. 6F, Supplementary Tables 3, 4). Indeed, CD34-MLL-AF9 and MV4–11 cells with SETD2-KD had a much higher tolerance to either Ara-C or Daunorubicin (Fig. 3e, f; Supplementary Fig. 6G–L). The results confirm that Setd2 mutations or SETD2 down-regulation confers resistance to chemotherapy in both murine and human leukemic cells, which is consistent with clinical findings.
Fig. 3.
Setd2 mutation leads to chemoresistance of Mll-Af9 AML cells. a–c Drug resistance assays in multiple clones from different individuals of both Mll-Af9 and Mll-Af9/Setd2F2478L/WT primary bone marrow leukemic populations treated with Ara-C (a), Daunorubicin (b), or Doxorubicin (c). Drug concentrations are indicated on the horizontal axis. Three biological replicates of each genotype are performed in triplicate and the data are presented as the mean ± SD values. d After transplantation of bone marrow cells from Mll-Af9 or Mll-Af9/ Setd2F2478L/WT mice to B6-SJL (CD45.1+) mice (n = 4 in each group, two independent biological replicates) for 3 weeks, the chemotherapy regimen was performed by intravenous (i.v.) injection of Doxorubicin and Ara-C. Days of survival of treated and nontreated mice in the Mll-Af9 and Mll-Af9/Setd2-mutant cohorts were recorded. e, f Drug resistance assays in Mll-Af9 human CD34+ cells (e) or the MV4–11 cell line (f) with shRNA-mediated SETD2 knockdown. Three biological replicates of each genotype are performed in triplicate, and the data are presented as the mean ± SD values
Checkpoint inhibition resensitizes resistant Mll-Af9/ Setd2-mutant AML to chemotherapy
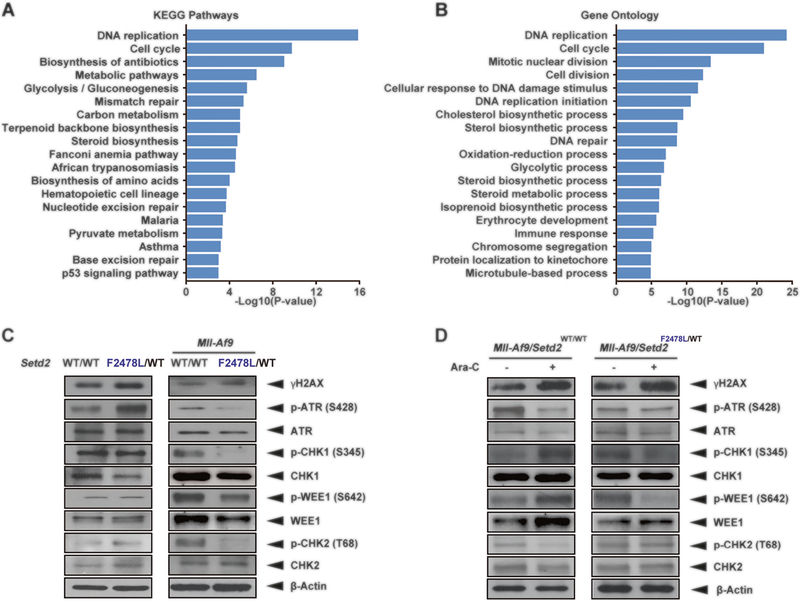
To understand the mechanism of chemoresistance caused by Setd2 LOF mutations, we took an unbiased wholegenome approach (RNA-seq) using Mll-Af9 cells with Setd2 knockdown to identify dysregulated pathways [8, 19]. Interestingly, we found enrichment of downregulated genes with roles in G2/M cell cycle progression, DNA replication, DNA repair (mismatch repair, Fanconi anemia, nucleotide excision repair, and base excision repair), and the p53 apoptosis pathway, all of which are involved in cellular DDR (Fig. 4a, b; Supplementary Table 2). Using immunoblotting assays, we further confirmed that the levels of the DNA damage marker γH2AX were increased in Setd2F2478L/WT compared with the Setd2 wild-type control (Fig. 4c). This indicates that the Setd2 mutation provokes an aberrant DNA damage signaling even without any exogenous stress. When the Setd2F2478L/WT mutation was introduced into the Mll-Af9 background, the levels of S and G2/M checkpoint kinases, phosphorylated ATR, phosphorylated CHK1, and WEE1 downstream, were dramatically reduced (Fig. 4c, right panel). This is consistent with increased γH2AX due to cell cycle progression without the completion of DNA repair. Moreover, phosphorylated CHK1 and WEE1 proteins were barely accumulated after Ara-C treatment in Mll-Af9/Setd2F2478L/WT cells compared with the untreated controls (Fig. 4d, right panel). These data indicate that Setd2 mutations confer the decreased S and G2/M checkpoint activity on AML cells. Activity of S and G2/M checkpoint function prevented cells with massive DNA damage from entering mitosis, arrested cells in the S phase, and induced cell death [20, 21]. Therefore, due to S and G2/M check-point defects, Mll-Af9/Setd2-mutant AML cells are able to survive by escaping from massive DNA damage-induced S-phase cell cycle arrest and cell death.
Fig. 4.
Checkpoint signaling is dysregulated in Mll-Af9/Setd2-mutant AML. a, b KEGG pathway analysis (a) and GO analysis (b) of RNA-seq data from Mll-Af9 and Mll-Af9/shSetd2 knockdown cell lines. c Detection of endogenous DNA damage and checkpoint-related proteins in untreated Setd2F2478L/WT and Mll-Af9/Setd2F2478L/WT cells using immunoblotting. d Detection of endogenous DNA damage and checkpoint-related proteins in Mll-Af9 and Mll-Af9/Setd2F2478L/WT cells following treatment with 100 nM Ara-C for 48 h using immunoblotting. The data are presented from one representative experiment with one of four independent clones’ replicates. The results were consistent across all biological replicates tested
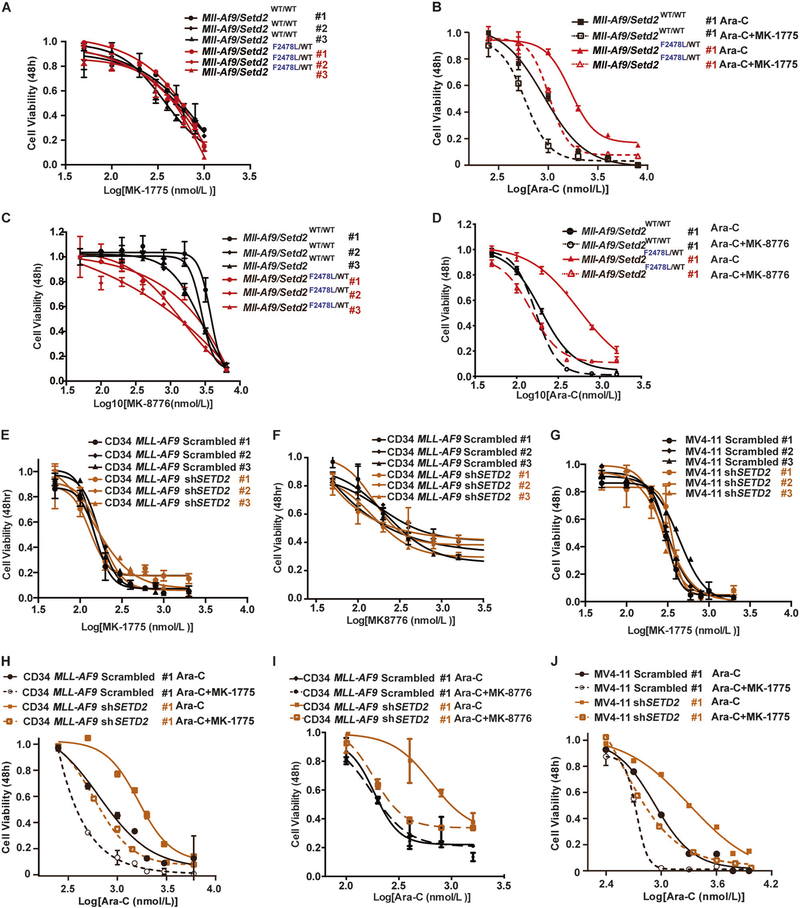
However, this advantage of Mll-Af9/Setd2-mutant AML cells also presents a potential unique vulnerability that can be targeted. When S and G2/M checkpoints are chemically inhibited, those cells can bypass both S and G2/M checkpoints and aberrantly enter the M phase (mitosis), in which those cells may undergo a mitotic catastrophe by DNA damage or incomplete chromosome segregation, ultimately leading to apoptosis [22, 23]. To test this hypothesis, Setd2-mutant leukemic cells were treated with Ara-C in combination with a WEE1 inhibitor, MK-1775 [24], in vitro. As shown in Fig. 5a, treatment of MK-1775 alone resulted in no significant difference in the viability of Mll-Af9 or Mll-Af9/Setd2F2478L/WT cells. However, when MK-1775 was added in combination with Ara-C, chemoresistant Mll-Af9/Setd2F2478L/WT cells were able to resensitize to Ara-C, while Mll-Af9 cells were also affected (Fig. 5b; Supplementary Fig. 7A, B). Similar results were found with the CHK1 inhibitor MK-8776 [25] (Fig. 5c, d; Supplementary Fig. 7C, D). Both drug combinations showed a synergistic effect with increased therapeutic response to Ara-C and reduced AML cell viabilities (Supplementary Fig. 8A–F). We further validated these results in human CD34-MLL-AF9 and MV4–11 cells with shRNA-mediated SETD2-KD, and observed a similar resensitization (Fig. 5e– j; Supplementary Fig. 7E–J) and synergistic effects of drug combinations (Supplementary Fig. 8G–O). These data indicate that S and G2/M cell cycle checkpoint inhibition can resensitize Setd2-mutant AML cells to chemotherapy.
Fig. 5.
Checkpoint inhibition resensitizes resistant Mll-Af9/Setd2 double-mutant AML cells to chemotherapy. a Mll-Af9 and Mll-Af9/ Setd2F2478L/WT primary bone marrow leukemic cells were treated with variable concentrations of the WEE1 inhibitor MK-1775 for 48 h. b Mll-Af9 and Mll-Af9/Setd2F2478L/WT primary bone marrow leukemic cells were treated with variable concentrations of the combination of Ara-C and MK-1775 for 48 h. c Mll-Af9 and Mll-Af9/Setd2F2478L/WT primary cells were treated with variable concentrations of the CHK1 inhibitor MK-8776 for 48 h. d Mll-Af9 and Mll-Af9/Setd2F2478L/WT primary cells were treated with variable concentrations of the combination of Ara-C and MK-8776 for 48 h. e–g shRNA-mediated Setd2 knockdown was performed in MLL-AF9 human CD34+ cells (e, f) and the MV4–11 cell line (g). After puromycin selection, the stable cell lines were treated with variable concentrations of the WEE1 inhibitor MK-1775 (e, g) or CHK1 inhibitor MK-8776 (f). h MLL-AF9 human CD34+ Scrambled and shSETD2 cells tested in the combination of Ara-C and MK-1775 for 48 h. i MLL-AF9 human CD34+ Scrambled and shSETD2 cells tested with the combination of Ara-C and MK-8776 for 48 h. j MV4–11 Scrambled and shSETD2 cells tested with the combination of Ara-C and MK-1775 for 48 h. The data in (a, c, e, f, g) are shown as three biological replicates of each genotype performed in triplicate and presented as the mean ± SD values. The data in (b, d, h, i, j) are shown as the mean ± SD values (n = 3 technical replicates) of one independent clone
Checkpoint inhibition increases mitotic catastrophe and promotes cell death in chemo-treated cells
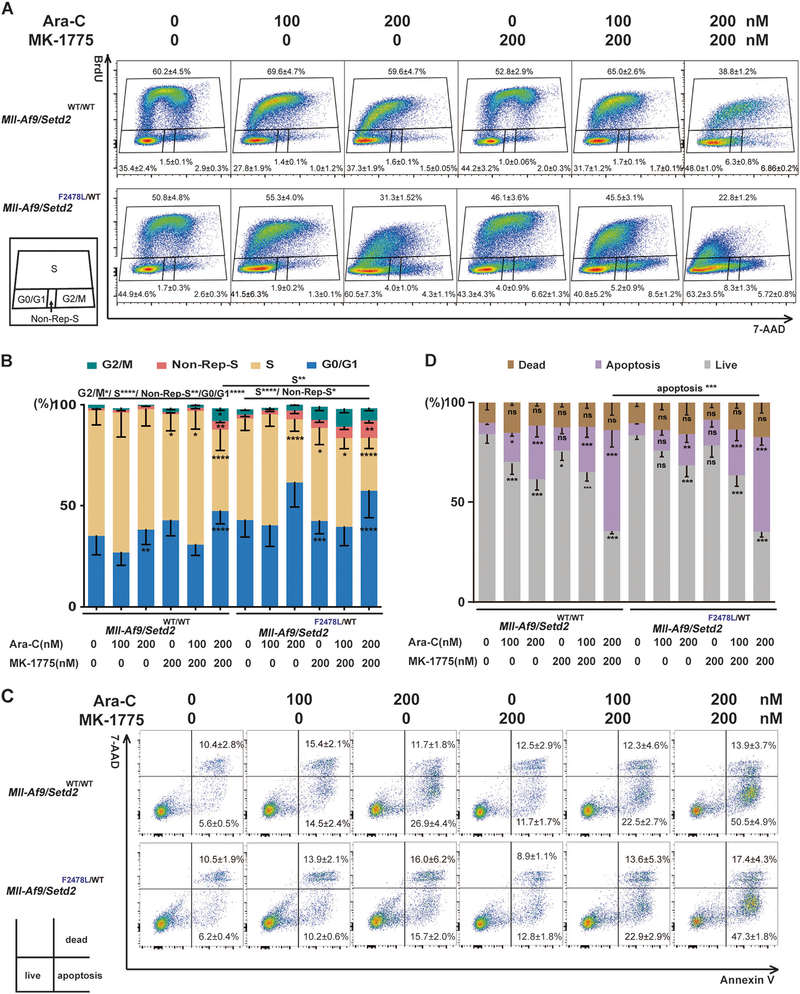
To understand the details of how cell cycle checkpoint inhibition is able to resensitize Setd2-mutant AML cells to chemotherapy, we first performed a detailed cell cycle analysis using BrdU and 7-AAD labeling. In the presence of Ara-C, Mll-Af9/Setd2F2478L/WT cells at the S phase were reduced. But the subpopulations at the G2/M phase and the non-replicating S phase (BrdUneg/7-AADint) were increased, as compared with Mll-Af9 cells (Fig. 6a, b). Non-replicating S-phase cells are cells with >2 N and <4 N DNA content, which have arrested in the S phase [26]. These suggested that a subpopulation of Mll-Af9/Setd2F2478L/WT cells were arrested in the S phase due to blockage of DNA synthesis upon 48 h of Ara-C treatment with BrdU labeling [27]. These cells could have subsequently resumed DNA synthesis and get along with Mll-Af9/Setd2F2478L/WT cells measured as BrdU+. BrdU+Mll-Af9/Setd2F2478L/WT cells may have been able to progress to the G2/M phase to enable DNA repair and mitotic entry, possibly because of an impaired G2/M checkpoint. Notably, in the presence of Ara-C and MK-1775, Mll-Af9/Setd2F2478L/WT cells showed significantly reduced S-phase cells and increased non-replicating S-phase cells after 48 h of treatment, as compared with the nontreated control. The reduction of S-phase cells was more significant in Mll-Af9/Setd2F2478L/WT cells than in Mll-Af9 cells after the treatment of Ara-C and MK-1775 (Fig. 6a, b). The reduced S-phase population might result from the increased non-replicating S phase and also increased premature entry into mitosis. Thus, with these two additive effects in the cell cycle, WEE1 inhibition could resensitize Mll-Af9/Setd2F2478L/WT cells to chemotherapy.
Fig. 6.
Checkpoint inhibition alters the cell cycle and promotes apoptosis in cells treated with chemotherapeutic agents. Mll-Af9 or Mll-Af9/Setd2F2478L/WT AML cells treated with Ara-C, MK-1775, or a combination, as indicated, for 24 h. a Cell cycle phase distributions were determined via BrdU incorporation for 40 min. b The percentage of cells at various stages of the cell cycle (G2/M, S, G0/G1, and non-Rep-S) in (a) was calculated and illustrated. Non-Rep-S represents the non-replicating S phase. c Apoptosis of the cells in quadrant 3 was evaluated by Annexin V/7-AAD staining. d Graphical representation of the percentage of apoptotic cells in (c). The stacked bar graphs indicate the mean percentage of viable (“live”), early apoptotic (“apoptosis”), and late apoptotic (“dead”) cells. Three biological replicates of each genotype are performed in triplicate and the data are presented as the mean ± SD values. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001
Next, to determine whether combination treatment with Ara-C and MK-1775 can enhance cell death, Annexin V and 7-AAD co-staining were performed to assess the percentage of cells undergoing early-stage apoptosis or becoming dead cells. As shown in Fig. 6c, d, when cells were treated with Ara-C only, they featured an increased early apoptosis population (Annexin V positive/7-AAD negative) in a drug dose-dependent manner as compared with the control group. Although MK-1775 has no effect on cell death by itself, the combination treatment of MK-1775 and Ara-C exhibited a much stronger cell death induction effect than Ara-C alone in both Mll-Af9 and Mll-Af9/ Setd2F2478L/WT cells. The Annexin V and 7-AAD double-positive populations containing both late apoptotic and necrotic cells are shown in Fig. 6d as “dead cells”.
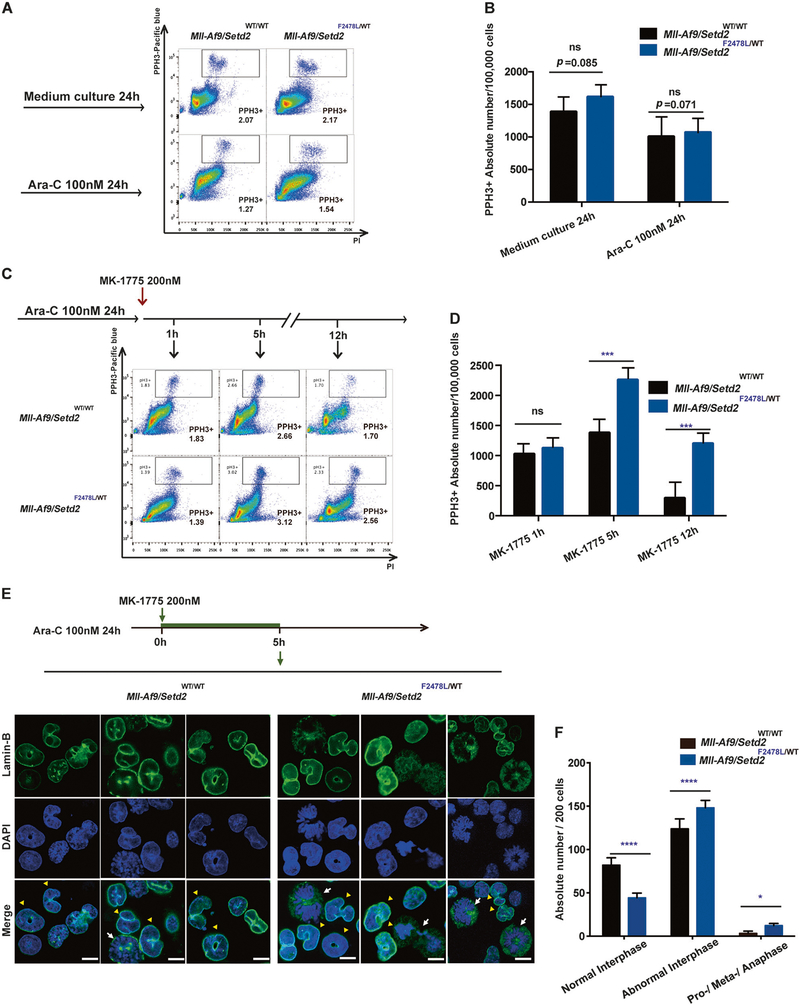
Furthermore, we attempted to specify the combinatory therapeutic effect on mitotic entry and mitotic catastrophe. Mll-Af9 and Mll-Af9/Setd2F2478L/WT cells were treated with Ara-C overnight followed by a combination with MK-1775. The mitotic cell numbers were measured with Phospho- Histone 3 (PHH3), a marker for mitosis [28], at different time points. Mll-Af9/Setd2F2478L/WT and Mll-Af9 populations showed no significant difference in the levels of PHH3+ cells, regardless of Ara-C treatment for 24 h (Fig. 7a, b), indicating that Setd2 mutation does not alter the mitotic activities of these leukemic cells. However, after MK-1775 was added following Ara-C treatment, the PHH3+ population was significantly increased in Mll-Af9/Setd2F2478L/WT cells within the intervals of 5 and 12 h of combinatory treatment, as compared with the same treatment of Mll-Af9 cells (Fig. 7c, d). These results indicate that the WEE1 inhibition has a stronger effect on bypassing the G2/M checkpoint and cell cycle progression to mitosis in Setd2- mutant leukemic cells.
Fig. 7.
Checkpoint inhibition increases mitotic catastrophe in chemotreated cells. a Flow cytometry analysis of the phospho-Histone H3 (PHH3)-positive (PHH3+) mitotic population (indicated by boxes) in Mll-Af9 and Mll-Af9/Setd2F2478L/WT primary cells after exposure to culture medium alone or Ara-C treatment for 24 h. b The total number of PHH3+ cells in Mll-Af9 and Mll-Af9/Setd2 F2478L/WT primary cells in (a) were calculated and illustrated. c After Mll-Af9 and Mll-Af9/ Setd2F2478L/WT primary cells were treated with Ara-C for 24 h, the WEE1 inhibitor MK-1775 was added and cells were collected at 1, 5, and 12 h from the same culture for pHH3+ analysis. d Total number of PHH3+ cells in Mll-Af9 and Mll-Af9/Setd2F2478L/WT primary populations in (c) were illustrated and calculated. e After Mll-Af9 and Mll- Af9/Setd2F2478L/WT primary cells were treated with Ara-C for 24 h, MK-1775 was added and the cells were collected at 5 h for confocal analysis. Mitotic catastrophe is visualized by micronucleation detected with an antibody against Lamin B (green). Nuclei or condensed chromosomes are shown by counterstaining with DAPI (blue). Scale bar indicates 10 μm. Yellow triangles are used to show abnormal interphase cells; white arrows are used to show pro-/meta-/anaphase cells. f Representative data of three independent experiments in (e) are shown. In total, 200 cells per slide were counted. Three biological replicates of each genotype are performed in triplicate and the data are presented as the mean ± SD values. *P < 0.05; ***P < 0.001; ****P < 0.0001
Finally, we sought to determine whether Mll-Af9/ Setd2F2478L/WT leukemic cells with DNA replication defects and forced mitotic entry lead to mitotic catastrophe after combinatory treatment [22, 23]. The nuclear envelope of either mononucleated (normal) or micronucleated cells, is detected with an antibody against Lamin B and is present only in interphase cells [29]. Untreated cells of both types were predominantly mononucleated (normal interphase), as indicated by Lamin B staining (Supplementary Fig. 9A). Next, Lamin B images were also collected at 5 h post MK-1775 treatment following the overnight pre-incubation of Ara-C. The percentages of micronucleation or abnormally shaped interphase nuclei (abnormal interphase), were increased in both Mll-Af9 and Mll-Af9/Setd2F2478L/WT populations treated with Ara-C and MK-1775, with Mll-Af9/Setd2F2478L/WT populations being significantly increased (Fig. 7e, f). Notably, WEE1 inhibition in Mll-Af9/ Setd2F2478L/WT cells also increased the number of cells in mitosis, which is consistent with difficulties in mitotic progression. Both abnormal interphase cells and cells in an impaired mitotic progression can lead to mitotic catastrophe characterized by incomplete chromosome segregation, and consequently induce cell death [22, 23] (Supplementary Fig. 9B).
Taken together, our results indicate that Setd2 mutations lead to impaired S and G2/M checkpoint regulation. Therefore, they confer chemoresistance on AML cells by altering cell cycle progression and decreasing cell death. Importantly, adding S and G2/M checkpoint inhibitors can resensitize chemoresistant AML cells bearing Setd2 mutations to standard doses of chemotherapy by multiple mechanisms: enhanced DNA replication defects, increased premature entry into mitosis, mitotic catastrophe, and cell death. Our findings present a promising therapeutic strategy for resensitizing chemoresistant AL with SETD2 or SETD2-like epigenetic mutations.
Discussion
Recently, epigenetic changes and mutations (e.g., SETD2 [8], DNMT3 [30], and EZH2 [31]) have been identified as regulators that confer chemoresistance in AL. To model these phenomena in vitro and in vivo, we have generated two genetically engineered mouse models with novel loss-of-function Setd2 mutation alleles (Setd2F2478L and Setd2Ex6-KO). The F2478L mutant in mice is equivalent to the F2505L loss-of-function mutant found in an AML patient [8]. We found that mice which were heterozygous for either mutant allele showed similar epigenetic, cellular, and growth retardation phenotypes (Fig. 1; Supplementary Fig. 1). Furthermore, the Setd2F2478L mutant allele cooperates with Mll-Af9 to accelerate AML. More importantly, Setd2-mutated AML shows chemoresistance to the standard DA (Daunorubicin and Ara-C) regimen, as compared with Mll-Af9 AML, which is chemosensitive (Fig. 3).
MLL/SETD2 leukemia, generated with our genetically engineered mouse models, helped us address several important questions: why does Setd2 loss accelerate leukemia and why does Setd2 loss confer chemoresistance? First, as shown in our previous study, Setd2 LOF results in activated gene expression in the mTOR and Jak–Stat signaling pathways, which are known to contribute directly to promoting leukemogenesis [8]. Second, we hypothesized that there are intrinsic drug-resistant mechanisms of Mll-Af9/Setd2F2478L cells, which are different from chemosusceptible (Mll-Af9) cells. Thus, we performed an unbiased whole-genome analysis and found downregulated genes, which are involved in the cellular DDR, downregulation of CHK1 activation (phosphorylation on Ser345), and of the total WEE1 protein in Mll-Af9/Setd2F2478L cells. These phenomena were independent of whether they were treated with chemotherapeutic agents or not (Fig. 4c, d). This indicates that pCHK1/WEE1 downregulation might lead to the intrinsic defects in DNA replication, and the S- and G2/ M-phase checkpoints caused by Setd2 mutations, instead of by extrinsic treatment with a chemotherapeutic agent. Chemotherapeutic agents often exert their cytotoxic effects by inducing DNA damage (based on increased levels of the DNA damage marker γH2AX, Fig. 4d) in the S phase of the cell cycle during DNA replication. Certainly, our finding that DNA replication proteins are downregulated in double-mutant cells could be a source of replication stress, particularly in the presence of chemotherapeutic agents, such as Ara-C. Further increased levels of γH2AX might be expected to emerge from such replication stress. Excess DNA damage triggers a DDR, including activation of checkpoints (such as ATR-CHK1 and WEE1 in the S phase and G2 phase). The current view of chemosensitive leukemic cells is that they can be arrested in the S phase and undergo massive cell death after chemotherapy (Supplementary Fig. 10A). In contrast, downregulation of pCHK1/ WEE1 will lead to an impaired S and G2/M checkpoints, resulting in exiting from the S phase and the increased cell cycle progression to G2 without completing DNA replication in Setd2 mutation AML cells (Fig. 4d, Fig. 6a, b). In this case, the leukemic cells may delay the G2 progression as they try to repair the damaged DNA in response to chemotherapeutic agents. Since the G2 checkpoint is still somewhat functional, leukemic cells therefore have an alternative option for DNA repair and are able to enter mitosis after resolving most of the DNA damage. This is recognized in general as the chemoresistance pathway (Supplementary Fig. 10B) [32].
AL, which is positive for mutations of epigenetic modifiers, such as SETD2, DNMT3A, and EZH2, also tends to be chemoresistant [10, 30, 31]. Similar to our findings, the DNMT3A-mutant murine AML cells and EZH2-deficient colorectal cancer cells also show the attenuated ATR-CHK1 phosphorylation [30, 33]. Thus, we hypothesize that impaired ATR-CHK1/WEE1-dependent S-phase and G2 checkpoints play a key role in chemoresistance in AL. This indicates that there may be a common mechanism by which epigenetic regulator mutations confer acquired resistance to chemotherapeutic agents on cancer cells. We propose a combinatory therapeutic strategy to target their unique vulnerability. Indeed, the WEE1 inhibitor MK-1775 resensitizes Mll-Af9/Setd2F2478L cells to chemotherapy.
SETD2 mutation is also reported to be associated with increased sensitivity to WEE1 inhibition in the human osteosarcoma cell line (U2OS) and kidney carcinoma cell lines (A498 and LB996-RCC), because of synergistic effects on the depletion of a ribonucleotide reductase subunit RRM2 by SETD2/H3K36me3 loss and WEE1 inhibition. Due to RRM2 depletion, SETD2/H3K36me3-deficient cells suffer from the DNA replication stress with critical dNTP depletion and arrest in the S phase, eventually undergoing apoptosis [34]. Consistent with this, we identified a similar downregulation of Rrm2 in Setd2-mutant leukemia by RNA-seq analysis (data not shown). This suggests that Setd2-mutant leukemia was sensitive to the WEE1 inhibitor, potentially through the same mechanism to enhance DNA replication stress and apoptosis. In addition, our study further suggests that WEE1 inhibition can resensitize Setd2-mutant leukemia to chemotherapy by additional mechanisms, including an increased non replicating S-phase population, enhanced premature entry into mitosis, and mitotic catastrophe (Supplementary Fig. 10C).
WEE1 inhibitors have been reported to enhance the antiproliferative effects of Ara-C in AML [10, 26, 35], pediatric Down syndrome AML [36], and T-cells acute lymphoblastic leukemia (T-ALL) [37, 38]. However, the status of SETD2–H3K36me3 was not examined in these studies. In addition to AL, the WEE1 inhibitor with other S-phase specific chemotherapies or radiation also showed a promising anticancer activity in advanced solid tumors [39]. WEE1 inhibitor (MK-1775) combined with carboplatin or cisplatin is currently under investigation in phase I or phase II clinical trials in patients with chemotherapy refractory ovarian cancer [40], or different types of advanced solid tumors [41, 42]. To date, the initial results show good tolerability and promising anticancer activity.
Although dual in vivo treatment with the same WEE1 inhibitor and Ara-C did not prolong survival in retroviral MLL-AF9 Setd2KO/WT leukemia mice, the leukemia burden was significantly reduced in this model [10]. Notably, the leukemia latency of this retroviral leukemia model is relatively short, indicating a correspondingly narrow therapeutic window to treat the leukemia. The pharmacokinetic variability, toxicity, and therapeutic efficacy may still need to be optimized in order to achieve the greatest therapeutic benefit without resulting in unacceptable side effects or toxicity [43].
SETD2 is not only mutated in AL but is also frequently mutated in a wide range of cancers [44], which are broadly treated with chemotherapy and radiotherapy. SETD2 or the related epigenetic dysregulations could similarly confer chemoresistance of solid tumors. Using cell cycle check-point inhibition in combination with standard chemotherapy, or radiotherapy, may effectively resensitize those other types of resistant cancer cells to the standard therapy. Thus, our new approach may represent a promising therapeutic strategy for a variety of cancer types and could benefit many patients. While beyond the scope of this work, this will be an important area for future study.
Supplementary Material
Acknowledgements
This work was supported by the Cincinnati Children’s Hospital Research Foundation (to P.R.A. and to G.H.), the Leukemia Research Foundation (to G.H.), the OCRA (to G.H.), the CFK (to G.H.), National Institutes of Health (NIH) (R21CA187276 to G.H.), the National Natural Science Foundation of China (NSFC) (grant 81470297 and grant 81770129 to G.H., and grant 81471911 to X.Z.), and the “Personalized Medicines–Molecular Signature-based Drug Discovery and Development”, Strategic Priority Research Program of the Chinese Academy of Sciences (grant XDA12020362 to Q.-f. Wang).
Footnotes
Supplementary information The online version of this article (https://doi.org/10.1038/s41375-019-0456-2) contains supplementary material, which is available to authorized users.
Conflict of interest The authors declare no potential conflicts of interest.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Estey E, Dohner H. Acute myeloid leukaemia. Lancet. 2006;368:1894–907. [DOI] [PubMed] [Google Scholar]
- 2.Short NJ, Ravandi F. Acute myeloid leukemia: past, present, and prospects for the future. Clin Lymphoma Myeloma Leuk. 2016;16 (Suppl):S25–9. [DOI] [PubMed] [Google Scholar]
- 3.Farkona S, Diamandis EP, Blasutig IM. Cancer immunotherapy: the beginning of the end of cancer? BMC Med. 2016;14:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagler E, Xavier MF, Frey N. Updates in immunotherapy for acute myeloid leukemia. Transl Cancer Res. 2017;6:86–92. [Google Scholar]
- 5.Hourigan CS. Editorial: targets for immunotherapy in acute leukemia. Curr Drug Targets. 2017;18:256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fesnak AD, June CH, Levine BL. Engineered T cells: the promise and challenges of cancer immunotherapy. Nat Rev Cancer. 2016;16:566–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. 2013;13:714–26. [DOI] [PubMed] [Google Scholar]
- 8.Zhu X, He F, Zeng H, Ling S, Chen A, Wang Y, et al. Identification of functional cooperative mutations of SETD2 in human acute leukemia. Nat Genet. 2014;46:287–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mar BG, Bullinger LB, McLean KM, Grauman PV, Harris MH, Stevenson K, et al. Mutations in epigenetic regulators including SETD2 are gained during relapse in paediatric acute lymphoblastic leukaemia. Nat Commun. 2014;5:3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mar BG, Chu SH, Kahn JD, Krivtsov AV, Koche R, Castellano CA, et al. SETD2 alterations impair DNA damage recognition and lead to resistance to chemotherapy in leukemia. Blood. 2017;130:2631–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao H, Wang LM, Luo Y, Lai X, Li C, Shi J, et al. Mutations in epigenetic regulators are involved in acute lymphoblastic leukemia relapse following allogeneic hematopoietic stem cell transplantation. Oncotarget. 2016;7:2696–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Yan X, Sashida G, Zhao X, Rao Y, Goyama S, et al. Stress hematopoiesis reveals abnormal control of self-renewal, lineage bias, and myeloid differentiation in Mll partial tandem duplication (Mll-PTD) hematopoietic stem/progenitor cells. Blood. 2012;120:1118–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li M, Phatnani HP, Guan Z, Sage H, Greenleaf AL, Zhou P. Solution structure of the Set2-Rpb1 interacting domain of human Set2 and its interaction with the hyperphosphorylated C-terminal domain of Rpb1. Proc Natl Acad Sci USA. 2005;102: 17636–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu M, Sun XJ, Zhang YL, Kuang Y, Hu CQ, Wu WL, et al. Histone H3 lysine 36 methyltransferase Hypb/Setd2 is required for embryonic vascular remodeling. Proc Natl Acad Sci USA. 2010;107:2956–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2003;23:4207–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker H, Rose-Zerilli MJ, Larrayoz M, Clifford R, Edelmann J, Blakemore S, et al. Genomic disruption of the histone methyltransferase SETD2 in chronic lymphocytic leukaemia. Leukemia. 2016;30:2179–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dobson CL, Warren AJ, Pannell R, Forster A, Lavenir I, Corral J, et al. The mll-AF9 gene fusion in mice controls myeloproliferation and specifies acute myeloid leukaemogenesis. EMBO J. 1999;18:3564–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wunderlich M, Chou FS, Link KA, Mizukawa B, Perry RL, Carroll M, et al. AML xenograft efficiency is significantly improved in NOD/SCID-IL2RG mice constitutively expressing human SCF, GM-CSF and IL-3. Leukemia. 2010;24:1785–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bu J, Chen A, Yan X, He F, Dong Y, Zhou Y, et al. SETD2-mediated crosstalk between H3K36me3 and H3K79me2 in MLL-rearranged leukemia. Leukemia. 2018;32:890–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;3:421–9. [DOI] [PubMed] [Google Scholar]
- 21.Schnerch D, Yalcintepe J, Schmidts A, Becker H, Follo M, Engelhardt M, et al. Cell cycle control in acute myeloid leukemia. Am J Cancer Res. 2012;2:508–28. [PMC free article] [PubMed] [Google Scholar]
- 22.Andreassen PR, Martineau SN, Margolis RL. Chemical induction of mitotic checkpoint override in mammalian cells results in aneuploidy following a transient tetraploid state. Mutat Res. 1996;372:181–94. [DOI] [PubMed] [Google Scholar]
- 23.Andreassen PR, Lacroix FB, Lohez OD, Margolis RL. Neither p21WAF1 nor 14–3-3sigma prevents G2 progression to mitotic catastrophe in human colon carcinoma cells after DNA damage, but p21WAF1 induces stable G1 arrest in resulting tetraploid cells. Cancer Res. 2001;61:7660–8. [PubMed] [Google Scholar]
- 24.Hirai H, Iwasawa Y, Okada M, Arai T, Nishibata T, Kobayashi M, et al. Small-molecule inhibition of Wee1 kinase by MK-1775 selectively sensitizes p53-deficient tumor cells to DNA-damaging agents. Mol Cancer Ther. 2009;8:2992–3000. [DOI] [PubMed] [Google Scholar]
- 25.Guzi TJ, Paruch K, Dwyer MP, Labroli M, Shanahan F, Davis N, et al. Targeting the replication checkpoint using SCH 900776, a potent and functionally selective CHK1 inhibitor identified via high content screening. Mol Cancer Ther. 2011;10:591–602. [DOI] [PubMed] [Google Scholar]
- 26.Porter CC, Kim J, Fosmire S, Gearheart CM, van Linden A, Baturin D, et al. Integrated genomic analyses identify WEE1 as a critical mediator of cell fate and a novel therapeutic target in acute myeloid leukemia. Leukemia. 2012;26:1266–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li FX, Zhu JW, Hogan CJ, DeGregori J. Defective gene expression, S phase progression, and maturation during hematopoiesis in E2F1/E2F2 mutant mice. Mol Cell Biol. 2003;23:3607–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hendzel MJ, Wei Y, Mancini MA, Van Hooser A, Ranalli T, Brinkley BR, et al. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–60. [DOI] [PubMed] [Google Scholar]
- 29.Chaudhary N, Courvalin JC. Stepwise reassembly of the nuclear envelope at the end of mitosis. J Cell Biol. 1993;122:295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guryanova OA, Shank K, Spitzer B, Luciani L, Koche RP, Garrett-Bakelman FE, et al. DNMT3A mutations promote anthracycline resistance in acute myeloid leukemia via impaired nucleosome remodeling. Nat Med. 2016;22:1488–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gollner S, Oellerich T, Agrawal-Singh S, Schenk T, Klein HU, Rohde C, et al. Loss of the histone methyltransferase EZH2 induces resistance to multiple drugs in acute myeloid leukemia. Nat Med. 2016;23:69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Speroni J, Federico MB, Mansilla SF, Soria G, Gottifredi V. Kinase-independent function of checkpoint kinase 1 (Chk1) in the replication of damaged DNA. Proc Natl Acad Sci USA. 2012;109: 7344–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Z, Lee ST, Qiao Y, Li Z, Lee PL, Lee YJ, et al. Polycomb protein EZH2 regulates cancer cell fate decision in response to DNA damage. Cell Death Differ. 2011;18:1771–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pfister SX, Markkanen E, Jiang Y, Sarkar S, Woodcock M, Orlando G, et al. Inhibiting WEE1 Selectively Kills Histone H3K36me3-Deficient Cancers by dNTP Starvation. Cancer Cell. 2015;28:557–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tibes R, Bogenberger JM, Chaudhuri L, Hagelstrom RT, Chow D, Buechel ME, et al. RNAi screening of the kinome with cytarabine in leukemias. Blood. 2012;119:2863–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caldwell JT, Edwards H, Buck SA, Ge Y, Taub JW. Targeting the wee1 kinase for treatment of pediatric Down syndrome acute myeloid leukemia. Pediatr Blood Cancer. 2014;61:1767–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ford JB, Baturin D, Burleson TM, Van Linden AA, Kim YM, Porter CC. AZD1775 sensitizes T cell acute lymphoblastic leukemia cells to cytarabine by promoting apoptosis over DNA repair. Oncotarget. 2015;6:28001–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia TB, Fosmire SP, Porter CC. Increased activity of both CDK1 and CDK2 is necessary for the combinatorial activity of WEE1 inhibition and cytarabine. Leuk Res. 2018;64:30–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamura K Development of cell-cycle checkpoint therapy for solid tumors. Jpn J Clin Oncol. 2015;45:1097–102. [DOI] [PubMed] [Google Scholar]
- 40.Leijen S, van Geel RM, Sonke GS, de Jong D, Rosenberg EH, Marchetti S, et al. Phase II study of WEE1 inhibitor AZD1775 plus carboplatin in patients with TP53-mutated ovarian cancer refractory or resistant to first-line therapy within 3 months. J Clin Oncol. 2016;34:4354–61. [DOI] [PubMed] [Google Scholar]
- 41.Mendez E, Rodriguez CP, Kao MC, Raju S, Diab A, Harbison RA, et al. A phase I clinical trial of AZD1775 in combination with neoadjuvant weekly docetaxel and cisplatin before definitive therapy in head and neck squamous cell carcinoma. Clin Cancer Res. 2018;24:2740–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leijen S, van Geel RM, Pavlick AC, Tibes R, Rosen L, Razak AR, et al. Phase I study evaluating WEE1 inhibitor AZD1775 as monotherapy and in combination with gemcitabine, cisplatin, or carboplatin in patients with advanced solid tumors. J Clin Oncol. 2016;34:4371–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Linden AA, Baturin D, Ford JB, Fosmire SP, Gardner L, Korch C, et al. Inhibition of Wee1 sensitizes cancer cells to antimetabolite chemotherapeutics in vitro and in vivo, independent of p53 functionality. Mol Cancer Ther. 2013;12:2675–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J, Duns G, Westers H, Sijmons R, van den Berg A, Kok K. SETD2: an epigenetic modifier with tumor suppressor functionality. Oncotarget. 2016;7:50719–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.