Abstract
Background/Objectives:
The salience network (SN) comprises brain regions that evaluate cues in the external environment in light of internal signals. We examined the SN response to meal intake and potential genetic and acquired influences on SN function.
Subjects/Methods:
Monozygotic (MZ; 40 pairs) and dizygotic (15 pairs) twins had body composition and plasma metabolic profile evaluated (glucose, insulin, leptin, ghrelin and GLP-1). Twins underwent resting-state functional magnetic resonance imaging (fMRI) scans before and after a standardized meal. The strength of SN connectivity was analyzed pre- and post-meal and the percentage change elicited by a meal was calculated. A multi-echo T2 MRI scan measured T2 relaxation time, a radiologic index of gliosis, in the mediobasal hypothalamus (MBH) and control regions. Statistical approaches included intraclass correlations (ICC) to investigate genetic influences and within-pair analyses to exclude genetic confounders.
Results:
SN connectivity was reduced by meal ingestion (β=−0.20; P<0.001). Inherited influences on both pre- and post-meal connectivity were present (ICC MZ twins 26%, P<0.05 and 47%, P<0.001, respectively), but not percentage change in response to the meal. SN connectivity in response to a meal did not differ between participants with obesity and of normal weight (χ2(1)=0.93; P=0.33). However, when participants were classified as having high or low signs of MBH gliosis, the high MBH gliosis group failed to reduce the connectivity in response to a meal (z=−1.32; P=0.19). Excluding genetic confounders, the percentage change in SN connectivity by a meal correlated to body fat percentage (r=0.24; P<0.01).
Conclusions:
SN connectivity was reduced by a meal, indicating potential participation of the SN in control of feeding. The strength of SN connectivity is inherited, but the degree to which SN connectivity is reduced by eating appears to be influenced by adiposity and the presence of hypothalamic gliosis.
Keywords: hypothalamus, gliosis, obesity, feeding behavior, twin study
Introduction
Resting state fMRI provides information on functional connectivity between anatomically distinct brain areas that form networks(1,2). Connectivity is strong when spontaneous fluctuations in neural activation (measured via blood oxygen level-dependent [BOLD] signal)(1,2) over time is found across the regions of a network. The salience network (SN) is a high order cognitive network(3,4) that detects information from the ambient environment(5) as well as from internal physiological functions (e.g. autonomic and endocrine signals)(6) to achieve a coordinated behavioral response.
To date, the SN’s involvement in feeding is not well established. Reduced connectivity in the satiated state was observed between individual SN regions such as the insula and hypothalamus(7), as well as caudate, insula, and anterior cingulate cortex(8,9), but these studies did not evaluate the SN as a whole. Contrary findings show increased insular connectivity with the hypothalamus after a breakfast(10) and no difference in SN connectivity after an oral glucose load(11).
In obesity, reports show both reduced connectivity within regions of the SN(10,12) and increased connectivity(7,13,14) or a positive correlation with BMI(15,16).
Recent findings of inflammation and reactive gliosis in the mediobasal hypothalamus (MBH) of diet-induced obese rodents(17,18) as well as human adults(17,19–21) and children(22) with obesity add another factor that could influence SN connectivity. Gliosis is an inflammatory reaction involving activation of glial cells(23). It’s presence in the MBH, a critical brain area controlling energy homeostasis(24), could impact brain function locally(17) but also distally due to direct and indirect inputs to multiple regions within the SN(25).
Connectivity in general(26,27) and sensory networks specifically exhibit inherited influences(26,28), but a study of the SN found strong environmental determinants(28). We therefore employed a twin study methodologies(29) to investigate the role of the SN in feeding and query inherited and environmental influences on basal SN connectivity and SN response to a meal.
Subjects and Methods
Participants.
Monozygotic (MZ, N=40 complete pairs, 81 participants) and dizygotic (DZ, N=15 complete pairs, 30 participants) same sex twin pairs were recruited from the community-based Washington State Twin Registry(30). Enrollment criteria were as previously described in a subsample from the current study(31) with additional pairs recruited with at least one twin presenting with obesity. Criteria included age 18–50 years, BMI: 18.5–50 kg/m2 and no contraindications to MRI, major medical problems, history of bariatric surgery, or current participation in weight-loss program. All study procedures were approved by the University of Washington Human Subjects Committee. Written informed consent was provided by all participants.
Study procedures.
Twin pairs completed all procedures on the same day 30 min apart (Figure 1A). The study protocol started at 08:00, after an overnight fast, with height and weight measurements, antecubital intravenous line placement, fasting labs, followed by a standardized breakfast representing 10% of the estimated daily caloric requirements(32) (standard activity factor of 1.3). Twins underwent the first MRI scan 3.5 h after breakfast followed by a standardized meal of macaroni and cheese consumed within 15 min (representing 20% of the individual’s estimated daily caloric needs(32)). Participants then underwent the second MRI scan. Further details in(31).
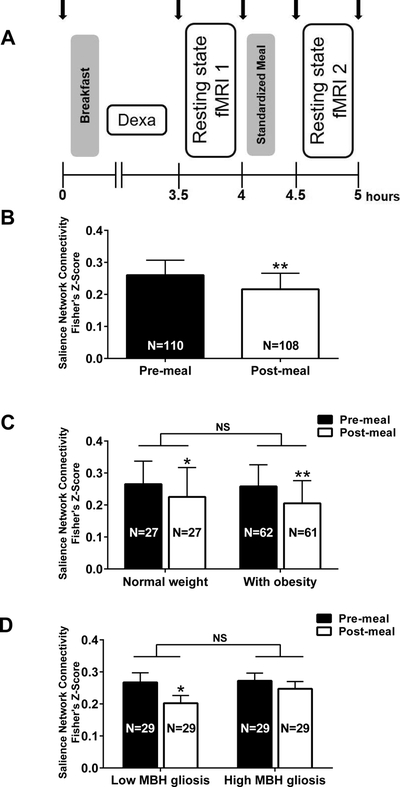
Figure 1.
Salience Network (SN) connectivity in response to a meal. (A) Study protocol: Twin pairs completed all the procedures on the same day, 30 minutes apart from each other. Blood samples were collected throughout the study visit (black arrows). Participants were given a breakfast and standardized meal, representing 10% and 20% respectively of their estimated daily caloric need. All twins underwent a DEXA scan for body composition measurement and two resting-state MRI sessions. (B) SN connectivity was reduced after the ingestion of a meal in all participants. (C) Overall SN connectivity did not differ by adiposity (P=0.33 main effect of group), however, consistent with panel B, it did differ by time (P<0.0001 main effect of time) and post-hoc tests confirmed a significant reduction in SN connectivity by a meal in both groups. (D) SN connectivity did not differ overall by gliosis group (P=0.31 main effect of gliosis group), but did differ by time (P=0.002 main effect of time) specifically that participants in the Low MBH gliosis group reduced SN connectivity after a meal, but this difference was attenuated in the High MBH gliosis group. Data are mean ± standard error of the mean of Fisher z-score log back-transformed. P-values determined by generalized estimating equation (B) or linear mixed models (C, D). *P<0.01 and **P<0.001 vs. Pre-meal.
Laboratory assessment.
Pre- and post-meal values are averages of samples taken before and after their respective MRI sessions, Figure 1A. Blood samples for plasma glucose, insulin, total ghrelin, leptin, and total glucagon-like peptide-1 (GLP-1) concentrations were collected and analyzed as in(33). HOMA-IR was calculated from fasting glucose and insulin levels(34).
Body composition.
Twins underwent a dual-energy X-ray absorptiometry (DEXA) scan (GE Lunar Prodigy or iDXA using a correction factor). Abdominal visceral and android fat mass were estimated by EnCore™ software 16.2 (GE Medical Systems).
MRI acquisition and processing.
All scans were acquired with a 3-Tesla Philips Achieva MR System using a 32-channel SENSE head coil, software 3.2.2 (Philips Medical Systems) and dual Quasar gradients (80mT/m at a slew rate of 110mT/m/s or 40mT/m at a slew rate of 220mT/m/s).
Resting state fMRI.
For both sessions, a 186-volume (7.5 min), T2*-weighted echo-planar imaging time series (44 axial slices; 2.75 × 2.75 × 3mm voxels; echo time [TE]: 30ms; repetition time [TR]: 2400ms, SENSE factor: 2) was acquired with eyes closed. A B0 field map (TE: minimum 2.8ms and Δ 1.0ms; TR: 10ms; flip angle: 10°) and a 3D Magnetization-Prepared Rapid Gradient-Echo image (176 sagittal slices; 1mm isotropic voxels; TE: 3.5ms; TR: 7.5ms; flip angle: 7°; SENSE factor: 2; matrix 256 × 256) were acquired for distortion correction and registration.
Pre-processing of functional data, using tools from Functional MRI of the Brain (FMRIB) Software Library (FSL 5.0.9), FreeSurfer 5.3, and Analysis of Functional NeuroImages, included: elimination of the first three volumes; simultaneous slice-timing and motion correction; removal of non-brain tissue and spike artifacts; spatial smoothing (full-width at half maximum=5mm); bandpass filtering at 0.008–0.15 Hz; removal of nuisance regressors(35).
The registration matrix and warp field for each timeseries to the participant’s skull-stripped structural scan were calculated using boundary-based registration in FSL(36,37). The structural scans were registered to standard space (MNI) through an intermediate template(38) (created by randomly selecting one individual from an equal number of MZ and DZ pairs and submitting their skull-stripped T1 images to an iterative template-building process) using the ANTs toolkit(38).
Regions of the SN (orbital frontoinsula, frontal and temporal pole, paracingulate, dorsal anterior cingulate cortex, supplementary motor area, superior temporal, parietal operculum, ventro and dorsolateral pre-frontal cortex, ventral striatum, thalamus, hypothalamus, amygdala, periaquedutal grey, substantia nigra/ventral tegmental area) were defined by placing spheres (4mm radius) around peaks as previously published by Seeley et al.(3). ROIs were warped into each subject’s functional space and mean timeseries extracted. Pairwise correlations were calculated among each pair of ROIs then transformed to Fisher’s Z-scores. An overall average Fisher’s Z-score was derived from pre- and post-meal values and used to calculate percentage change by a meal.
Quantitative T2 parametric map.
A quantitative multi-slice/multi-echo T2-weighted sequence with 16 echoes was obtained (TR/TE/NSA: 2000/20–170/1, interecho space: 10ms). Slices (N=12 per subject) were acquired coronally between the optic chiasm and the mammillary bodies (slice thickness: 2.5mm; interslice gap: 0.2mm; voxel size: 1.313mm3). The T2 parametric map was constructed based on the signal decay curve from the 16 echoes on a voxel-by-voxel basis, from which the T2 relaxation time (ms) was obtained.
For analyses, the coronal slice directly posterior to the optic chiasm was located(19) and the bilateral MBH was delimited, along with the bilateral reference regions (amygdala and putamen). All ROIs were transferred to the T2 parametric map of the corresponding slice and the T2 relaxation time (mean and SD) was obtained using OsiriX Imaging Software 5.6. Eleven DZ and 28 MZ pairs completed scans for both MRI techniques (functional and structural).
Statistics.
For descriptive statistics mean(SD) are reported. Chi-squared tests were used to test proportions of categorical variables. In overall analyses including all twins, generalized estimating equation (GEE) regression models (to take into consideration the non-independence of the twins(39)) or linear mixed models with the restricted maximum likelihood estimation were used.
In twin analyses, intraclass correlations (ICC) were calculated by linear mixed models. In twin association analyses using within-pair differences to investigate influences on SN connectivity controlling for genetic confounding, a within-pair analysis approach restricted to the MZ twins was used. Twins within each pair were ordered according to their body fat mass and the difference between values obtained from each twin was calculated. Simple and multiple linear regressions were applied(39). Pearson r values were calculated for descriptive purposes.
Non-normally distributed variables were log transformed for analyses, then log-back transformed for graphical presentation. Statistics were conducted using STATA 15.1 and graphing completed with GraphPad Prism 6.0.
Results
Twin Characteristics
Table 1 shows characteristics among all participants and stratified into MZ and DZ groups. Due to planned oversampling for obesity during recruitment, 57% of all participants were classified with obesity, 24% as normal weight and 19% as overweight. Most DZ pairs were BMI discordant, with 80% having >5kg/m2 difference, whereas only 22% of MZ pairs were BMI discordant (>5kg/m2 difference, Pearson’s χ2(1)=31.07; P<0.001). MZ and DZ groups were similar, except for pre-meal levels of GLP-1 (Table 1).
Table 1.
Twin characteristics for all participants and stratified by zygosity.
| All participants | Dizygotic | Monozygotic | ||
|---|---|---|---|---|
| Female, % | 53.2 | 53.3 | 53.1 | |
| Age, y | 29.7±9.0 | 29.3±8.3 | 29.9±9.3 | |
| BMI, kg/m2 | 30.5±6.0 | 31.8±6.1 | 30.0±5.9 | |
| Body fat, % | 37.3±9.1 | 39.0±8.3 | 36.7±9.4 | |
| Lean mass, % | 60.6±8.5 | 59.0±7.7 | 61.1±8.8 | |
| Fasting leptin, ng/mL | 28.4±23.1 | 33.4±21.8 | 26.6±23.4 | |
| HOMA-IR | 2.4±1.7 | 2.4±1.7 | 2.4±1.7 | |
| Glucose, mg/dL | Pre-meal | 88.8±6.6 | 88.9±7.5 | 88.7±6.2 |
| Post-meal | 99.1±9.7 | 98.8±9.0 | 99.2±10.0 | |
| Insulin, μU/mL | Pre-meal | 8.1±4.9 | 7.9±5.0 | 8.1±4.8 |
| Post-meal | 24.1±15.3 | 25.5±14.4 | 23.5±15.7 | |
| Ghrelin, pg/mL | Pre-meal | 902.9±324.8 | 887.6±247.3 | 908.5±350.3 |
| Post-meal | 854.2±303.6 | 837.9±252.2 | 860.2±321.8 | |
| GLP-1, pmol/L | Pre-meal | 8.2±9.6 | 6.2±3.8 | 9.0±10.9* |
| Post-meal | 8.7±9.0 | 7.1±4.2 | 9.3±10.2 | |
| SN connectivity, % change | −9.3±46.6 | −5.1±60.3 | −10.8±40.7 | |
Data are mean ± standard deviation unless otherwise indicated. Group comparisons (dizygotic vs monozygotic) by generalized estimating equations. N=111 (Dizygotic N=30, except for SN connectivity [N=29]; Monozygotic N=81, except for Ghrelin, post-meal [N=80] and SN connectivity [N=79]). Pre- and post-meal plasma measures reflect the average of the 2 samples collected during the 30 minutes before (−30, 0, flanking the pre-meal MRI) and after a standardized meal (30, 60, flanking the post-meal MRI). HOMA-IR, homeostatic model assessment of insulin resistance; GLP-1, glucagon like peptide-1. SN, Salience network.
P=0.04 vs dizygotic.
Overall Analyses Mimicking Studies of Unrelated Individuals
SN connectivity in response to a meal.
Considering twins as individuals, the strength of SN connectivity did not vary based on age (Pre-meal: β=−0.002; P=0.82; Post-meal: β=−0.006, P=0.36) or sex (Pre-meal: β=0.08; P=0.41; Post-meal: β=0.06, P=0.59), however SN connectivity was significantly reduced by a meal (Figure 1B).
Relation of SN connectivity to circulating hormone concentrations.
SN connectivity was not associated with fasting leptin concentration (Pre-meal: β=−0.001, P=0.65; Post-meal: β=−0.002, P=0.48) or HOMA-IR (Pre-meal: β=−0.02, P=0.40; Post-meal: β=−0.05, P=0.09). Moreover, insulin, glucose, GLP-1 and ghrelin average concentrations during each MRI scan were unrelated to the strength of SN connectivity either pre- or post-meal (Suppl. Table 1). Furthermore, the percentage change in SN connectivity evoked by a meal was not associated to the degree of change in concentrations by a meal of insulin, glucose, GLP-1 or ghrelin (Suppl. Table 1). For gut-peptides, the postprandial peak for GLP-1 (30 min) and nadir for Ghrelin (60 min) were significantly different from pre-meal concentrations (GLP-1: β=1.03, P=0.004; Ghrelin: β=−102.40, P<0.001). The percentage change from pre-meal values to GLP-1 peak and Ghrelin nadir were also not correlated to the percentage change in SN connectivity (GLP-1: β=0.11, P=0.29; Ghrelin: β=−0.23, P=0.52).
Relationship of obesity to SN connectivity in response to a meal.
Still considering twins as individuals, no main effect of obesity on the strength of SN connectivity was found (χ2(1)=0.93, P=0.33; Figure 1C) nor was there a significant interaction between obesity and SN connectivity pre- and post-meal (χ2(1)=−0.0002; P=0.99). However, there was a significant main effect of time (χ2(1)=19.08; P<0.0001) and a significant reduction in SN connectivity by a meal in both groups (Figure 1C). The percentage change in SN connectivity by a meal also did not differ between BMI categories (Normal weight [N=27]:−11.3±42.2%; vs. with obesity [N=61]:−12.6±41.5%; P=0.94).
Relationship of MBH gliosis to SN connectivity in response to a meal.
There is currently no diagnostic cutoff for the presence of gliosis. Therefore, among all participants, we categorized the bottom and top tertiles of mean MBH T2 relaxation time into low and high MBH gliosis groups (N=29 per group; Suppl. Table 2). Mean BMI tended to be higher between the low and high gliosis groups (29.9±6.2 vs. 31.5±5.8kg/m2, β=0.02, P=0.08). The groups differed in markers for metabolic risk such as visceral fat percentage (β=0.22, P=0.02) and HOMA-IR (β=0.08, P=0.03; Suppl. Table 2). A main effect of time was again present (χ2(1)=9.50; P=0.002) for SN connectivity, but there was no main effect for the presence of MBH gliosis (χ2(1)=1.05; P=0.31) or interaction (χ2(1)=1.47; P=0.23) as the groups were quite similar pre-meal. Nonetheless, post-meal SN connectivity was significantly lower than pre-meal connectivity in the low (z=−3.04, P=0.002; Figure 1D), but not in the high gliosis group (z=−1.32; P=0.19). Results were not altered when body fat percentage was included in the model (effect of time: χ2(1)=9.50, P=0.002; MBH gliosis: χ2(1)=1.10; P=0.30; interaction: χ2(1)=1.47; P=0.23). Low and high MBH gliosis groups were not significantly different in the percentage change in SN connectivity by a meal, although a trend was present toward greater reductions among the low MBH gliosis group (−18.4±32.4% vs. high MBH gliosis:4.3±67.8%; P=0.10).
Twin Analyses – Within-Pair Correlations
Inherited and familial factors in SN connectivity.
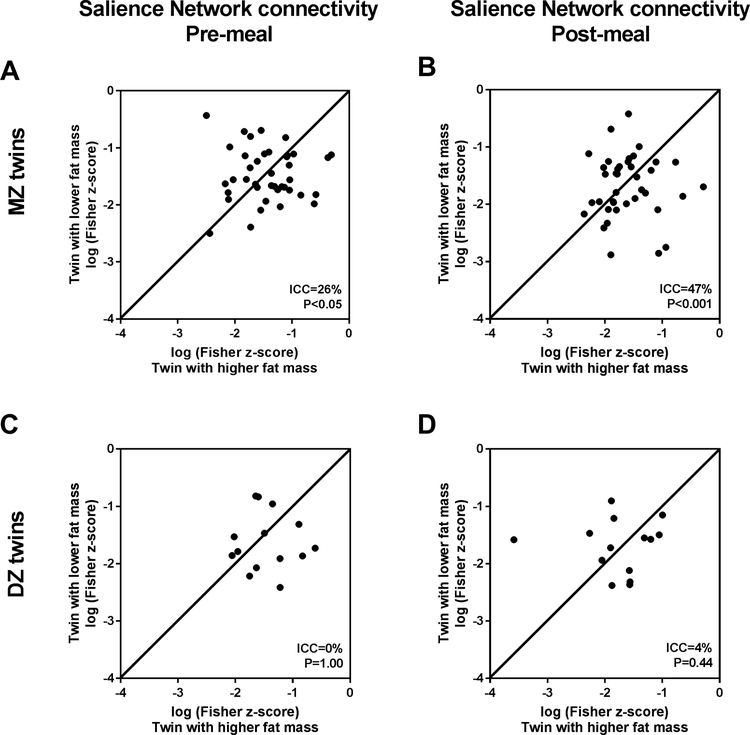
We then considered the potential effects of genetics by testing if twins were more similar to each other than to unrelated individuals in their SN connectivity. MZ twins share 100% of their genetic background and DZ twins share ~50%. Therefore, if correlations are higher for MZ than DZ twins, a genetic influence is suggested for that analyzed variable. Among MZ twins, the strength of SN connectivity was significantly more similar within the pairs both before (ICC=26%, P<0.05; Figure 2A) and after a meal (ICC=47%, P<0.001; Figure 2B). In contrast, DZ twins were not more similar to each other than to unrelated individuals (Figure 2C and D), providing evidence that genetic and not familial factors influence the basal strength of SN connectivity, pre- and post-meal. In contrast, for the percentage change in SN connectivity by a meal, there was no evidence for genetic or familial influences (MZ [N=40 pairs] ICC 8.5%, P=0.30 vs. DZ [N=15 pairs] ICC 5.4%, P=0.42), suggesting that SN response to food intake is a malleable response affected by environmental or acquired factors.
Figure 2.
Inherited factors in Salience Network (SN) connectivity. (A) Intraclass correlations (ICC) analyzed for monozygotic (MZ) twin pairs for pre-meal SN connectivity (N=41) and for (B) post-meal connectivity (N=40). (C) ICC for dizygotic (DZ) twin pairs for pre-meal SN connectivity (N=15) and for (D) post-meal connectivity (N=15). Line in each graph represents ICC of 100%.
Twin Analyses – Twin Association Study Using Within-Pair Differences
Relationships of adiposity to SN connectivity independent of inherited factors.
Given the above evidence for inherited influences on SN connectivity and recognized inherited influence on adiposity, we next employed a Twin Association Study approach(40) among the MZ twin pairs to control for genetic confounding. We calculated the within-pair differences in continuous variables (higher fat mass twin minus lower fat mass twin) to control for inherited and familial factors; these analyses are also inherently controlled for age and sex. Characteristics for the higher fat mass vs. lower fat mass members of twin pairs are presented in Table 2. A significant relationship of within-pair differences in adiposity to within-pair differences in SN connectivity can be interpreted as supporting an effect of adiposity on SN connectivity that is related to adiposity itself independent of genetic predispositions, family background, age, sex, and numerous unmeasured factors.
Table 2.
Characteristics of monozygotic twins with higher fat mass and lower fat mass
| Twin with higher fat mass | Twin with lower fat mass | ||
|---|---|---|---|
| BMI | 31.4±6.2* | 28.8±5.4 | |
| Fasting leptin, ng/mL | 30.8±26.4* | 22.4±19.9 | |
| HOMA-IR | 2.8±1.7* | 2.0±1.6 | |
| Glucose, mg/dL | Pre-meal | 88.6±6.6 | 88.9±5.9 |
| Post-meal | 100.1±11.0 | 98.6±8.9 | |
| Insulin, μU/mL | Pre-meal | 9.1±5.3* | 7.1±4.2 |
| Post-meal | 26.4±17.3* | 20.9±13.7 | |
| Ghrelin, pg/mL | Pre-meal | 881.7±308.0 | 934.1±394.3 |
| Post-meal | 828.9±270.5 | 892.3±370.3 | |
| GLP-1, pmol/L | Pre-meal | 11.2±14.4 | 6.9±5.0 |
| Post-meal | 11.0±13.2 | 7.8±5.8 | |
| MBH T2 relaxation time, ms | 95.4±5.1 | 93.3±5.3 | |
Twins ordered according to body fat mass (kg); Data are mean ± standard deviation. Group comparisons by generalized estimating equations. Number of participants in each group N=40, except for T2 relaxation time: Twin with higher fat mass (N=32), Twin with lower fat mass (N=31). Pre- and post-meal plasma measures reflect the average of the 2 samples collected during the 30 minutes before (−30, 0, flanking the pre-meal MRI) and after a standardized meal (30, 60, flanking the post-meal MRI). HOMA-IR, homeostatic model assessment of insulin resistance; GLP-1, glucagon like peptide-1; MBH, mediobasal hypothalamus.
P<0.01.
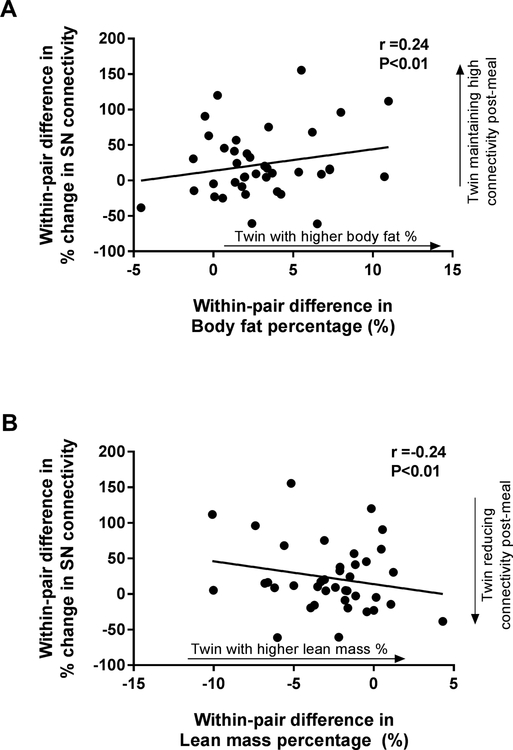
Pre-meal, higher body fat percentage was associated with lower strength of SN connectivity (i.e., the twin with higher fat mass had lower pre-meal SN connectivity, Table 3), but no other component of body composition was related. Post-meal, there were no relationships between difference in the strength of SN connectivity and adiposity or lean mass. However, differences in the percentage change in SN connectivity by a meal positively correlated with differences in body fat percentage such that the heavier twin did not reduce SN connectivity after a meal (Table 3, Figure 3A). The reciprocal relationship was also present such that the twin with higher lean mass percentage demonstrated a more effective reduction in SN connectivity post-meal (Figure 3B). These associations were also present for BMI, total body fat mass, and visceral fat mass, but not for total lean mass (Table 3). We then used sequential regression modeling to explore potential mediators of the significant relationships between percentage change in SN connectivity and measures of body composition. Still considering only MZ pairs (N=40), all data available for each variable was used which included a subset of MZ pairs (N=28) with both functional and structural MRI. The significance of the models was not reduced by the addition of within-pair differences in MBH T2 relaxation time, in fact, including radiologic evidence of MBH gliosis in the models raised the adjusted R2 (adjR2) in the significant models by 0.03–0.05, representing increases of 19–55% in the amount of variance explained (Table 3). In contrast, the addition of within-pair differences in fasting insulin concentrations to the models reduced both the P-values (0.01–0.14) and the adjR2 (0.07–0.18) as did including the differences in fasting leptin concentrations (adjR2:0.07–0.19; P=0.01–0.12), suggesting that insulin and leptin concentrations explained some of the effect of adiposity, but that radiologic evidence of MBH gliosis might be an independent contributor to changes in SN connectivity by a meal.
Table 3.
Unadjusted and adjusted associations of within monozygotic twin pair differences in Salience Network connectivity and body composition
| Within-pair differences in Salience Network Connectivity | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pre-meal | Post-meal | Percentage change | ||||||||||||
| Unadjusted | Adjusted for MBH gliosis | |||||||||||||
| N | β | P | N | β | P | N | β | P | adjR2 | N | β | P | adjR2 | |
| BMI, kg/m2 | 40 | −0.009 | 0.19 | 39 | 0.001 | 0.92 | 39 | 6.51 | 0.01 | 0.16 | 28 | 6.86 | 0.01 | 0.2 |
| Fat mass, kg | 40 | −0.005 | 0.13 | 39 | 0 | 0.91 | 39 | 3.26 | <0.01 | 0.2 | 28 | 3.5 | <0.01 | 0.25 |
| Lean mass, kg | 40 | −0.004 | 0.55 | 39 | 0 | 0.94 | 39 | 2.96 | 0.16 | 0.03 | 28 | 3.41 | 0.17 | 0.02 |
| Visceral fat, kg | 38 | −0.004 | 0.44 | 37 | 0 | 0.7 | 37 | 0.03 | 0.04 | 0.09 | 27 | 0.03 | 0.03 | 0.13 |
| Fat mass, % | 40 | −0.012 | <0.05 | 39 | −0.001 | 0.87 | 39 | 5.44 | <0.01 | 0.17 | 28 | 6.37 | 0.01 | 0.22 |
| Lean mass, % | 40 | 0.013 | 0.05 | 39 | 0.001 | 0.89 | 39 | −5.84 | 0.01 | 0.17 | 28 | −6.88 | 0.01 | 0.22 |
| Visceral fat, % | 38 | −0.049 | 0.42 | 37 | 0.015 | 0.76 | 37 | 32.79 | 0.09 | 0.05 | 27 | 40.68 | 0.09 | 0.08 |
All covariates represent within-pair differences (higher fat mass twin − lower fat mass twin); analyses are inherently adjusted for age, sex and genetics. Pre- and post-meal within-pair connectivity Fischer’s z-scores are log-transformed. N=number of twin pairs. P-values determined by simple linear regression and multiple linear regressions. β, regression coefficient; adjR2, adjusted R2; MBH, mediobasal hypothalamus.
Figure 3.
Correlations of within-pair differences in adiposity and the percentage change in Salience Network (SN) connectivity by a meal. Within-pair differences were calculated for each measure by subtracting the value for the twin with lower fat mass from that of the twin with higher fat mass. Scatterplots and regression lines shown for association of within-pair differences in (A) fat mass and (B) lean mass with differences in the percentage change in SN connectivity by a meal. Pearson’s correlation coefficients were calculated for descriptive purposes. P-values determined by simple linear regression. N=39 monozygotic twin pairs.
Discussion
We found that SN connectivity was reduced by meal ingestion. There was no evidence that circulating hormone concentrations were associated with basal connectivity or with meal-induced changes in connectivity. Instead, we demonstrated inherited, but not familial, influences on the strength of SN connectivity, both pre- and post-meal. When considered as individuals, participants with obesity and normal weight controls equivalently reduced SN connectivity post-meal. However, participants with evidence of MBH gliosis had an attenuated reduction in SN connectivity. Further analyses that controlled for genetic confounding showed that twins with higher adiposity than their identical co-twin tended to not reduce connectivity after a meal. The presence of MBH gliosis explained additional variance in this change in SN connectivity above and beyond the presence of excess adiposity or any genetic predispositions. These findings provide initial data supporting a role for the SN in feeding and suggest that obesity and/or obesity-associated MBH gliosis may be acquired factors related to an attenuated reduction in SN response to a meal.
The SN is a high order cognition network(3) and includes brain regions that process autonomic signs(41) and sensory information from the ambient environment(5). The SN has the capacity to then marshal affective, reward, and cognitive processes to select and execute the most appropriate behavioral response to interoceptive signals(3,6). As such, it logically might play an important role in drawing attention and decisional capacity toward identifying and procuring food when energy is insufficient and, conversely, relinquishing its hold on cognitive resources once homeostatically driven needs for nutrition are met. Measured in the resting-state, SN connectivity was significantly reduced from before to after ingestion of a meal. Test-retest analyses have shown that the SN has a consistent connectivity in humans and a good reliability for intrasession repetitions (<45min between scans)(42) and also long-term repetitions (up to 1y)(43). These prior reliability results strengthen our interpretation that the reduction in SN connectivity is a consequence of meal intake. These data augment general descriptions of SN function bridging salient stimuli and internal body information(3,6) with the final purpose of maintaining body homeostasis(4) and support the SN’s involvement in feeding and energy balance. The findings echo prior studies showing reduced connectivity between individual regions of the SN by nutrient intake(7–9). Regions of the SN (e.g., hypothalamus, ventral tegmental area) participate in downstream pathways stimulated by satiety signals (acting via vagal afferents synapsing in the hindbrain) and have sites of action for circulating peptides such as ghrelin and insulin(review in(44)). Nonetheless, circulating hormone concentrations were unrelated to SN connectivity or SN response to the meal. In sum, the reduction in salience of securing and consuming food may be reflected after a meal by reducing connectivity between SN regions, a shift that could potentially redirect cognitive resources toward alternative tasks, such as executive function(45).
Although a stable resting state network in humans(1,4), the SN can be functionally disrupted in pathological states like behavioral variant frontotemporal dementia and autism(46,47). In frontotemporal dementia, loss of satiety and binge eating(48) accompany its pathognomonic feature of loss of SN connectivity(46). Findings are mixed in individuals with obesity(10,13,49). Kullman et al.(10) showed reduced SN connectivity in a group with obesity when compared to normal weight controls whereas others(13,15,16) suggest increased connectivity among subjects with obesity. In addition, distinct regions have been implicated as the source of difference among groups including the insula(10,14), anterior cingulate cortex(50), and left putamen(13). Our study uncovers possible sources of variability in published findings. In our analyses that mimicked prior studies of unrelated individuals, we found no differences in SN connectivity, before or after a meal or in the degree of change by a meal, when comparing our participants solely based on the presence of obesity. However, in twin analyses that controlled for genetic confounding, a negative association between greater body fat and pre-meal connectivity was revealed as well as a failure to reduce SN connectivity after eating, implicating that genetic confounding may have affected prior discrepant results. Conflicting findings of SN connectivity in obesity could be also due to methodological differences such as 1) applying connectivity analyses to distinct fMRI task paradigm-derived data (visual food cues(14), taste cues(7,49), glucose ingestion(11) or liquid meal(8,9); 2) divergent definitions for the SN(10,13) and, likely; 3) genetical variability in the degree of SN connectivity, uncontrolled in most studies(26,27).
The current study attempted to address these differences, and thus facilitate comparison and reproducibility(51) by utilized a resting-state acquisition, applied an independent previously-defined SN template(3) and accounted for genetics(26,27,40).
These data provide evidence that the strength of SN connectivity has a polygenic inheritance component, which we identified consistently in both pre- and post-meal functional responses. In consequence, when we took genetic confounders into account via within-pair analyses(39,40) our results differed somewhat from when twins were considered as individuals. Twins with greater body adiposity than their identical co-twin maintained higher SN connectivity after the meal. This suggests that, in addition to genetic influences(12,52), acquired increases in body fat, presumably due to individual environmental factors affecting one twin more than the other, play a role in SN brain response to feeding. In contrast to basal connectivity, we did not detect inherited influence on the change in SN connectivity evoked by a meal. Interestingly, one prior study showed strong environmental influences on SN connectivity, but the state of satiety was uncontrolled in the sample(28) which could have masked inherited similarities. Together, these findings suggest inherited influence on SN connectivity, but also illuminate novel potential mechanisms whereby the central nervous system response to nutrient intake retains sufficient flexibility to react to environmental challenges, such as nutrient intake, or shifts in energy balance, such as added fat mass.
Besides adiposity, gliosis is another acquired factor that could potentially influence connectivity via effects on hypothalamic brain structure(17). Hypothalamic gliosis was first identified in diet-induced obese rodents in the arcuate nucleus of the MBH(17) and has been detected in humans by quantitative MRI methods in association with obesity in both adults(17,19,20) and children(22). MBH gliosis correlates positively with BMI(19) and visceral fat content(22). The data herein suggest that MBH gliosis, independent of obesity, might affect meal-related response in SN connectivity, but not necessarily basal connectivity. Participants with signs of MBH gliosis had an attenuated reduction in SN connectivity after a meal, as compared to the group without signs of MBH gliosis. That leads us to hypothesize that the group with high signs of MBH gliosis, by maintaining high SN connectivity after a meal, may be at risk for increased susceptibility to food stimuli. This surmise is based in a rodent literature showing that gliosis is integral to the hyperphagia and weight gain seen in diet-induced obesity(17,18,53). Our exploratory mediation analysis further indicated that MBH gliosis may have additive effects to those of adiposity alone, potentially promoting additional functional compromise in appetite control. Due to our twin design, we can state that the relationship between body fat and a lesser reduction in SN connectivity evoked by a meal is not rooted in genetic predispositions, age or sex. Instead, it suggests adaptation that follows an increase in body adiposity and, hence, could be speculated to reflect changes in brain processing of food intake needed to sustain a new, higher “set point” for body adiposity(24), perhaps even beyond that predicted by inheritance alone.
The current study’s design does not allow establishing cause-effect between SN connectivity and acquired components like MBH gliosis – longitudinal studies may help to address that limitation. The twin study design including MZ and DZ twins is a strength in our work by allowing evaluation of genetic, familial, and acquired influences separately. However, it only permits us to make conclusions on polygenic inheritance, not specific genetic markers. Furthermore, we found no sex difference in SN connectivity response to a meal which is in contrast with previous works that have suggested the effect of sex on general connectivity(54,55) as well as for SN specifically(56), but have not been confirmed by all(57). Regardless, all within-pair analyses were inherently controlled for sex. In addition, twin pairs in our study completed the study visit on the same day, which limited our possibility to control for menstrual cycle phase and potential effects of sex hormones fluctuation in the brain’s functional communication(54). Also, participants received a standardized meal which did not allow exploration of whether distinct portion sizes of food modulate changes in SN connectivity. Finally, brain control over feeding behavior is complex and likely involves other networks besides the SN(58). Future work should consider feeding effects on connectivity within and between resting state networks(2).
In conclusion, the findings support the involvement of the SN in feeding behavior as well as genetic influences on SN connectivity. Moreover, SN connectivity is not consistently disrupted based on the presence of obesity. Other factors such as genetic predispositions and/or structural compromise in the hypothalamus represented by MBH gliosis may either overshadow or augment the effect of obesity itself. Finally, the degree of change in SN connectivity evoked by a meal appears to be a malleable response reflecting flexibility within the regulatory system to adapt to shifts in environmental or homeostatic conditions.
Supplementary Material
Acknowledgements:
This work was supported by funding provided by the National Institutes of Health (DK089036, DK098466) and by the American Diabetes Association (ADA 1–17-ICTS-085). Additional assistance was provided by the University of Washington’s Nutrition Obesity Research Center (P30 DK035816), Diabetes Research Center (P30 DK017047), and the Institute of Translational Health Sciences (UL1 TR000423). LES was funded by Sao Paulo Research Foundation Post-Doctoral Fellowship (FAPESP 2017/00657–0).
Footnotes
Conflict of interest: The authors declare that they have no conflict of interest.
Supplementary information is available at International Journal of Obesity’s website.
References
- 1.Damoiseaux JS, Rombouts SARB, Barkhof F, Scheltens P, Stam CJ, Smith SM, et al. Consistent resting-state networks across healthy subjects. Proc Natl Acad Sci. 2006;103(37):13848–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA, et al. Functional Network Organization of the Human Brain. Neuron. 2011;72(4):665–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seeley WW, Menon V, Schatzberg AF, Keller J, Glover GH, Kenna H, et al. Dissociable Intrinsic Connectivity Networks for Salience Processing and Executive Control. J Neurosci. 2007;27(9):2349–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bressler SL, Menon V. Large-scale brain networks in cognition: emerging methods and principles. Trends Cogn Sci. 2010;14(6):277–90. [DOI] [PubMed] [Google Scholar]
- 5.Rolls ET. Reward Systems in the Brain and Nutrition. Annu Rev Nutr. 2016;36(1):435–70. [DOI] [PubMed] [Google Scholar]
- 6.Menon V Salience Network. In: Brain Mapping. Elsevier; 2015. p. 597–611. [Google Scholar]
- 7.Lips MA., Wijngaarden MA, Van Der Grond J, Van Buchem MA, De Groot GH, Rombouts S a RB, et al. Resting-state functional connectivity of brain regions involved in cognitive control, motivation, and reward is enhanced in obese females. Am J Clin Nutr. 2014;100:524–31. [DOI] [PubMed] [Google Scholar]
- 8.Ryan JP, Karim HT, Aizenstein HJ, Helbling NL, Toledo FGS. Insulin sensitivity predicts brain network connectivity following a meal. Neuroimage. 2018;171:268–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paolini BM, Laurienti PJ, Norris J, Jack Rejeski W. Meal replacement: Calming the hot-state brain network of appetite. Front Psychol. 2014;5:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kullmann S, Heni M, Veit R, Ketterer C, Schick F, Häring HU, et al. The obese brain: Association of body mass index and insulin sensitivity with resting state network functional connectivity. Hum Brain Mapp. 2012;33(5):1052–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Opstal AM, Hafkemeijer A, van den Berg-Huysmans AA, Hoeksma M, Blonk C, Pijl H, et al. Brain activity and connectivity changes in response to glucose ingestion. Nutr Neurosci. 2018;0(0):1–8. [DOI] [PubMed] [Google Scholar]
- 12.Doucet GE, Rasgon N, McEwen BS, Micali N, Frangou S. Elevated Body Mass Index is Associated with Increased Integration and Reduced Cohesion of Sensory-Driven and Internally Guided Resting-State Functional Brain Networks. Cereb Cortex. 2018;28(3):988–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García-García I, Jurado MÁ, Garolera M, Segura B, Sala-Llonch R, Marqués-Iturria I, et al. Alterations of the salience network in obesity: a resting-state fMRI study. Hum Brain Mapp. 2013. November;34(11):2786–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nummenmaa L, Hirvonen J, Hannukainen JC, Immonen H, Lindroos MM, Salminen P, et al. Dorsal striatum and its limbic connectivity mediate abnormal anticipatory reward processing in obesity. PLoS One. 2012;7(2):e31089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Figley CR, Asem JSA, Levenbaum EL, Courtney SM. Effects of Body Mass Index and Body Fat Percent on Default Mode, Executive Control, and Salience Network Structure and Function. Front Neurosci. 2016;10:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta A, Mayer EA, Hamadani K, Bhatt R, Fling C, Alaverdyan M, et al. Sex differences in the influence of body mass index on anatomical architecture of brain networks. Int J Obes (Lond). 2017;41(8):1185–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thaler JP, Yi C, Schur EA, Guyenet SJ, Hwang BH, Dietrich MO, et al. Obesity is associated with hypothalamic injury in rodents and humans. J Clin Invest. 2012;122(1):153–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Douglass JD, Dorfman MD, Fasnacht R, Shaffer LD, Thaler JP. Astrocyte IKKβ/NF-κB signaling is required for diet-induced obesity and hypothalamic inflammation. Mol Metab. 2017;6(4):366–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schur EA, Melhorn SJ, Oh S-K, Lacy JM, Berkseth KE, Guyenet SJ, et al. Radiologic evidence that hypothalamic gliosis is associated with obesity and insulin resistance in humans. Obesity (Silver Spring). 2015;23(11):2142–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kreutzer C, Peters S, Schulte DM, Fangmann D, Türk K, Wolff S, et al. Hypothalamic Inflammation in Human Obesity Is Mediated by Environmental and Genetic Factors. Diabetes. 2017;66(9):2407–15. [DOI] [PubMed] [Google Scholar]
- 21.Baufeld C, Osterloh A, Prokop S, Miller KR, Heppner FL. High-fat diet-induced brain region-specific phenotypic spectrum of CNS resident microglia. Acta Neuropathol. 2016;132(3):361–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sewaybricker LE, Schur EA, Melhorn SJ, Campos BM, Askren MK, Nogueira GAS, et al. Initial evidence for hypothalamic gliosis in children with obesity by quantitative T2 MRI and implications for blood oxygen-level dependent response to glucose ingestion. Pediatr Obes. 2019;14(2):e12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burda JE, Sofroniew MV. Reactive Gliosis and the Multicellular Response to CNS Damage and Disease. Neuron. 2014;81(2):229–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwartz MW, Seeley RJ, Zeltser LM, Drewnowski A, Ravussin E, Redman LM, et al. Obesity Pathogenesis: An Endocrine Society Scientific Statement. Endocr Rev. 2017;38(4):267–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelley AE, Baldo B a., Pratt WE, Will MJ. Corticostriatal-hypothalamic circuitry and food motivation: Integration of energy, action and reward. Physiol Behav. 2005;86(5):773–95. [DOI] [PubMed] [Google Scholar]
- 26.Fu Y, Ma Z, Hamilton C, Liang Z, Hou X, Ma X, et al. Genetic influences on resting-state functional networks: A twin study. Hum Brain Mapp. 2015;36(10):3959–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glahn DC, Winkler AM, Kochunov P, Almasy L, Duggirala R, Carless MA, et al. Genetic control over the resting brain. Proc Natl Acad Sci. 2010;107(3):1223–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Z, Zuo X-N, McMahon KL, Craddock RC, Kelly C, de Zubicaray GI, et al. Genetic and Environmental Contributions to Functional Connectivity Architecture of the Human Brain. Cereb Cortex. 2016;26(5):2341–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boomsma D, Busjahn A, Peltonen L. Classical twin studies and beyond. Nat Rev Genet. 2002;3(11):872–82. [DOI] [PubMed] [Google Scholar]
- 30.Strachan E, Hunt C, Afari N, Duncan G, Noonan C, Schur E, et al. University of Washington Twin registry: Poised for the next generation of twin research. Twin Res Hum Genet. 2013;16(1):455–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melhorn SJ, Mehta S, Kratz M, Tyagi V, Webb MF, Noonan CJ, et al. Brain regulation of appetite in twins. Am J Clin Nutr. 2016;103(2):314–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mifflin MD, St Jeor ST, Hill L a, Scott BJ, Daugherty S a, Koh YO. A new predictive equation for resting energy expenditure in healthy individuals. Am J Clin Nutr. 1990;51(2):241–7. [DOI] [PubMed] [Google Scholar]
- 33.Melhorn SJ, Askren MK, Chung WK, Kratz M, Bosch TA, Tyagi V, et al. FTO genotype impacts food intake and corticolimbic activation. Am J Clin Nutr. 2018;107(2):145–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28(7):412–9. [DOI] [PubMed] [Google Scholar]
- 35.Behzadi Y, Restom K, Liau J, Liu TT. A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. Neuroimage. 2007;37(1):90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fast Jenkinson M., automated, N-dimensional phase-unwrapping algorithm. Magn Reson Med. 2003;49(1):193–7. [DOI] [PubMed] [Google Scholar]
- 37.Greve DN, Fischl B. Accurate and robust brain image alignment using boundary-based registration. Neuroimage. 2009;48(1):63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54(3):2033–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carlin JB, Gurrin LC, Sterne JAC, Morley R, Dwyer T. Regression models for twin studies: A critical review. Int J Epidemiol. 2005;34(5):1089–99. [DOI] [PubMed] [Google Scholar]
- 40.Schur E, Carnell S. What Twin Studies Tell Us About Brain Responses to Food Cues. Curr Obes Rep. 2017;6(4):371–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Craig ADB. How do you feel--now? The anterior insula and human awareness. Nat Rev Neurosci. 2009;10(1):59–70. [DOI] [PubMed] [Google Scholar]
- 42.Shehzad Z, Kelly AMC, Reiss PT, Gee DG, Gotimer K, Uddin LQ, et al. The resting brain: Unconstrained yet reliable. Cereb Cortex. 2009;19(10):2209–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo CC, Kurth F, Zhou J, Mayer EA, Eickhoff SB, Kramer JH, et al. One-year test-retest reliability of intrinsic connectivity network fMRI in older adults. Neuroimage. 2012;61(4):1471–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andermann ML, Lowell BB. Toward a Wiring Diagram Understanding of Appetite Control. Neuron. 2017;95(4):757–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Menon V, Uddin LQ. Saliency, switching, attention and control: a network model of insula function. Brain Struct Funct. 2010;214(5–6):655–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD. Neurodegenerative Diseases Target Large-Scale Human Brain Networks. Neuron. 2009;62(1):42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uddin LQ, Menon V. The anterior insula in autism: under-connected and under-examined. Neurosci Biobehav Rev. 2009;33(8):1198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woolley JD, Gorno-Tempini ML, Seeley WW, Rankin K, Lee SS, Matthews BR, et al. Binge eating is associated with right orbitofrontal-insular-striatal atrophy in frontotemporal dementia. Neurology. 2007;69(14):1424–33. [DOI] [PubMed] [Google Scholar]
- 49.Frank S, Linder K, Kullmann S, Heni M, Ketterer C, Çavuşoǧlu M, et al. Fat intake modulates cerebral blood flow in homeostatic and gustatory brain areas in humans. Am J Clin Nutr. 2012;95(6):1342–9. [DOI] [PubMed] [Google Scholar]
- 50.Martin LE, Holsen LM, Chambers RJ, Bruce AS, Brooks WM, Zarcone JR, et al. Neural mechanisms associated with food motivation in obese and healthy weight adults. Obesity (Silver Spring). 2010;18(2):254–60. [DOI] [PubMed] [Google Scholar]
- 51.Poldrack RA, Baker CI, Durnez J, Gorgolewski KJ, Matthews PM, Munafò MR, et al. Scanning the horizon: Towards transparent and reproducible neuroimaging research. Nat Rev Neurosci. 2017;18(2):115–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Doornweerd S, van Duinkerken E, de Geus EJ, Arbab-Zadeh P, Veltman DJ, Ijzerman RG. Overweight is associated with lower resting state functional connectivity in females after eliminating genetic effects: A twin study. Hum Brain Mapp. 2017;38(10):5069–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valdearcos M, Douglass JD, Robblee MM, Dorfman MD, Stifler DR, Bennett ML, et al. Microglial Inflammatory Signaling Orchestrates the Hypothalamic Immune Response to Dietary Excess and Mediates Obesity Susceptibility. Cell Metab. 2017;26(1):185–197.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tomasi D, Volkow ND. Gender differences in brain functional connectivity density. Hum Brain Mapp. 2012;33(4):849–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Biswal BB, Mennes M, Zuo X-N, Gohel S, Kelly C, Smith SM, et al. Toward discovery science of human brain function. Proc Natl Acad Sci. 2010;107(10):4734–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jamadar SD, Sforazzini F, Raniga P, Ferris NJ, Paton B, Bailey MJ, et al. Sexual Dimorphism of Resting-State Network Connectivity in Healthy Ageing. J Gerontol B Psychol Sci Soc Sci. 2018;00(00):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weissman-Fogel I, Moayedi M, Taylor KS, Pope G, Davis KD. Cognitive and default-mode resting state networks: Do male and female brains “rest” differently? Hum Brain Mapp. 2010;31(11):1713–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rossi MA, Stuber GD. Overlapping Brain Circuits for Homeostatic and Hedonic Feeding. Cell Metab. 2018;27(1):42–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.