Abstract
The goal of this study was to provide quantitative data on the catechin contents and underlying molecular regulatory mechanisms in cucumber during fruit development. The dynamic changes in the total catechin contents and RNA-seq-based transcriptome profiling of the flesh and peel of the cucumber cultivar ‘YanBai’, which is strongly astringent, were examined at three key developmental stages 3, 6 and 9 days post-pollination. The total catechin content decreased as cucumber fruit developed and was significantly lower in the flesh than in the peel. In total, 5092 and 4004 genes were found to be differently expressed in the peel and flesh, respectively. Based on a functional annotation, eight structural genes encode enzymes involved in the catechin biosynthesis pathway. Three genes encoding 4-coumarate-CoA ligases, two genes encoding chalcone isomerases, two genes encoding dihydroflavonol-4-reductase and one gene each encoding a phenylalanine ammonia-lyase, flavanone 3-hydroxylase and cinnamate 4-hydroxylase were identified as affecting the catechin content of cucumber. The transcriptome data also revealed the significance of transcription factors, including WD40-repeat proteins, MYB and bHLH, in regulating catechin biosynthesis. These findings help increase our understanding of the molecular mechanisms controlling catechin biosynthesis and astringency development in cucumber fruit.
Electronic supplementary material
The online version of this article (10.1007/s13205-019-1922-2) contains supplementary material, which is available to authorized users.
Keywords: Cucumber, Astringency, Catechins, RNA-seq
Introduction
Cucumber (Cucumis sativus L.) is an important cultivated vegetable, which is typically harvested when immature and consumed as fleshy fruit in China (Ando et al. 2012). For consumers, the oral sensory properties (astringency, sweetness, sourness, bitterness, spiciness and aroma) of the fleshy fruit greatly affect their consumption and acceptability (Troszyńska et al. 2011; He et al. 2015). Astringency is a key fruit oral sensory quality that produces a tactile sensation, which has been summarized as a feeling of dryness, as well as the puckering and tightening of the mouth, resulting from the interaction between polyphenolic compounds and salivary proteins (Soares et al. 2016). However, astringency can be irritating to consumers because of the unpleasant oral sensation it causes.
Catechins are main contributors to astringency (Li et al. 2011; Xu et al. 2018). They are biosynthesized by glucose metabolic pathways, including the shikimate, phenylpropanoid and flavonoid pathways (Eungwanichayapant and Popluechai 2009). In recent decades, because of the potential antioxidant properties of catechins, their biosynthesis has been widely studied (Grzesik et al. 2018). Catechin biosynthesis enzyme-encoding genes have been previously cloned and analyzed in Polygonum hydropiper (Furukawa et al. 2002), grapevine (Castellarin et al. 2007), strawberry fruit (Severo et al. 2011) and tea plant (Rani et al. 2012; Xiong et al. 2013). The enzymes catalyzing the biosynthesis of catechins in plants include phenylalanine ammonia-lyase (PAL, EC4.3.1.24), cinnamate 4-hydroxylase (C4H, EC1.14.13.11), 4-coumarate-CoA ligase (4CL, EC6.2.1.12), chalcone synthase (CHS, EC2.3.1.74), chalcone isomerase (CHI, EC5.5.1.6), flavonoid 3′-hydroxylase (F3′H, EC1.14.13.21), dihydroflavonols 4-reductase (DFR, EC1.1.1.219), anthocyanidin synthase (ANS, EC1.14.11.19), anthocyanidin reductase (ANR, EC1.3.1.77), leucocyanidin reductase (LAR, EC1.17.1.3) and flavan-3-ol gallate synthase (FGS, EC number not found) (Xiong et al. 2013; Liu et al. 2015). The traditional way of identifying key structural genes that contribute to catechin biosynthesis is through a correlation analysis of the transcript abundances of catechin biosynthetic enzyme-encoding genes and the catechin contents (Rani et al. 2012; Zhang et al. 2018). Using such a correlation study, Zhang et al. (2016) demonstrated that ANS may control earlier stages in the synthesis of tea catechin, while both ANR and LAR are critical for the conversion of galloylated catechins. However, in plant genomes, almost all of the enzymes that are responsible for the biosynthesis of catechins are encoded by multiple genes. Only some genes that encode catechin biosynthetic enzymes have been studied.
Presently, there are limited reports on cucumber catechins and their underlying molecular regulatory mechanisms. To increase the daily cucumber consumption and understand the potential health benefits, it is necessary to evaluate the catechin contents in cucumber fruit. Recent breakthroughs in high-throughput RNA sequencing technology has made the rapid detection of gene expression levels possible on a genome-wide scale (Xu et al. 2015). Transcriptome analyses will allow us to uncover key structure genes responsible for catechin biosynthesis and provide information regarding their roles during cucumber fruit development. Consequently, we analyzed the transcriptomic changes in cucumber fruit during three key development stages at 3, 6 and 9 days post-pollination (dpp). The results shed light on the molecular mechanisms regulating cucumber catechin biosynthesis.
Materials and methods
Plant materials and sampling
The strongly astringent cucumber variety ‘YanBai’ was used to analyze the catechins content and perform RNA-seq. The seedlings were cultivated in a greenhouse of Yangzhou University (Yangzhou, China). All the female flowers were manually self-pollinated. To avoid competition for nutrients and to ensure the full growth of the fruit, we retained one well-shaped ovary per 5–10 plant nodes and removed all the other ovaries before pollination. Peel (exocarp) and flesh (mesocarp) samples at 3, 6, and 9 dpp were collected separately using a razor blade. Independent samples from at least ten fruits were pooled and then separated into two portions: one for the high-performance liquid chromatography (HPLC) analysis and the other for the RNA-seq study.
Total catechin extraction and HPLC quantification
The total catechin was extracted according to Liu et al. (2015) with some modifications. In total, 5 g of fresh materials was ground in a pestle and extracted with 4 mL of 40% (v/v) methanol for 20 min at 60 °C. The mixture was centrifuged at 13,000×g for 12 min at 4 °C. The supernatant was further filtered through a 0.22-μm organic membrane and then subjected to the HPLC analysis. The HPLC-grade standard solutions of gallocatechin (GC), catechin (C), epigallocatechin, epicatechin, epigallocatechin gallate (EGCG) and epicatechin gallate (Sigma, St Louis, MO, USA), were prepared by dissolving the appropriate amount of each standard in 40% (v/v) methanol to generate stock solutions containing the six catechins at 100 mg/mL, 50 mg/mL, 25 mg/mL, 12.5 mg/mL and 6.25 mg/mL, respectively. Samples (extracted and standard) were stored at 0 °C and protected from light until quantification.
The HPLC analysis was performed using a Trace DSQII detector (Thermo Scientific, Waltham, MN, USA) with a C18 ODS column (4.6 mm × 250 mm, 5 μm) maintained in an oven at 35 °C. Mobile phases A and B were methanol and 0.1% formic acid in water, respectively. The gradient elution procedure was set from 20% mobile phase B to 65% mobile phase B by a linear gradient that increased during the first 30 min and then decreased to 20% mobile phase B for an additional 5 min. The samples were eluted at a flow rate of 1 mL/min. The detector wavelength was set as 280 nm. The sample injection volume was 10 mL. The peaks were identified by comparing the retention times for the extracted samples to those of standard solutions. The total catechin contents (TCs) were calculated by the summation of the six individual catechin fractions.
RNA extraction and sequencing
Flesh and peel samples were independently ground to a fine powder in liquid nitrogen, and total RNA was extracted using RNAiso Plus (Takara, Dalian, China). We assessed the RNA quality using a 2100 Bioanalyzer (Agilent Technologies, PaloAlto, CA, USA). The concentration of the total RNA was measured using a Qubit 2.0 fluorometer (Life Technologies, Carlsbad, CA, USA). Transcriptome libraries were prepared from each sample (100 ng of total RNA) using a TruSeq RNA Sample Prep Kit V2 (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. The transcriptome libraries were sequenced on an Illumina HiSeq 4000 platform. After removing the low overall quality reads containing sequencing primers or exhibiting a low mass, the clean sequence reads were subjected to a BLAST analyses using TopHat v2 (Trapnell et al. 2012). The 9930 draft genome assembly (version 2, http://www.icugi.org/cgi-bin/ICuGI/index.cgi) was used as the cucumber reference genome. Cufflinks was applied to assembly mapped reads and calculate the fragments per kilobase of exon model per million mapped reads (FPKM) of each transcript (Trapnell et al. 2010, 2012).
qRT-PCR analysis
The expression levels of genes related to astringency development were validated using qRT-PCR. Total RNA was extracted from each sample using a TaKaRa MiniBEST Plant RNA Extraction Kit (TaKaRa, Dalian, China), and first-strand cDNA was synthesized using total RNA and a PrimeScript RT reagent kit (TaKaRa). The primers were designed with Primer 3 (http://bioinfo.ut.ee/primer3-0.4.0/; Online Resource 1). qRT-PCR was performed using a SYBR PrimeScript RT-PCR kit (TaKaRa) under the following conditions: 30 s at 95 °C, 35 cycles of 5 s at 95 °C and 1 min at 60 °C, and a final step of 30 s at 72 °C. The relative transcriptional levels were calculated using the 2−ΔΔCt method. The specificity levels of the PCR products were verified using a melting curve analysis.
Results
TCs in the peel and flesh of cucumber fruits
In our previous work, the astringency substances in different cucumber accessions were investigated, and the main astringency substances in cucumber fruit were determined to be catechins (Tian 2015). Thus, to evaluate the dynamic changes of astringency during cucumber fruit development and the differences in various fruit parts, we analyzed the TCs in the cucumber fruit peel and flesh harvested at 3, 6 and 9 dpp by HPLC. A typical chromatogram of the six individual catechins separated under the described settings are shown in Online Resource 2a. The separation of the six components was completed within 18.5 min. A total of three peaks, representing GC, C and EGCG, were detected when using cucumber samples (Online Resource 2b). Among these, GC was the dominant component (Table 1; Online Resource 2b). We calculated the TC through the summation of GG, C and EGCG. As shown in Table 1, the TCs were significantly higher than in the peels than in the flesh at 3, 6 and 9 dpp, suggesting that the flesh was less astringent than the peel. We also found that the TCs decreased with time in both the peel and flesh after pollination, indicating that the degree of astringency decreased with fruit development. To investigate the molecular details that occurred during astringency changes, the peel and flesh of ‘YanBai’ at 3, 6 and 9 dpp were dissected separately for the RNA-seq analysis.
Table 1.
Total catechin contents of the peel and flesh of ‘YanBai’ fruit at 3, 6 and 9 days post-pollination (dpp)
| 3 dpp | 6 dpp | 9 dpp | ||||
|---|---|---|---|---|---|---|
| Peel (mg/g FW) | Flesh (mg/g FW) | Peel (mg/g FW) | Flesh (mg/g FW) | Peel (mg/g FW) | Flesh (mg/g FW) | |
| GC | 0.284 ± 0.023 | 0.075 ± 0.014 | 0.143 ± 0.032 | 0.041 ± 0.005 | 0.075 ± 0.011 | 0.022 ± 0.004 |
| EGCG | 0.043 ± 0.002 | 0.057 ± 0.008 | 0.028 ± 0.003 | 0.032 ± 0.008 | 0.023 ± 0.003 | 0.023 ± 0.002 |
| C | 0.031 ± 0.003 | 0.017 ± 0.003 | 0.022 ± 0.004 | 0.014 ± 0.003 | 0.019 ± 0.005 | 0.013 ± 0.001 |
| TC | 0.358 ± 0.017 | 0.149 ± 0.013 | 0.193 ± 0.018 | 0.087 ± 0.006 | 0.117 ± 0.012 | 0.058 ± 0.004 |
Error bars represent ± SD of three biological replicates. Only three catechin fractions, gallocatechin (GC), catechin (C) and epigallocatechin gallate (EGCG) were detected; therefore, the total catechin contents (TC) were calculated by the summation of the GC, EGCG and C fractions
Extensive transcriptomic reprogramming during astringency changes
At least 35.1 million clean reads per sample were obtained after removal of the low-quality reads (Table 2). These clean reads were then mapped to the 9930 reference genome, and the mapping ratios ranged from 88.70 to 89.39%. Hierarchical clustering revealed differential expression profiles of genes in cucumber peels and flesh at different developmental stages (Fig. 1). The heat map patterns for the peel and flesh transcripts were different from each other at all of the examined time points during fruit development. The expression profiles of peel harvested at 6 dpp (P-6) and (peel harvested at 3 dpp (P-3) were in the same cluster, but that of peel harvested at 9 dpp (P-9) was in a different subgroup. A similar trend also existed for expression profiles of the flesh samples. Those of flesh harvested at 6 dpp (F-6) and flesh harvested at 3 dpp (F-3) were in the same cluster, while the profile of flesh harvested at 9 dpp (F-9) was distinctly different. The results suggested an alteration in gene expression patterns during the late cucumber fruit enlargement process.
Table 2.
Sequencing and assembly statistics for the six samples from YanBai cucumber at the three examined developmental stages
| Sample ID | Total reads | Mapped reads | Mapped paired Reads | Mapped unique reads | Mapped multi-reads | Mapping ratio (%) |
|---|---|---|---|---|---|---|
| P-3 | 54,327,636 | 48,336,925 | 46,795,704 | 46,946,106 | 1,390,819 | 88.97 |
| F-3 | 60,826,806 | 53,955,227 | 52,225,786 | 52,529,414 | 1,425,813 | 88.70 |
| P-6 | 50,530,048 | 45,045,315 | 43,614,748 | 43,761,856 | 1,283,459 | 89.15 |
| F-6 | 59,933,106 | 53,576,530 | 51,846,688 | 52,054,172 | 1,522,358 | 89.39 |
| P-9 | 40,847,258 | 36,233,269 | 34,988,548 | 35,118,367 | 1,114,902 | 88.70 |
| F-9 | 50,766,798 | 45,360,755 | 43,877,166 | 44,216,247 | 1,144,508 | 89.35 |
P-3 peel harvested at 3 days post-pollination (dpp), P-6 peel harvested at 6 dpp, P-9 peel harvested at 9 dpp, F-3 flesh harvested at 3 dpp, F-6 flesh harvested at 6 dpp, F-9 flesh harvested at 9 dpp
Fig. 1.
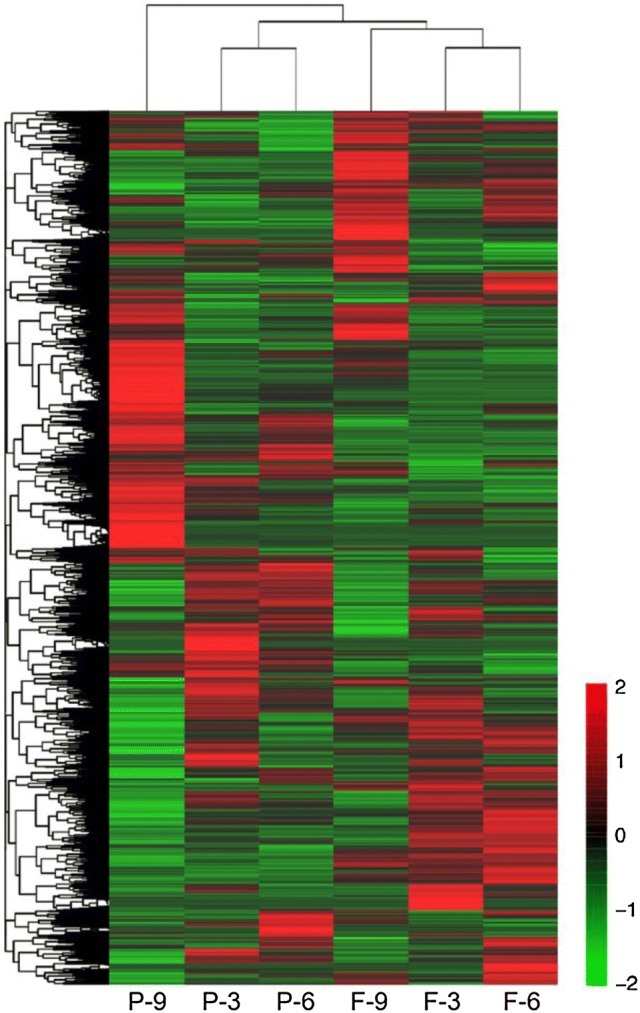
Hierarchical cluster analysis of peel and flesh transcripts in cucumber fruits at 3, 6 and 9 days post-pollination (dpp). The maps were drawn by Cluster 3.0 and Java tree view software. P-3: peel harvested at 3 dpp; P-6: peel harvested at 6 dpp; P-9: peel harvested at 9 dpp; F-3: flesh harvested at 3 dpp; F-6: flesh harvested at 6 dpp; F-9: flesh harvested at 9 dpp
Genes related to the catechin biosynthetic pathway
We found 2528 down-regulated genes that had FPKM values greater than 5 and were highest in P-3, but lowest in P-9; (P-3 > P-6 > P-9) and 2562 up-regulated genes that had FPKM values greater than 5 and were highest in P-9, but lowest in P-3 (P-3 < P-6 < P-9) in the peel (Online Resource 3). However, there were only 1646 down-regulated genes that had FPKM values greater than 5 and were highest in F-3, but lowest in F-9 (F-3 > F-6 > F-9) and 2358 up-regulated genes that had FPKM values greater than 5 and were highest in F-9, but lowest in F-3 (F-3 < F-6 < F-9) in the flesh (Online Resource 4). In total, 1723 genes (776 up-regulated and 947 down-regulated) were commonly regulated in the two organs. In addition, 2930 genes (1477 up-regulated and 1453 down-regulated) were only identified in the peel, and 1844 genes (707 up-regulated and 1137 down-regulated) were only found in the flesh. In total, 274 genes were up-regulated in the peel but down-regulated in the flesh, while 162 genes were up-regulated in the flesh but down-regulated in the peel (Fig. 2).
Fig. 2.
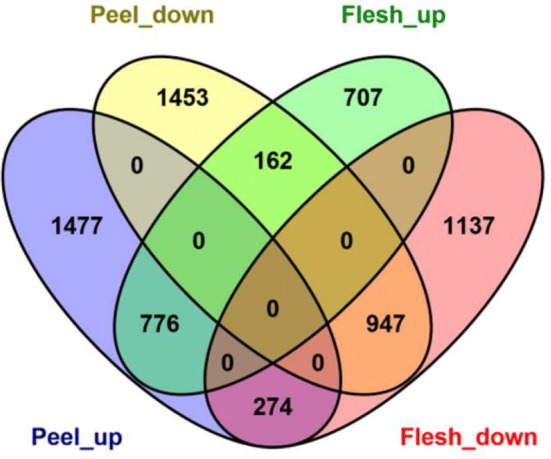
Venn diagram analyses of the differentially expressed genes that were identified in this study. Peel_up: FPKM values were highest in the peel at 9 days-post pollination (dpp), but lowest at 3 dpp; Peel_down: FPKM values were highest in the peel at 3 dpp, but lowest at 9 dpp; Flesh_up: FPKM values were highest in the flesh at 9 dpp, but lowest at 3 dpp; Flesh_down: FPKM values were highest in the flesh at 3 dpp, but lowest at 9 dpp
Based on functional annotation, eight genes, three down-regulated (Csa1G050290, Csa6G077400 and Csa7G388440) and five up-regulated (Csa1G050280, Csa1G590300, Csa4G622760, Csa6G108510 and Csa6G133710) in the peel were determined to be involved in catechin biosynthesis (Table 3). However, only two up-regulated genes (Csa6G133710 and Csa7G388440) and two down-regulated genes (Csa6G133710 and Csa7G388440) in the flesh were related to catechin biosynthesis (Table 3). Csa1G050280, Csa1G050290 and Csa1G108800 encode 4CL; Csa7G388440 and Csa4G622760 encode CHI; Csa1G590300 encodes PAL; Csa6G108510 encodes F3′H; Csa6G133710 encodes C4H; and Csa6G077400 and Csa3G098550 encode DFR. Considering the continuous decrease in the TC content at the three tested time points (Table 2) in peel and flesh, these genes may be critical for the de-astringency process, which makes them candidates for further studies.
Table 3.
Comparisons of the fragments per kilobase of exon model per million mapped reads (FPKM) values of genes involved in the catechin biosynthetic pathway
| Gene ID | Function annotation | P-3 FPKM |
P-6 FPKM |
P-9 FPKM |
F-3 FPKM |
F-6 FPKM |
F-9 FPKM |
|---|---|---|---|---|---|---|---|
| Csa1G050280 | 4-Coumarate-CoA ligase | 17.9 | 22.4 | 88.4 | 19.3 | 24.7 | 15.7 |
| Csa1G050290 | 4-Coumarate-CoA ligase | 37.3 | 19.3 | 9.1 | 5.6 | 12.6 | 8.2 |
| Csa1G108800 | 4-Coumarate-CoA ligase | 25.2 | 21.9 | 26.0 | 12.4 | 16.9 | 22.2 |
| Csa7G388440 | Chalcone isomerase | 41.7 | 31.7 | 18.7 | 32.2 | 23.4 | 18.0 |
| Csa4G622760 | Chalcone isomerase | 23.0 | 27.7 | 35.7 | 21.5 | 19.2 | 21.9 |
| Csa1G590300 | Phenylalanine ammonia-lyase | 8.7 | 9.0 | 12.6 | 8.2 | 8.7 | 7.7 |
| Csa6G108510 | Flavanone 3-hydroxylase | 13.0 | 19.6 | 29.8 | 18.7 | 15.8 | 26.7 |
| Csa6G133710 | Cinnamate 4-hydroxylase | 17.4 | 19.3 | 163.5 | 43.4 | 33.2 | 14.0 |
| Csa6G077400 | Dihydroflavonol-4-reductase | 21.8 | 18.1 | 6.8 | 37.3 | 55.2 | 28.8 |
| Csa3G098550 | Dihydroflavonol-4-reductase | 30.7 | 90.2 | 31.2 | 27.2 | 34.0 | 46.3 |
P-3 peel harvested at 3 days post-pollination (dpp), P-6 peel harvested at 6 dpp, P-9 peel harvested at 9 dpp, F-3 flesh harvested at 3 dpp, F-6 flesh harvested at 6 dpp, F-9 flesh harvested at 9 dpp
Detection of transcription factors (TFs) related to catechin biosynthesis
Transcription factors play important roles in the regulation of secondary metabolism (Xu et al. 2014). In total, 449 TFs were identified and divided into 52 gene families (Online Resource 5). Of these, 99 TFs were commonly regulated in peel and flesh, while 164 TFs were regulated in peel only (P-3 > P-6 > P-9 or P-3 < P-6 < P-9), and 86 TFs were regulated in flesh only (F-3 > F-6 > F-9 or F-3 < F-6 < F-9). WD40 proteins (100), MYB (37), bHLH (27) and WRKY (23) were the most frequently identified TFs (Fig. 3). Some TFs, such as GRAS (14), Trihelix (10), SBP (6) and WOX (3) family members, which were not previously reported to regulate catechin biosynthesis, were also continuously induced or repressed. R2R3MYB, bHLH and WD40 proteins are generally thought to be involved in catechin biosynthesis (Guo et al. 2017). We found 17 MYB genes, 11 bHLH genes and 8 WD40 protein-encoding genes were up-regulated in both the peel (P-3 < P-6 < P-9) and flesh (F-3 < F-6 < F-9), and 14 MYB genes, 9 bHLH genes and 10 WD40 protein-encoding genes were down-regulated in both the peel (P-3 > P-6 > P-9) and flesh (F-3 > F-6 > F-9).
Fig. 3.
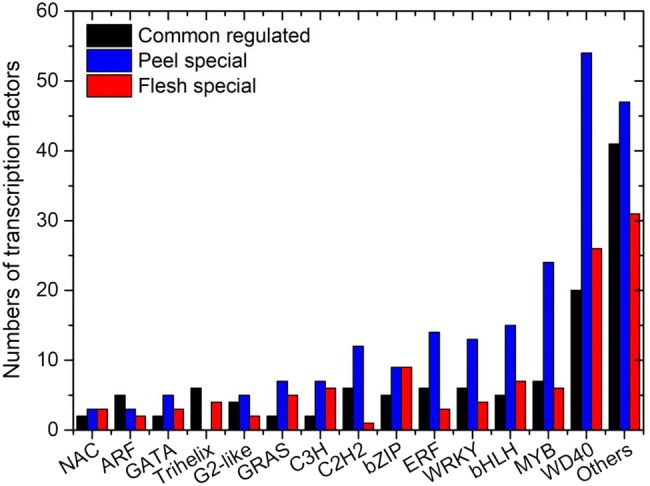
Graphical representations of transcription factors (TFs) related to catechin biosynthesis based on their assigned protein families. Peel special: TFs regulated in peel only (P-3 > P-6 > P-9 or P-3 < P-6 < P-9); Flesh special: TFs regulated in flesh only (F-3 > F-6 > F-9 or F-3 < F-6 < F-9)
Validation of gene expression associated with catechin biosynthesis as assessed by qRT-PCR
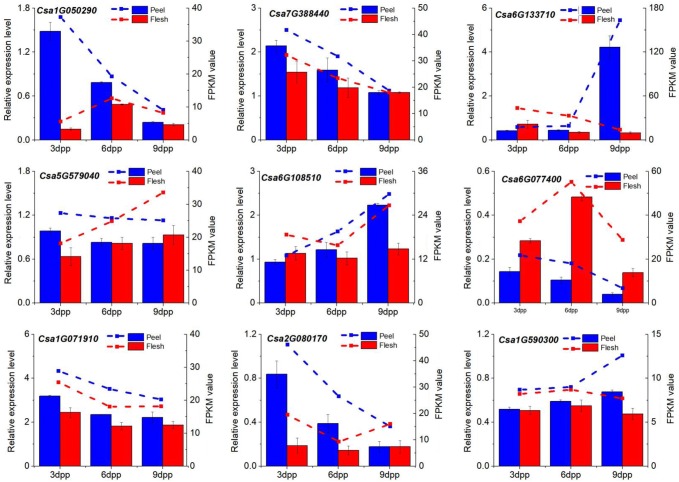
To validate the expression patterns obtained from RNA-seq, six catechin synthetic genes (Csa1G050290, Csa7G388440, Csa1G590300, Csa6G108510, Csa6G133710 and Csa6G077400) and three TFs (Csa2G080170, Csa5G579040 and Csa1G071910) were selected and verified by qPCR in all six samples. As shown in Fig. 4, all nine genes showed the same expression patterns in the RNA-seq data as in the qRT-PCR assay, indicating that the RNA-seq data were credible.
Fig. 4.
Comparison of quantitative real-time reverse transcription-PCR (qPCR) analyses of selected genes (left y-axis, histograms) and fragments per kilobase of exon model per million mapped reads (FPKM) values as assessed by RNA-seq (right y-axis, dotted lines). The cucumber β-actin gene (GenBank AB010922) was used as an internal control to normalize the expression data. Each value denotes the mean relative level of expression of three biological replicates
Discussion
A positive correlation between the TCs and age of tea leaves was found, with the new buds having the lowest concentration and the third leaves having the highest concentration (Pang et al. 2007). Contrary to tea, we found that the TCs decreased gradually during the maturation of cucumber fruit, being highest at 3 dpp (ovary stage) and lowest at 9 dpp (immature stage) (Table 1). In pear and apple, large quantities of catechins are generated in the early fruit and then decrease rapidly during the following progressive growth stages until the reach a steady level that is retained at maturity (Mosel and Herrmann 1974). Catechins produced in strawberry leaves may act as protective agents against fungal infections (Yamamoto et al. 2000). However, Ghassempour et al. (2011) found that Puccinia triticina-infected wheat leaves had lower catechin contents than healthy leaves.
In plants, PAL and 4CL are two crucial enzymes in the phenylpropanoid pathway (Baldi et al. 2017). A significant correlation between catechin abundance and PAL expression was reported previously in tea leaves at maturity (Singh et al. 2009; Samanta et al. 2017). However, the correlation is not always positive. Xiong et al. (2013) found that PAL expression was negatively correlated with the catechin contents in albino tea plants (AnJiBaiCha), but the PAL activity and TC concentration were positively correlated. Here, our RNA-seq results showed that PAL (Csa1G590300) expression was significant higher in peel than in fruit flesh at 3, 6 and 9 dpp, but was negatively correlated with the TC content in the cucumber peel, which was similar to the findings in albino tea plants (AnJiBaiCha) (Table 3). The Arabidopsis genome has four PAL members (AtPAL1–4) (Huang et al. 2010). AtPAL1 and AtPAL2 are specialized to flavonoid synthesis, AtPAL4 is involved in lignin synthesis, and the relative expression of AtPAL3 is low (Huang et al. 2010). An in-depth functional analysis of these PAL family members is needed to reveal their exact roles in catechin synthesis. Multiple genes encoding 4CLs have been studied in several plant species, including Arabidopsis (Ehlting et al. 1999), soybean (Lindermayr et al. 2002), rice (Gui et al. 2011) and Plagiochasma appendiculatum (Gao 2015). The 4CL isoenzymes direct metabolic flux toward the biosynthesis of different compounds, including flavonoids, isoflavonoids, monolignols and coumarins (Baldi et al. 2017). Our RNA-seq data suggested that at least three 4CLs (Csa1G050280, Csa1G050290 and Csa1G108800) may be involved in the fine-tuning of cucumber catechin biosynthesis. Csa1G050280 and Csa1G050290 are tandemly duplicated and show highly similar (84.7%) amino acid sequences, but they had opposite expression patterns during cucumber peel development (Table 3). The variations in gene expression patterns indicate the functional diversification of the 4CLs after divergence during the evolutionary process.
CHI, which catalyzes the cyclization of chalcone into flavanone in the cytoplasm of plant cells, is thought to be a key enzyme of catechin biosynthesis (Nishihara et al. 2005; Xiong et al. 2013). The overexpression of petunia CHI in tomato fruit resulted in a higher flavonol contents than the control, while the suppression of tobacco CHI (AB213651) by RNAi resulted in reduced pigmentation and a change in the flavonoid components of flower petals (Nishihara et al. 2005). Of the two CHI genes Csa4G622760 and Csa7G388440 identified in our transcriptome data, the latter was commonly downregulated, but the expression level was higher in the peel than in the flesh, indicating the importance of the gene in cucumber catechin biosynthesis (Table 3). Therefore, in future studies, more attention should be paid to Csa7G388440.
DFR catalyzes the reduction of dihydroflavonols to leucoanthocyanins, which occurs at a later stage prior to the biosynthesis of catechin (Singh et al. 2009). DFR has been studied in several plant species mainly as a single gene (e.g. Arabidopsis thaliana, tomato, barley, rice and grape), but species with multiple genes (e.g. Populus trichocarpa and Medicago truncatula) also exist (Mayr et al. 1997; Khanizadeh et al. 2007; Singh et al. 2009; Huang et al. 2012). Our transcriptome-wide analysis showed that at least two DFRs (Csa6G077400 and Csa3G098550) exist in the cucumber genome. Interestingly, we found that Csa6G077400 was downregulated in the peel (P-3 > P-6 > P-9), and Csa3G098550 was upregulated in the flesh (F-3 < F-6 < F-9). Csa6G077400 and Csa3G098550 showed a low similarity (32.9%) between amino acid sequences. The amino acid residue 134 of DFR plays a critical role in substrate specificity and can be divided into three types: Asp-, Asn- and non-Asn/Asp-type DFRs (Johnson et al. 2001). By searching the cucumber genome database, we found that Csa6G077400 (residue 134 is Met) and Csa3G098550 (residue 134 is Ser) are non-Asn/Asp-type DFRs. Lotus japonicus DFR1 belongs to the non-Asn/Asp-type, having a Ser residue at position 134, but it has no DFR activity (Hua et al. 2013). Thus, we speculated that Csa3G098550 expression in cucumber flesh may not contribute to catechin biosynthesis.
In A. thaliana, the expression levels of the flavonoid biosynthetic genes are specifically induced by a ternary MYB–bHLH–WD40 protein complex, comprising an R2R3-MYB (AT5G35550), a bHLH (At4g09820) and a WD40 repeat protein (AT5G24520) (Baudry et al. 2004). Conducting a correlation analysis between TFs and TCs is an effective method for identifying key candidate TFs (Fang et al. 2016; Guo et al. 2017). Using this strategy, we found two R2R3MYBs (Csa5G579040 and Csa7G452880), four bHLHs (Csa1G007910, Csa2G080170, Csa3G810530 and Csa6G516730) and 10 WD40-repeat proteins (Csa1G042240, Csa1G071910, Csa1G153530, Csa3G002310, Csa3G088960, Csa4G001690, Csa4G638360, Csa4G643110, Csa6G524070 and Csa7G4529500) that were significantly correlated with the TC contents in the peel. In contrast, only one bHLH (Csa1G051760), six WD40-repeat proteins (Csa3G061010, Csa3G166250, Csa3G416130, Csa3G775290, Csa4G325540 and Csa6G152320), and no R2R3MYBs were significantly correlated with the TC content in the flesh. These data suggested the basic transcriptional control of catechin biosynthesis was regulated by MYB–bHLH–WD40 complexes.
Conclusion
A comparison between metabolite and transcriptional profiles revealed that PAL (Csa1G590300), 4CLs (Csa1G050280, Csa1G050290 and Csa1G108800), CHI (Csa7G388440), F3H (Csa6G108510), C4H (Csa6G133710) and DFRs (Csa6G077400 and Csa3G098550) may be critical for de-astringency during cucumber fruit development. Furthermore, TFs, such as WD40-repeat proteins, MYB and bHLH, may be involved in the regulatory process.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource 2 (a) HPLC chromatogram of the catechin components in standard solutions. (b) A representative HPLC chromatogram of catechin components in a tested sample (peel of “YanBai” harvested at 6 dpp). (JPEG 194 kb)
Acknowledgements
This work was supported by the National Basic Research Program of China (973 Program; no. 2012CB113900), the Research Innovation Program for College Graduates of Jiangsu Province (no. KYLX15_1374) and the Jiangsu Science and Technology Project (BE2012326). We thank Lesley Benyon, PhD, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Author contributions
XC designed the study; XX, MH and HT carried out the experiments; XX, JP, QX and XQ analyzed the date; XX, JP and MH wrote the paper. All of the authors reviewed and approved the final manuscript.
Data availability
The data generated or analyzed during this study are included in this published article, its supplementary information files, and publicly available repositories. The raw RNA-seq reads have been deposited in NCBI Gene Expression Omnibus under accession GSE112666.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
References
- Ando K, Carr K, Grumet R. Transcriptome analyses of early cucumber fruit growth identifies distinct gene modules associated with phases of development. BMC Genom. 2012;13:518. doi: 10.1186/1471-2164-13-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldi P, Moser M, Brilli M, Vrhovsek U, Pindo M, Si-Ammour A. Fine-tuning of the flavonoid and monolignol pathways during apple early fruit development. Planta. 2017;245:1021–1035. doi: 10.1007/s00425-017-2660-5. [DOI] [PubMed] [Google Scholar]
- Baudry A, Heim M, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004;39:366–380. doi: 10.1111/j.1365-313X.2004.02138.x. [DOI] [PubMed] [Google Scholar]
- Castellarin S, Pfeiffer A, Sivilotti P, Degan M, Peterlunger E, Di Gaspero G. Transcriptional regulation of anthocyanin biosynthesis in ripening fruits of grapevine under seasonal water deficit. Plant Cell Environ. 2007;30:1381–1399. doi: 10.1111/j.1365-3040.2007.01716.x. [DOI] [PubMed] [Google Scholar]
- Ehlting J, Büttner D, Wang Q, Douglas C, Somssich I, Kombrink E. Three 4-coumarate: coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J. 1999;19:9–20. doi: 10.1046/j.1365-313x.1999.00491.x. [DOI] [PubMed] [Google Scholar]
- Eungwanichayapant P, Popluechai S. Accumulation of catechins in tea in relation to accumulation of mRNA from genes involved in catechin biosynthesis. Plant Physiol Biochem. 2009;47:94–97. doi: 10.1016/j.plaphy.2008.11.002. [DOI] [PubMed] [Google Scholar]
- Fang Z, Zhou D, Ye X, Jiang C, Pan S. Identification of candidate anthocyanin-related genes by transcriptomic analysis of ‘Furongli’ plum (Prunus salicina Lindl.) during fruit ripening using RNA-seq. Front Plant Sci. 2016;7:1338. doi: 10.3389/fpls.2016.01338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T, Eshima A, Kouya M, Takio S, Takano H, Ono K. Coordinate expression of genes involved in catechin biosynthesis in Polygonum hydropiper cells. Plant Cell Rep. 2002;21:385–389. [Google Scholar]
- Gao S. Cloning and functional characterization of a 4-coumarate CoA ligase from liverwort Plagiochasma appendiculatum. Phytochemistry. 2015;111:48–58. doi: 10.1016/j.phytochem.2014.12.017. [DOI] [PubMed] [Google Scholar]
- Ghassempour A, Mollayi S, Farzaneh M, Sharifi-Tehrani A, Aboul-Enein H. Variation of Catechin, epicatechin and their enantiomers concentrations before and after wheat cultivar-Puccinia triticina infection. Food Chem. 2011;125:1287–1290. [Google Scholar]
- Grzesik M, Naparło K, Bartosz G, Sadowska-Bartosz I. Antioxidant properties of catechins: comparison with other antioxidants. Food Chem. 2018;241:480–492. doi: 10.1016/j.foodchem.2017.08.117. [DOI] [PubMed] [Google Scholar]
- Gui J, Shen J, Li L. Functional characterization of evolutionarily divergent 4-coumarate: coenzyme A ligases in rice. Plant Physiol. 2011;157:574–586. doi: 10.1104/pp.111.178301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Guo Y, Wang P, Wang Y, Ni D. Transcriptional profiling of catechins biosynthesis genes during tea plant leaf development. Planta. 2017;246:1139–1152. doi: 10.1007/s00425-017-2760-2. [DOI] [PubMed] [Google Scholar]
- He M, Tian H, Luo X, Qi X, Chen X. Molecular progress in research on fruit astringency. Molecules. 2015;20:1434–1451. doi: 10.3390/molecules20011434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua C, Linling L, Shuiyuan C, Fuliang C, Feng X, Honghui Y, Conghua W. Molecular cloning and characterization of three genes encoding dihydroflavonol-4-reductase from Ginkgo biloba in anthocyanin biosynthetic pathway. PLoS One. 2013;8:e72017. doi: 10.1371/journal.pone.0072017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Gu M, Lai Z, Fan B, Shi K, Zhou Y, Yu J, Chen Z. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010;153:1526–1538. doi: 10.1104/pp.110.157370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Gou J, Jia Z, Yang L, Sun Y, Xiao X, Song F, Luo K. Molecular cloning and characterization of two genes encoding dihydroflavonol-4-reductase from Populus trichocarpa. PLoS One. 2012;7:e30364. doi: 10.1371/journal.pone.0030364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E, Ryu S, Yi H, Shin B, Cheong H, Choi G. Alteration of a single amino acid changes the substrate specificity of dihydroflavonol 4-reductase. Plant J. 2001;25:325–333. doi: 10.1046/j.1365-313x.2001.00962.x. [DOI] [PubMed] [Google Scholar]
- Khanizadeh S, Tsao R, Rekika D, Yang R, De Ell J. Phenolic composition and antioxidant activity of selected apple genotypes. J Food Agric Environ. 2007;5:61–66. [Google Scholar]
- Li P, Du G, Ma F. Phenolics concentration and antioxidant capacity of different fruit tissues of astringent versus non-astringent persimmons. Sci Hortic. 2011;129:710–714. [Google Scholar]
- Lindermayr C, Möllers B, Fliegmann J, Uhlmann A, Lottspeich F, Meimberg H, Ebel J. Divergent members of a soybean (Glycine max L.) 4-coumarate:coenzyme A ligase gene family. Eur J Biochem. 2002;269:1304–1315. doi: 10.1046/j.1432-1033.2002.02775.x. [DOI] [PubMed] [Google Scholar]
- Liu M, Tian H, Wu J, Cang R, Wang R, Qi X, Xu Q, Chen X. Relationship between gene expression and the accumulation of catechin during spring and autumn in tea plants (Camellia sinensis L.) Hortic Res. 2015;2:15011. doi: 10.1038/hortres.2015.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr U, Michalek S, Treutter D, Feucht W. Phenolic compounds of apple and their relationship to scab resistance. J Phytopathol-Phytopathol Z. 1997;145:69–75. [Google Scholar]
- Mosel H, Herrmann K. Changes in catechins and hydroxycinnamic acid derivatives during development of apples and pears. J Sci Food Agric. 1974;25:251–256. doi: 10.1002/jsfa.2740250304. [DOI] [PubMed] [Google Scholar]
- Nishihara M, Nakatsuka T, Yamamura S. Flavonoid components and flower color change in transgenic tobacco plants by suppression of chalcone isomerase gene. FEBS Lett. 2005;579:6074–6078. doi: 10.1016/j.febslet.2005.09.073. [DOI] [PubMed] [Google Scholar]
- Pang Y, Peel G, Wright E, Wang Z, Dixon R. Early steps in proanthocyanidin biosynthesis in the model legume Medicago truncatula. Plant Physiol. 2007;145:601–615. doi: 10.1104/pp.107.107326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani A, Singh K, Ahuja P, Kumar S. Molecular regulation of catechins biosynthesis in tea [Camellia sinensis (L.) O. Kuntze] Gene. 2012;495:205–210. doi: 10.1016/j.gene.2011.12.029. [DOI] [PubMed] [Google Scholar]
- Samanta T, Kotamreddy J, Ghosh B, Mitra A. Changes in targeted metabolites, enzyme activities and transcripts at different developmental stages of tea leaves: a study for understanding the biochemical basis of tea shoot plucking. Acta Physiol Plant. 2017;39:11. [Google Scholar]
- Severo J, Tiecher A, Chaves F, Silva J, Rombaldi C. Gene transcript accumulation associated with physiological and chemical changes during developmental stages of strawberry cv. Camarosa. Food Chem. 2011;126(3):995–1000. [Google Scholar]
- Singh K, Kumar S, Rani A, Gulati A, Ahuja P. Phenylalanine ammonia-lyase (PAL) and cinnamate 4-hydroxylase (C4H) and catechins (flavan-3-ols) accumulation in tea. Funct Integr Genomic. 2009;9:125. doi: 10.1007/s10142-008-0092-9. [DOI] [PubMed] [Google Scholar]
- Soares S, Ferrer-Galego R, Brandão E, Silva M, Mateus N, Freitas V. Contribution of human oral cells to astringency by binding salivary protein/tannin complexes. J Agric Food Chem. 2016;64:7823–7828. doi: 10.1021/acs.jafc.6b02659. [DOI] [PubMed] [Google Scholar]
- Tian H (2015) The relationship between cucumber astrengency form and catechins metabolism, and the study of molecular basis. Master’s degree dissertation in Yangzhou University 20–24
- Trapnell C, Williams B, Pertea G, Mortazavi A, Kwan G, Van Baren M, Salzberg S, Wold B, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotech. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley D, Pimentel H, Salzberg S, Rinn J, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troszyńska A, Estrella I, Lamparski G, Hernández T, Amarowicz R, Pegg R. Relationship between the sensory quality of lentil (Lens culinaris) sprouts and their phenolic constituents. Food Res Int. 2011;44:3195–3201. [Google Scholar]
- Xiong L, Li J, Li Y, Yuan L, Liu S, Huang J, Liu Z. Dynamic changes in catechin levels and catechin biosynthesis-related gene expression in albino tea plants (Camellia sinensis L.) Plant Physiol Bioch. 2013;71:132–143. doi: 10.1016/j.plaphy.2013.06.019. [DOI] [PubMed] [Google Scholar]
- Xu W, Lepiniec L, Dubos C. New insights toward the transcriptional engineering of proanthocyanidin biosynthesis. Plant Signal Behav. 2014;9:e28736. doi: 10.4161/psb.28736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Xu R, Zhu B, Yu T, Qu W, Lu L, Xu Q, Qi X, Chen X. A high-density genetic map of cucumber derived from Specific Length Amplified Fragment sequencing (SLAF-seq) Front Plant Sci. 2015;5:768. doi: 10.3389/fpls.2014.00768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zhang Y, Chen J, Wang F, Du Q, Yin J. Quantitative analyses of the bitterness and astringency of catechins from green tea. Food Chem. 2018;258:16–24. doi: 10.1016/j.foodchem.2018.03.042. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Nakatsuka S, Otani H, Kohmoto K, Nishimura S. (+)-Catechin acts as an infection-inhibiting factor in strawberry leaf. Phytopathology. 2000;90:595–600. doi: 10.1094/PHYTO.2000.90.6.595. [DOI] [PubMed] [Google Scholar]
- Zhang L, Wei K, Cheng H, Wang L, Zhang C. Accumulation of catechins and expression of catechin synthetic genes in Camellia sinensis at different developmental stages. Bot Stud. 2016;57:31. doi: 10.1186/s40529-016-0143-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wei K, Li H, Wang L, Ruan L, Pang D, Cheng H. Identification of key genes involved in catechin metabolism in tea seedlings based on transcriptomic and HPLC analysis. Plant Physiol Biochem. 2018;133:107–115. doi: 10.1016/j.plaphy.2018.10.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Online Resource 2 (a) HPLC chromatogram of the catechin components in standard solutions. (b) A representative HPLC chromatogram of catechin components in a tested sample (peel of “YanBai” harvested at 6 dpp). (JPEG 194 kb)
Data Availability Statement
The data generated or analyzed during this study are included in this published article, its supplementary information files, and publicly available repositories. The raw RNA-seq reads have been deposited in NCBI Gene Expression Omnibus under accession GSE112666.