Abstract
Cancer consists of heterogeneous cells that contain a small population of cells that possess stem cell properties; these cells, referred to as cancer stem cells (CSCs) or tumor-initiating cells, are involved in tumor progression and metastasis. Using a sphere-forming assay, canine mammary CSCs were found to be similar to human breast CSCs. Metabolic reprogramming has been recognized as a hallmark of various cancers. However, the significance of cellular metabolism in CSCs remains unclear. The aim of this study was to define the metabolic characteristics of CSCs derived from canine mammary tumors and gain an understanding of the maintenance of stemness. We identified metabolite profiles of canine mammary adenocarcinoma cell lines using gas chromatography-mass spectrometry. Metabolites were extracted from both adherent and sphere-forming cells derived from three cell lines. Sphere-forming cells were separated from adherent cells using an orthogonal, partial least-squares discriminant analysis. Sphere-forming cells were found to contain high levels of the amino acids alanine, glycine and proline compared with adherent cells. They also had high levels of palmitoleate, palmitate and dihomo-gamma-linolenic acid compared with adherent cells. In a sphere-forming assay, palmitate increased the number of spheres for all cell lines. These results indicate that the sphere-forming cells derived from canine mammary adenocarcinoma cell lines have specific metabolic profiles that may be useful for the development of CSC-specific therapies targeting metabolic pathways and potential stemness biomarkers; these results also clarify the maintenance of stemness in canine mammary CSCs.
Keywords: cancer stem cell, gas chromatography-mass spectrometry, mammary adenocarcinoma, metabolite
Cancer consists of heterogeneous cells that contain a subset of tumor cells with stemness features, including the ability to self-renew and differentiate and high tumorigenicity, with low sensitivities for chemotherapy and radiotherapy [5, 21]. Such tumor cells, termed cancer stem cells (CSCs) or tumor-initiating cells (TICs), are involved in tumor initiation, recurrence and metastasis [5, 21]. In veterinary medicine, there is recent evidence for the presence of CSCs in various solid cancers, including osteosarcoma, mammary adenocarcinoma, hepatocellular carcinoma, rhabdomyosarcoma and pulmonary adenocarcinoma [13, 17,18,19, 23, 29, 36]. In canine mammary cancers, CSCs were identified using a sphere-forming assay, ALDEFLUOR™ assay, side population analysis and the expression of surface markers, including CD44 and CD24 [9, 17, 18, 23]. Moreover, sphere-forming cells derived from canine mammary cancer cell lines possess high tumorigenicity in immunodeficient mice, and exhibit resistance to anticancer drugs and increased expression of stem cell-related genes, such as CD133 and multidrug resistance [18, 23]. Thus, sphere-forming cells possess stem cell-like properties.
Recently, tumorigenesis-associated metabolic reprogramming has received attention in cancer research. The hallmarks of cancer metabolism include the deregulation of glucose and amino acid uptake, use of opportunistic modes of nutrient acquisition, use of glycolysis/TCA cycle intermediates for biosynthesis and NADPH production, increased demand for nitrogen, alterations in metabolite-driven gene regulation, and metabolic interactions with the microenvironment [24]. Cancer cells can generate ATP via glycolysis, even under normoxic conditions (Warburg effect), rather than by mitochondrial oxidative phosphorylation (OXPHOS), and can facilitate the synthesis of amino acids, lipids, and nucleotides as well as lactate production [10, 35]. In contrast to cancer cells, CSCs adapt to various environmental factors, such as hyponutrition, therapeutic toxicity and oxidant stress, compared with non-CSCs; the metabolic properties of CSCs differ from those of non-CSCs [37]. Moreover, the metabolic phenotypes of CSCs depend on the specific cancer type, such as breast, ovarian and pancreatic cancers, and use either the glycolytic or OXPHOS pathway in humans [25]. However, the metabolic reprogramming of CSCs in dogs remains unknown.
In humans, metabolomic analyses have been performed on many different tissue types, including solid tissue as well as on serum, plasma and urine specimens [1]. In veterinary medicine, gas chromatography-mass spectrometry (GC-MS) analysis has been used to analyze plasma specimens from dogs with cancer, including mammary adenocarcinoma, brain cancer, lymphoma and melanoma [3, 12, 28, 30]. The concentrations of plasma amino acids are altered in dogs with cancer compared with healthy dogs. For example, the levels of threonine, proline and serine are increased in dogs with melanoma, and alanine, proline and isoleucine are increased in dogs with brain tumors compared with healthy dogs [12, 28]. Moreover, increased levels of methionine, serine, asparagine, glycine and alanine have been observed in dogs with carcinoma (no metastasis) and decreased levels of aspartate and ornithine have been observed in dogs with metastatic carcinoma [3]. Changes in various amino acids have also been associated with cancer progression in dogs. However, the metabolic alterations of the cancer cells and CSCs themselves remain largely unknown.
Mammary adenocarcinoma is the most common cancer in female dogs and often exhibits distinct metastases [20]. Cancer treatments, such as chemotherapy, radiation and their combination except for surgery, are not very effective in preventing recurrence [16]. Therefore, it is essential to develop novel therapeutic strategies targeting CSCs to eradicate mammary adenocarcinoma in dogs. In this study, we metabolically profiled sphere-forming cells derived from canine mammary adenocarcinoma cell lines using GC-MS to clarify the mechanism underlying the maintenance of stemness mediated by metabolic changes.
MATERIALS AND METHODS
Cell lines and culture
Three canine mammary adenocarcinoma cell lines (CHMp, CNMp and CTBp) [31] used in the present study were kindly provided by Dr. N. Sasaki, the University of Tokyo. The cell lines were maintained in Dulbecco’s modified Eagle medium and nutrient F-12 (DMEM/F12, Invitrogen, Carlsbad, CA, U.S.A.) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT, U.S.A.) and antibiotics (Nacalai Tesque, Kyoto, Japan) at 37°C in an atmosphere containing 5% CO2.
Sphere-forming assay
The sphere-forming assay was performed as described previously [18]. In brief, singly suspended cells were plated at a density of 1 × 105 viable cells per ultralow attachment 100-mm dish (Corning Inc., Corning, NY, U.S.A.). The cells were grown in serum-free DMEM/F12 supplemented with 10 ng/ml basic fibroblast growth factor (Invitrogen), 10 ng/ml epidermal growth factor (Invitrogen), 4 mg/ml heparin (Sigma-Aldrich, St. Louis, MO, U.S.A.) and 1 × NeuroBrew-21 (Miltenyi Biotech, Tokyo, Japan) (GF+ medium) or without ones (GF−medium) for 7 days. A colony of cells with a diameter of greater than 50 µm was considered a sphere. Spheres were counted under low magnification, collected, and then used for the western blotting and GC-MS analyses.
Western blotting
The cells were lysed in ice-cold Mammalian Lysis Buffer (Promega, Madison, WI, U.S.A.) containing Protease Inhibitor Cocktail (Promega) and incubated for 15 min at 4°C. Insoluble fragments were removed by centrifugation at 16,000 × g for 10 min at 4°C, and the supernatant was collected. Protein concentrations were determined using the Bicinchoninate Protein Assay kit (Nacalai Tesque). Further, cell lysates containing 10 µg of total protein were prepared for western blotting analysis by boiling samples in sodium dodecyl sulfate (SDS) sample buffer (50 mM Tris, 2% SDS, 5% glycerol, 5% 2-mercaptoethanol, pH6.8).The prepared lysates were resolved on a 5–12.5% Tris-glycine polyacrylamide gradient gel using a Perfect Cell B from DRC (Tokyo, Japan) (ΔV=150 V, 50 min at 25°C), transferred to a PVDF membrane using a standard semi-dry apparatus from Bio-Rad (Hercules, CA, U.S.A.) (ΔV=15 V, 1 hr at 25°C). After blocking with 10% skim milk, 6% glycine in TBS containing 0.1% Tween-20, the membrane was incubated with the following primary antibodies: mouse anti-ALDH1 (44/ALDH, 1:1,000, BD Biosciences, Franklin Lakes, NJ, U.S.A.) and rabbit anti-β-actin (PM053, 1:2,000; MBL, Nagoya, Japan). Horseradish peroxidase-conjugated secondary antibodies and Ez WestLumi plus (ATTO, Tokyo, Japan) were used for the detection of antibody-bound proteins.
GC-MS analyses of amino acids and fatty acids
The metabolites from adherent and sphere-forming cells were extracted using a method as described previously [26]. For amino acid analysis, sample preparation was performed using EZ:faast (Phenomenex, Torrance, CA, U.S.A.) according to the manufacturer’s instructions. Norvaline was used as an internal standard. The following amino acids were analyzed: alanine, glycine, alpha-aminobutyric acid, valine, leucine, allo-isoleucine, threonine, serine, proline, asparagine, thioproline, aspartic acid, methionine, 4-hydroxyproline (4-HG), glutamic acid, phenylalanine, ornithine, lysine, histidine, tyrosine and tryptophan. For fatty acid analysis, samples of fatty acids from both adherent and sphere-forming cells were methylated using a fatty acid methylation kit (Nacalai Tesque), and extracted using a fatty acid methyl ester purification kit (Nacalai Tesque). Ethyl arachidonate was used as an internal standard. The following fatty acids were analyzed: C12:0 (laurate), C14:0 (myristate), C16:1n-7 (palmitoleate), C16:0 (palmitate), C17:0 (margarate), C18:2n-6 (linoleate), C18:1n-9 (oleate), C18:1n-7 (cis-vaccenate), C18:0 (stearate), C20:4n-6 (arachidonate), C20:5n-3 (eicosapentaenoate, EPA), C20:3n-6 (dihomo-gamma-linolenic acid, DGLA), C22:6n-3 (docosahexaenoate, DHA) and C22:5n-3 (docosapentaenoate, DPA). Ethyl arachidonate was used as an internal standard. The GC-MS analysis was performed using a GC/MS-QP2010Plus instrument (Shimadzu, Milan, Italy). The correlation of peak retention times with the retention time of a standard alkane series mixture was performed using the Automatic Adjustment of Retention Time function of the GC/MS solution software (Shimadzu). Chromatogram acquisition and compound identification using a mass spectral library search were performed using the Shimadzu GC-MS solution software.
Sphere-forming assay with fatty acids treatment
Sensitivity to palmitate was examined for spheres obtained from CTBp, CHMp and CNMp cells. Palmitate (Sigma-Aldrich) and DHA (Cayman chemical, Ann Arbor, MI, U.S.A.) was dissolved in hexane (Wako, Osaka, Japan) and dimethyl sulfoxide (Sigma-Aldrich), respectively. Singly suspended cells were seeded at a density of 5 × 103 cells per well on 6-well ultralow attachment plates (Corning), and cultured in a GF+ medium containing five different doses (final conc. 1 nM, 10 nM, 100 nM, 1 µM and 10 µM) from the beginning or GF−medium. The total number of spheres was counted after 5 days. All experiments were repeated thrice in each cell lines.
Statistical analysis
Multivariate analysis was performed using principal component analysis (PCA) and orthogonal, partial least-squares discriminant analysis (OPLS-DA) in the SIMCA 13.0 software (Umetrics, Umea, Sweden). Statistical significance was analyzed using Welch’s t-test, ordinary one-way ANOVA, and Bonferroni’s multiple comparisons test for PCA and OPLS-DA, or Dunnett’s test for the sphere-forming assay with palmitate treatment. A P value less than 0.05 was considered significant.
RESULTS
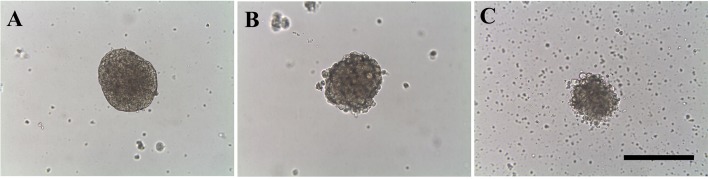
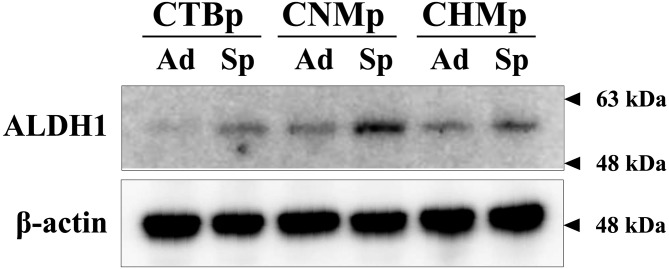
To identify the amino acid and fatty acid profiles of adherent and sphere-forming cells from canine mammary adenocarcinoma cell lines, we used the sphere-forming assay. All spheres appeared large, round and sharp (Fig. 1). To examine the expression of the stem cell marker ALDH1, sphere-forming cells were analyzed by western blotting compared with the corresponding adherent cells. ALDH1 expression was higher in all sphere-forming cells than in the adherent cells (Fig. 2).
Fig. 1.
Characterization of spheres formed from CTBp (A), CNMp (B), and CHMp cell lines (C). Bar, 100 µm.
Fig. 2.
Western blotting for detecting the stem cell marker ALDH1 in sphere-forming cells derived from canine mammary adenocarcinoma cell lines. Ad, adherent cells; Sp, sphere-forming cells.
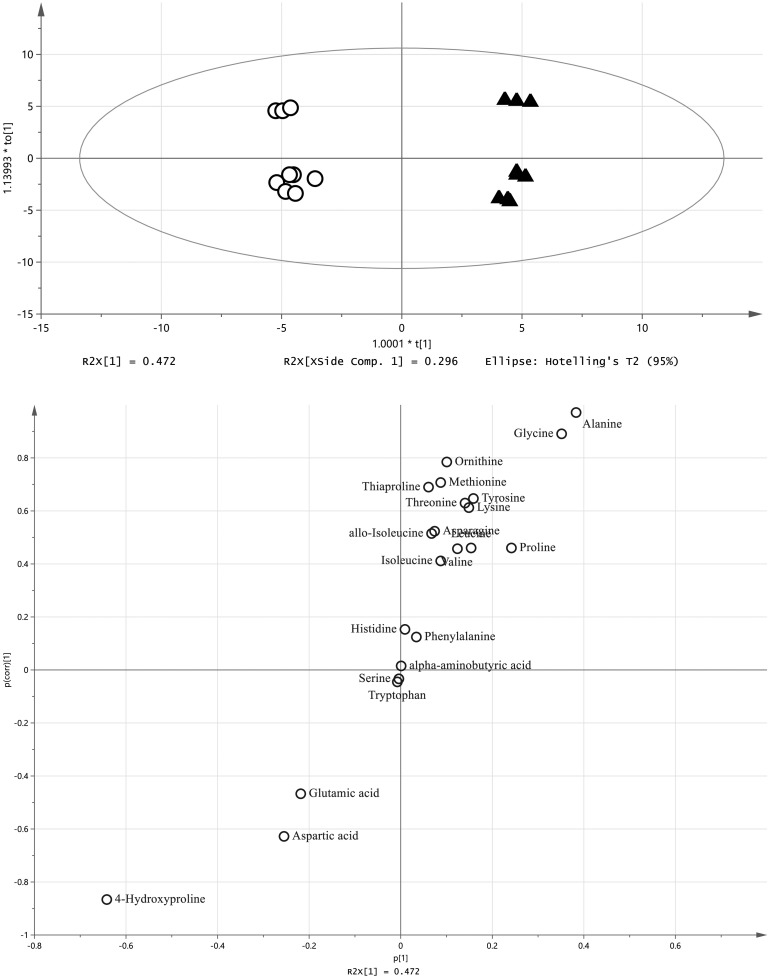
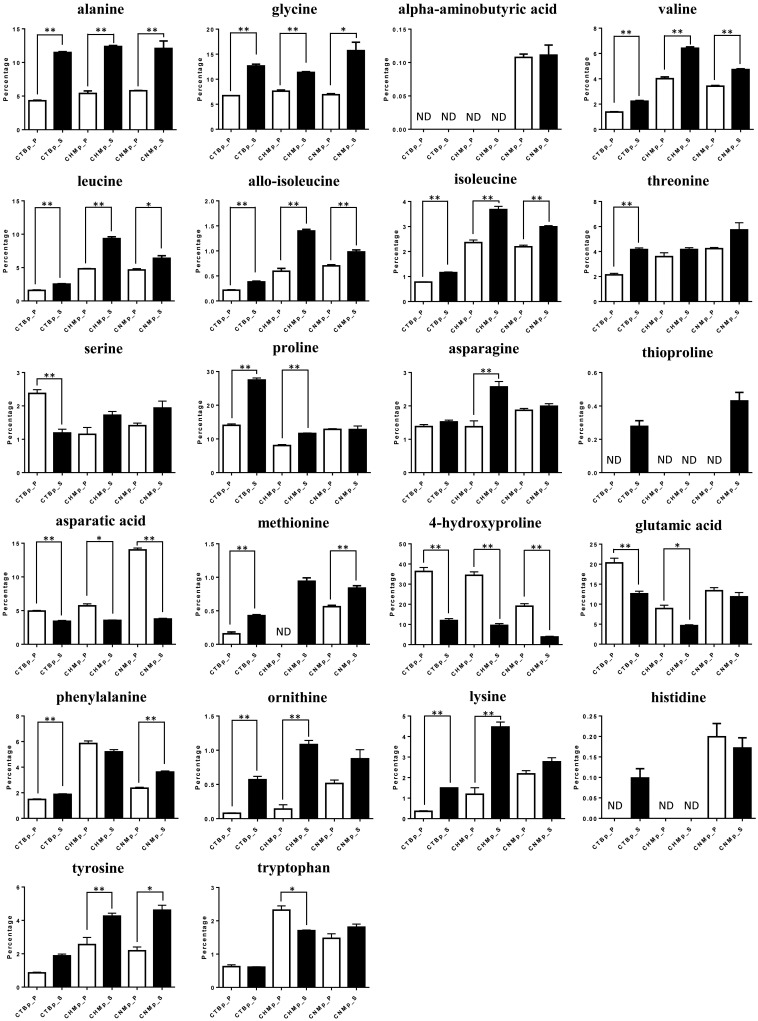
For the analysis of the amino acids, each cell population was plotted in a different area by PCA (Fig. 3). In OPLS-DA, sphere-forming cells were separated from adherent cells, where higher levels of alanine, glycine and proline, and lower levels of aspartic acid, glutamic acid and 4-HG were identified in the sphere-forming cells (Fig. 4). Levels of six amino acids (alanine, glycine valine, leucine, allo-isoleucine and isoleucine) were significantly increased in all sphere-forming cells, whereas aspartic acid and 4-HG were significantly decreased compared with adherent cells (Fig. 5).
Fig. 3.
Principal component analysis (PCA) of amino acids derived from canine mammary adenocarcinoma cell lines. Upper, score plot. Bottom, loading plot. Ad, adherent cells; Sp, sphere-forming cells.
Fig. 4.
Orthogonal, partial least squares discriminant analysis (OPLS-DA) of amino acids derived from canine mammary adenocarcinoma cell lines. Upper, score plot. Bottom, s-plot. White circles, adherent cells. Black triangles, sphere-forming cells.
Fig. 5.
Percentage of amino acids in parental cells (white) and sphere-forming cells (black) derived from canine mammary adenocarcinoma cell lines. Data are based on three independent experiments. ND, not detected. *P<0.05, **P<0.01.
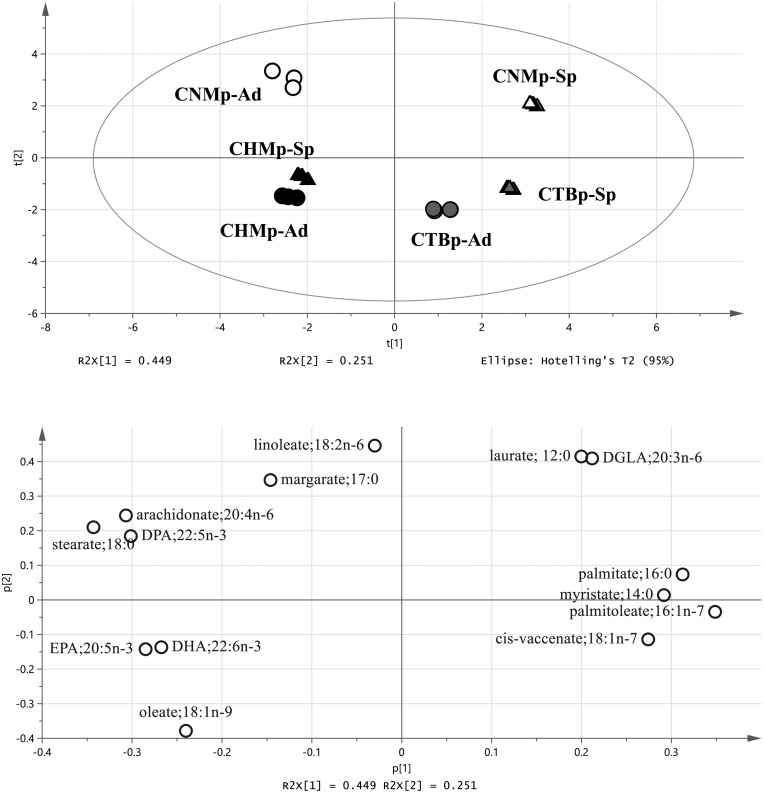
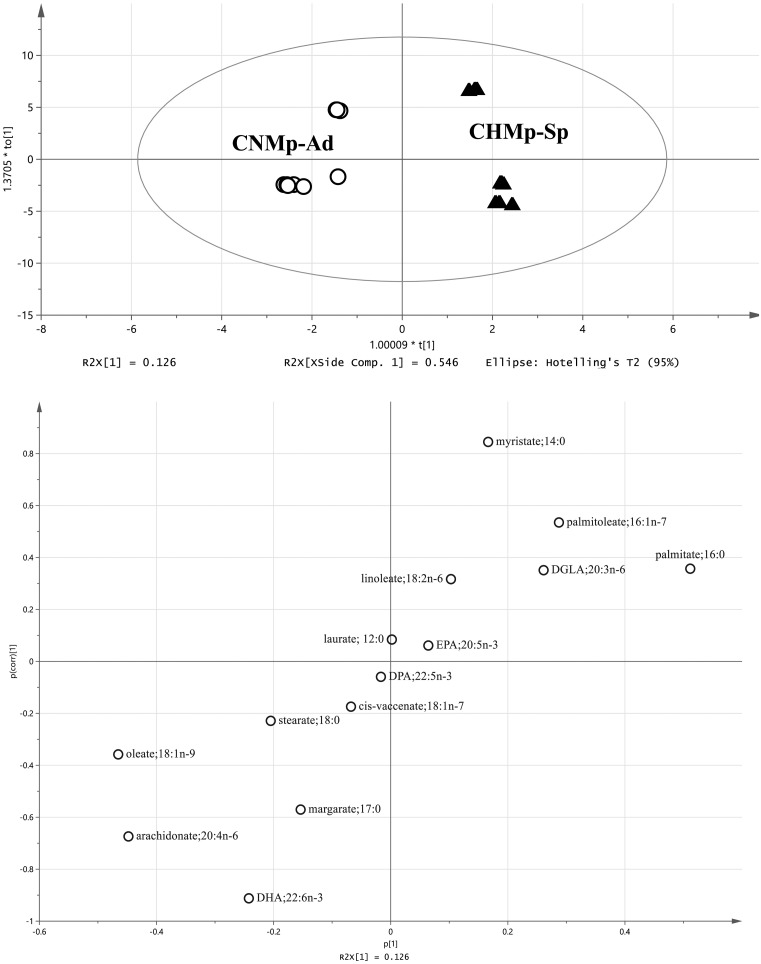
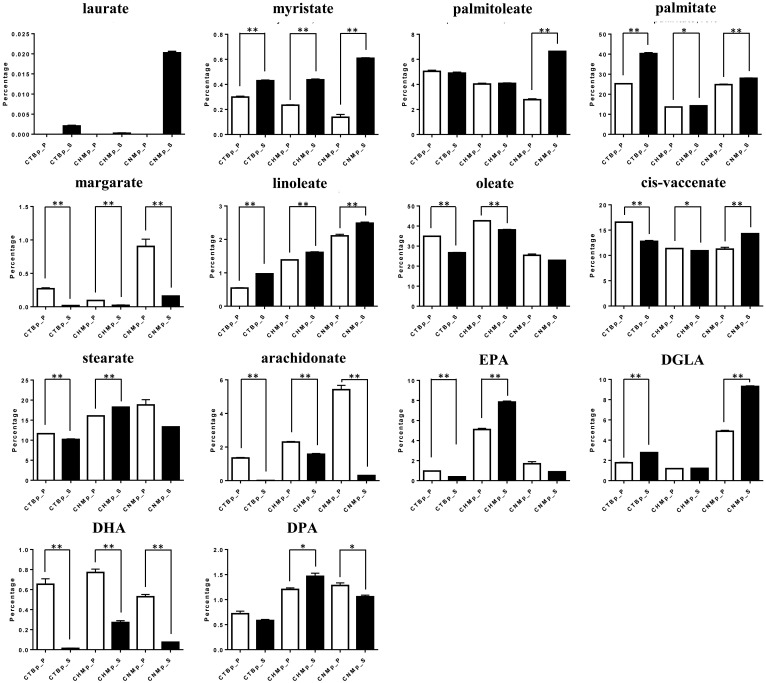
For the analysis of the fatty acids, each cell population was plotted in a different area by PCA (Fig. 6). In OPLS-DA, higher levels of palmitoleate, palmitate and DGLA, and lower levels of stearate, oleate, arachidonate and DHA were found in the sphere-forming cells compared with adherent cells (Fig. 7). Three fatty acids (myristate, palmitate and linoleate) were significantly increased in the sphere-forming cells, whereas margarate, arachidonate, and DHA were significantly decreased compared with adherent cells (Fig. 8).
Fig. 6.
Principal component analysis (PCA) of fatty acids derived from canine mammary adenocarcinoma cell lines. Upper, score plot. Bottom, loading plot. Ad, adherent cells; DGLA, dihomo-gamma-linolenic acid; DHA, docosahexaenoate; DPA, docosapentaenoate; EPA, eicosapentaenoate; Sp, sphere-forming cells.
Fig. 7.
Orthogonal, partial least squares discriminant analysis (OPLS-DA) of fatty acids derived from canine mammary adenocarcinoma cell lines. Upper, score plot. Bottom, s-plot. White circles, adherent cells. Black triangles, sphere-forming cells. DGLA, dihomo-gamma-linolenic acid; DHA, docosahexaenoate; DPA, docosapentaenoate; EPA.
Fig. 8.
Percentage of fatty acids in adherent cells (white) and sphere-forming cells (black) derived from canine mammary adenocarcinoma cell lines. Data are based on three independent experiments. *P<0.05, **P<0.01. DGLA, dihomo-gamma-linolenic acid; DHA, docosahexaenoate; DPA, docosapentaenoate; EPA, eicosapentaenoate.
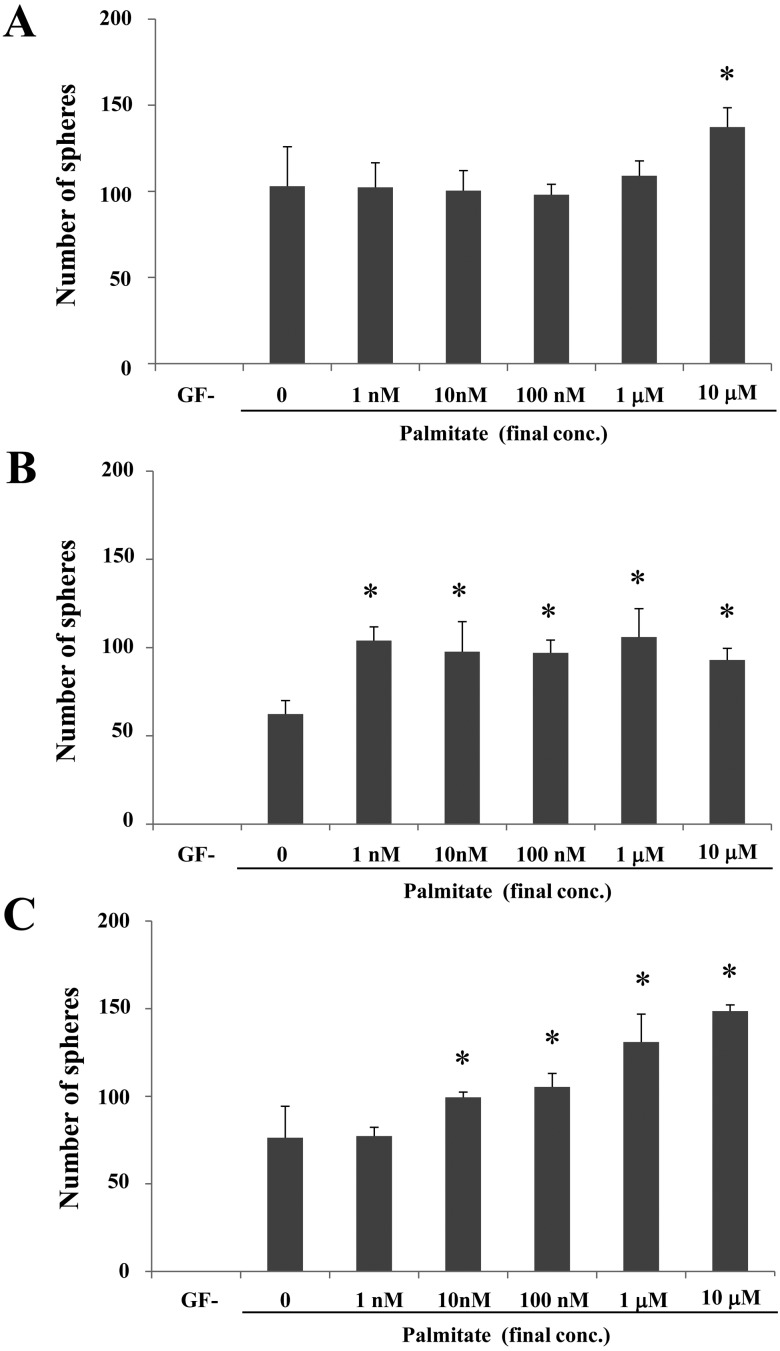
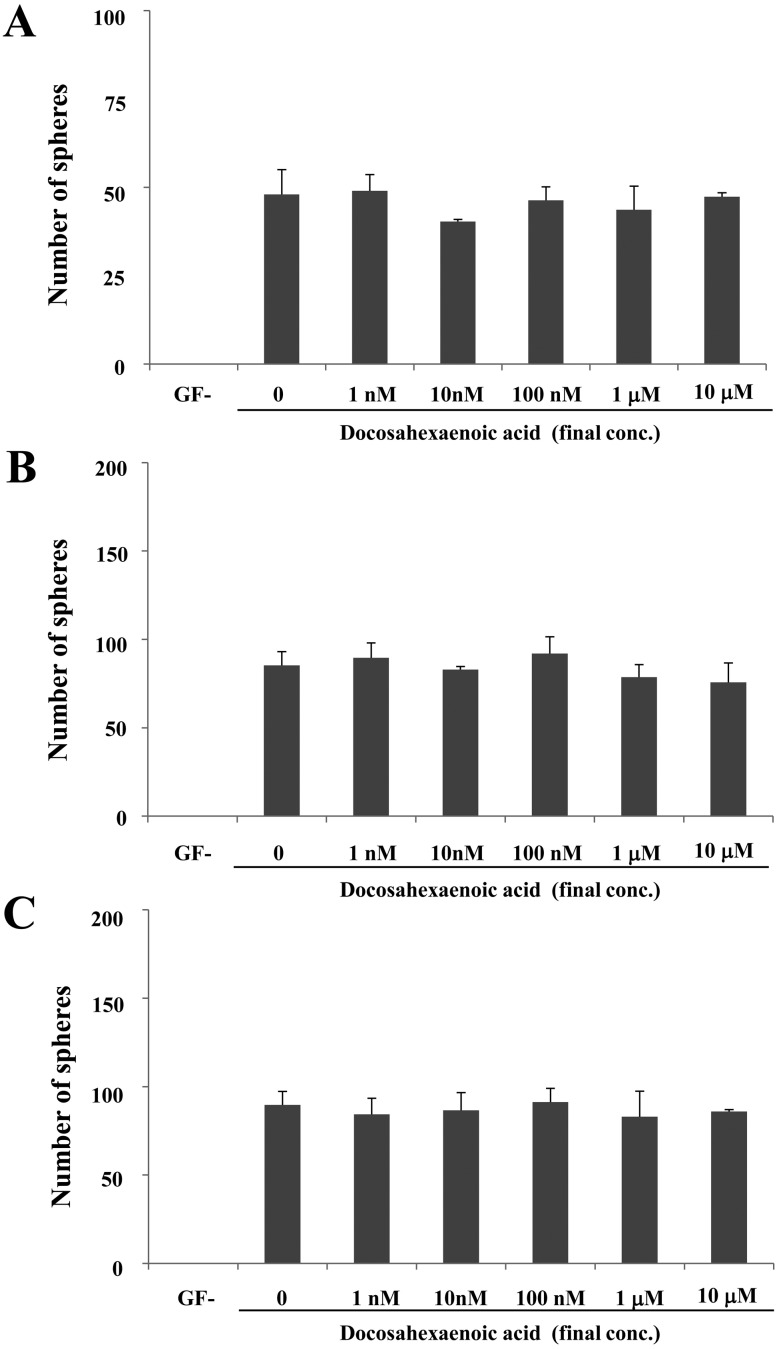
Next, we examined the effect of palmitate on sphere formation. All cell lines exhibited sphere formation in the GF+ medium containing palmitate or DHA, whereas no sphere formation was observed on GF−medium (Figs. 9 and 10). Notably, the sharp outline of spheres did not different among the cells cultures in the GF+ medium on the presence/absence of palmitate or DHA. In CTBp, a significant increased number of spheres was observed only at a final palmitate concentration of 10 mM than other concentrations (Fig. 9A), whereas in CNMp, the number of spheres did not significantly increase as the concentration of palmitate increased, although significant difference was observed on GF+ medium without palmitate (Fig. 9B). In CHMp, the number of spheres increased in a palmitate dose-dependent manner (Fig. 9C). On the other hand, in all cell lines, the number of spheres did not significantly increase as the concentration of DHA increased (Fig. 10).
Fig. 9.
Effect of palmitate on sphere-forming cells derived from the canine mammary adenocarcinoma cell lines CTBp (A), CNMp (B) and CHMp (C). Statistical analysis was performed using Dunnett’s test. Results shown are representative of three independent experiments. *P<0.05.
Fig. 10.
Effect of docosahexaenoic acid on sphere-forming cells derived from the canine mammary adenocarcinoma cell lines CTBp (A), CNMp (B) and CHMp (C). Statistical analysis was performed using Dunnett’s test. Results shown are representative of three independent experiments. *P<0.05.
DISCUSSION
In this study, we identified the metabolic profiles of sphere-forming cells derived from canine mammary adenocarcinoma cell lines using GC-MS analysis. Sphere-forming cells exhibited higher expression of ALDH1 and concentrations of alanine, glycine and proline as well as higher levels of palmitoleate, palmitate and DGLA compared with adherent cells, indicating that these cells possess specific metabolites that may be associated with the maintenance of stemness.
Recently, several metabolic analyses of amino acids and fatty acids demonstrated that specific metabolic phenotypes regulate the stemness of both normal stem cells and CSCs [27, 34, 40]. Methionine and threonine metabolism is regulated during the maintenance of quiescence and differentiation in human pluripotent stem cells, including embryonic stem (ES) and induced pluripotent stem (iPS) cells [27, 33]. Higher levels of valine are associated with the progression of myeloid leukemia, which is mediated by enhanced uptake into leukemic stem cells and the proliferation of undifferentiated cells in the embryonic liver [11, 14]. Branched-chain amino acids, such as valine, leucine and isoleucine, suppress growth of CSCs via the mammalian target of rapamycin pathway, following enhanced sensitivity to chemotherapy [22]. The amino acids found in CSCs are independent of the higher expression of metabolic enzymes, including glycine decarboxylase, branched-chain amino acid aminotransferase 1 and threonine dehydrogenase [11, 33, 40]. Lysine metabolism decreases the level of reactive oxygen species, which suppress the proliferation and progression of cancer, following enhanced self-renewal and Wnt signal activation in CSCs [38]. In this study, sphere-forming cells exhibited higher levels of alanine, glycine and proline compared with adherent cells. Zhang et al. [40] reported that glycine metabolism is necessary to promote tumorigenesis, supporting the idea that glycine decarboxylase activity drives TICs. Arnold et al. [2] also demonstrated increased levels of alanine and glycine in ES and iPS cells. Proline metabolism via proline dehydrogenase also plays an important role in the self-renewal capacity of human breast CSCs [6]. Canine mammary CSCs are characterized by an increase in alanine, glycine and proline, and may be associated with the maintenance of stemness. Cancer-specific metabolic reprogramming may exist in solid cancers in dogs, as the levels of amino acids in CSCs differ according to cancer type. Moreover, eight plasma metabolites, including alanine and proline, appear to be useful diagnostic markers for different subtypes of human breast cancer [8]. Therefore, amino acid profiling may be useful for the development of CSC markers as well as for diagnostic and prognostic markers.
Both ES and iPS cells have a higher capacity to produce palmitate and oleate [34]. The stemness of CSCs is also regulated by unsaturated fatty acids [15]. In this study, sphere-forming cells exhibited higher levels of palmitoleate, palmitate and DGLA, and lower levels of DHA, stearate, oleate and arachidonate compared with adherent cells. Moreover, palmitate increased the number of spheres formed, suggesting that palmitate is enhanced a self-renewal in sphere-forming cells derived from canine mammary adenocarcinoma cell lines. In human ovarian cancer, CSCs contain higher levels of unsaturated fatty acids, such as palmitoleate, arachidonate, oleate and DHA compared with non-CSCs, suggesting their contribution to the maintenance of stemness [15]. Palmitate is associated with the facilitation of stemness and tumorigenic capacity [4]. On the other hand, both EPA and DHA act not only to suppress self-renewal capacity but also to decrease viability by inducing CSC apoptosis [7, 39]. In human breast cancer, the levels of oleate and palmitate tend to be decreased in CSCs compared with non-CSCs [32]. In this study, DHA levels were low and variable in sphere-forming cells. These findings suggest a function in the proliferation of cancer cells rather than the maintenance of stemness in CSCs. However, the function of unsaturated fatty acids in CSCs remains controversial and may differ by animal species and cancer type/subtype. Therefore, further studies are necessary to determine the essential functions of unsaturated fatty acids in CSCs. The metabolic signatures and pathways identified will provide insights into mammary cancer pathogenesis and the development of novel therapies for canine mammary cancers.
REFERENCES
- 1.Armitage E. G., Barbas C.2014. Metabolomics in cancer biomarker discovery: current trends and future perspectives. J. Pharm. Biomed. Anal. 87: 1–11. doi: 10.1016/j.jpba.2013.08.041 [DOI] [PubMed] [Google Scholar]
- 2.Arnold J. M., Choi W. T., Sreekumar A., Maletić-Savatić M.2015. Analytical strategies for studying stem cell metabolism. Front. Biol. (Beijing) 10: 141–153. doi: 10.1007/s11515-015-1357-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azuma K., Osaki T., Tsuka T., Imagawa T., Minami S., Okamoto Y.2012. Plasma free amino acid profiles of canine mammary gland tumors. J. Vet. Sci. 13: 433–436. doi: 10.4142/jvs.2012.13.4.433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beyaz S., Mana M. D., Roper J., Kedrin D., Saadatpour A., Hong S. J., Bauer-Rowe K. E., Xifaras M. E., Akkad A., Arias E., Pinello L., Katz Y., Shinagare S., Abu-Remaileh M., Mihaylova M. M., Lamming D. W., Dogum R., Guo G., Bell G. W., Selig M., Nielsen G. P., Gupta N., Ferrone C. R., Deshpande V., Yuan G. C., Orkin S. H., Sabatini D. M., Yilmaz Ö. H.2016. High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature 531: 53–58. doi: 10.1038/nature17173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke M. F., Dick J. E., Dirks P. B., Eaves C. J., Jamieson C. H., Jones D. L., Visvader J., Weissman I. L., Wahl G. M.2006. Cancer stem cells-perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 66: 9339–9344. doi: 10.1158/0008-5472.CAN-06-3126 [DOI] [PubMed] [Google Scholar]
- 6.Elia I., Broekaert D., Christen S., Boon R., Radaelli E., Orth M. F., Verfaillie C., Grünewald T. G. P., Fendt S. M.2017. Proline metabolism supports metastasis formation and could be inhibited to selectively target metastasizing cancer cells. Nat. Commun. 8: 15267. doi: 10.1038/ncomms15267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erickson K. L., Hubbard N. E.2010. Fatty acids and breast cancer: the role of stem cells. Prostaglandins Leukot. Essent. Fatty Acids 82: 237–241. doi: 10.1016/j.plefa.2010.02.019 [DOI] [PubMed] [Google Scholar]
- 8.Fan Y., Zhou X., Xia T. S., Chen Z., Li J., Liu Q., Alolga R. N., Chen Y., Lai M. D., Li P., Zhu W., Qi L. W.2016. Human plasma metabolomics for identifying differential metabolites and predicting molecular subtypes of breast cancer. Oncotarget 7: 9925–9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferletta M., Grawé J., Hellmén E.2011. Canine mammary tumors contain cancer stem-like cells and form spheroids with an embryonic stem cell signature. Int. J. Dev. Biol. 55: 791–799. doi: 10.1387/ijdb.113363mf [DOI] [PubMed] [Google Scholar]
- 10.Gatenby R. A., Gillies R. J.2004. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 4: 891–899. doi: 10.1038/nrc1478 [DOI] [PubMed] [Google Scholar]
- 11.Hattori A., Tsunoda M., Konuma T., Kobayashi M., Nagy T., Glushka J., Tayyari F., McSkimming D., Kannan N., Tojo A., Edison A. S., Ito T.2017. Cancer progression by reprogrammed BCAA metabolism in myeloid leukaemia. Nature 545: 500–504. doi: 10.1038/nature22314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawabe M., Baba Y., Tamai R., Yamamoto R., Komori M., Mori T., Takenaka S.2015. Profiling of plasma metabolites in canine oral melanoma using gas chromatography-mass spectrometry. J. Vet. Med. Sci. 77: 1025–1028. doi: 10.1292/jvms.14-0641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kishimoto T. E., Yashima S., Nakahira R., Onozawa E., Azakami D., Ujike M., Ochiai K., Ishiwata T., Takahashi K., Michishita M.2017. Identification of tumor-initiating cells derived from two canine rhabdomyosarcoma cell lines. J. Vet. Med. Sci. 79: 1155–1162. doi: 10.1292/jvms.16-0412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koike H., Zhang R. R., Ueno Y., Sekine K., Zheng Y. W., Takebe T., Taniguchi H.2017. Nutritional modulation of mouse and human liver bud growth through a branched-chain amino acid metabolism. Development 144: 1018–1024. doi: 10.1242/dev.143032 [DOI] [PubMed] [Google Scholar]
- 15.Li J., Condello S., Thomes-Pepin J., Ma X., Xia Y., Hurley T. D., Matei D., Cheng J. X.2017. Lipid desaturation is a metabolic marker and therapeutic target of ovarian cancer stem cells. Cell Stem Cell 20: 303–314.e5. doi: 10.1016/j.stem.2016.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacEwen E. G.1990. Spontaneous tumors in dogs and cats: models for the study of cancer biology and treatment. Cancer Metastasis Rev. 9: 125–136. doi: 10.1007/BF00046339 [DOI] [PubMed] [Google Scholar]
- 17.Michishita M., Akiyoshi R., Suemizu H., Nakagawa T., Sasaki N., Takemitsu H., Arai T., Takahashi K.2012. Aldehyde dehydrogenase activity in cancer stem cells from canine mammary carcinoma cell lines. Vet. J. 193: 508–513. doi: 10.1016/j.tvjl.2012.01.006 [DOI] [PubMed] [Google Scholar]
- 18.Michishita M., Akiyoshi R., Yoshimura H., Katsumoto T., Ichikawa H., Ohkusu-Tsukada K., Nakagawa T., Sasaki N., Takahashi K.2011. Characterization of spheres derived from canine mammary gland adenocarcinoma cell lines. Res. Vet. Sci. 91: 254–260. doi: 10.1016/j.rvsc.2010.11.016 [DOI] [PubMed] [Google Scholar]
- 19.Michishita M., Ezaki S., Ogihara K., Naya Y., Azakami D., Nakagawa T., Sasaki N., Arai T., Shida T., Takahashi K.2014. Identification of tumor-initiating cells in a canine hepatocellular carcinoma cell line. Res. Vet. Sci. 96: 315–322. doi: 10.1016/j.rvsc.2014.01.004 [DOI] [PubMed] [Google Scholar]
- 20.Misdorp W., Else R. W., Hellmen E., Lipscomb T. P.1999. Histological Classification of Mammary Tumors of the dog and the cat. 2nd ser. Vol. VII. Armed Forces Institute of Pathology, Wahington D.C. [Google Scholar]
- 21.Nguyen L. V., Vanner R., Dirks P., Eaves C. J.2012. Cancer stem cells: an evolving concept. Nat. Rev. Cancer 12: 133–143. doi: 10.1038/nrc3184 [DOI] [PubMed] [Google Scholar]
- 22.Nishitani S., Horie M., Ishizaki S., Yano H.2013. Branched chain amino acid suppresses hepatocellular cancer stem cells through the activation of mammalian target of rapamycin. PLoS One 8: e82346. doi: 10.1371/journal.pone.0082346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pang L. Y., Cervantes-Arias A., Else R. W., Argyle D. J.2011. Canine mammary cancer stem cells are radio- and chemo-resistant and exhibit an epithelial-mesenchymal transition phenotype. Cancers (Basel) 3: 1744–1762. doi: 10.3390/cancers3021744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pavlova N. N., Thompson C. B.2016. The emerging hallmarks of cancer metabolism. Cell Metab. 23: 27–47. doi: 10.1016/j.cmet.2015.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sancho P., Barneda D., Heeschen C.2016. Hallmarks of cancer stem cell metabolism. Br. J. Cancer 114: 1305–1312. doi: 10.1038/bjc.2016.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheikh K. D., Khanna S., Byers S. W., Fornace A., Jr., Cheema A. K.2011. Small molecule metabolite extraction strategy for improving LC/MS detection of cancer cell metabolome. J. Biomol. Tech. 22: 1–4. [PMC free article] [PubMed] [Google Scholar]
- 27.Shiraki N., Shiraki Y., Tsuyama T., Obata F., Miura M., Nagae G., Aburatani H., Kume K., Endo F., Kume S.2014. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab. 19: 780–794. doi: 10.1016/j.cmet.2014.03.017 [DOI] [PubMed] [Google Scholar]
- 28.Tamai R., Furuya M., Hatoya S., Akiyoshi H., Yamamoto R., Komori Y., Yokoi S., Tani K., Hirano Y., Komori M., Takenaka S.2014. Profiling of serum metabolites in canine lymphoma using gas chromatography mass spectrometry. J. Vet. Med. Sci. 76: 1513–1518. doi: 10.1292/jvms.14-0043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanabe A., Deguchi T., Sato T., Nemoto Y., Maruo T., Madarame H., Shida T., Naya Y., Ogihara K., Sahara H.2016. Radioresistance of cancer stem-like cell derived from canine tumours. Vet. Comp. Oncol. 14: e93–e101. doi: 10.1111/vco.12110 [DOI] [PubMed] [Google Scholar]
- 30.Utsugi S., Azuma K., Osaki T., Murahata Y., Tsuka T., Ito N., Imagawa T., Okamoto Y.2017. Analysis of plasma free amino acid profiles in canine brain tumors. Biomed. Rep. 6: 195–200. doi: 10.3892/br.2016.825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uyama R., Nakagawa T., Hong S. H., Mochizuki M., Nishimura R., Sasaki N.2006. Establishment of four pairs of canine mammary tumour cell lines derived from primary and metastatic origin and their E-cadherin expression. Vet. Comp. Oncol. 4: 104–113. doi: 10.1111/j.1476-5810.2006.00098.x [DOI] [PubMed] [Google Scholar]
- 32.Waki M., Ide Y., Ishizaki I., Nagata Y., Masaki N., Sugiyama E., Kurabe N., Nicolaescu D., Yamazaki F., Hayasaka T., Ikegami K., Kondo T., Shibata K., Hiraide T., Taki Y., Ogura H., Shiiya N., Sanada N., Setou M.2014. Single-cell time-of-flight secondary ion mass spectrometry reveals that human breast cancer stem cells have significantly lower content of palmitoleic acid compared to their counterpart non-stem cancer cells. Biochimie 107 Pt A: 73–77. doi: 10.1016/j.biochi.2014.10.003 [DOI] [PubMed] [Google Scholar]
- 33.Wang J., Alexander P., Wu L., Hammer R., Cleaver O., McKnight S. L.2009. Dependence of mouse embryonic stem cells on threonine catabolism. Science 325: 435–439. doi: 10.1126/science.1173288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L., Zhang T., Wang L., Cai Y., Zhong X., He X., Hu L., Tian S., Wu M., Hui L., Zhang H., Gao P.2017. Fatty acid synthesis is critical for stem cell pluripotency via promoting mitochondrial fission. EMBO J. 36: 1330–1347. doi: 10.15252/embj.201695417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warburg O.1956. On the origin of cancer cells. Science 123: 309–314. doi: 10.1126/science.123.3191.309 [DOI] [PubMed] [Google Scholar]
- 36.Wilson H., Huelsmeyer M., Chun R., Young K. M., Friedrichs K., Argyle D. J.2008. Isolation and characterisation of cancer stem cells from canine osteosarcoma. Vet. J. 175: 69–75. doi: 10.1016/j.tvjl.2007.07.025 [DOI] [PubMed] [Google Scholar]
- 37.Wolf D. A.2014. Is reliance on mitochondrial respiration a “chink in the armor” of therapy-resistant cancer? Cancer Cell 26: 788–795. doi: 10.1016/j.ccell.2014.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu Z., Wei D., Gao W., Xu Y., Hu Z., Ma Z., Gao C., Zhu X., Li Q.2015. TPO-induced metabolic reprogramming drives metastasis of colorectal cancer CD110+ tumor-initiating cells. Cell Stem Cell 17: 47–59. doi: 10.1016/j.stem.2015.05.016 [DOI] [PubMed] [Google Scholar]
- 39.Yang T., Fang S., Zhang H. X., Xu L. X., Zhang Z. Q., Yuan K. T., Xue C. L., Yu H. L., Zhang S., Li Y. F., Shi H. P., Zhang Y.2013. N-3 PUFAs have antiproliferative and apoptotic effects on human colorectal cancer stem-like cells in vitro. J. Nutr. Biochem. 24: 744–753. doi: 10.1016/j.jnutbio.2012.03.023 [DOI] [PubMed] [Google Scholar]
- 40.Zhang W. C., Shyh-Chang N., Yang H., Rai A., Umashankar S., Ma S., Soh B. S., Sun L. L., Tai B. C., Nga M. E., Bhakoo K. K., Jayapal S. R., Nichane M., Yu Q., Ahmed D. A., Tan C., Sing W. P., Tam J., Thirugananam A., Noghabi M. S., Pang Y. H., Ang H. S., Mitchell W., Robson P., Kaldis P., Soo R. A., Swarup S., Lim E. H., Lim B.2012. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell 148: 259–272. doi: 10.1016/j.cell.2011.11.050 [DOI] [PubMed] [Google Scholar]