Abstract
Background
Nonsyndromic cleft lip with or without cleft palate (NSCL/P) is a common birth defect with complex etiology. One strategy for studying the genetic risk factors of NSCL/P is to consider gene–gene interaction (G × G) among gene pathways having a role in craniofacial development. The present study aimed to investigate the G × G among cell adhesion gene pathway.
Methods
We carried out an interaction analysis of eight genes involved in cell adherens junctions among 806 NSCL/P Chinese case‐parent trios originally recruited for a genome‐wide association study (GWAS). Regression‐based approach was used to test for two‐way G × G interaction, while machine learning algorithm was run for exploring both two‐way and multi‐way interaction that may affect the risk of NSCL/P.
Results
A two‐way ACTN1 × CTNNB1 interaction reached the adjusted significance level. The single nucleotide polymorphisms pair composed of rs17252114 (CTNNB1) and rs1274944 (ACTN1) yielded a p value of .0002, and this interaction was also supported by the logic regression algorithm. Higher order interactions involving ACTN1, CTNNB1, and CDH1 were picked out by logic regression, suggesting a potential role in NSCL/P risk.
Conclusion
This study suggests for the first time evidence of both two‐way and multi‐way G × G interactions among cell adhesion genes contributing to the NSCL/P risk.
Keywords: adherens junctions, case‐parent trios, cell adhesion, gene–gene interaction, nonsyndromic cleft lip with or without cleft palate
1. INTRODUCTION
Nonsyndromic cleft lip with or without cleft palate (NSCL/P) is a common congenital malformation worldwide. This disorder is a result of facial tissues not joining properly during development, with the reason of which, in most cases, is unknown (Leslie & Marazita, 2013). NSCL/P has a complex and heterogeneous etiology, and the well‐recognized casual factors so far are still limited despite tens of suspicious genetic variants and environmental risk factors.
Craniofacial development is an ordered sequence of events involving for instance structure growth, elevation, attachment, and fusion (Marazita, 2012). Dysfunction of cell adhesion during embryo development can prevent the necessary process and give rise to a cleft (Meng, Bian, Torensma, & Von den Hoff, 2009). Adherens junctions are key structures for cell‐cell adhesion (Harris & Tepass, 2010). At adherens junctions, cadherin and nectins serve as essential cell adhesion molecules (CAMs), associated with cytoskeleton to physically link cells, and with intracellular signaling molecules to transduce signals (KEGG map04520). In this context, genes encoding proteins involved in the formation of adherens junctions may influence the risk of NSCL/P. Although a few studies conducted in a candidate framework suggested significant association between cell adhesion genes and oral clefts (Oner & Tastan, 2016; Rafighdoost et al., 2013; Song et al., 2017), none of the genes showed up in any genome‐wide association studies (GWASs). We assume there could be other forms of genetic effect which are statistically detectable in this pathway.
For a complex disease like NSCL/P, candidate genes may interact with one another in biological pathways which can be identified through statistical tests (Eichler et al., 2010). Logistic regression is the standard way of modeling two‐way gene–gene (G × G) interaction. For higher order interaction though, parametric model can easily become daunting as the number of single nucleotide polymorphisms (SNPs) increase. New methods are emerging and being applied to genetic studies with the purpose of testing G × G interaction, among which machine learning approaches represent a major category (Cordell, 2009). Machine learning algorithms have the superior feature of exploring potential nonlinear relationships, and the core idea of data‐driven decision enables these methods to approach the realistic biological system (Cordell, 2009; Upstill‐Goddard, Eccles, Fliege, & Collins, 2012). Logic regression (LR) is a machine learning method to model an outcome of interest with interaction between binary covariates (Kooperberg, Ruczinski, LeBlanc, & Hsu, 2001; Ruczinski, Kooperberg, & LeBlanc, 2003). LR has been applied successfully in the studies with preselected candidate genes for various phenotypes, such as bladder cancer (Andrew et al., 2008), schizophrenia (Nicodemus et al., 2010), and cardiovascular disease (Enquobahrie et al., 2008). Recently, researchers applied LR to NSCL/P case‐parent trios and found convincing evidence of interaction between markers in WNT pathway genes and in GWAS‐confirmed regions (Li et al., 2015).
Here, we reported a G × G interaction study of genes involved in adherens junctions using both regression‐based method and machine learning algorithm among 806 Chinese case‐parent trios, with the particular purpose of evaluating both two‐way and multi‐way interactions that may affect the risk of NSCL/P.
2. MATERIAL AND METHODS
2.1. Editorial policies and ethical considerations
Research protocols were approved by Institutional Review Boards (IRBs) at Johns Hopkins Bloomberg School of Public Health. Informed consent was obtained from parents of each participating family.
2.2. Samples
We used data from an established consortium where a GWAS of NSCL/P was conducted (Beaty et al., 2010). A total of 806 Chinese case‐parent trios were included in the present study. Detailed information was discussed previously (Beaty et al., 2010). The datasets analyzed in the current study are available from dbGaP: phs000094.v1.p1.
2.3. Genotyping and quality control
The genome‐wide genotyping was performed by The Center for Inherited Disease Research (CIDR) at the Johns Hopkins University using Illumina Human610‐Quad v.1_B Bead Chip. From the original set of GWAS SNPs, we extracted 254 SNPs in or near eight genes having a function related to adherens junctions (Table 1, Figure S1, Table S1). A SNP was dropped during data quality control if its missing rate was >5%, or its minor allele frequency (MAF) was <0.1, or Mendelian errors occurred >5% of all trios, or it substantially deviated from Hardy–Weinberg equilibrium (HWE, p < .001). Single nucleotide polymorphisms with MAF <.1 were dropped due to limited power for detecting their interaction. Eighty SNPs and one SNP were dropped for unsatisfactory MAF and HWE, respectively, leaving 173 SNPs qualified for further analysis.
Table 1.
Genes involved in cell adherens junctions
| Chromosome | Gene full name | Gene symbol | Number of SNPs passing QC |
|---|---|---|---|
| 16q22 | Cadherin 1 | CDH1 | 15 |
| 3p22 | Catenin beta 1 | CTNNB1 | 45 |
| 11q23 | Nectin cell adhesion molecule 1 | NECTIN1 | 13 |
| 19q13 | Nectin cell adhesion molecule 2 | NECTIN2 | 10 |
| 3q13 | Nectin cell adhesion molecule 3 | NECTIN3 | 34 |
| 14q24 | Actinin alpha 1 | ACTN1 | 39 |
| 10q22 | Vinculin | VCL | 11 |
| 4q25 | Lymphoid enhancer binding factor 1 | LEF1 | 6 |
3. STATISTICAL ANALYSES
3.1. Conditional logistic regression
A modified form of conditional logistic regression, as proposed by Cordell (Cordell, 2002; Cordell, Barratt, & Clayton, 2004), is the standard method of testing two‐way G × G interaction in case‐parent trio data, which compares the genotypes at two loci in a case and its matched pseudocontrols generated from untransmitted alleles under all parental mating types. Only interactions between two SNPs from different genes were analyzed. p values were generated from a 4df likelihood ratio test comparing the likelihood of the full model to the likelihood of the reduced model without interaction terms. Since Cordell's method is technically unable to estimate the effect size, we applied another model, Logit(p) = β0 + β1 SNP1+β2 SNP2 + β3 SNP1 × SNP2, under an additive inheritance pattern to estimate relative risks (RR). To appreciate the exploratory nature of this interaction analysis, we relaxed the Bonferroni threshold (.05 divided by number of test) for the conditional logistic regression to a more liberal threshold, .05 divided by the number of SNPs included (.05/173), so that potential signals will have a chance to be taken into further studies.
3.2. Logic regression
Logic regression (LR) is a method of delineating higher order interactions especially suitable for SNP data (Kooperberg et al., 2001; Ruczinski et al., 2003). Basically, it was derived to find Boolean combinations of binary predictors that are associated with an outcome, as embedded in a regression framework. Single nucleotide polymorphisms are connected using “AND,” “OR,” “NOT” in these combinations, also referred to as logic expression, that can be interpreted as ‘‘risk factors.” One particular SNP is denoted by two binary variable SNPD and SNPR, coding for the dominant and recessive effect, respectively. To be specific, homozygous genotype with two copies of minor allele at a SNP corresponds to SNPD = SNPR = 1 and genotype with two copies of major allele corresponds to SNPD = SNPR = 0. Heterozygous genotype is specified by SNPD = 1 and SNPR = 0. The model search of LR is carried out using a stochastic search algorithm called simulated annealing, where the “goodness‐of‐fit” of a model is evaluated by a score function suited for the underlying model type.
LR was extended to studies of case‐parent trios with affected probands to detect disease‐associated SNP interactions of any order (Li et al., 2010). Conditional logistic regression is embedded as the underlying model, with model performance measured by the deviance from the conditional regression. In one run of LR, the logic expression fitting in the model with the lowest deviance (referred to as “score” in the text below) is picked up to be the optimal expression. Two types of permutation are implemented in trio LR. The global permutation test checks whether there is a signal in the data, and the conditional permutation helps to determine the best model with optimal size, that is, the number of predictors allowed to be in the logic expression.
The LR cannot fully differentiate linkage disequilibrium (LD) from interaction among markers (Kooperberg & Ruczinski, 2005; Li et al., 2015). We generated a pruned set of SNPs with reduced LD in Haploview v4.2. These tag SNPs were determined in a way that alleles to be captured are correlated with one tag SNP at r 2 > .1 as estimated based on the founder population. Single nucleotide polymorphisms reaching the significance level in conditional logistic regressions were forced into the list of tag SNPs. This procedure generated 24 tag SNPs with all pairwise r 2 < .1 except for one pair with r 2 = .3 (Figure S2). We specified those two SNPs to be in a LD block so that LR would not pick them up together.
Using this set of tag SNP, we first run the global permutation test allowing a maximum of model size 8. Then the optimal model size was decided via sequential permutation tests conditional on increasing model size from 1 with an increment of 1. Due to the computational burden of this procedure, 500 permutations were performed for each model. Both conditional logistic regression and trio LR are implemented in the “trio” R package (version 3.8.0, available at http://www.bioconductor.org).
4. RESULTS
4.1. Conditional logistic regression
Beaty et al., 2010 have shown that none of the SNPs included in the current study had significant main effect with NSCL/P. Using the Cordell's method, we identified ACTN1 × CTNNB1 interaction reaching the prespecified significance level. The pair composed of rs17252114 (CTNNB1) and rs1274944 (ACTN1) gave a p‐value of .0002. We calculated the size of interaction effect under the additive model described above and showed a negative interaction. For individuals carrying the reference genotype (CC) at rs17252114, the RR of carrying one risk allele (C) at rs1274944 was 2.04 (95% CI: 1.38 to 3.02), while the RR of rs1274944 decreased to 1.42 (95% CI: 1.16 to 1.74) for individuals carrying one copy of risk allele (T) at rs17252114. No other SNP × SNP interaction was significant after correction for multiple tests.
4.2. Trio LR
In the global permutation test, only 1 of 500 permutations yielded a score that was better (“better” means provide a better goodness‐of‐fit) than that of the best scoring model on the original data, which indicated G × G signals in the data. See Figure S3 for the histogram of the 500 global permutation scores. Figure 1 showed the results from conditional permutation tests. The proportion of permutation scores better than the score of the overall best model was claimed as a p value, as shown in the top right corner of each panel, indicating evidence against the null hypothesis that any improvement of the quality of the fit beyond the current model size is due to noise. According to the sequential permutation procedure, 4‐SNP or 5‐SNP model was suggested to be optimal for this data. The best Boolean combinations of SNPs picked by LR algorithm under different model sizes were displayed in Table 2. Since LR identified the combinations of SNPs that were associated with increased or decreased disease risk, the best 4‐SNP logic expression, for example, allowed then for an explanation that an individual was at lower risk of NSCL/P if he or she carried at least one minor allele at rs409228(CTNNB1) and at least one minor allele at s1274944(ACTN1) and at the most one minor allele at rs4783676(CDH1), or at least one minor allele at rs10490822(CTNNB1).
Figure 1.
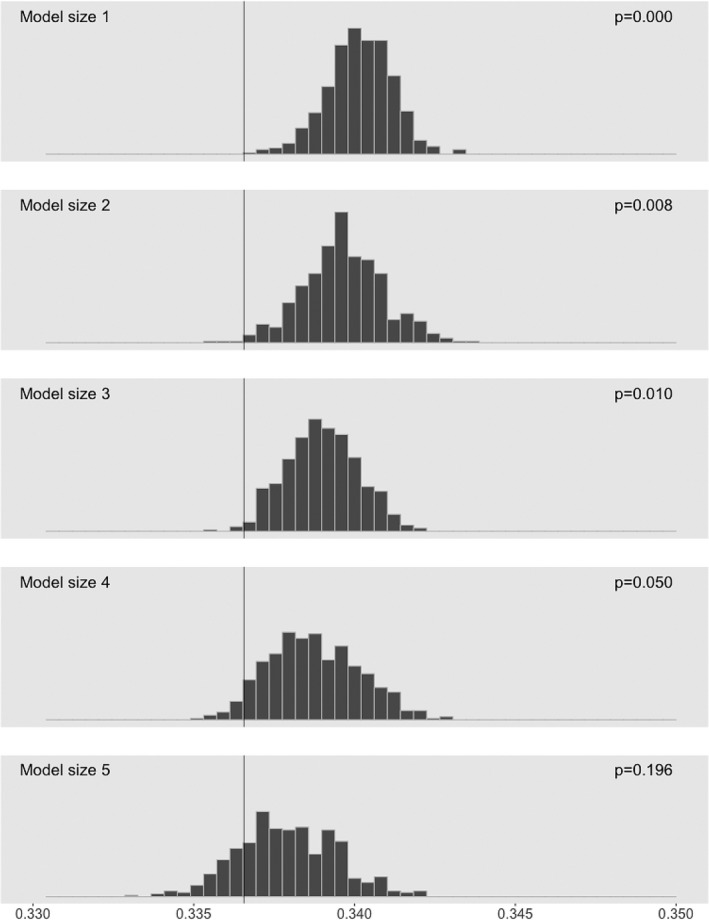
Histograms of permutation scores from conditional permutation tests for model size determination. Smaller scores corresponded to better performance of the model. Empirical p values of the permutation tests are showed in the top right. The solid line at score 0.3366 indicated the overall best score on the nonpermuted data. Since the overall best score, 0.3366, was unlikely sampled from the distribution until we condition on the model of size 4 or 5, we considered a model with 4 or 5 SNPs to be optimal for the current data
Table 2.
The optimal logic expressions identified by LR among 806 Chinese trios
| Model size | Optimal logic term |
|---|---|
| 2 | {NOT rs6802872R (CTNNB1)} AND rs10490822D (CTNNB1) |
| 3 |
{rs17252114D
(CTNNB1) AND rs1274944D
(ACTN1)} OR rs10490822D (CTNNB1) |
| 4 |
{rs409228D
(CTNNB1) AND rs1274944D
(ACTN1) AND {NOT rs4783676R
(CDH1)}} OR rs10490822D (CTNNB1) |
| 5 |
{NOT rs10490822D
(CTNNB1)} AND {{{NOT rs409228D (CTNNB1)} AND {NOT rs174213D (ACTN1)}} OR {rs4783676R (CDH1) OR {NOT rs1274944D (ACTN1)}}} |
Superscript D denoted that at least one minor allele existed at a particular SNP, and superscript R meant a particular SNP had two copies of minor allele.
5. DISCUSSION
For human complex diseases, testing for G × G interaction can make better use of the genomic structure and shed more light on the disease etiology. We evaluated the G × G interaction within cell adhesion‐related genes among 806 Chinese trios where the probands were affected with NSCL/P. Our conditional logistic regression revealed a two‐way ACTN1 × CTNNB1 interaction, which was also supported by the logic regression algorithm. Besides, higher order interactions involving ACTN1, CTNNB1, and CDH1 were suggested to confer elevated or reduced risk for NSCL/P.
For statistically identified interactions between candidate genes, proving functional basis remains a significant challenge. Pathway‐based analysis is therefore biologically appealing. We preselected a group of genes having molecular cross talk and related function—genes bound up with adherens junctions. Development of the lip and palate involves spatiotemporally coordinated growth and morphogenesis of initially separate primordia (Meng et al., 2009). This highly complicated process requires regulatable, dynamic cell adhesion to surrounding, namely series of the formation and disruption of adhesion at the cell front or rear (Meng et al., 2009). Any disturbance in these processes may result in human clefting. Cell adhesion also interplays with WNT signaling pathway which was reported to be associated with human oral cleft (Heuberger & Birchmeier, 2010; Liu & Millar, 2010).
E‐cadherins (CDH1) are primary CAMs within adherens junctions. In human embryos, E‐cadherin was found to be highly expressed at the fourth and the fifth weeks in the frontonasal prominence, and at the sixth week in the lateral and medial nasal prominences (Frebourg et al., 2006). A few studies showed that pathogenic mutations in CDH1 played a role in the etiology of oral clefts (Brito et al., 2015; Bureau et al., 2014). Alpha‐actinin (ACTN1) and beta‐catenin (CTNNB1) are integral elements involved in regulation and coordination of cell adhesion. So far, little evidence exists for a relation between these two genes and oral clefts. At adherens junctions, E‐cadherins have physical interplay with CTNNB1 (Harris & Tepass, 2010), and one possible biological explanation of the statistical interaction between these two molecules would be that a change in one protein may somehow compensate for the defective function of a second mutant protein. CTNNB1 and ACTN1 were not in direct cross talk, and the mechanism of epistasis could be that the substitution of alleles occurring in either of these two genes alone is enough to block the pathway so that mutant alleles at both loci would not have an additive effect. Three‐way and higher order interactions may involve even more complex biological mechanism with various kinds of potential relationship interweaving together. It is not surprising that all the SNPs picked out either by Cordell's method or LR algorithm were noncoding variants which are less likely to give rise to a different gene product. But chances are these SNPs or the surrounding SNPs in high LD may still have some regulatory function on gene expression. For example, the Genotype‐Tissue Expression (GTEx) project identified rs4783676 (appeared in the best 4‐ and 5‐SNP logic expressions) as a cis‐eQTL for CDH1 in the whole blood tissue (GTEx Consortium, 2015). See Table S2 for the potential regulatory function of all SNPs included in Table 2 annotated by HaploReg v4.1 (Ward & Kellis, 2012). The interactions among CTNNB1, ATCN1, and CDH1 suggested in our study would therefore be a reasonable starting point for functional assay testing epistasis.
G × G interaction is being revealed to be a crucial player in the etiology of oral clefts. Some reported interactions include BHMT/BHMT2 × DMGDH (Wang et al., 2018), WNT5B × MAFB (Li et al., 2015), IRF6 × MAFB (Xiao et al., 2016), MSX1 × TGFB3 (Suazo, Luis Santos, Colombo, & Pardo, 2018), BMP4 × IRF6 (Blanco, Colombo, Pardo, & Suazo, 2017), and so on. Interaction analyses are mostly considered in a candidate framework so far, with the candidates often being genes near GWAS hits. No SNPs in this study, however, showed a significant main effect, yet the analyses here still captured the potential interaction signals, which shows the advantage of regularly incorporating interaction analysis into association study even without significant main genetic effect. We showed that two SNPs in CTNNB1 and ACTN1 interact with each other in an antagonistic manner where the presence of the risk allele at one SNP alleviates the risk‐increasing effect of the risk allele at the other SNP, with the RR dropped moderately from 2.04 to 1.42. This type of interaction effect for oral clefts was reported before. For example, Wang et al even reported reversed direction of association of one SNP when stratified by the genotype at the other SNP (Wang et al., 2018). The two SNPs involved in CTNNB1 × ACTN1 are both noncoding variants, and how the alleles affect gene products and behave antagonistically are worth exploring in future function study.
Two types of statistical methods were employed to investigate G × G interactions in our study. A significant interaction identified by conditional logistic regression represents departure from expected effects under a multiplicative model for alleles at two genes. LR implies interactions in a way that certain combination of SNPs is associated with a higher or lower disease risk. The results of these two approaches were to some extent in agreement with each other. Marker rs17252114 (CTNNB1) and/or rs1274944 (ACTN1), exhibiting significant interactive effect in the logistic regression, appeared in the best 3‐, 4‐, and 5‐SNP logic expressions as well (Table 2). In particular, these two SNPs were connected by the Boolean operator AND in the best model of size 3, which fitted in well with the understanding of “traditional” interaction that the co‐existence of two factors confers altered risk, as what underlies the interpretation of regression‐recognized interaction. Hopefully, in future studies these findings can be taken further to increase our understanding of the molecular mechanism of oral clefts.
One desirable property of LR algorithm when compared with other machine learning methods is its ease of interpretation of selected models, without putting so much emphasis on prediction (Ruczinski, Kooperberg, & LeBlanc, 2004). Traditional regression‐based approaches are limited in its ability to deal with high‐dimensional data. In this context, our study presented an example of multi‐way interaction analysis workflow with computational burden fairly acceptable. LR also provides an advance over traditional statistical methods in that it can search for more biologically plausible forms of interaction. Thus, it gives more confidence for our study in suggesting the future study of ACTN1, CTNNB1, and CDH1 under experimental design. Moreover, multiple testing is not a big issue in LR, whereas it is of distressing concern in regression‐based methods. The number of potential hypotheses and the incurred multiple tests burden can greatly limit the statistical power (Cordell, 2002), even though our study included relatively large number of trios. We relaxed the Bonferroni threshold for the conditional logistic regression in which 0.05 was divided by the number of SNPs included instead of the number of tests undertaken, as we do not want to miss any potential signal. The major drawbacks of LR in the context of the present study are its incapability to give p value and effect size estimate for individual combination of SNPs, which are of the most interest to researchers, and the challenge of LD that forced the usage of a pruned list of SNPs.
In summary, our study demonstrated both two‐way and multi‐way G × G interactions contributing to NSCL/P risk among ACTN1, CTNNB1, and CDH1, which are involved in cell adhesion. The interactions were not previously reported, and are in need of independent replication. The next challenge also relates to verifying the genotype‐phenotype relationship and providing solid justification that these statistically identified interactions are of importance in a functional perspective.
CONFLICTS OF INTEREST
The authors declare no conflict of interests.
Supporting information
ACKNOWLEDGMENTS
We sincerely thank all participating families at each recruitment site who made this work possible. Every effort of the dedicated clinical, field, and laboratory staff are appreciated. We thank the Smile Train Foundation for supporting cleft research in China. The International Cleft Consortium including genotyping and analysis was supported by the National Institute for Dental and Craniofacial Research through U01‐DE‐018993; ‘‘International Consortium to Identify Genes &Interactions Controlling Oral Clefts'', 2007–2009; We acknowledge the invaluable contributions of X. Ye (Wuhan University), S. Huang (Peking Union Medical College), V. Yeow (KKWCH), S. S. Chong (National University of Singapore), Y. H. Wu‐Chou (Chang Gung Memorial Hospital), P. K. Chen (Chang Gung Memorial Hospital), B. Shi (Sichuan University), and S. H. Jee (Yonsei University). This research was also supported by the National Natural Science Foundation of China (81102178, 81573225, PI: Tao Wu) and Beijing Natural Science Foundation of China (7172115, PI: Tao Wu).
Liu D, Wang M, Yuan Y, et al. Gene–gene interaction among cell adhesion genes and risk of nonsyndromic cleft lip with or without cleft palate in Chinese case‐parent trios. Mol Genet Genomic Med. 2019;7:e872 10.1002/mgg3.872
Funding information
The study was supported by the National Institute for Dental and Craniofacial Research through U01‐DE‐018993; “International Consortium to Identify Genes &Interactions Controlling Oral Clefts”, 2007–2009. This research was also supported by the National Natural Science Foundation of China (81102178, 81573225, PI: Tao Wu) and Beijing Natural Science Foundation of China (7172115, PI: Tao Wu).
REFERENCES
- Andrew, A. S. , Karagas, M. R. , Nelson, H. H. , Guarrera, S. , Polidoro, S. , Gamberini, S. , … Matullo, G. (2008). DNA repair polymorphisms modify bladder cancer risk: A multi‐factor analytic strategy. Human Heredity, 65, 105–118. 10.1159/000108942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty, T. H. , Murray, J. C. , Marazita, M. L. , Munger, R. G. , Ruczinski, I. , Hetmanski, J. B. , … Scott, A. F. (2010). A genome‐wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nature Genetics, 42, 525–529. 10.1038/ng.580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco, R. , Colombo, A. , Pardo, R. , & Suazo, J. (2017). Haplotype‐based gene‐gene interaction of bone morphogenetic protein 4 and interferon regulatory factor 6 in the etiology of non‐syndromic cleft lip with or without cleft palate in a Chilean population. European Journal of Oral Sciences, 125, 102–109. 10.1111/eos.12332 [DOI] [PubMed] [Google Scholar]
- Brito, L. A. , Yamamoto, G. L. , Melo, S. , Malcher, C. , Ferreira, S. G. , Figueiredo, J. , … Passos‐Bueno, M. R. (2015). Rare variants in the epithelial cadherin gene underlying the genetic etiology of nonsyndromic cleft lip with or without cleft palate. Human Mutation, 36, 1029–1033. 10.1002/humu.22827 [DOI] [PubMed] [Google Scholar]
- Bureau, A. , Parker, M. M. , Ruczinski, I. , Taub, M. A. , Marazita, M. L. , Murray, J. C. , … Beaty, T. H. (2014). Whole exome sequencing of distant relatives in multiplex families implicates rare variants in candidate genes for oral clefts. Genetics, 197, 1039–1044. 10.1534/genetics.114.165225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell, H. J. (2002). Epistasis: What it means, what it doesn't mean, and statistical methods to detect it in humans. Human Molecular Genetics, 11, 2463–2468. 10.1093/hmg/11.20.2463 [DOI] [PubMed] [Google Scholar]
- Cordell, H. J. (2009). Detecting gene‐gene interactions that underlie human diseases. Nature Review Genetics, 10, 392–404. 10.1038/nrg2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell, H. J. , Barratt, B. J. , & Clayton, D. G. (2004). Case/pseudocontrol analysis in genetic association studies: A unified framework for detection of genotype and haplotype associations, gene‐gene and gene‐environment interactions, and parent‐of‐origin effects. Genetic Epidemiology, 26, 167–185. 10.1002/gepi.10307 [DOI] [PubMed] [Google Scholar]
- Eichler, E. E. , Flint, J. , Gibson, G. , Kong, A. , Leal, S. M. , Moore, J. H. , & Nadeau, J. H. (2010). Missing heritability and strategies for finding the underlying causes of complex disease. Nature Review Genetics, 11, 446–450. 10.1038/nrg2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enquobahrie, D. A. , Smith, N. L. , Bis, J. C. , Carty, C. L. , Rice, K. M. , Lumley, T. , … Psaty, B. M. (2008). Cholesterol ester transfer protein, interleukin‐8, peroxisome proliferator activator receptor alpha, and toll‐like receptor 4 genetic variations and risk of incident nonfatal myocardial infarction and ischemic stroke. American Journal of Cardiology, 101, 1683–1688. 10.1016/j.amjcard.2008.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frebourg, T. , Oliveira, C. , Hochain, P. , Karam, R. , Manouvrier, S. , Graziadio, C. , … Seruca, R. (2006). Cleft lip/palate and CDH1/E‐cadherin mutations in families with hereditary diffuse gastric cancer. Journal of Medical Genetics, 43, 138–142. 10.1136/jmg.2005.031385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium (2015). Human genomics. The Genotype‐Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science, 348, 648–660. 10.1126/science.1262110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, T. J. , & Tepass, U. (2010). Adherens junctions: From molecules to morphogenesis. Nature Reviews Molecular Cell Biology, 11, 502–514. 10.1038/nrm2927 [DOI] [PubMed] [Google Scholar]
- Heuberger, J. , & Birchmeier, W. (2010). Interplay of cadherin‐mediated cell adhesion and canonical wnt signaling. Cold Spring Harbor Perspectives in Biology, 2, a002915 10.1101/cshperspect.a002915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooperberg, C. , & Ruczinski, I. (2005). Identifying interacting SNPs using Monte Carlo logic regression. Genetic Epidemiology, 28, 157–170. 10.1002/gepi.20042 [DOI] [PubMed] [Google Scholar]
- Kooperberg, C. , Ruczinski, I. , LeBlanc, M. L. , & Hsu, L. (2001). Sequence analysis using logic regression. Genetic Epidemiology, 21, S626–S631. 10.1002/gepi.2001.21.s1.s626 [DOI] [PubMed] [Google Scholar]
- Leslie, E. J. , & Marazita, M. L. (2013). Genetics of cleft lip and cleft palate. American Journal of Medical Genetics, 163, 246–258. 10.1002/ajmg.c.31381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q. , Fallin, M. D. , Louis, T. A. , Lasseter, V. K. , McGrath, J. A. , Avramopoulos, D. , … Ruczinski, I. (2010). Detection of SNP‐SNP interactions in trios of parents with schizophrenic children. Genetic Epidemiology, 34, 396–406. 10.1002/gepi.20488 [DOI] [PubMed] [Google Scholar]
- Li, Q. , Kim, Y. , Suktitipat, B. , Hetmanski, J. B. , Marazita, M. L. , Duggal, P. , … Bailey‐Wilson, J. E. (2015). Gene‐gene interaction among WNTGenes for Oral Cleft in Trios. Genetic Epidemiology, 39, 385–394. 10.1002/gepi.21888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, F. , & Millar, S. E. (2010). Wnt/beta‐catenin signaling in oral tissue development and disease. Journal of Dental Research, 89, 318–330. 10.1177/0022034510363373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazita, M. L. (2012). The evolution of human genetic studies of cleft lip and cleft palate. Annual Review of Genomics and Human Genetics, 13, 263–283. 10.1146/annurev-genom-090711-163729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, L. , Bian, Z. , Torensma, R. , & Von den Hoff, J. W. (2009). Biological mechanisms in palatogenesis and cleft palate. Journal of Dental Research, 88, 22–33. 10.1177/0022034508327868 [DOI] [PubMed] [Google Scholar]
- Nicodemus, K. K. , Callicott, J. H. , Higier, R. G. , Luna, A. , Nixon, D. C. , Lipska, B. K. , … Weinberger, D. R. (2010). Evidence of statistical epistasis between DISC1, CIT and NDEL1 impacting risk for schizophrenia: Biological validation with functional neuroimaging. Human Genetics, 127, 441–452. 10.1007/s00439-009-0782-y [DOI] [PubMed] [Google Scholar]
- Oner, D. A. , & Tastan, H. (2016). Identification of novel variants in the pvrl1 gene in patients with nonsyndromic cleft lip with or without cleft palate. Genetic Testing and Molecular Biomarkers, 20, 269–272. 10.1089/gtmb.2015.0276 [DOI] [PubMed] [Google Scholar]
- Rafighdoost, H. , Hashemi, M. , Narouei, A. , Eskanadri‐Nasab, E. , Dashti‐Khadivaki, G. , & Taheri, M. (2013). Association between CDH1 and MSX1 gene polymorphisms and the risk of nonsyndromic cleft lip and/or cleft palate in a southeast Iranian population. The Cleft Palate‐Craniofacial Journal, 50, e98–e104. 10.1597/12-144 [DOI] [PubMed] [Google Scholar]
- Ruczinski, I. , Kooperberg, C. , & LeBlanc, M. L. (2003). Logic regression. Journal of Computational and Graphical Statistics, 12, 475–511. 10.1198/1061860032238 [DOI] [Google Scholar]
- Ruczinski, I. , Kooperberg, C. , & LeBlanc, M. L. (2004). Exploring interactions in high‐dimensional genomic data: An overview of Logic Regression, with applications. Journal of Multivariate Analysis, 90, 178–195. 10.1016/j.jmva.2004.02.010 [DOI] [Google Scholar]
- Song, H. , Wang, X. , Yan, J. , Mi, N. A. , Jiao, X. , Hao, Y. , … Gao, Y. (2017). Association of single‐nucleotide polymorphisms of CDH1 with nonsyndromic cleft lip with or without cleft palate in a northern Chinese Han population. Medicine, 96, e5574 10.1097/MD.0000000000005574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suazo, J. , Luis Santos, J. , Colombo, A. , & Pardo, R. (2018). Gene‐gene interaction for nonsyndromic cleft lip with or without cleft palate in Chilean case‐parent trios. Genetic Testing and Molecular Biomarkers, 91, 91–95. 10.1016/j.archoralbio.2018.04.009 [DOI] [PubMed] [Google Scholar]
- Upstill‐Goddard, R. , Eccles, D. , Fliege, J. , & Collins, A. (2012). Machine learning approaches for the discovery of gene‐gene interactions in disease data. Briefings in Bioinformatics, 14, 251–260. 10.1093/bib/bbs024 [DOI] [PubMed] [Google Scholar]
- Wang, P. , Wu, T. , Schwender, H. , Wang, H. , Shi, B. , Wang, Z. Q. , … Beaty, T. H. (2018). Evidence of interaction between genes in the folate/homocysteine metabolic pathway in controlling risk of non‐syndromic oral cleft. Oral Diseases, 24, 820–828. 10.1111/odi.12831 [DOI] [PubMed] [Google Scholar]
- Ward, L. D. , & Kellis, M. (2012). HaploReg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Research, 40, D930–D934. 10.1093/nar/gkr917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, Y. , Taub, M. A. , Ruczinski, I. , Begum, F. , Hetmanski, J. B. , Schwender, H. , … Beaty, T. H. (2016). Evidence for SNP‐SNP interaction identified through targeted sequencing of cleft case‐parent trios. Genetic Epidemiology, 41, 1–7. 10.1002/gepi.22023 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


