Abstract
Many different species of gram-negative bacteria are associated with infection in the lung, causing exacerbations of chronic obstructive pulmonary disease, cystic fibrosis (CF), and ventilator-associated pneumonias. These airway pathogens must adapt to common host clearance mechanisms that include killing by antimicrobial peptides, antibiotics, oxidative stress, and phagocytosis by leukocytes. Bacterial adaptation to the host is often evident phenotypically, with increased extracellular polysaccharide production characteristic of some biofilm-associated organisms. Given the relatively limited repertoire of bacterial strategies to elude airway defenses, it seems likely that organisms sharing the same ecological niche might also share common strategies to persistently infect the lung. In this review, we will highlight some of the major factors responsible for the adaptation of Pseudomonas aeruginosa to the lung, addressing how growth in biofilms enables persistent infection, relevant to, but not limited to, the pathogenesis of infection in CF. In contrast, we will discuss how carbapenem-resistant Klebsiella pneumoniae evade immune clearance, an organism often associated with ventilator-associated pneumonia and health-care-acquired pneumonias, but not a typical pathogen in CF.
Keywords: Pseudomonas aeruginosa, Klebsiella pneumoniae, Biofilm, Bacterial adaptation, Immune evasion, Cystic fibrosis, Bacterial infection, Inflammasome, Pathogen-associated molecular patterns, Reactive oxygen species
Introduction
The gram-negative airway pathogens, Pseudomonas aeruginosa and Klebsiella pneumoniae, are opportunists that are inadvertently inhaled and, upon finding a conducive environment, rapidly adjust gene expression to proliferate. This entails major changes: the upregulation of genes involved in the acquisition of newly available carbon sources, the scavenging of essential nutrients, such as iron, and the positive selection for clones resistant to immune clearance mechanisms. Mutations in genes that potentially impact both major processes would likely be under significant positive selection. A substantial literature has been amassed detailing how many organisms form biofilms in pulmonary and endovascular infections as well as a mode of infection on abiotic surfaces. Many features of biofilms contribute to bacterial persistence and the survival of the bacterial community within the lung. These processes are highly relevant to numerous forms of infection, including colonization of endotracheal tubes, airways, and intravascular and urinary catheters. In addition, recent findings suggest host immunity to play a key role during bacterial adaptation, which, in conjunction with the ability of these pathogens to adapt a biofilm lifestyle, are a matter of discussion in this review.
Gram-Negative Bacteria and Lung Inflammation: Cystic Fibrosis
Interactions of bacterial pathogens and innate immune signaling have been well studied but not fully understood in cystic fibrosis (CF) [1]. CF is a genetic disease initially linked to the Caucasian population and now recognized in recent studies in much of the rest of the world population [2, 3, 4, 5]. CF is produced by mutations in the CF transmembrane conductance regulator (CFTR), a chloride channel at the surface of epithelial and immune cells [1]. As hydration and mucin viscosity in the airways is dependent on the ionic equilibrium across the epithelial barrier, lack of CFTR function reduces airway mucus fluidity, trapping inhaled bacteria [6]. Although CFTR mutations also have negative effects on the function of other organs, such as the pancreas, CF patients receiving nutritional supplementation are able to manage these limitations [7]. However, the inflammation and chronic infection associated with CF lung disease limits quality of life and ultimately longevity [8].
During childhood, most CF patients are colonized by both P. aeruginosa and Staphylococcus aureus. However, later in adulthood, the primary pathogen causing pulmonary infection in CF is Pseudomonas [1]. Many studies have addressed why P. aeruginosa is the major pathogen in CF. The most commonly accepted hypothesis suggests that physiological abnormalities linked to CFTR mutations confer advantages to P. aeruginosa over others pathogens. These advantages include mucus viscosity, excessive production of reactive oxygen species (ROS), impaired autophagy, reduced airway acidity, and accumulation of ceramides, all factors that contribute to the genomic and metabolic plasticity of P. aeruginosa and its ability to colonize the airways [9]. Whether the exaggerated inflammatory response seen in CF facilitates the colonization and then chronicity of P. aeruginosa infection over other bacteria is not well understood, although recent findings suggest that mutant CFTR directly influences inflammatory signaling, and this favors P. aeruginosa over other respiratory pathogens, such as S. aureus and K. pneumoniae [10].
In this work, we review the recent literature detailing how P. aeruginosa adapts to the CF lung through interactions with host immunity and the metabolic environment of the airways. We also highlight how the mechanisms of patho-adaptation that are operative for P. aeruginosa are conserved among lung pathogens by reviewing the pathways by which K. pneumoniae, another clinically relevant multidrug-resistant (MDR) gram-negative pathogen, causes airway invasion.
P. aeruginosa Adaptation to the Airways through Biofilm Formation
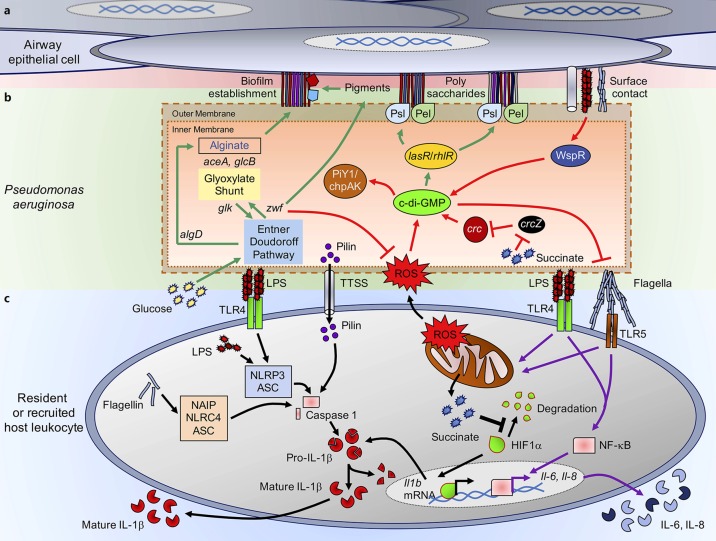
Biofilm formation is a highly conserved mechanism of bacterial adaptation, which has been well studied in the setting of CF, and endovascular and foreign body infections, and common mechanisms pertain to many clinical settings. P. aeruginosa responds to a surface through the activation of WspR and the cyclic (c)-di-GMP cascade [11, 12, 13] (Fig. 1). The c-di-GMP cascade controls expression of over 500 genes [11, 14]. Initial probiofilm stimuli can be chemotactic or metabolic. These include operons that regulate twitching motility through the upregulated expression of type IV pili, as well as by decreasing swimming motility through suppression of flagellin gene expression. WspR signaling is critical in the expression of pslA and pelA, genes that encode the structural components that lead to biofilm formation [15] (Fig. 1a, b). In the process of pulmonary colonization and chronic infection, there is selection for both small colony variants and mucoid variants. Organisms generally produce either a Pel or Psl predominant biofilm [16]. The Pel-directed biofilm, which refers to the formation of Pseudomonas pellicles on the surface of cultures, is a polymer of N-acetyl-galactosamine and N-acetyl-glucosamine [15, 17]. The Psl operon results in the formation of a biofilm matrix composed of mannose, rhamnose, and glucose [15, 18]. P. aeruginosa can also become mucoid, a property conferred by the activation of the alg operon, which in the presence of the mucA mutation activates gene expression to cause production of copious extracellular polysaccharides composed of repeating subunits of guluronic and mannuric acids [19, 20]. Mucoid phenotypes are related with infectious chronicity [21].
Fig. 1.
Pseudonomas aeruginosa biofilm production and interactions with host immunity. a Airway epithelial cells contact P. aeruginosa during infection triggering chemotactic signaling and biofilm production. Polysaccharide structures generated by bacterial metabolism are anchored to the extracellular matrix of these cells. bP. aeruginosa metabolic pathways provide biofilm components. Intracellular pathways that direct the metabolism of glucose (green arrows) through the Entner-Doudoroff (central loci zwf and glk) and glyoxylate shunt (mediated by aceA and glcB) pathways. Glucose flux through these pathways allows the production of carbohydrates that form extracellular polysaccharides for the attachment to the airway cells. Upon contact with the host cell, surface recognition and activation of WspR and cyclic (c)-di-GMP production (red arrows) result in stimulation of lasR and rhlR loci and production of PslA and PelA, which support formation of extracellular polysaccharides. c-di-GMP coordinates biofilm formation and bacterial motility, switching from flagellar- to a pilus-dependent twitching motility through activation of the PiY1/chpAK complex. Both endogenous/exogenous reactive oxygen species (ROS) sources and catabolite repressor succinate collaborate in the synthesis of c-di-GMP. Intracellular ROS and redox balance are regulated by NADPH generated by zwf activity. cP. aeruginosa interaction with host immunity. Pathogen-associated molecular patterns, such as lipopolysaccharide (LPS), flagella, pilus, or type three secretion system components (TTSS), activate proinflammatory surface receptors in immune cells. Both TLR4/MD2-LPS and TLR5-flagella surface interactions trigger activation of transcription factor NF-κB and mitochondrial destabilization (purple arrows). NF-κB induces production and secretion of inflammatory cytokines, such as IL-6 and IL-8. Mitochondrial collapse is accompanied by accumulation of ROS and succinate release and hypoxia-induced factor-1α (HIF1α) stabilization, which acts as a transcription factor that mediates production of pro- IL-1β (black arrows). Pro-IL-1β is cleaved to its mature form by caspase-1 produced by three different inflammasome systems: (1) NAIP/NLRC4/ASC activated by cytoplasmic uptake of flagellin; (2) NLRP3/ASC activated by cytoplasmic uptake of LPS; and (3) pilin secreted by P. aeruginosa.
Biofilms and Oxidant Stress
Several stimuli for the production of such biofilms have been proposed, most importantly oxidant stress. It is apparent that endogenous oxidant stress is sufficient, as organisms exposed to H2O2 spin off mucoid variants in vitro [22], a process that is blocked in the presence of an antioxidant such as N-acetyl cysteine. The extracellular polysaccharides themselves can also contribute to protection from ROS [16]. In addition to the oxidant stress generated by bacterial metabolism, which for P. aeruginosa is preferentially oxidative, there are numerous sources of oxidant stress in vivo. In a clinical setting such as CF, organisms are continually exposed to antimicrobial agents, which, despite their specific bacterial targets, wind up killing through the production of toxic oxidants [23]. Low-level exposure to aminoglycosides, as well as the fluoroquinolones that induce DNA damage, presents a major inducer of oxidant stress within the bacteria and inhibits bacterial growth. The recruitment of immune cells and their release of oxidants, such a mitochondrial-related anion superoxide (O2–) are highly toxic oxidants as well. The organisms isolated from clinical settings often have upregulated expression of DNA repair mechanisms and are observed to have increased mutation rates in response to oxidant stress encountered from the host response.
Bacterial metabolism must be very significantly altered to support the bioenergetic demands of biofilm formation. This adaptation has been termed “metabolic adaptation” [24, 25]. The metabolic changes provide a mechanism to diminish oxidant production and to enhance the production of NADPH, which promotes the formation of antioxidants, as opposed to NADH, which is prooxidant. This can be accomplished through increased Entner-Doudoroff activity, a catabolic route for glucose, which increases NADPH production and diverts the energetic program away from ATP generation through the more oxidant tricarboxylic acid (TCA) cycle activity [26]. In order to optimize extensive polysaccharide production, key components of the alginate and/or biofilm matrix, metabolic adaptation can also be associated with increased use of the glyoxylate shunt to generate even more carbohydrate substrates [27]. Thus, the generation of extracellular polysaccharide matrices may well be primarily a response to oxidant stress encountered in vivo.
c-di-GMP signaling also results in the suppression of motility, and, with it, decreased expression of gene products that can activate inflammasomes including flagellin [14]. In addition, there is suppression of the type three secretion system (TTSS) components which activate the inflammasome. TTSS toxins themselves alter the integrity of the epithelial tight junctions [28] and cause widespread damage in the lung through a patatin-like activity [29, 30]. The consequences of this global downregulation of immunostimulatory activity have been interpreted as a mechanism to avoid immune clearance.
Biofilms and Innate Immune Clearance Mechanisms
The major driving force for biofilm formation in vivo has been thought to be resistance to innate immune clearance. The biofilm community, overall, affords physical protection to a substantial amount of the biomass, as the organisms are protected from phagocytes, antibody, and complement [16]. The biomass also provides a diversity of microenvironments exposed to different levels of oxygenation and nutrients, diversity that overall may enhance the viability of the biofilm community. In vitro studies are less clear-cut. The extracellular polysaccharides associated with biofilms do not appear to constitute a significant barrier to antimicrobial penetration [31], as they form a loose polysaccharide mesh. Depending upon the nature of the biofilm matrix, whether formed of alginate, or PelA or PslA polymers, there may or may not be an effect of charge on antibiotic diffusion.
Whether biofilms arise as a response to phagocytic clearance is not definitively established. While the prevailing dogma suggests that mucoid or biofilm-producing P. aeruginosa are “resistant” to phagocytic clearance, supporting data are limited, although there are few, if any, murine model systems that faithfully reproduce what happens in humans. Moreover, confusion as to the predominance of the Pel or Psl biofilm components, as opposed to the production of alginate, has made interpretation of the importance of each of these genes in pathogenesis difficult [15, 17]. Models of infection performed with P. aeruginosa mutants lacking specific biofilm components and/or the ability to produce alginate reflect the relative contribution of these extracellular polysaccharides to the pathogenesis of pulmonary infection [32]. While an immunostimulatory activity and complement resistance could be ascribed to the expression of Pel, a mucoid strain expressing Pel was readily cleared from the murine lung in an acute infection model, suggesting that phagocytic clearance is not at all adversely affected by the presence of these extracellular polysaccharides [32]. While the presence of alginate or biofilm-specific antibodies to promote clearance of these strains has been proposed, experiments done in infection-naïve mice indicate that this is not the case, at least in the murine model.
The ability to proliferate in both planktonic as well as biofilm modes of growth is undoubtedly a major factor in the success of P. aeruginosa as an opportunistic pathogen. While it is unclear exactly how the bacterial population shifts to biofilm formation in vivo, it seems likely that some critical number of organisms must reach the epithelial surface to activate the bacterial chemosensors that would initiate c-di-GMP signaling and biofilm formation [14]. The planktonic organisms that are presumably the initial sources of infection are generally motile, express immunostimulatory lipopolysaccharide (LPS) and other pathogen-associated molecular patterns (PAMPs) and should activate the numerous protective responses in the airways. Thus, the infecting organisms must have specific mechanisms to evade this initial innate immune response.
P. aeruginosa Resistance to Antimicrobial Peptides
The airway epithelium, through the effects of IL-22 and other cytokines and interferons, produces a large number of antimicrobial peptides [33]. Many organisms, including P. aeruginosa, are able to undergo mutation to acquire resistance to antimicrobial peptides, which would enhance their ability to persistently infect epithelial surfaces. An important site of P. aeruginosa infection is the eye, where these organisms are an important cause of keratitis [34, 35]. A major therapeutic strategy against these biofilm infections is the development of optimized antimicrobial peptides delivered topically [36]. There is substantial interest in the development of more active antimicrobial peptides that could be used topically to treat organisms growing at various anatomical sites, whether in a planktonic or biofilm format.
Resistance to Inflammasome-Mediated Clearance
P. aeruginosa can be especially virulent pathogens, expressing potent toxins, both proteases and the more complex TTSS that target specific eukaryotic receptors [37]. They express multiple PAMPs associated with the activation of inflammasomes (Fig. 1c): intracellular flagellin can be detected by the NLRC4/NAIP-dependent inflammasome, which causes caspase-1 maturation and pro-IL-1β cleavage, producing its secretion [38, 39, 40]. Flagellin can also activate surface TLR5 receptors to induce NF-κB-dependent inflammation [41]. LPS, which is recognized by the TLR4/MD2 surface complex, triggers NLRP3/ASC-dependent inflammasome activation and cleavage of caspase-1, which processes pro-IL-1β into mature IL-1β [38, 39]. Surface TLR4/MD2 engagement also primes NF-κB signaling [42]. Intracellular LPS can be recognized by procaspase-11 (mouse) or procaspase-4 to -5 (in humans) and mount the noncanonical NLRP3/ASC inflammasome response to produce and secrete more IL-1β [38]. The type IV pilus-derived pilin, which directly activates caspase-1 without priming either NLRC4, NLRP3, or ASC, causes pro-IL-1β cleavage and its secretion [43]. As noted above, the switch to a biofilm mode of growth through the induction of c-di-GMP signaling limits the expression of PAMPs such as flagellin or PcrV, both NLRC4/NAIP inflammasome activators. This diminished expression of PAMPs is usually interpreted as bacterial strategy to avoid the activation of the inflammasome and resulting production of IL-1β and recruitment of phagocytes [28, 44]. However, several studies fail to demonstrate that inflammasome activation promotes P. aeruginosa clearance [39, 40, 45]. Mice lacking inflammasome components, or those that had been treated with anakinra, which blocks IL-1 signaling, in fact had significantly increased ability to clear P. aeruginosa [39]. These studies do not support the hypothesis that decreased expression of PAMPs is a bacterial mechanism to increase survival within the lung by limiting inflammation. It is suggested that the intense proinflammatory response elicited by the NLRC4-NLRP3 inflammasomes actually interferes with bacterial clearance, such as by activating autophagy-mediated P. aeruginosa protection inside macrophages [46], although other effects of inflammasome activation on the local milieu may also be involved. In addition, these studies do not take into consideration whether the same properties play a role in the clearance or conservation of long-term strains that cause chronic infections, which due to their adaptation and reduction of immunogenicity must use other mechanisms to enhance their survival.
Host Immunity and Catabolite Repression-Mediated Biofilm Production
Catabolite repression is a signaling pathway present in several pathogens, which in P. aeruginosa is mediated by the crc locus [47]. It is activated by succinate, a keto acid and intermediate of the TCA cycle, which produces ATP under O2 respiration and internal ROS. Succinate restricts glucose and other nutrient utilization, such as amino acids and mannitol, committing P. aeruginosa metabolism to the TCA cycle to generate energy, as opposed to the bioenergetically more favorable Entner-Doudoroff and glyoxylate shunt pathways [48, 49]. Indeed, zwf, the gene that codes for the glucose 6-phosphate dehydrogenase enzyme (G6PDH) and that initiates the Entner-Doudoroff and pentose phosphate pathway, is negatively regulated by crc [47, 50]. Lack of zwf signaling compromises the production of NADPH, the antioxidant molecule in charge of controlling excessive endogenous and exogenous ROS [26, 50]. Catabolite repression through Crc increases the accumulation of intracellular oxidative species, which, in addition to the ROS produced by the TCA cycle operation, boosts the probability of damaging DNA [22, 51]. As defenses against free radicals and ROS require upregulation of intracellular signaling such as zwf, selection of mutants that fulfill these requirements also promotes the generation of a biofilm lifestyle.
Acute pneumonia causes enormous airway damage, allowing cells leaking out their cytoplasmic contents. Many inflammatory reactions, such as the inflammasome, activate pyroptosis and cell death with the release of intracellular contents [52]. These host-derived nutrients become available for Pseudomonas during acute pneumonia. Yet, the impact of these carbon sources on bacterial metabolism and pathogenicity remains to be established, influencing the predominance of glucose- or amino-acid-based metabolism of P. aeruginosa. Utilization of substrates for the TCA cycle and endogenous ROS production could promote the selection of mutants adapted to the newly available nutrients [22]. This ROS-mediated selection of oxidant-friendly clones and biofilm production is further supported by studies showing that either scavenging ROS with N-acetyl-L-cysteine [53] or deleting the crc locus [47], which primes c-di-GMP synthesis, modulates pilin formation and results in biofilm disruption [14]. Thus, organisms such as P. aeruginosa can readily alter their metabolism to a particular environment, even in the presence of recruited immune cells and phagocytes.
Augmented expression of Entner-Doudoroff (zwf) and glyoxylate shunt (aceA and glcB) regulatory genes in mucoid CF-related P. aeruginosa has been reported [54, 55, 56], which conferred advantages for sputum-mediated clearance. Of interest, zwf, aceA, and glcB were constitutively upregulated in mucoid CF isolates, suggesting that adaptation in late-stage CF may include the rewiring of the regulatory networks that control preferential use of metabolites. Whether defense against host immune clearance is increased by altered use of these metabolic pathways is not established; it is clear that the biofilm lifestyle acquired by chronic strains along their transformation into the mucoid phenotype is in response to the hostile surroundings produced by both the host and the bacterial metabolism itself.
Overall, despite decades of research using clinical isolates as well as many different in vivo and in vitro models, exactly how P. aeruginosa adapts to the milieu of the airways and overcomes innate immune clearance at other sites remains incompletely understood. Ongoing analyses of whole-genome sequencing data might provide a more encompassing view of the overall adaptive process, with the airway surface, the production of biofilms, and their interaction with the host immunity. The adaptive responses to common innate immune clearance mechanisms exploited by opportunistic bacteria are predicted to be similar. The opportunistic gram-negative bacterial genomes would be thought to have roughly the same repertoire of genes and metabolic capabilities. These organisms must all adapt to the same innate immune effectors, antimicrobial peptides, phagocytes, and oxidant stress. Nonetheless, it appears that there may be highly species-specific strategies employed by different pathogens, particularly in the setting of pneumonia.
K. pneumoniae Resistance to Innate Immune Clearance
P. aeruginosa and K. pneumoniae share a common ecological niche as major causes of health-care-associated infections, particularly pneumonia and sepsis [57]. As gram-negative pathogens with the ability to acquire exogenous DNA to confer antibiotic resistance, both can adapt to the milieu of human airways. K. pneumoniae in general produces extracellular polysaccharides that are associated with human colonization [58]. K. pneumoniae strains are relatively heterogeneous, with different phenotypes and genotypes associated with infection. Whereas the typical virulent serotypes, as typified by ATCC 43816 (KPPR1), have been widely studied, the carbapenem-resistant ST258 isolates, while less immunostimulatory in murine models, are increasingly prevalent and are associated with high rates of morbidity and mortality in humans [59]. Most notorious are the hypermucovirulent isolates often associated with liver abscess formation that are important causes of sepsis but in relatively limited geographic locales [60]. Carbapenem-resistant (CRKP) and even colistin-resistant strains have become more endemic, colonizing healthy humans [61]. As an opportunist, these MDR organisms can cause severe disease that is often difficult to treat, associated with persistent bacteremia and high mortality, and disproportionately affecting patients with comorbidities and critical illness (> 50%) [62].
K. pneumoniae Surface Structures Important in Pathogenesis: Production of Biofilm and Fimbriae
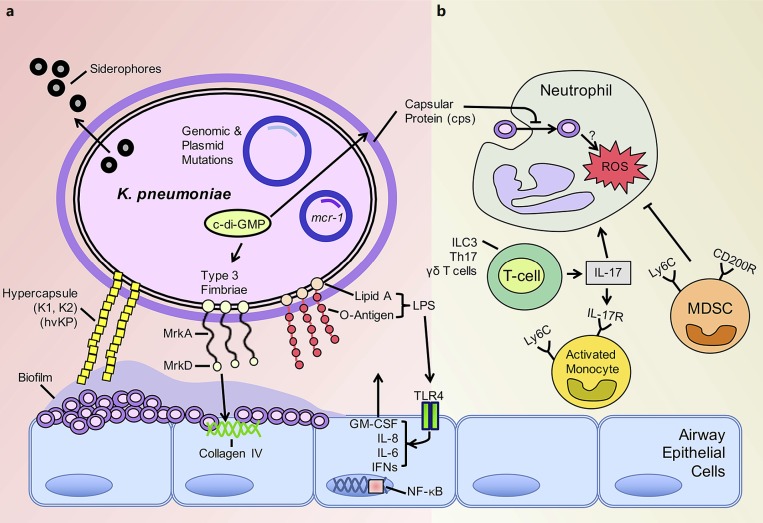
Biofilm formation is a major characteristic of K. pneumoniae strains, seen especially in MDR and extensively drug-resistant isolates [63] (Fig. 2a). Mechanisms of biofilm formation such as c-di-GMP network regulation are common to gram-negative organisms, while capsular protein variability is unique to K. pneumoniae. Across characterized isolates, fimbriae and capsular protein are variably expressed reflecting their genetic diversity and differing contributions to pathogenicity [63, 64]. Like P. aeruginosa, K. pneumoniae utilizes c-di-GMP, a critical second messenger restricted to bacterial metabolism [65], to regulate type 1 and 3 fimbrial production and subsequent biofilm formation. Redundant and conserved components of the c-di-GMP network are found across clinical isolates of K. pneumoniae suggesting that c-di-GMP regulation plays a major role in virulence and/or colonization [66]. This has been corroborated in murine models of K. pneumoniae infection with levels of c-di-GMP influencing the extent of pathology. Elevated c-di-GMP levels are associated with attenuated lung pathology and inflammation but increased fimbrial production [67]. Increased expression of fimbriae has been observed in the organisms that express the extended-spectrum β- lactamase (ESBL) [68] likely contributing to their ability to cause infection.
Fig. 2.
Klebsiella pneumoniae biofilm production and evasion of host immunity. aK. pneumoniae avidly forms biofilms on epithelial cells and abiotic surfaces. Induction of the cyclic (c)-di-GMP system increases production of type 3 fimbriae and subsequent biofilm formation. The hypervirulent K. pneumoniae (hvKP) forms biofilm enhanced by the abundant production of capsular protein. The composition of lipid A, a major component of lipopolysaccharide (LPS) and specific target of colistin, is altered in K. pneumoniae harboring the mcr-1 plasmid. Siderophores are utilized by nearly all K. pneumoniae to scavenge iron for bacterial growth. b Neutrophils readily take up ST258 isolates intracellularly although intracellular killing is impaired. Neutrophil function is inhibited by recruitment of immunosuppressive myeloid-derived suppressor cells (MDSC). IL-17 is produced predominantly by ILC3s, Th17 cells, and γδ T cells in response to K. pneu moniae infection. Monocytes are then activated by IL-17, with the potential to be microbicidal. Heterogeneity in IL-17 levels as well as monocyte plasticity in response to carbapenem-resistant K. pneumoniae isolates may explain the variability in the phenotype of these cells recruited to the site of infection.
The major function of K. pneumoniae fimbriae is to mediate attachment to the surfaces, a critical property of biofilm formation on biotic and abiotic surfaces [69, 70]. As this is a critical component in pathogenesis, fimbrial biogenesis has been considered a potential target to interrupt K. pneumoniae biofilm formation. The regulation of type 3 fimbrial production in K. pneumoniae by c-di-GMP has been well studied. MrkH acts as the upstream regulator of the mrkABCDF operon [71] to increase c-di-GMP-dependent expression of mrkHI and resulting biofilm formation [72, 73]. This c-di-GMP- dependent process leads to increased amounts of polymerized fimbrial shaft (MrkA) and fimbrial adhesion protein (MrkD) [73, 74]. These purified fimbrial proteins (MrkD) are a potential antigen for vaccine development [75] given the protective immunity conferred by their immunogenicity [76]. A downstream regulator of type 3 fimbrial expression, MrkJ, is a phosphodiesterase that reduces c-di-GMP levels to suppress fimbrial expression and subsequent biofilm formation [77]. Modulation of the c-di-GMP system could possibly enhance clearance in the chronically colonized host [78] but could also lead to increased inflammation leading to worse clinical outcomes [67].
Iron Scavenging and K. pneumoniae
A mechanism for iron acquisition is another shared property of K. pneumoniae and P. aeruginosa. For both pathogens, the presence of human lactoferrin in the airways interferes with biofilm formation [79, 80]. For K. pneumoniae, the production of several siderophores, including enterobactin (Ent+) and yersinobactin (Ybt+), provides a mechanism to compete for iron. Siderophores contribute significantly to the success of K. pneumoniae as a pathogen given the overrepresentation of organisms with increased expression of the major K. pneumoniae siderophores, the Ent+Ybt+ isolates, in the ST258 clade [81]. As a scavenger of iron, lactoferrin is a potential therapeutic that will inhibit the growth of K. pneumoniae [80]. However, the expression of capsular protein (cps) also plays a role in limiting interactions of the lactoferrin and the bacterial surface restricting its effect [82].
Emergence of Hypervirulent K. pneumoniae
While the production of extracellular polysaccharides and proteins is a general feature of K. pneumoniae isolates, a subgroup of “hypervirulent” (hvKP) strains produce huge amounts of extracellular material [83]. This Klebsiella has been typically associated with bloodstream infections and liver abscesses leading to high mortality [84]. The epidemiology of the hypermucovirulent isolates and their related resistance mechanisms have been recently reviewed [85]. These organisms are usually of the capsular type K2, and whole-genome sequencing suggests several unique lo ci, distinct from those of other pathogenic K. pneumoniae strains [86]. In murine models, they cause lethality at 102 cfu inocula, which is in stark contrast to previously characterized CRKP isolates that require inocula 5–6 orders of magnitude to cause illness in murine models [87, 88]. rmpA mutations are thought to be responsible for the hypermucoviscous phenotype by increasing expression of capsular protein and thereby biofilm formation [89, 90].
Recent ST11 isolates of K. pneumoniae isolated from patients with ventilator-associated pneumonia have the hypervirulent phenotype and are also carbapenem resistant [91]. This alarming development is not unexpected given the proclivity of Klebsiella for acquiring extracellular DNA, and the hypervirulent phenotype is associated with the acquisition of a 170-kbp plasmid [91]. These strains have also been isolated from patients without predisposing conditions [61]. The acquisition of colistin resistance in these hypervirulent strains has also been reported, which appears to generate a fitness cost [60]. In vitro studies of both antibiotic-resistant and -susceptible K. pneumoniae strains indicate that, like P. aeruginosa, the organisms tend to revert to the biofilm mode of growth in stressful laboratory conditions [92], suggesting a common bacterial response to the oxidant stress of human airways and the resulting immune response.
K. pneumoniae Evasion of Host Immune Clearance
The genetic flexibility of K. pneumoniae that has enabled their acquisition of antimicrobial resistance determinants [93] has also contributed to resistance to innate immune clearance (Fig. 2b). Transcriptomic analyses suggest that organisms that have acquired carbapenem resistance in vivo have a significantly altered pattern of gene expression [60]. The role of immune cells in the clearance of K. pneumoniae infection in the lung has been well characterized with the laboratory reference strain ATCC 43816 (KPPR1) recently reviewed [94]. However, current clinical isolates including the carbapenem-resistant ST258 clones [93] and the hvKP have many other genotypic and phenotypic changes.
Surface Properties of K. pneumoniae and Immune Recognition
Modification of surface PAMPs and capsular material is a well-recognized mechanism of immune evasion in Klebsiella. Sialic acid, a common feature of eukaryotic cell glycoproteins, is abundant in the expression of capsular protein of K. pneumoniae K1 isolates [95]. Sialic acid contributes to the hypermucovicous phenotype and directly impedes interaction with and uptake by immune cells. Indirectly, binding of sialic acid to Siglec-9 on the surface of neutrophils interferes with neutrophil bactericidal activity [95]. On the other hand, other cell surface moieties, such as mannose residues, enhance the recognition of these organisms by immune cells [68, 96].
The modification of LPS is another widely conserved target for immune adaptation in many gram-negative organisms. This is an especially important site of mutation in the Klebsiella isolates with resistance to colistin, an antimicrobial that specifically targets the lipid A moiety of LPS. The colistin-resistant K. pneumoniae strains have altered LPS structures that limit the binding of the drug to the bacterial membrane [97]. Recent characterization of LPS from infected patients showed the rapid acquisition of genomic- and plasmid-mediated mutations, leading to alterations in the composition of lipid A and colistin resistance [98]. Perhaps the most concerning of these genetic elements is the emergence of the mobilized colistin resistance (mcr-1), a mobile plasmid rapidly shared amongst all members of the Enterobacteriaceae [99]. Whether or not these mutations carry a high fitness cost is not well established in humans. In a Galleria mellonella infection model, MCR-1-mediated resistance was associated with decreased fitness and attenuated cytokine production [100]. How the alterations in the structure of LPS affect immune cell recognition and subsequent immunogenicity has not been published to date.
K. pneumoniae Resistance to Phagocytic Killing
A major factor in the pathogenesis of K. pneumoniae infections that is replicated in murine models is the remarkable resistance of these organisms, particularly the carbapenem-resistant K. pneumoniae to phagocytic clearance. The exact mechanism by which these CRKP isolates avoid intracellular killing, particularly by neutrophils, has yet to be elucidated. In vitro studies have established that ST258 isolates, in contrast to the KPPR1 strain, are resistant to neutrophil killing ex vivo [101, 102]. Differences in the production of respiratory burst and ROS generation [103, 104] have been observed; however, no specific bacterial genes have been identified that correlate with this phenotype. The mechanisms appear to involve resistance to Ca2+-dependent killing [102]. It is clear that neutrophils and/or monocytes are critical to the host in clearance of ST258 isolates, as mice treated with anti-Ly6G antibodies have increased bacterial burden [102, 105]. One recent study utilized an anti-LPS-O-antigen antibody to increase opsonophagocytic killing capacity of neutrophils without loss of TLR4 signaling, likely from enhanced bacterial recognition by neutrophils [106]. However, this may not be universal as the ST258 isolates appear to be readily phagocytosed [87] but not killed [102], suggesting a mechanism by which intracellular killing processes are inhibited.
Responses of Macrophages and Monocytes to K. pneumoniae Infection
In addition to the limited killing capacity of neutrophils, in vivo the CRKP isolates have acquired additional mechanisms to thwart phagocytic clearance. Alveolar macrophages, the resident responders to pathogens in the airways, are critical to the clearance of K. pneumoniae pneumonia [107, 108]. The activation state and polarization of these macrophages are important in this process, as macrophages in the M2b or anti-inflammatory state were more susceptible to infection [109, 110]. Treating mice with GM-CSF to polarize macrophages to an inflammatory phenotype appeared to improve bacterial clearance [111]. However, a recent study with a CRKP isolate found that the removal of alveolar macrophages with clodronate liposomes does not change bacterial clearance [102]. Therefore, the role of resident alveolar macrophages in pulmonary infection with CRKP isolates of K. pneumoniae may be less essential.
Some studies with the classic K. pneumoniae KPPR1 have highlighted the key role of recruited monocytes in the pathogenesis of K. pneumoniae pneumonia [62, 112]. The function of monocytes in response to infection with the genetically and phenotypically diverse CRKP isolates is less clear. Monocyte-derived suppressor cell recruitment has been identified as an important component in the resolution phase of K. pneumoniae KPPR1 infection [113]. These cells function in the clearance of infected and apoptotic neutrophils in an IL-10-dependent manner. However, the premature influx of these immunosuppressive monocytes (Ly6C+) in ST258 K. pneumoniae pneumonia interfered with neutrophil function and contributed to persistent infection [102]. Other groups have shown the central role of these Ly6C monocytes in the coordination and activation of Th17 cells in response to infection [114]. Subsequent bacterial clearance was dependent on the activation of these cells via the production of intracellular ROS. These discordant findings can most likely be explained by the observation that mononuclear cells infected with isolates from different clonal lineages of ST258 induced variable levels of Th17 expression in exposed T cells [115]. Again, these findings provide yet another example of the variability in the phenotype of these highly genetically variable organisms.
Conclusions
The success of certain gram-negative bacteria as human pathogens is due mainly to the genetic flexibility of these organisms, as they adapt to the milieu of the human airways and other sites of infection. Both K. pneumoniae and P. aeruginosa use c-di GMP signaling to assume the biofilm lifestyle which provides protection from phagocytes, antibiotic exposure, and oxidant stress. The biomass of the biofilm provides a sufficient population to support mutation in response to antibiotic and innate immune selective pressures. These pathogens, such as in the case of K. pneumoniae, also readily acquire exogenous DNA conferring many important phenotypes relevant to human disease, including both colistin resistance and the hypervirulent phenotype. The use of whole-genome sequencing highlights the substantial diversity of clinically prevalent isolates but may also lead to the identification of shared targets in both of these species, which could be exploited therapeutically.
The presence of different metabolites in the airways and their influence on the adaptive programs required to produce biofilm is less well understood. Whereas the consumption of glucose and synthesis of polysaccharides to construct the extracellular matrix is well accepted, utilization of keto acids or amino acids secreted by host cells is not established. As they may help “turning on” the switch of adaptation, they also arise as possible targets for therapy. There are scattered reports supporting this hypothesis; the use of specific carbon substrates to deregulate biofilm production is attractive as an alternative to antibiotics. Future research must also focus on comprehensive analyses of bacterial metabolomics to better understand the biochemistry behind host-pathogen adaptation and identify metabolic targets to disrupt bacterial colonization.
Disclosure Statement
Authors declare no conflict of interest exist.
Acknowledgments
A.P. is supported by NIH 1R35HL135800. S.A.R. is supported by a CFF Postdoctoral Fellowship (CFF RIQUEL 17F0/PG008837). D.A. is supported by NIH 1K08HL138289.
References
- 1.Elborn JS. Cystic fibrosis. Lancet. 2016;388:2519–2531. doi: 10.1016/S0140-6736(16)00576-6. [DOI] [PubMed] [Google Scholar]
- 2.Gutierrez HH, Sanchez I, Schidlow DV. Cystic fibrosis care in Chile. Curr Opin Pulm Med. 2009;15:632–637. doi: 10.1097/MCP.0b013e328330db7a. [DOI] [PubMed] [Google Scholar]
- 3.Silva Filho LV, Castanos C, Ruiz HH. Cystic fibrosis in Latin America - improving the awareness. J Cyst Fibros. 2016;15:791–793. doi: 10.1016/j.jcf.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 4.Masekela R, Zampoli M, Westwood AT, White DA, Green RJ, Olorunju S, Kwofie-Mensah M. Phenotypic expression of the 3120 + 1G>A mutation in non-Caucasian children with cystic fibrosis in South Africa. J Cyst Fibros. 2013;12:363–366. doi: 10.1016/j.jcf.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Westwood T, Henderson B, Ramsay M, Medical and Scientific Advisory Committee of the South African Cystic Fibrosis Association Diagnosing cystic fibrosis in South Africa. S Afr Med J. 2006;96:304–306. [PubMed] [Google Scholar]
- 6.Wine JJ, Hansson GC, Konig P, Joo NS, Ermund A, Pieper M. Progress in understanding mucus abnormalities in cystic fibrosis airways. J Cyst Fibros. 2018;17:S35–S39. doi: 10.1016/j.jcf.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 7.Barbas AS, Dib MJ, Al-Adra DP, Goldaracena N, Sapisochin G, Waddell TK, Keshavjee S, Selzner N, Chaparro C, Cattral MS. Combined lung-liver-pancreas transplantation in a recipient with cystic fibrosis. J Cyst Fibros. 2018;17:e1–e4. doi: 10.1016/j.jcf.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Lowery EM, Adams W, Grim SA, Clark NM, Edwards L, Layden JE. Increased risk of PTLD in lung transplant recipients with cystic fibrosis. J Cyst Fibros. 2017;16:727–734. doi: 10.1016/j.jcf.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 9.Cohen TS, Prince A. Cystic fibrosis: a mucosal immunodeficiency syndrome. Nat Med. 2012;18:509–519. doi: 10.1038/nm.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riquelme SA, Hopkins BD, Wolfe AL, DiMango E, Kitur K, Parsons R, Prince A. Cystic fibrosis transmembrane conductance regulator attaches tumor suppressor PTEN to the membrane and promotes anti Pseudomonas aeruginosa immunity. Immunity. 2017;47:1169–1181. doi: 10.1016/j.immuni.2017.11.010. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo Y, Zhao K, Baker AE, Kuchma SL, Coggan KA, Wolfgang MC, Wong GC, O'Toole GA. A hierarchical cascade of second messengers regulates Pseudomonas aeruginosa surface behaviors. MBio. 2015;6:e02456–14. doi: 10.1128/mBio.02456-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De N, Navarro MV, Raghavan RV, Sondermann H. Determinants for the activation and autoinhibition of the diguanylate cyclase response regulator WspR. J Mol Biol. 2009;393:619–633. doi: 10.1016/j.jmb.2009.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huangyutitham V, Guvener ZT, Harwood CS. Subcellular clustering of the phosphorylated WspR response regulator protein stimulates its diguanylate cyclase activity. MBio. 2013;4:e00242–13. doi: 10.1128/mBio.00242-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valentini M, Filloux A. Biofilms and cyclic di-GMP (c-di-GMP) signaling: lessons from Pseudomonas aeruginosa and other bacteria. J Biol Chem. 2016;291:12547–12555. doi: 10.1074/jbc.R115.711507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colvin KM, Irie Y, Tart CS, Urbano R, Whitney JC, Ryder C, Howell PL, Wozniak DJ, Parsek MR. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ Microbiol. 2012;14:1913–1928. doi: 10.1111/j.1462-2920.2011.02657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mishra M, Byrd MS, Sergeant S, Azad AK, Parsek MR, McPhail L, Schlesinger LS, Wozniak DJ. Pseudomonas aeruginosa Psl polysaccharide reduces neutrophil phagocytosis and the oxidative response by limiting complement-mediated opsonization. Cell Microbiol. 2012;14:95–106. doi: 10.1111/j.1462-5822.2011.01704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jennings LK, Storek KM, Ledvina HE, Coulon C, Marmont LS, Sadovskaya I, Secor PR, Tseng BS, Scian M, Filloux A, Wozniak DJ, Howell PL, Parsek MR. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc Natl Acad Sci USA. 2015;112:11353–11358. doi: 10.1073/pnas.1503058112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irie Y, Roberts AEL, Kragh KN, Gordon VD, Hutchison J, Allen RJ, Melaugh G, Bjarnsholt T, West SA, Diggle SP. The Pseudomonas aeruginosa PSL polysaccharide is a social but noncheatable trait in biofilms. MBio. 2017;8:e00374–17. doi: 10.1128/mBio.00374-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie ZD, Hershberger CD, Shankar S, Ye RW, Chakrabarty AM. Sigma factor-anti-sigma factor interaction in alginate synthesis: inhibition of AlgT by MucA. J Bacteriol. 1996;178:4990–4996. doi: 10.1128/jb.178.16.4990-4996.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathee K, McPherson CJ, Ohman DE. Posttranslational control of the algT (algU)-encoded sigma22 for expression of the alginate regulon in Pseudomonas aeruginosa and localization of its antagonist proteins MucA and MucB (AlgN) J Bacteriol. 1997;179:3711–3720. doi: 10.1128/jb.179.11.3711-3720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deretic V, Schurr MJ, Yu H. Pseudomonas aeruginosa, mucoidy and the chronic infection phenotype in cystic fibrosis. Trends Microbiol. 1995;3:351–356. doi: 10.1016/s0966-842x(00)88974-x. [DOI] [PubMed] [Google Scholar]
- 22.Boles BR, Singh PK. Endogenous oxidative stress produces diversity and adaptability in biofilm communities. Proc Natl Acad Sci USA. 2008;105:12503–12508. doi: 10.1073/pnas.0801499105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meylan S, Porter CBM, Yang JH, Belenky P, Gutierrez A, Lobritz MA, Park J, Kim SH, Moskowitz SM, Collins JJ. Carbon sources tune antibiotic susceptibility in Pseudomonas aeruginosa via tricarboxylic acid cycle control. Cell Chem Biol. 2017;24:195–206. doi: 10.1016/j.chembiol.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Behrends V, Bell TJ, Liebeke M, Cordes-Blauert A, Ashraf SN, Nair C, Zlosnik JE, Williams HD, Bundy JG. Metabolite profiling to characterize disease-related bacteria: gluconate excretion by Pseudomonas aeruginosa mutants and clinical isolates from cystic fibrosis patients. J Biol Chem. 2013;288:15098–15109. doi: 10.1074/jbc.M112.442814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Behrends V, Ryall B, Zlosnik JE, Speert DP, Bundy JG, Williams HD. Metabolic adaptations of Pseudomonas aeruginosa during cystic fibrosis chronic lung infections. Environ Microbiol. 2013;15:398–408. doi: 10.1111/j.1462-2920.2012.02840.x. [DOI] [PubMed] [Google Scholar]
- 26.Ma JF, Hager PW, Howell ML, Phibbs PV, Hassett DJ. Cloning and characterization of the Pseudomonas aeruginosa zwf gene encoding glucose-6-phosphate dehydrogenase, an enzyme important in resistance to methyl viologen (paraquat) J Bacteriol. 1998;180:1741–1749. doi: 10.1128/jb.180.7.1741-1749.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahn S, Jung J, Jang IA, Madsen EL, Park W. Role of glyoxylate shunt in oxidative stress response. J Biol Chem. 2016;291:11928–11938. doi: 10.1074/jbc.M115.708149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jain M, Ramirez D, Seshadri R, Cullina JF, Powers CA, Schulert GS, Bar-Meir M, Sullivan CL, McColley SA, Hauser AR. Type III secretion phenotypes of Pseudomonas aeruginosa strains change during infection of individuals with cystic fibrosis. J Clin Microbiol. 2004;42:5229–5237. doi: 10.1128/JCM.42.11.5229-5237.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pankhaniya RR, Tamura M, Allmond LR, Moriyama K, Ajayi T, Wiener-Kronish JP, Sawa T. Pseudomonas aeruginosa causes acute lung injury via the catalytic activity of the patatin-like phospholipase domain of ExoU. Crit Care Med. 2004;32:2293–2299. doi: 10.1097/01.ccm.0000145588.79063.07. [DOI] [PubMed] [Google Scholar]
- 30.Salacha R, Kovacic F, Brochier-Armanet C, Wilhelm S, Tommassen J, Filloux A, Voulhoux R, Bleves S. The Pseudomonas aeruginosa patatin-like protein PlpD is the archetype of a novel type V secretion system. Environ Microbiol. 2010;12:1498–1512. doi: 10.1111/j.1462-2920.2010.02174.x. [DOI] [PubMed] [Google Scholar]
- 31.Drenkard E, Ausubel FM. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature. 2002;416:740–743. doi: 10.1038/416740a. [DOI] [PubMed] [Google Scholar]
- 32.Jones CJ, Wozniak DJ. Psl produced by mucoid Pseudomonas aeruginosa contributes to the establishment of biofilms and immune evasion. MBio. 2017;8:e00864–17. doi: 10.1128/mBio.00864-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sonnenberg GF, Fouser LA, Artis D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat Immunol. 2011;12:383–390. doi: 10.1038/ni.2025. [DOI] [PubMed] [Google Scholar]
- 34.McClellan SA, Jerome A, Suvas S, Hazlett LD. NLRC4 regulates caspase-1 and IL-1β production in a CD11blowLy6Glow population of cells required for resistance to Pseudomonas aeruginosa keratitis. PLoS One. 2017;12:e0185718. doi: 10.1371/journal.pone.0185718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ross BX, Gao N, Cui X, Standiford TJ, Xu J, Yu FX. IL-24 promotes Pseudomonas aeruginosa keratitis in C57BL/6 mouse corneas. J Immunol. 2017;198:3536–3547. doi: 10.4049/jimmunol.1602087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li SA, Liu J, Xiang Y, Wang YJ, Lee WH, Zhang Y. Therapeutic potential of the antimicrobial peptide OH-CATH30 for antibiotic-resistant Pseudomonas aeruginosa keratitis. Antimicrob Agents Chemother. 2014;58:3144–3150. doi: 10.1128/AAC.00095-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huber P, Basso P, Reboud E, Attree I. Pseudomonas aeruginosa renews its virulence factors. Environ Microbiol Rep. 2016 doi: 10.1111/1758-2229.12443. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 38.Guo H, Callaway JB, Ting JP. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat Med. 2015;21:677–687. doi: 10.1038/nm.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iannitti RG, Napolioni V, Oikonomou V, De Luca A, Galosi C, Pariano M, Massi-Benedetti C, Borghi M, Puccetti M, Lucidi V, Colombo C, Fiscarelli E, Lass-Florl C, Majo F, Cariani L, Russo M, Porcaro L, Ricciotti G, Ellemunter H, Ratclif L, De Benedictis FM, Talesa VN, Dinarello CA, van de Veerdonk FL, Romani L. IL-1 receptor antagonist ameliorates inflammasome-dependent inflammation in murine and human cystic fibrosis. Nat Commun. 2016;7:10791. doi: 10.1038/ncomms10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cohen TS, Prince AS. Activation of inflammasome signaling mediates pathology of acute P.aeruginosa pneumonia. J Clin Invest. 2013;123:1630–1637. doi: 10.1172/JCI66142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pena J, Fu Z, Schwarzer C, Machen TE. Pseudomonas aeruginosa inhibition of flagellin-activated NF-κB and interleukin-8 by human airway epithelial cells. Infect Immun. 2009;77:2857–2865. doi: 10.1128/IAI.01355-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andreakos E, Sacre SM, Smith C, Lundberg A, Kiriakidis S, Stonehouse T, Monaco C, Feldmann M, Foxwell BM. Distinct pathways of LPS-induced NF-κB activation and cytokine production in human myeloid and nonmyeloid cells defined by selective utilization of MyD88 and Mal/TIRAP. Blood. 2004;103:2229–2237. doi: 10.1182/blood-2003-04-1356. [DOI] [PubMed] [Google Scholar]
- 43.Arlehamn CS, Evans TJ. Pseudomonas aeruginosa pilin activates the inflammasome. Cell Microbiol. 2011;13:388–401. doi: 10.1111/j.1462-5822.2010.01541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huus KE, Joseph J, Zhang L, Wong A, Aaron SD, Mah TF, Sad S. Clinical isolates of Pseudomonas aeruginosa from chronically infected cystic fibrosis patients fail to activate the inflammasome during both stable infection and pulmonary exacerbation. J Immunol. 2016;196:3097–3108. doi: 10.4049/jimmunol.1501642. [DOI] [PubMed] [Google Scholar]
- 45.Schultz MJ, Rijneveld AW, Florquin S, Edwards CK, Dinarello CA, van der Poll T. Role of interleukin-1 in the pulmonary immune response during Pseudomonas aeruginosa pneumonia. Am J Physiol Lung Cell Mol Physiol. 2002;282:L285–L290. doi: 10.1152/ajplung.00461.2000. [DOI] [PubMed] [Google Scholar]
- 46.Deng Q, Wang Y, Zhang Y, Li M, Li D, Huang X, Wu Y, Pu J, Wu M. Pseudomonas aeruginosa triggers macrophage autophagy to escape intracellular killing by activation of the NLRP3 inflammasome. Infect Immun. 2015;84:56–66. doi: 10.1128/IAI.00945-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O'Toole GA, Gibbs KA, Hager PW, Phibbs PV, Jr, Kolter R. The global carbon metabolism regulator Crc is a component of a signal transduction pathway required for biofilm development by Pseudomonas aeruginosa. J Bacteriol. 2000;182:425–431. doi: 10.1128/jb.182.2.425-431.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Collier DN, Hager PW, Phibbs PV., Jr Catabolite repression control in the Pseudomonads. Res Microbiol. 1996;147:551–561. doi: 10.1016/0923-2508(96)84011-3. [DOI] [PubMed] [Google Scholar]
- 49.Rojo F. Carbon catabolite repression in Pseudomonas: optimizing metabolic versatility and interactions with the environment. FEMS Microbiol Rev. 2010;34:658–684. doi: 10.1111/j.1574-6976.2010.00218.x. [DOI] [PubMed] [Google Scholar]
- 50.Kim J, Jeon CO, Park W. Dual regulation of zwf-1 by both 2-keto-3-deoxy-6-phosphogluconate and oxidative stress in Pseudomonas putida. Microbiology. 2008;154:3905–3916. doi: 10.1099/mic.0.2008/020362-0. [DOI] [PubMed] [Google Scholar]
- 51.Linares JF, Moreno R, Fajardo A, Martinez-Solano L, Escalante R, Rojo F, Martinez JL. The global regulator Crc modulates metabolism, susceptibility to antibiotics and virulence in Pseudomonas aeruginosa. Environ Microbiol. 2010;12:3196–3212. doi: 10.1111/j.1462-2920.2010.02292.x. [DOI] [PubMed] [Google Scholar]
- 52.Martin-Sanchez F, Diamond C, Zeitler M, Gomez AI, Baroja-Mazo A, Bagnall J, Spiller D, White M, Daniels MJ, Mortellaro A, Penalver M, Paszek P, Steringer JP, Nickel W, Brough D, Pelegrin P. Inflammasome-dependent IL-1β release depends upon membrane permeabilisation. Cell Death Differ. 2016;23:1219–1231. doi: 10.1038/cdd.2015.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao T, Liu Y. N-acetylcysteine inhibit biofilms produced by Pseudomonas aeruginosa. BMC Microbiol. 2010;10:140. doi: 10.1186/1471-2180-10-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Silo-Suh L, Suh SJ, Phibbs PV, Ohman DE. Adaptations of Pseudomonas aeruginosa to the cystic fibrosis lung environment can include deregulation of zwf, encoding glucose-6-phosphate dehydrogenase. J Bacteriol. 2005;187:7561–7568. doi: 10.1128/JB.187.22.7561-7568.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hagins JM, Scoffield J, Suh SJ, Silo-Suh L. Malate synthase expression is deregulated in the Pseudomonas aeruginosa cystic fibrosis isolate FRD1. Can J Microbiol. 2011;57:186–195. doi: 10.1139/W10-118. [DOI] [PubMed] [Google Scholar]
- 56.Hagins JM, Scoffield JA, Suh SJ, Silo-Suh L. Influence of RpoN on isocitrate lyase activity in Pseudomonas aeruginosa. Microbiology. 2010;156:1201–1210. doi: 10.1099/mic.0.033381-0. [DOI] [PubMed] [Google Scholar]
- 57.Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, Lynfield R, Maloney M, McAllister-Hollod L, Nadle J, Ray SM, Thompson DL, Wilson LE, Fridkin SK, Emerging Infections Program Healthcare- Associated Infections and Antimicrobial Use Prevalence Survey Team Multistate point-prevalence survey of health care-associated infections. N Engl J Med. 2014;370:1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lawlor MS, Handley SA, Miller VL. Comparison of the host responses to wild-type and cpsB mutant Klebsiella pneumoniae infections. Infect Immun. 2006;74:5402–5407. doi: 10.1128/IAI.00244-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Diago-Navarro E, Chen L, Passet V, Burack S, Ulacia-Hernando A, Kodiyanplakkal RP, Levi MH, Brisse S, Kreiswirth BN, Fries BC. Carbapenem-resistant Klebsiella pneumoniae exhibit variability in capsular polysaccharide and capsule associated virulence traits. J Infect Dis. 2014;210:803–813. doi: 10.1093/infdis/jiu157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choi MJ, Ko KS. Loss of hypermucoviscosity and increased fitness cost in colistin-resistant Klebsiella pneumoniae sequence type 23 strains. Antimicrob Agents Chemother. 2015;59:6763–6773. doi: 10.1128/AAC.00952-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4:107–118. doi: 10.4161/viru.22718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Satlin MJ, Chen L, Patel G, Gomez-Simmonds A, Weston G, Kim AC, Seo SK, Rosenthal ME, Sperber SJ, Jenkins SG, Hamula CL, Uhlemann AC, Levi MH, Fries BC, Tang YW, Juretschko S, Rojtman AD, Hong T, Mathema B, Jacobs MR, Walsh TJ, Bonomo RA, Kreiswirth BN. Multicenter clinical and molecular epidemiological analysis of bacteremia due to carbapenem-resistant Enterobacteriaceae (CRE) in the CRE Epicenter of the United States. Antimicrob Agents Chemother. 2017;61:e02349–16. doi: 10.1128/AAC.02349-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vuotto C, Longo F, Pascolini C, Donelli G, Balice MP, Libori MF, Tiracchia V, Salvia A, Varaldo PE. Biofilm formation and antibiotic resistance in Klebsiella pneumoniae urinary strains. J Appl Microbiol. 2017;123:1003–1018. doi: 10.1111/jam.13533. [DOI] [PubMed] [Google Scholar]
- 64.Lawlor MS, Hsu J, Rick PD, Miller VL. Identification of Klebsiella pneumoniae virulence determinants using an intranasal infection model. Mol Microbiol. 2005;58:1054–1073. doi: 10.1111/j.1365-2958.2005.04918.x. [DOI] [PubMed] [Google Scholar]
- 65.Tamayo R, Pratt JT, Camilli A. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu Rev Microbiol. 2007;61:131–148. doi: 10.1146/annurev.micro.61.080706.093426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cruz DP, Huertas MG, Lozano M, Zarate L, Zambrano MM. Comparative analysis of diguanylate cyclase and phosphodiesterase genes in Klebsiella pneumoniae. BMC Microbiol. 2012;12:139. doi: 10.1186/1471-2180-12-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rosen DA, Twentyman J, Hunstad DA. High levels of cyclic di-GMP in Klebsiella pneumoniae attenuate virulence in the lung. Infect Immun. 2018;86:e00647–17. doi: 10.1128/IAI.00647-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sahly H, Navon-Venezia S, Roesler L, Hay A, Carmeli Y, Podschun R, Hennequin C, Forestier C, Ofek I. Extended-spectrum β-lactamase production is associated with an increase in cell invasion and expression of fimbrial adhesins in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2008;52:3029–3034. doi: 10.1128/AAC.00010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sebghati TA, Korhonen TK, Hornick DB, Clegg S. Characterization of the type 3 fimbrial adhesins of Klebsiella strains. Infect Immun. 1998;66:2887–2894. doi: 10.1128/iai.66.6.2887-2894.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sebghati TA, Clegg S. Construction and characterization of mutations within the Klebsiella mrkD1P gene that affect binding to collagen type V. Infect Immun. 1999;67:1672–1676. doi: 10.1128/iai.67.4.1672-1676.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wilksch JJ, Yang J, Clements A, Gabbe JL, Short KR, Cao H, Cavaliere R, James CE, Whitchurch CB, Schembri MA, Chuah ML, Liang ZX, Wijburg OL, Jenney AW, Lithgow T, Strugnell RA. MrkH, a novel c-di-GMP-dependent transcriptional activator, controls Klebsiella pneumoniae biofilm formation by regulating type 3 fimbriae expression. PLoS Pathog. 2011;7:e1002204. doi: 10.1371/journal.ppat.1002204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tan JW, Wilksch JJ, Hocking DM, Wang N, Srikhanta YN, Tauschek M, Lithgow T, Robins-Browne RM, Yang J, Strugnell RA. Positive autoregulation of mrkHI by the cyclic di-GMP-dependent MrkH protein in the biofilm regulatory circuit of Klebsiella pneumoniae. J Bacteriol. 2015;197:1659–1667. doi: 10.1128/JB.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu CC, Lin CT, Cheng WY, Huang CJ, Wang ZC, Peng HL. Fur-dependent MrkHI regulation of type 3 fimbriae in Klebsiella pneumoniae CG43. Microbiology. 2012;158:1045–1056. doi: 10.1099/mic.0.053801-0. [DOI] [PubMed] [Google Scholar]
- 74.Johnson JG, Murphy CN, Sippy J, Johnson TJ, Clegg S. Type 3 fimbriae and biofilm formation are regulated by the transcriptional regulators MrkHI in Klebsiella pneumoniae. J Bacteriol. 2011;193:3453–3460. doi: 10.1128/JB.00286-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li Y, Li ZJ, Han WY, Lei LC, Sun CJ, Feng X, Du CT, Du TF, Gu JM. Identification and characterization of Th cell epitopes in MrkD adhesin of Klebsiella pneumoniae. Microb Pathog. 2010;49:8–13. doi: 10.1016/j.micpath.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 76.Lavender H, Jagnow JJ, Clegg S. Klebsiella pneumoniae type 3 fimbria-mediated immunity to infection in the murine model of respiratory disease. Int J Med Microbiol. 2005;295:153–159. doi: 10.1016/j.ijmm.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 77.Johnson JG, Clegg S. Role of MrkJ, a phosphodiesterase, in type 3 fimbrial expression and biofilm formation in Klebsiella pneumoniae. J Bacteriol. 2010;192:3944–3950. doi: 10.1128/JB.00304-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Karaolis DK, Newstead MW, Zeng X, Hyodo M, Hayakawa Y, Bhan U, Liang H, Standiford TJ. Cyclic di-GMP stimulates protective innate immunity in bacterial pneumonia. Infect Immun. 2007;75:4942–4950. doi: 10.1128/IAI.01762-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Singh PK, Parsek MR, Greenberg EP, Welsh MJ. A component of innate immunity prevents bacterial biofilm development. Nature. 2002;417:552–555. doi: 10.1038/417552a. [DOI] [PubMed] [Google Scholar]
- 80.Morici P, Florio W, Rizzato C, Ghelardi E, Tavanti A, Rossolini GM, Lupetti A. Synergistic activity of synthetic N-terminal peptide of human lactoferrin in combination with various antibiotics against carbapenem-resistant Klebsiella pneumoniae strains. Eur J Clin Microbiol Infect Dis. 2017;36:1739–1748. doi: 10.1007/s10096-017-2987-7. [DOI] [PubMed] [Google Scholar]
- 81.Bachman MA, Oyler JE, Burns SH, Caza M, Lepine F, Dozois CM, Weiser JN. Klebsiella pneumoniae yersiniabactin promotes respiratory tract infection through evasion of lipocalin 2. Infect Immun. 2011;79:3309–3316. doi: 10.1128/IAI.05114-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Campos MA, Vargas MA, Regueiro V, Llompart CM, Alberti S, Bengoechea JA. Capsule polysaccharide mediates bacterial resistance to antimicrobial peptides. Infect Immun. 2004;72:7107–7114. doi: 10.1128/IAI.72.12.7107-7114.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. 2016;80:629–661. doi: 10.1128/MMBR.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ko WC, Paterson DL, Sagnimeni AJ, Hansen DS, Von Gottberg A, Mohapatra S, Casellas JM, Goossens H, Mulazimoglu L, Trenholme G, Klugman KP, McCormack JG, Yu VL. Community-acquired Klebsiella pneumoniae bacteremia: global differences in clinical patterns. Emerg Infect Dis. 2002;8:160–166. doi: 10.3201/eid0802.010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee CR, Lee JH, Park KS, Jeon JH, Kim YB, Cha CJ, Jeong BC, Lee SH. Antimicrobial resistance of hypervirulent Klebsiella pneumoniae: epidemiology, hypervirulence-associated determinants, and resistance mechanisms. Front Cell Infect Microbiol. 2017;7:483. doi: 10.3389/fcimb.2017.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang X, Xie Y, Li G, Liu J, Li X, Tian L, Sun J, Ou HY, Qu H. Whole-genome-sequencing characterization of bloodstream infection-causing hypervirulent Klebsiella pneumoniae of capsular serotype K2 and ST374. Virulence. 2018;9:510–521. doi: 10.1080/21505594.2017.1421894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tzouvelekis LS, Miriagou V, Kotsakis SD, Spyridopoulou K, Athanasiou E, Karagouni E, Tzelepi E, Daikos GL. KPC-producing, multidrug-resistant Klebsiella pneumoniae sequence type 258 as a typical opportunistic pathogen. Antimicrob Agents Chemother. 2013;57:5144–5146. doi: 10.1128/AAC.01052-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chiang TT, Yang YS, Yeh KM, Chiu SK, Wang NC, Lin TY, Huang LY, Chang FY, Siu LK, Lin JC, Chen JH. Quantification and comparison of virulence and characteristics of different variants of carbapenemase-producing Klebsiella pneumoniae clinical isolates from Taiwan and the United States. J Microbiol Immunol Infect. 2016;49:83–90. doi: 10.1016/j.jmii.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 89.Lai YC, Peng HL, Chang HY. RmpA2, an activator of capsule biosynthesis in Klebsiella pneumoniae CG43, regulates K2 cps gene expression at the transcriptional level. J Bacteriol. 2003;185:788–800. doi: 10.1128/JB.185.3.788-800.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cheng HY, Chen YS, Wu CY, Chang HY, Lai YC, Peng HL. RmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43. J Bacteriol. 2010;192:3144–3158. doi: 10.1128/JB.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Du P, Zhang Y, Chen C. Emergence of carbapenem-resistant hypervirulent Klebsiella pneumoniae. Lancet Infect Dis. 2018;18:23–24. doi: 10.1016/S1473-3099(17)30629-1. [DOI] [PubMed] [Google Scholar]
- 92.Anes J, Hurley D, Martins M, Fanning S. Exploring the genome and phenotype of multi-drug resistant Klebsiella pneumoniae of clinical origin. Front Microbiol. 2017;8:1913. doi: 10.3389/fmicb.2017.01913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gomez-Simmonds A, Uhlemann AC. clinical implications of genomic adaptation and evolution of carbapenem-resistant Klebsiella pneumoniae. J Infect Dis. 2017;215:S18–S27. doi: 10.1093/infdis/jiw378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Parker D, Ahn D, Cohen T, Prince A. Innate immune signaling activated by MDR bacteria in the airway. Physiol Rev. 2016;96:19–53. doi: 10.1152/physrev.00009.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee CH, Chang CC, Liu JW, Chen RF, Yang KD. Sialic acid involved in hypermucoviscosity phenotype of Klebsiella pneumoniae and associated with resistance to neutrophil phagocytosis. Virulence. 2014;5:673–679. doi: 10.4161/viru.32076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kabha K, Nissimov L, Athamna A, Keisari Y, Parolis H, Parolis LA, Grue RM, Schlepper-Schafer J, Ezekowitz AR, Ohman DE, et al. Relationships among capsular structure, phagocytosis, and mouse virulence in Klebsiella pneumoniae. Infect Immun. 1995;63:847–852. doi: 10.1128/iai.63.3.847-852.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Olaitan AO, Morand S, Rolain JM. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol. 2014;5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Leung LM, Cooper VS, Rasko DA, Guo Q, Pacey MP, McElheny CL, Mettus RT, Yoon SH, Goodlett DR, Ernst RK, Doi Y. Structural modification of LPS in colistin-resistant, KPC-producing Klebsiella pneumoniae. J Antimicrob Chemother. 2017;72:3035–3042. doi: 10.1093/jac/dkx234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 100.Yang Q, Li M, Spiller OB, Andrey DO, Hinchliffe P, Li H, MacLean C, Niumsup P, Powell L, Pritchard M, Papkou A, Shen Y, Portal E, Sands K, Spencer J, Tansawai U, Thomas D, Wang S, Wang Y, Shen J, Walsh T. Balancing mcr-1 expression and bacterial survival is a delicate equilibrium between essential cellular defence mechanisms. Nat Commun. 2017;8:2054. doi: 10.1038/s41467-017-02149-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kobayashi SD, Porter AR, Dorward DW, Brinkworth AJ, Chen L, Kreiswirth BN, DeLeo FR. Phagocytosis and killing of carbapenem-resistant ST258 Klebsiella pneumoniae by human neutrophils. J Infect Dis. 2016;213:1615–1622. doi: 10.1093/infdis/jiw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ahn D, Penaloza H, Wang Z, Wickersham M, Parker D, Patel P, Koller A, Chen EI, Bueno SM, Uhlemann AC, Prince A. Acquired resistance to innate immune clearance promotes Klebsiella pneumoniae ST258 pulmonary infection. JCI Insight. 2016;1:e89704. doi: 10.1172/jci.insight.89704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sahly H, Aucken H, Benedi VJ, Forestier C, Fussing V, Hansen DS, Ofek I, Podschun R, Sirot D, Sandvang D, Tomas JM, Ullmann U. Impairment of respiratory burst in polymorphonuclear leukocytes by extended-spectrum β-lactamase-producing strains of Klebsiella pneumoniae. Eur J Clin Microbiol Infect Dis. 2004;23:20–26. doi: 10.1007/s10096-003-1047-7. [DOI] [PubMed] [Google Scholar]
- 104.Demirel I, Kinnunen A, Onnberg A, Soderquist B, Persson K. Comparison of host response mechanisms evoked by extended spectrum βlactamase (ESBL)–and non- ESBL-producing uropathogenic E. coli. BMC Microbiol. 2013;13:181. doi: 10.1186/1471-2180-13-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xiong H, Carter RA, Leiner IM, Tang YW, Chen L, Kreiswirth BN, Pamer EG. Distinct contributions of neutrophils and CCR2+ monocytes to pulmonary clearance of different Klebsiella pneumoniae strains. Infect Immun. 2015;83:3418–3427. doi: 10.1128/IAI.00678-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cohen TS, Pelletier M, Cheng L, Pennini ME, Bonnell J, Cvitkovic R, Chang CS, Xiao X, Cameroni E, Corti D, Semenova E, Warrener P, Sellman BR, Suzich J, Wang Q, Stover CK. Anti-LPS antibodies protect against Klebsiella pneumoniae by empowering neutrophil-mediated clearance without neutralizing TLR4. JCI Insight. 2017:2. doi: 10.1172/jci.insight.92774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bettina A, Zhang Z, Michels K, Cagnina RE, Vincent IS, Burdick MD, Kadl A, Mehrad B. M-CSF mediates host defense during bacterial pneumonia by promoting the survival of lung and liver mononuclear phagocytes. J Immunol. 2016;196:5047–5055. doi: 10.4049/jimmunol.1600306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Broug-Holub E, Toews GB, van Iwaarden JF, Strieter RM, Kunkel SL, Paine R, 3rd, Standiford TJ. Alveolar macrophages are required for protective pulmonary defenses in murine Klebsiella pneumonia: elimination of alveolar macrophages increases neutrophil recruitment but decreases bacterial clearance and survival. Infect Immun. 1997;65:1139–1146. doi: 10.1128/iai.65.4.1139-1146.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tsuchimoto Y, Asai A, Tsuda Y, Ito I, Nishiguchi T, Garcia MC, Suzuki S, Kobayashi M, Higuchi K, Suzuki F. M2b monocytes provoke bacterial pneumonia and gut bacteria-associated sepsis in alcoholics. J Immunol. 2015;195:5169–5177. doi: 10.4049/jimmunol.1501369. [DOI] [PubMed] [Google Scholar]
- 110.Ohama H, Asai A, Ito I, Suzuki S, Kobayashi M, Higuchi K, Suzuki F. M2b macrophage elimination and improved resistance of mice with chronic alcohol consumption to opportunistic infections. Am J Pathol. 2015;185:420–431. doi: 10.1016/j.ajpath.2014.09.022. [DOI] [PubMed] [Google Scholar]
- 111.Standiford LR, Standiford TJ, Newstead MJ, Zeng X, Ballinger MN, Kovach MA, Reka AK, Bhan U. TLR4-dependent GM-CSF protects against lung injury in Gram-negative bacterial pneumonia. Am J Physiol Lung Cell Mol Physiol. 2012;302:L447–L454. doi: 10.1152/ajplung.00415.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Herold S, Tabar TS, Janssen H, Hoegner K, Cabanski M, Lewe-Schlosser P, Albrecht J, Driever F, Vadasz I, Seeger W, Steinmueller M, Lohmeyer J. Exudate macrophages attenuate lung injury by the release of IL-1 receptor antagonist in Gram-negative pneumonia. Am J Respir Crit Care Med. 2011;183:1380–1390. doi: 10.1164/rccm.201009-1431OC. [DOI] [PubMed] [Google Scholar]
- 113.Poe SL, Arora M, Oriss TB, Yarlagadda M, Isse K, Khare A, Levy DE, Lee JS, Mallampalli RK, Chan YR, Ray A, Ray P. STAT1-regulated lung MDSC-like cells produce IL-10 and efferocytose apoptotic neutrophils with relevance in resolution of bacterial pneumonia. Mucosal Immunol. 2013;6:189–199. doi: 10.1038/mi.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xiong H, Keith JW, Samilo DW, Carter RA, Leiner IM, Pamer EG. Innate lymphocyte/Ly6Chi monocyte crosstalk promotes Klebsiella pneumoniae clearance. Cell. 2016;165:679–689. doi: 10.1016/j.cell.2016.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Clemente AM, Castronovo G, Antonelli A, D'Andrea MM, Tanturli M, Perissi E, Paccosi S, Parenti A, Cozzolino F, Rossolini GM, Torcia MG. Differential Th17 response induced by the two clades of the pandemic ST258 Klebsiella pneumoniae clonal lineages producing KPC-type carbapenemase. PLoS One. 2017;12:e0178847. doi: 10.1371/journal.pone.0178847. [DOI] [PMC free article] [PubMed] [Google Scholar]