Abstract
Purpose
Metastatic uveal melanoma (UM) has a very poor prognosis and no effective therapy. Despite remarkable advances in treatment of cutaneous melanoma, UM remains recalcitrant to chemotherapy, small-molecule kinase inhibitors, and immune-based therapy.
Methods
We assessed two sets of oxidative phosphorylation (OxPhos) genes within 9858 tumors across 31 cancer types. An OxPhos inhibitor was used to characterize differential metabolic programming of highly metastatic monosomy 3 (M3) UM. Seahorse analysis and global metabolomics profiling were done to identify metabolic vulnerabilities. Analyses of UM TCGA data set were performed to determine expressions of key OxPhos effectors in M3 and non-M3 UM. We used targeted knockdown of succinate dehydrogenase A (SDHA) to determine the role of SDHA in M3 UM in conferring resistance to OxPhos inhibition.
Results
We identified UM to have among the highest median OxPhos levels and showed that M3 UM exhibits a distinct metabolic profile. M3 UM shows markedly low succinate levels and has highly increased levels of SDHA, the enzyme that couples the tricarboxylic acid cycle with OxPhos by oxidizing (lowering) succinate. We showed that SDHA-high M3 UM have elevated expression of key OxPhos molecules, exhibit abundant mitochondrial reserve respiratory capacity, and are resistant to OxPhos antagonism, which can be reversed by SDHA knockdown.
Conclusions
Our study has identified a critical metabolic program within poor prognostic M3 UM. In addition to the heightened mitochondrial functional capacity due to elevated SDHA, M3 UM SDHA-high mediate resistance to therapy that is reversible with targeted treatment.
Keywords: uveal melanoma, SDHA, OxPhos, mitochondrial metabolism, monosomy 3
Uveal melanoma (UM), which arises from melanocytes in the uveal tract, is the most common intraocular cancer of the adult eye.1 Although primary disease in the eye can be effectively treated, half of UM patients develop distant metastatic disease, often to the liver, with a median survival of less than 12 months.2,3 The loss of one copy of chromosome 3 (Chr3) in a primary UM tumor, referred to as monosomy 3 (M3), is associated with metastasis and poor prognosis.4 More than 90% of metastatic UM is M3. In contrast, primary UM with a normal complement of two copies of Chr3, referred to as disomy 3 (D3), rarely metastasizes. In some cases, the single Chr3 in M3 UM undergoes a duplication, termed isodisomy 3 (iD3). Dysregulated mitochondrial metabolism is a hallmark of cancer.5 Specifically, cancer cells show remarkable metabolic reprogramming through alterations in the tricarboxylic acid (TCA) cycle and/or oxidative phosphorylation (OxPhos) within mitochondria.6 The enzyme succinate dehydrogenase A (SDHA), a component of Complex II of the electron transport chain (ETC), is a fundamental link between the TCA cycle and OxPhos. As part of the TCA cycle, SDHA oxidizes succinate, which reduces FAD to FADH2, liberating two electrons. As a component of the ETC, SDHA ensures that these electrons are shuttled through Ubiquinone to Cytochrome C (CYC1), facilitating OxPhos. Recent studies show that SDHA is a major source of OxPhos reserve respiratory capacity to support cell survival in noncancer cells.7 Loss-of-function mutations in SDHA, or other alterations in Complex II, result in elevated succinate, which serves as an oncometabolite. Thus, SDHA plays a fundamental role in normal and cancer-related metabolism. In this study, we explored the metabolic features that underlie metastasis-prone UMs and identified critical features that may serve as precise therapeutic targets for UM.
Materials and Methods
Reagents for OxPhos Inhibition, Mito Stress Test, and Antibodies
IACS-010759, a small-molecule inhibitor of Complex I of OxPhos, was a generous gift of the Institute of Applied Cancer Science (IACS) at UT MD Anderson Cancer Center.8 The SDHA antibody was obtained from ABCAM (#ab14715; Abcam, Cambridge, MA, USA). The Mito Stress Kit was obtained from Agilent Technologies (Seahorse XF Cell Mito Stress Test Kit, #103015–100; Agilent, Santa Clara, CA, USA).
Cell Lines
Cell line 39 (official name: Mel20–06–039, RRID:CVCL_8473),9,10 70 (official name: Mel20–07–070, RRID:CVCL_8475),9,10 and 196 (official name Mel20–09–196, RRID:CVCL_D715)10 were obtained from Dr. Tara A. McCannel. Cell line OMM-1 (RRID:CVCL_6939),11 Mel202 (RRID:CVCL_C301),11 92–1 (RRID:CVCL_8607),11 and Mel270 (RRID:CVCL_C302)11 were kindly provided by Drs. Martine Jager and Bruce Ksander. Cell line MP41 (RRID: CVCL_4D12) and MP46 (RRID:CVCL_4D13)12 were generous gifts from Dr. Laurence Desjardins. Additional UM cell line information is provided in the references.13–16
Cell Culture
UM cells were cultured in RPMI media with 10% FBS, glutamine, penicillin-streptomycin and insulin supplement, under ambient oxygen at 37°C. Hypoxic culture conditions (1% O2) were enforced when required in the MCO-170M IncuSafe Multigas Incubator (Panasonic, Kadoma, Japan). Treatment with IACS-010759 was done in normal culture media.
Cell Line Validation
Cell lines were validated by short random repeat (STR) DNA fingerprinting techniques and mutational analysis, by the MDACC Cancer Center Support Grant (CCSG)-supported Characterized Cell Line Core, using the AmpFLSTR Identifier Kit (Applied Biosystems, Foster City, CA, USA), according to manufacturer's instructions. The STR profiles were compared with known ATCC fingerprints (ATCC.org), and with the Cell Line Integrated Molecular Authentication database (CLIMA) version 0.1.200808 (http://bioinformatics.istge.it/clima/). The STR profiles matched known DNA fingerprints or were unique (Supplementary Table S1).
Chromosome Copy Number Analysis of UM Cells
To determine global copy number status of each UM cell line, next-generation sequencing was performed at the MD Anderson Institute of Personalized Cancer Therapy Genomic Laboratory (IPCT Lab).17 Genomic DNA was quantified by Picogreen (Invitrogen, Carlsbad, CA, USA) and quality checked using a TapeStation (Agilent). DNA was sheared by ultrasonication and after library preparation, samples were analyzed on a TapeStation. Equimolar amounts of DNA from each sample were pooled, and biotin-labeled probes from Roche Nimblegen (Basel, Switzerland) used for capturing global copy number regions. Targeted regions were recovered using streptavidin beads, streptavidin-biotin-probe-target complex was washed, and another round of PCR amplification performed. Captured sample was analyzed on a TapeStation using the DNA High Sensitivity kit, and the enrichment assessed by quantitative PCR (qPCR) using specific primers designed by Roche Nimblegen. The cutoff for the enrichment was a minimum 50-fold. Captured libraries were sequenced on a HiSeq 4000 (Illumina Inc., San Diego, CA, USA) on a version 3 TruSeq paired-end flowcell at a cluster density of between 700 and 1000 K clusters/mm2. The resulting BCL files were converted into “.fastq.gz” files and individual libraries within the samples were demultiplexed, allowing no mismatches, using CASAVA 1.8.2. The captured sequencing data was aligned17 to human reference assembly GRCh37 (ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp/technical/reference/phase2_reference_assembly_sequence/hs37d5.fa.gz) using BWA (version 0.6.2) and duplicated reads removed using Picard (version 1.80). In preliminary data analysis we called single nucleotide variants and small indels using an in-house developed analysis pipeline, which classifies variants into three categories: somatic, germline, and loss-of-heterozygosity, based on variant allele frequencies. We compared these variants with dbSNP 138, COSMIC 70, and TCGA databases, and annotated them using the VEP (version 2.7), Annovar (version 2014–07–14), and CanDrA (version 1.1) programs. Specific copy number alterations and B allele frequency (BAF) were analyzed using Nexus Copy Number Discovery Version 9.0 (BioDiscovery, Inc., El Segundo, CA, USA). Previously aligned BAM files of tumor samples (from the targeted sequencing pipeline) were loaded as input, and pseudo-matched paired analyses were carried out using a nontumor sequencing database from IPCT that contained data from more than 5000 normal samples. For copy number analysis, log2-ratio cutoffs of 0.18 and 0.6 were used for amplified and highly amplified status, and −0.18 and −1.0 were used for deletion and homozygous-deletion status, respectively. For BAF analysis, only SNP loci in the general population were used. Reads with alignment quality score less than 30, and loci with sequencing quality score less than 20 or read depth less than 20, were filtered out. For each locus, the number of alleles with largest count was divided by the total count of all alleles, then subtracted from 1 to obtain BAF.
Western Blotting
Cells were lysed in a buffer containing 50 mM Tris (pH 7.9), 150 mM NaCl, 1% NP40, 1 mM EDTA, 10% glycerol, 1 mM sodium vanadate, and a protease inhibitor cocktail (Roche Pharmaceuticals, Nutley, NJ, USA). Proteins were separated by SDS-PAGE with 4% to 20% gradient gels (Bio-Rad Laboratories, Hercules, CA, USA), transferred to a Hybond-ECL nitrocellulose membrane (GE Healthcare Biosciences, Piscataway, NJ, USA) and blocked in 5% dry milk in PBS. The membrane was then incubated with primary and secondary antibodies, and target proteins were detected with ECL detection reagent (GE Healthcare Biosciences).
Small Interfering RNA (siRNA) Transfection of UM Cells
siRNA directed to SDHA and nontargeting siRNA were purchased from Santa Cruz Biotechnology (Dallas, TX, USA) (sc-61834, sc-37007). UM cells were plated in six-well plates at a density of 1.5 × 105 cells/well. Transfection of SDHA siRNA and control siRNA was performed the next day using Oligofectamine reagent (Invitrogen). In each case, the specified concentration of siRNA was transfected with 3 μL of Oligofectamine. Cells were treated with IACS-010759 2 days post transfection, tested with Western blots or MTT assays.
Cell Viability Assays
MTT-based cell viability assays were used for estimating cell survival. UM cells were plated at a density of 1 × 104 cells per well in triplicate in a 24-well plate with 1 mL culture media per well. To assess cell viability, MTT reagent (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) (Sigma-Aldrich, St. Louis, MO, USA), dissolved in PBS, was added to a final concentration of 1 mg/mL. After 3 hours, the precipitate formed was dissolved in dimethyl sulfoxide, and the color intensity estimated in a MRX Revelation microplate absorbance reader (Dynex Technologies, Chantilly, VA, USA) at 570 nm.
Mito Stress Test: Seahorse Analysis
UM cell lines were seeded on poly-Lysine coated 96-well Seahorse plates (Agilent Technologies); 24 hours later the cells were treated with IACS-010759 at 1.5 nm (the lowest effective dose tested) for the next 24 hours. The level of oxygen consumption rate (OCR) was measured in a Seahorse Mito stress test using the Agilent Seahorse XF Cell Mito Stress Test Kit. OCR, as well as reserve capacity, were measured following the protocol described in the kit. For normalization purposes, after Seahorse testing, cells were stained with Hoechst and propidium iodide, and scanned in Operetta and cell numbers utilized for normalization.
Mitotracker Red Staining, Imaging, and Quantitation
MitoTracker Red from Invitrogen (#M22425) was used to stain mitochondria from M3 and D3 UM cells. Cells were grown on chamber slides with the appropriate culture medium. When cells reached the desired confluence, the medium was removed from the chambers and prewarmed (37°C) growth medium containing the Mito tracker red dye was added. After the cells were incubated for 30 minutes under appropriate growth conditions, the loading solution was replaced with PBS. The cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) for 2 to 5 minutes (a stock of 5 mg/mL in dimethylformamide was diluted 1:10,000 in PBS to reach a working concentration) and observed using Olympus (Tokyo, Japan) FV1000 confocal microscope, after mounting with Dako (Glostrup, Denmark) Cytomation Fluorescent Mounting Medium (Dako Cytomation #S3023).
Images captured on the Olympus FV1000 (four fields per condition) were analyzed using Bitplane's Imaris software. Specifically, ImarisCell was used to mask the cell (cytoplasmic, nuclei, and vesicles [mitochondria]). DAPI staining was used to identify the nuclear area, the mitochondria label along with DAPI and autofluorescence was used to identify the cell body. The vesicle function was then used to identify the foci representing the mitochondria. A creation wizard was then created and used to batch analyze all four datasets using ImarisArena. These data were then presented in ImarisVantage, which represents the number of mitochondria counted per cell in each cell line.
Metabolic Profile Analysis by Mass Spectrometry
A profile analysis of 200 metabolites in D3 or M3 UM cells was performed using mass spectrometric methods at BIDMC (Boston, MA, USA), by liquid chromatography–mass spectrometry analysis.18 Culture media was changed the day before sample harvesting (at 50%–80% cell confluency). To collect adherent cells, culture media was aspirated, cells washed with cold PBS, and 80% methanol (−80 °C) added; cells were then incubated for 15 minutes or longer on dry ice. Cells were scraped and transferred to tubes. All the after-wash solution was added to tubes, to capture all metabolites. Cells were centrifuged at 2000g for 8 minutes at 4 °C to pellet cell debris and proteins. Supernatants were collected and the matching cell pellets were used for protein normalization. Supernatants were centrifugally evaporated and stored at −80 °C until they were used for the experiments.
Molecular and Survival Analysis of UM Tumor Samples
For TCGA cohort samples, data for normalized RSEM gene-level expression and for copy number were downloaded from www.firebrowse.org, and clinical data was downloaded from the publication website as Supplementary Table S1.19 For the validation cohort samples, data were obtained from GEO as GSE22138.20 To compare gene expression levels, normalized RSEM expression values were used. To determine pairwise statistical associations between mRNA transcript expression and M3 status across the entire UM TCGA cohort (n = 80), Regulome Explorer was used.19 Specifically, associations were filtered to include only the somatic copy number status of Chr3 (both 3p and 3q Gistic Arms) and the expression of all genes. The results of each pairwise test includes the number of samples used in the test, the Spearman correlation, and the −log10 (p) and −log10 (p) adjusted for multiple testing. Kaplan-Meier analyses were performed using the ‘survival' R package, with low and high expression defined as below and above the median expression. UM-specific metastasis was calculated as the time interval from primary UM diagnosis to development of distant UM metastasis.
Expression of OxPhos Gene Signatures and SDHA Across Cancers
We assessed two sets of oxidative phosphorylation genes. The first was a Hallmark set of 200 genes (derived from MSigDB), in which we changed “ECI1|1632,” to “DCI|1632” to be compatible with the RSEM gene names|Entrez IDs. The second was a set of 113 genes from the TCGA CHOL publication.21 We did not assess the gene set for the KEGG oxidative phosphorylation pathway (hsa00190), because TCGA RSEM expression data contained no genes expressed from mitochondrial DNA. All analyses were done in R 3.5.1.
We used two files from the PanCancer TCGA cell-of-origin publication22: (1) Table S3, which lists the sample barcodes and cancer types for the mRNA unsupervised clusters, and (2) batch-corrected mRNA sequencing (mRNA-seq) gene-level RSEM normalized expression data for 20531 genes × 11069 samples (https://gdc.cancer.gov/node/977). For most cancer types, we used only primary tumors (sample type code 01, https://gdc.cancer.gov/resources-tcga-users/tcga-code-tables/sample-type-codes). However, for SKCM, consistent with the TCGA publication,23 we retained both primary (01) and metastatic (06) tumors. For OV, we retained primary tumor samples corresponding to the 300 RNA-seq datasets that had passed the data-generating center's QC metrics for use in expression analyses. To focus the results on RNA-seq data from solid tissue cancers, we excluded samples from LAML and DLBC, TCGA's two blood cancers. This filtering resulted in 9858 tumor samples that were present in the batch-corrected mRNA-seq data file.
For each OxPhos gene set, we extracted a subset of the full mRNA-seq data set with that gene set and the 9858 tumor samples. For each tumor sample we calculated the median normalized RSEM expression value for that OxPhos gene set. We generated a box-whisker plot of the per-sample OxPhos gene set medians for each cancer type, overlaying beeswarm plots, and ordering the boxplots, left-to-right, by increasing median (i.e., median of per-sample signature medians) for each cancer type.
For SDHA expression analysis we extracted the SDHA expression record from the 200 gene × 9858 tumor sample Hallmark RSEM expression data and used the TCGA UM publication's supplemental master table19 to identify case IDs for n = 42 D3 and n = 38 M3 samples. We added SDHA expression data for the UVM-D3 and UVM-M3 subtypes, increasing the total number of tumor samples to 9938. We generated a boxplot similar to those for the signature medians above, for the resulting SDHA normalized RSEM expression results, for 9938 samples, 31 cancer types, and two subtypes.
Results
Chr 3-Aberrant UM Cells Are Highly Resistant to Oxidative Phosphorylation Inhibition
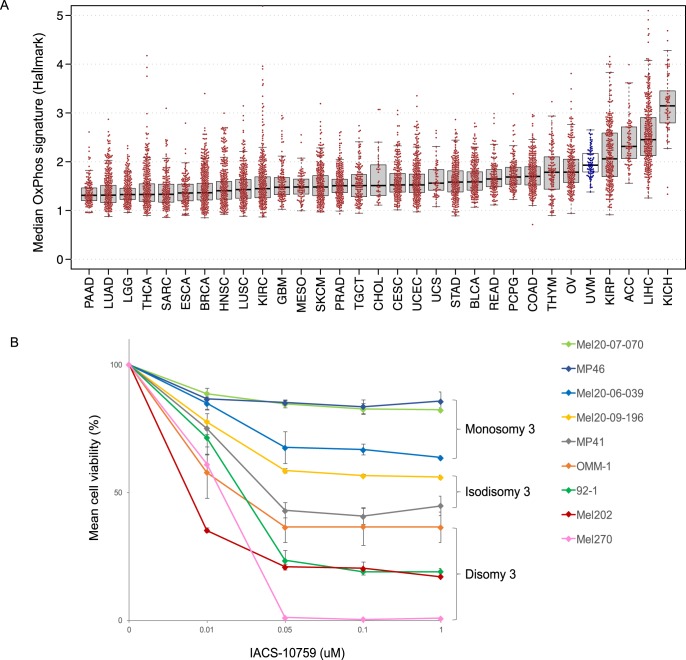
To determine the OxPhos activity in UM tumors, relative to other cancers, we used two distinct OxPhos signatures (Supplementary Table S2) to analyze 31 different tumor types. In each case, UM ranked among the tumors with the highest OxPhos signature profile (Fig. 1a; Supplementary Fig. S1). We then sought to evaluate the effect of targeting mitochondrial OxPhos in genetically well-annotated UM cells (i.e., Chr3-aberrant [M3 and iD3] versus Chr3-normal [D3] UM) (Supplementary Fig. S2A; Supplementary Table S3).9,11,12 UM cells were treated with increasing concentrations of the recently reported OxPhos antagonist IACS-010759, which directly inhibits Complex I of the ETC.8 M3 UM cells were observed to be highly resistant to OxPhos antagonism compared with D3 UM cells (21.9% versus 79.5% decrease in mean cell viability at 50 nM, respectively), with iD3 UM cells exhibiting an intermediate decrease (49.3%) (Fig. 1b). Notably, the effect of antagonizing OxPhos on cell viability was concentration-dependent at low doses, and the maximal effect on cell viability was reached at low nanomolar concentrations, consistent with the high sensitivity and specificity of the OxPhos inhibitor.8
Figure 1.
Chr3-aberrant UM cells are highly resistant to oxidative phosphorylation inhibition. (A) TCGA cancers ranked by median expression of the Hallmark OxPhos signature gene set (UVM = uveal melanoma samples, indicated in blue). Each dot represents the median normalized RSEM expression, in 1000s, of the set of signature genes in a sample. Box plots show median-of-median values (horizontal lines), and the 25th to 75th percentile range in the medians, that is, the interquartile range (IQR). Whiskers extend 1.5 times the IQR. (B) Effect of OxPhos antagonism on viability in UM cell lines. Cells were treated with the OxPhos inhibitor (IACS-010759) and cell viability was determined at 4 days. The cell lines MEL 202, MEL270, 92.1, MEL-20–07–070 and MEL20–09–196 have chr 8q gain, whereas cell lines MEL 20–06–039, MP41 and MP46 have chr 8q loss and 8p gain.
Chr3-Aberrant UM Cells Have a Higher Mitochondrial Reserve Capacity and Greater Number of Active Mitochondria
To characterize the key parameters of mitochondrial function that differentiate Chr3-aberrant from Chr3-normal cells, we measured the OCR in UM cells with different Chr3 genotypes, using the Seahorse Cell Mito Stress Test (Fig. 2a, Supplementary Fig. S2B and S2C). Although basal respiration was similar in all UM cell lines, the maximal and reserve spare respiration capacity was significantly elevated in M3, as compared with iD3 or D3 UM cells, revealing M3 UM cells to have an increased capacity to respond to a heightened energy demand. In micrographs of M3 UM cells, mitochondrial activity was more widely distributed within the cytoplasm, whereas in D3 UM cells, mitochondrial activity was concentrated in a perinuclear pattern (Fig. 2b; Supplementary Fig. S3).
Figure 2.
Chr3-aberrant UM cells have a higher mitochondrial reserve capacity and greater number of active mitochondria. (A) Mito stress test analysis shows M3 UM cells to have a higher mitochondrial reserve capacity than the D3 or iD3 cells at 1.5 nM IACS-010759. Between the two graphs, the dashed gray line indicates the maximum capacity, and the solid gray line indicates reserve the capacity. (B) M3 UM cells have more mitochondria than D3 cells. Mitotracker red dye staining shows that the M3 UM cell lines Mel20–06–039 (lower left) and Mel20–07–070 (lower right) have more mitochondria spread throughout the entire cell, while the D3 cell lines Mel202 (upper left) and 92–1 (upper right) have fewer mitochondria, concentrated around the nuclear membrane.
Monosomy 3 UM Cells Exhibit a Distinct Metabolic Profile
To better characterize the metabolic features underlying the mitochondrial phenotype that we observed between M3 and D3 UM cells, we performed comprehensive profiling of 200 key small-molecule metabolites using a mass spectrometry–based approach.18 A comparative assessment showed the most significantly diminished and elevated metabolite levels to be succinate (∼100–1000× lower) and malate (∼10,000× higher) in M3 than in D3 UM cells, respectively (Fig. 3a; Supplementary Table S4). Succinate and malate are key metabolites in the TCA cycle (Fig. 3b). Importantly, SDHA, the enzyme that catalyzes the oxidation of succinate in the TCA cycle, is also a subunit in ETC Complex II.
Figure 3.
Monosomy 3 UM cells exhibit a distinct metabolic profile. (A) Global metabolite analysis comparing the metabolomic profile of cell line Mel20–06–039 (M3) versus cell line Mel202 (D3). Gray: top 10 metabolites with increased levels in M3 versus D3; Blue: top 10 metabolites with decreased levels in M3 versus D3. (B) Schematic of the TCA cycle coupled with mitochondrial ETC. (C) Circos plot showing the most highly expressed genes associated with M3 status and corresponding summary.
Monosomy 3 UM Tumors Display a Heightened Gene Expression of Specific ETC Complex II Genes
To determine the underlying gene expression profile associated with the metabolic observations, we identified the gene transcripts that were most highly correlated (positively or negatively) with M3 UM status in the TCGA data set (42 M3 versus 38 D3 UM tumors).19 As expected, most genes with the lowest expression levels (Spearman rank correlation coefficient ρ > 0.80) localized to Chr3 (Supplementary Table S5), whereas the most highly expressed genes (ρ < −0.80) did not localize to a specific chromosomal region (Fig. 3c). Notably, two of the most highly expressed transcripts are well-described genes of metabolism: succinate dehydrogenase complex flavoprotein subunit A (SDHA) and coenzyme Q6, monooxygenase (COQ6). The role of SDHA in the TCA cycle is to oxidize succinate (Fig. 3b); thus, the increased SDHA in M3 UM tumors is consistent with the marked reduction in succinate and elevation in its downstream product, malate, observed in the metabolic profile of M3 UM (Figs. 3a–c). The strong association between elevated SDHA expression and M3 status was verified in an independent set of 50 UM tumors (Fig. 4a),20 consistent with our observation in the TCGA cohort (Fig. 4b). A comparative analysis showed M3 UM to have the second highest median level of SDHA gene expression across 31 cancer types, second only to the highly metabolic chromophobe kidney cancer (Supplementary Fig. S4; Supplementary Table S6).
Figure 4.
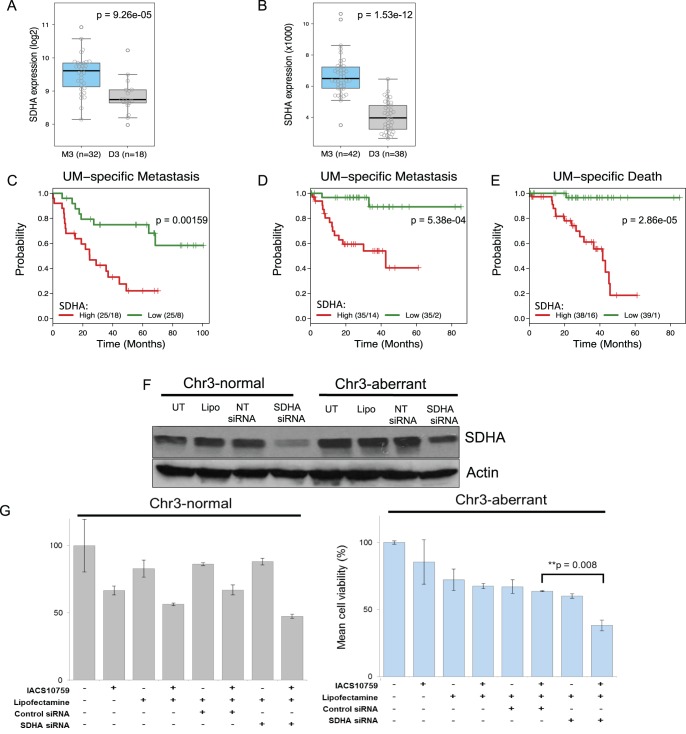
Monosomy 3 UM tumors display a heightened gene expression of specific ETC complex II genes. (A) SDHA gene expression in M3 (n = 32) versus D3 (n = 18) UM tumors.20 SDHA expression was significantly higher in M3 than in D3 UM tumors (P = 9.3·10−5, Kruskal-Wallis test). Box plots show median values and the 25th to 75th percentile range in the data (i.e., the IQR). Whiskers extend 1.5 times the IQR. Circles show all data values. (B) SDHA gene expression of M3 (n = 42) versus D3 (n = 38) UM tumors from UM TCGA data. SDHA expression (normalized RSEM mRNA expression) was significantly higher in M3 tumors than in D3 tumors (P = 1.5·10−12, Kruskal-Wallis test). (C–E) Kaplan-Meier analyses with log-rank tests for SDHA expression groups that were above and below the median. For each graph, numbers give the cases and events in each group. UM-specific metastasis for (C), Laurent cohort (n = 50), (D) UM TCGA cohort (n = 70). (E) UM-specific death for the UM TCGA cohort (n = 77). (F) Western blot analysis showing successful knockdown of SDHA protein resulting from SDHA siRNA transfection in both the Chr3-aberrant cell line Mel20–06–039 and the Chr3-normal Mel202 cell line. UT, untransfected; Lipo, lipofectamine-treated; NT, nontargeting. (H) Restoration of OxPhos inhibitor IACS-010759 sensitivity in Chr3-aberrant UM cells by SDHA knockdown. Right: siRNA-based knockdown of SDHA in cell line Mel20–06–039 (M3). Left: Mel202 cells (D3).
Kaplan-Meier analysis of both cohorts showed high SDHA expression to be strongly associated with a poor clinical outcome. In the Laurent cohort, for which only UM-specific metastasis data were available, SDHA expression was significantly associated with UM metastasis (median time to– UM-specific metastasis, high SDHA group = 24.5 months, low SDHA group = not reached, log-rank P = 1.6·10−3) (Fig. 4c). Likewise, patients from the UM TCGA cohort with SDHA-high UM tumors showed a markedly shorter time to UM-specific metastasis (median time to UM-specific metastasis, 42.6 months, low SDHA group = not reached, log-rank P = 5.4·10−4) (Fig. 4d). Further, patients with SDHA-high UM tumors had a significantly shorter time to UM-specific death (median survival time, 41.6 months, low SDHA group = not reached, log-rank P = 2.9·10−5) (Fig. 4e).
The oxidation of succinate by Complex II SDHA liberates two electrons, which, along with electrons generated by Complex I, are further shuttled between the remaining Complexes in the ETC by soluble mediators, ubiquinone (UBQ) and cytochrome c (CYC1). COQ6, a cofactor required for the biosynthesis of UBQ, is among the most highly expressed genes in the TCGA UM cohort (P = 1.1·10−13, Kruskal-Wallis test), as well as in the Laurent cohort in M3 UM (P = 5.8·10−5) (Supplementary Figs. S5A, S5B). Given that CYC1 is localized to Chr 8q, which has greater increases in copy number in M3 versus D3 UM, we observed a statistically significant elevation of CYC1 in M3 UM in both cohorts (P = 1.64·10−8 for TCGA UM, P = 2.0·10−3 for Laurent cohort, respectively) (Supplementary Figs. S5A, S5B). Elevated expression of COQ6 and CYC1 were also both associated with poor clinical outcome in both cohorts (Supplementary Figs. S5C–E).
Normalization of SDHA in M3 UM Restores Sensitivity to OxPhos Inhibition
Given the clear association between elevated SDHA and OxPhos antagonist resistance in M3 UM, we assessed whether normalization of SDHA levels would be sufficient to improve the sensitivity of M3 UM cells to OxPhos antagonism. Thus, M3 UM cells with targeted lowering of SDHA, to levels similar to D3 UM cells (Fig. 4f), were treated with IACS-010759 (Fig. 4g, Supplementary Fig. S6). Sensitivity to the OxPhos inhibitor was increased in M3 UM cells with normalized SDHA levels (Fig. 4g, Supplementary Fig. S6), indicating that elevated SDHA expression confers resistance to OxPhos antagonism.
Discussion
Patients with M3 UM have a very high risk of developing metastases and dying from their disease. Although inhibition of growth signaling pathways shows preclinical promise,24,25 no clearly effective therapies have been realized in clinical trials.26 Thus, there is a critical need to better understand the molecular pathology of M3 UM and identify potential novel treatment approaches.
SDHA is an essential metabolic enzyme that functionally couples the flux of the TCA cycle with the ETC electron transfer associated with OxPhos. Loss-of-function mutations in the SDH genes (SDHA, SDHB, SDHC and SDHD), which comprise Complex II of the ETC, increase the propensity for cellular transformation and the development of tumors.27 Mutations in SDH genes and in fumarate hydratase (FH), which catalyzes in the TCA cycle, result in the accumulation of succinate or fumarate, respectively. The build-up of these dicarboxylic acids is akin to the accumulation of D-2-hydroxyglutarate (D-2HG) that results from mutations in isocitrate dehydrogenase-1 or -2 (IDH-1 or -2). The term “oncometabolites” has emerged to describe the products of these (SDH, FH, IDH1, and IDH2) metabolic genes when they are mutated. Each of these interfere with dioxygenase function and promote tumorigenesis.28
Our data support a distinctly different cancer metastasis mechanism in which elevated, nonmutant SDHA drives pathological metabolism in M3 UM. Our molecular studies revealed high SDHA levels to be central to a distinct metabolic program with a markedly elevated maximal and reserve OxPhos respiration capacity, and resistance to OxPhos antagonism. M3 UM tumors also showed significantly increased expression of COQ6, the cofactor for the biosynthesis of ubiquinone, which shuttles electrons from Complex II to Complex III of the ETC. M3 UM tumors also had increased expression of CYC1, which shuttles electrons from Complex III to IV. Notably, M3 UM tumors are highly associated with chromosome 8q copy number gain,29 and CYC1 localizes to Chr 8q24.3. Thus, M3 UM cells are metabolically reprogrammed with heightened levels of the key TCA enzyme, SDHA, and also with ETC electron-shuttling molecules to respond to cellular states with high energy demands that require heightened OxPhos. Clinically, elevated SDHA in UM tumors was associated with significantly shorter times to metastasis and death.
Two recent studies have implicated BAP1, on chromosome 3, as playing a role in metabolic function, although the conclusions of the studies are somewhat contradictory. In Baughman et al.,30 loss of BAP1, and its de-ubiquitinating role, resulted in mitochondrial protein depletion, due to hyperubiquitination. In Hebert et al.,31 reconstitution of BAP1wt in a BAP1−/− mesothelioma cell line (NCI-H226) resulted in a modest decrease in the level of 20 mitochondrial proteins across each of the five ETC complexes, and an elevated reserve respiratory capacity. Consistent with our study, SDHA was one of the 20 proteins identified, and our work shows that normalization of elevated SDHA is sufficient to sensitize M3 UM cells to OxPhos antagonism, supporting its key role in UM cellular pathology.
Targeting the metabolic substrates and ETC features required for pathological OxPhos regulation has emerged as a potential anticancer strategy for some tumor types.32 Modulating particular members of specific ETC complexes appears to be a feasible approach, although cancer cell versus normal cell metabolic dependencies and context-dependent differences among ETC pathway members need to be taken into account. To translate our current findings to the clinical setting, studies should be aimed at better understanding the substrate reliance(s), divergent pathway responses, and efficacy of ETC complex member-specific inhibitors in M3 UM.
Supplementary Material
Acknowledgments
The authors thank Marina Protopopova for help with Seahorse Analysis, the Flow Cytometry and Cellular Imaging Facility for assistance with mitotracker imaging, and Jennifer McGee for her expert graphical design work.
Supported by Miriam and Sheldon Adelson AMRF (CC, JR, EAG), MELANOMA SPORE MDACC (CC, EAG, SEW), MDACC Melanoma Moonshot (SEW, JO), Melanoma Research Alliance Young Investigator Award, University of Texas System Rising Star Award, NCI SPORE in Melanoma (P50 CA093459) Developmental Research Project Award, NIH/NCI Cancer Center Support Grant P30 CA016672 Faculty Award (SEW), the George E. and Ruth Moss Trust, Cancer Prevention and Research Institute of Texas (SEW, JO), and an unrestricted grant from Research to Prevent Blindness (TMC).
Disclosure: C. Chattopadhyay, None; J. Oba, None; J. Roszik, None; J.R. Marszalek, None; K. Chen, None; Y. Qi, None; K. Eterovic, None; A.G. Robertson, None; J.K. Burks, None; T.A. McCannel, None; E.A. Grimm, None; S.E. Woodman, None
References
- 1.McLaughlin CC, Wu XC, Jemal A, Martin HJ, Roche LM, Chen VW. Incidence of noncutaneous melanomas in the U.S. Cancer. 2005;103:1000–1007. doi: 10.1002/cncr.20866. [DOI] [PubMed] [Google Scholar]
- 2.Jager MJ, Dogrusoz M, Woodman SE. Uveal melanoma: identifying immunological and chemotherapeutic targets to treat metastases. Asia Pac J Ophthalmol (Phila) 2017;6:179–185. doi: 10.22608/APO.201782. [DOI] [PubMed] [Google Scholar]
- 3.Chattopadhyay C, Kim DW, Gombos DS, et al. Uveal melanoma: from diagnosis to treatment and the science in between. Cancer. 2016;122:2299–2312. doi: 10.1002/cncr.29727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ewens KG, Kanetsky PA, Richards-Yutz J, et al. Chromosome 3 status combined with BAP1 and EIF1AX mutation profiles are associated with metastasis in uveal melanoma. Invest Ophthalmol Vis Sci. 2014;55:5160–5167. doi: 10.1167/iovs.14-14550. [DOI] [PubMed] [Google Scholar]
- 5.Vander Heiden MG, DeBerardinis RJ. Understanding the intersections between metabolism and cancer biology. Cell. 2017;168:657–669. doi: 10.1016/j.cell.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci Adv. 2016;2:e1600200. doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfleger J, He M, Abdellatif M. Mitochondrial complex II is a source of the reserve respiratory capacity that is regulated by metabolic sensors and promotes cell survival. Cell Death Dis. 2015;6:e1835. doi: 10.1038/cddis.2015.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Molina JR, Sun Y, Protopopova M, et al. An inhibitor of oxidative phosphorylation exploits cancer vulnerability. Nat Med. 2018;24:1036–1046. doi: 10.1038/s41591-018-0052-4. [DOI] [PubMed] [Google Scholar]
- 9.Burgess BL, Rao NP, Eskin A, Nelson SF, McCannel TA. Characterization of three cell lines derived from fine needle biopsy of choroidal melanoma with metastatic outcome. Mol Vis. 2011;17:607–615. [PMC free article] [PubMed] [Google Scholar]
- 10.von Euw E, Atefi M, Attar N, et al. Antitumor effects of the investigational selective MEK inhibitor TAK733 against cutaneous and uveal melanoma cell lines. Mol Cancer. 2012;11:22. doi: 10.1186/1476-4598-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griewank KG, Yu X, Khalili J, et al. Genetic and molecular characterization of uveal melanoma cell lines. Pigment Cell Melanoma Res. 2012;25:182–187. doi: 10.1111/j.1755-148X.2012.00971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amirouchene-Angelozzi N, Nemati F, Gentien D, et al. Establishment of novel cell lines recapitulating the genetic landscape of uveal melanoma and preclinical validation of mTOR as a therapeutic target. Mol Oncol. 2014;8:1508–1520. doi: 10.1016/j.molonc.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jager MJ, Magner JA, Ksander BR, Dubovy SR. Uveal melanoma cell lines: where do they come from? (An American Ophthalmological Society Thesis) Trans Am Ophthalmol Soc. 2016;114:T5. [PMC free article] [PubMed] [Google Scholar]
- 14.Ksander BR, Rubsamen PE, Olsen KR, Cousins SW, Streilein JW. Studies of tumor-infiltrating lymphocytes from a human choroidal melanoma. Invest Ophthalmol Vis Sci. 1991;32:3198–3208. [PubMed] [Google Scholar]
- 15.De Waard-Siebinga I, Blom DJ, Griffioen M, et al. Establishment and characterization of an uveal-melanoma cell line. Int J Cancer. 1995;62:155–161. doi: 10.1002/ijc.2910620208. [DOI] [PubMed] [Google Scholar]
- 16.Verbik DJ, Murray TG, Tran JM, Ksander BR. Melanomas that develop within the eye inhibit lymphocyte proliferation. Int J Cancer. 1997;73:470–478. doi: 10.1002/(sici)1097-0215(19971114)73:4<470::aid-ijc3>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 17.Chen K, Meric-Bernstam F, Zhao H, et al. Clinical actionability enhanced through deep targeted sequencing of solid tumors. Clin Chem. 2015;61:544–553. doi: 10.1373/clinchem.2014.231100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dettmer K, Aronov PA, Hammock BD. Mass spectrometry-based metabolomics. Mass Spectrom Rev. 2007;26:51–78. doi: 10.1002/mas.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robertson AG, Shih J, Yau C, et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell. 2017;32:204–220.e215. doi: 10.1016/j.ccell.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laurent C, Valet F, Planque N, et al. High PTP4A3 phosphatase expression correlates with metastatic risk in uveal melanoma patients. Cancer Res. 2011;71:666–674. doi: 10.1158/0008-5472.CAN-10-0605. [DOI] [PubMed] [Google Scholar]
- 21.Farshidfar F, Zheng S, Gingras MC, et al. Integrative genomic analysis of cholangiocarcinoma identifies distinct IDH-mutant molecular profiles. Cell Rep. 2017;18:2780–2794. doi: 10.1016/j.celrep.2017.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoadley KA, Yau C, Hinoue T, et al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell. 2018;173:291–304.e296. doi: 10.1016/j.cell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cancer Genome Atlas N. Genomic classification of cutaneous melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chattopadhyay C, Grimm EA, Woodman SE. Simultaneous inhibition of the HGF/MET and Erk1/2 pathways affect uveal melanoma cell growth and migration. PLoS One. 2014;9:e83957. doi: 10.1371/journal.pone.0083957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khalili JS, Yu X, Wang J, et al. Combination small molecule MEK and PI3K inhibition enhances uveal melanoma cell death in a mutant GNAQ- and GNA11-dependent manner. Clin Cancer Res. 2012;18:4345–4355. doi: 10.1158/1078-0432.CCR-11-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J, Manson DK, Marr BP, Carvajal RD. Treatment of uveal melanoma: where are we now? Ther Adv Med Oncol. 2018;10:1758834018757175. doi: 10.1177/1758834018757175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bezawork-Geleta A, Rohlena J, Dong L, Pacak K, Neuzil J. Mitochondrial complex II: at the crossroads. Trends Biochem Sci. 2017;42:312–325. doi: 10.1016/j.tibs.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sciacovelli M, Frezza C. Oncometabolites: unconventional triggers of oncogenic signalling cascades. Free Radic Biol Med. 2016;100:175–181. doi: 10.1016/j.freeradbiomed.2016.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson CP, Kim IK, Esmaeli B, et al. Systematic genomic and translational efficiency studies of uveal melanoma. PLoS One. 2017;12:e0178189. doi: 10.1371/journal.pone.0178189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baughman JM, Rose CM, Kolumam G, et al. NeuCode proteomics reveals Bap1 regulation of metabolism. Cell Rep. 2016;16:583–595. doi: 10.1016/j.celrep.2016.05.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hebert L, Bellanger D, Guillas C, et al. Modulating BAP1 expression affects ROS homeostasis, cell motility and mitochondrial function. Oncotarget. 2017;8:72513–72527. doi: 10.18632/oncotarget.19872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kluckova K, Bezawork-Geleta A, Rohlena J, Dong L, Neuzil J. Mitochondrial complex II, a novel target for anti-cancer agents. Biochim Biophys Acta. 2013;1827:552–564. doi: 10.1016/j.bbabio.2012.10.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.