Abstract
Transposable elements in prokaryotes are found in many forms and therefore a robust nomenclature system is needed in order to allow researchers to describe and search for them in publications and databases. Here we provide an update on The Transposon Registry which allocates numbers to any prokaryotic transposable element. Additionally, we present the completion of registry records for all transposons assigned Tn numbers from Tn1 onwards where sequence data or publications exist.
Keywords: Transposon, Tn, Registry, Conjugative transposon, Composite transposon, Integrative conjugative transposon, Antimicrobial resistance, Accessory function, Horizontal gene transfer, Plasmid
Introduction and background
A diverse array of transposable elements (TEs) can be found in the genomes of most prokaryotes where their evolutionary strategies for replication and mobility are often inextricably linked with fundamental roles in the evolution and adaptation of their hosts. Defined as “specific DNA segments that can repeatedly insert into one or more sites in one or more genomes” [1] an increasing variety of TEs are being discovered, facilitated by the normalisation of rapid whole genome sequencing and analysis. The opportunity for a researcher to be able to use a number-based, characteristic-free (in terms of the TE genome), nomenclature system to assign a unique name to a new TE means that the sequence of, and associated publications on, the newly described TE will be searchable and the nomenclature system can deal with any manner of genetic variation within TEs and their hosts.
The Transposon Registry is a nomenclature system for the assignment of Tn numbers for bacterial and archaeal autonomous TEs, including unit transposons, composite transposons, conjugative transposons (CTns)/Integrative Conjugative Elements (ICEs), Mobilisable transposons (MTns)/Integrative mobilisable elements (IMEs) and mobile genomic islands. The online registry is well established and has been utilised extensively over the last decade assigning over 700 Tn numbers to researchers to date. It excludes insertion sequences (ISs), which are managed by ISfinder database (www-is.biotoul.fr) and other TEs such as introns and inteins for which other databases already exist, and non-autonomous TEs such as integron cassettes and MITES. It is also worth noting that alternative nomenclature guidelines have been proposed for the ICEs and IMEs which uses “ICE” or “IME” followed by a two or three letter acronym of the host and a sequential number [2]. There is however cross-over between these two nomenclature systems as the “Tn” part of a newly designated Tn number from The Transposon Registry can be written as “CTn” (for conjugative transposon) or “ICE” or indeed “MTn” or “IME” as preferred [1] which enables researchers to cope with findings from metagenomes where host identification may not be possible. In this short review we provide a historical perspective, thoughts on future directions for TE annotation and nomenclature and an update on The Transposon Registry.
What’s in a name?
Why should researchers contemplate naming a newly identified TE? It is useful to name a mobile genetic element such as a TE so the discoverer and other researchers can specifically refer to it. This is becoming increasingly important as our understanding about the influence of TEs upon their hosts becomes more apparent. Epidemiological and evolutionary studies throughout biology require names in order to refer to any mobile and / or evolving entity, whether this is a macro-scale population or a molecular scale unit such as DNA. This is particularly appropriate when describing mobile genetic elements due to their recombinatorial nature, and their ability to spread prolifically between different hosts, and within their hosts around the planet. Using higher order names such as composite transposon or resistance plasmid does not allow for the fine scale details to be described following comparative analysis within the increasingly large DNA sequence databases and the proliferation of microbial genome sequences.
A historical perspective on Tn numbers
The nomenclature of transposable elements was first discussed in a meeting on DNA Insertions at Cold Spring Harbor in 1976. A set of rules for the nomenclature were modified based on the proposal from D.E. Berg and W. Szybalski, which was subsequently revised in 1979, due to development of early DNA sequencing techniques [3, 4]. Insertion sequences and transposable elements were named separately by having IS and Tn as a prefix, respectively, followed by a sequential number in italics such as IS1, IS2 and Tn1, Tn2, etc. The administration and allocation of numbers were carried out by the late Dr. Esther Lederberg from Stanford University Medical School, CA, USA. The names and locations of registrants for Tn1 to Tn4685 were published previously [5, 6]. The allocations were continued up to Tn5500 and above but were not published as a list and allocation ceased when Dr. Lederberg stopped running the plasmid reference centre.
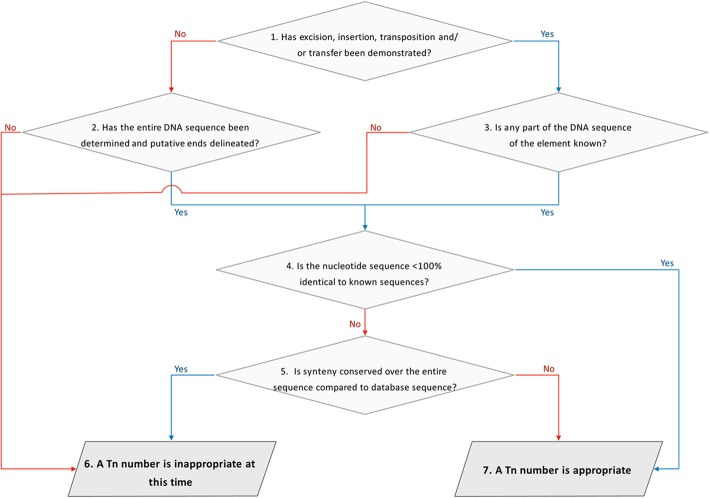
Subsequently, a variety of nomenclature systems were adopted by different research groups, especially for novel types of TE, due to the discontinuation of the sequential numbering system and the need to name mobile genetic elements being studied as outlined above. To fill this gap the “Tn Registry” was launched in 2006, hosted by University College London, London, UK and an accompanying description published in 2008 [1]. It began assigning Tn numbers from Tn6000 to avoid any duplicative assignations as there are published records within the 5000 range (see Additional file 1: Table S1). A set of criteria, was also proposed to determine whether a new Tn number is appropriate [1], summarized in Fig. 1.
Fig. 1.
Flow chart for the determining whether a transposon suitable for a new Tn number based on the guidelines published previously [1]
The Transposon Registry
In 2017 the Tn registry moved to Liverpool School of Tropical Medicine (LSTM), Liverpool, UK, (https://transposon.lstmed.ac.uk/) and was upgraded, updated and renamed “The Transposon Registry”. The registry is now fully searchable and can be updated by users as and when their sequences are deposited and when their publications arise by linking their individual records to accession numbers and digital object identifiers respectively. In order to obtain a Tn number, users simply register and request as many as they need when they need them; there is no reason to request surplus numbers as we are not going to run out. Subsequent to its relocation to LSTM, we have collated and backfilled The Transposon Registry with all published records from Tn1 to Tn5999, where either a publication or a sequence is available. It is worth noting that many records assigned previously do not have a sequence or publication associated with them. This we think is in part because researchers were assigned blocks of numbers by traditional postal methods to use as and when they were needed, something which is no longer necessary due to the online nature of The Transposon Registry. We have also updated the records from Tn6000 onwards where users have not yet had the opportunity to do so.
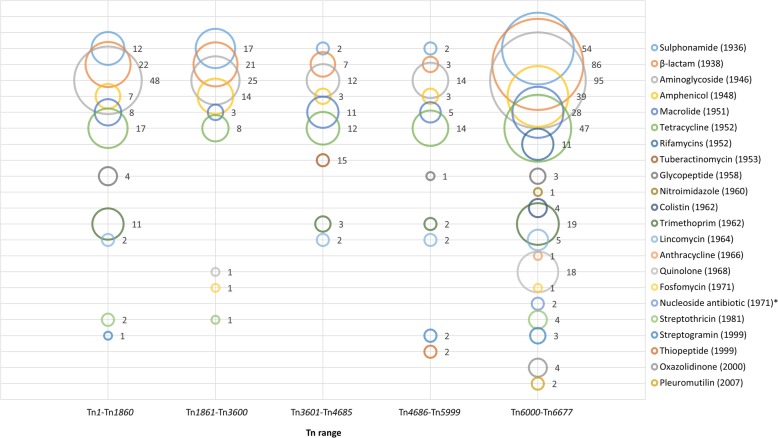
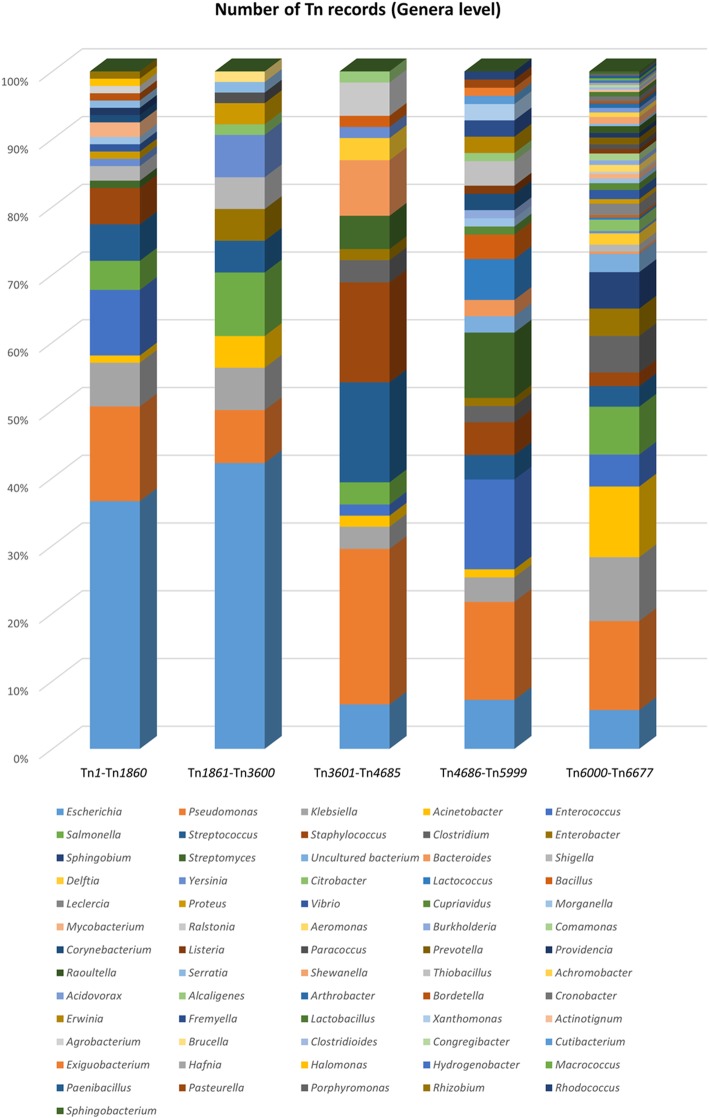
The current, complete information on all available records in The Transposon Registry is summarized in Table 1, including available metadata on the types, size, original host, accession numbers, accessory functions and references (Table 1). The accessory functions assigned to cargo genes primarily include antimicrobial resistance and show that antibiotic resistance genes against all major antibiotic classes are increasingly found to be associated with transposons and an increase in the numbers of antibiotics to which resistance is conferred by transposon located genes (Fig. 2 and Additional file 2: Table S2). Also noted are antiseptic resistance, heavy metal resistance, efflux functions, metabolic capability and virulence factor and CRISPR functions. It is interesting to note the trends observable with respect to the hosts of transposons being reported (Fig. 3). It is clear that more transposons are being reported in Acinetobacter and Klebsiella and that the diversity of transposon hosts is increasing, presumably as sequencing becomes more common place. As the identification of the host species is not a requirement for the nomenclature system, transposable elements identified from metagenomic studies are also included, e.g. Tn6032 and Tn6300. Details of all the entries within the Transposon Registry are provided in the Additional file 1: Table S1.
Table 1.
Summary of all available records in The Transposon Registry based on the data in Additional file 1
| Characteristics | Number | ||
|---|---|---|---|
| Available records in The Transposon Registry (Records) | Tn1-Tn1860 [3] | 102 | |
| Tn1861-Tn3600 [5] | 64 | ||
| Tn3601-Tn4685 [6] | 62 | ||
| Tn4686-Tn5999(Not published) | 83 | ||
| Tn6000-Tn6677(This work) | 299 | ||
| Total | 610 | ||
| Types and Size of transposons | Unit transposons | Number of records | 313 |
| Average (range) in kb | 12.4 (2.4–86) | ||
| Composite transposons | Number of records | 136 | |
| Average (range) in kb | 11.7 (1.5–65) | ||
| Conjugative transposons | Number of records | 97 | |
| Average (range) in kb | 45.7 (9.6–120) | ||
| Composite transposon and unit transposon | Number of records | 23 | |
| Average (range) in kb | 36.6 (8.4–111.2) | ||
| Not specified | Number of records | 41 | |
| Overall | Number of records | 610 | |
| Average (range) in kb | 19.1 (1.5–120) | ||
| Accessory functions (Records) | AMR | 399 | |
| Antiseptic resistance | 18 | ||
| Efflux functions | 31 | ||
| Heavy metal resistance | 32 | ||
| Metabolism | 15 | ||
| Virulence determinants | 127 | ||
| CRISPR | 1 | ||
| Other / not defined | 97 | ||
| Accession numbers (Records) | Available in GenBank | 448 | |
| Not available in GenBank | 162 | ||
| Publication (Records) | Tn with available publication | 583 | |
| No publication | 27 | ||
Fig. 2.
Bubble chart depicting the number of Tn records containing resistance genes against different types of antibiotic classes. Antibiotic classes were sorted according to the year of introduction on the Y-axis. Tn records were grouped into 5 groups on the X-axis according to the previously published lists [3–5], and before and after the allocation by the Tn registry (starting at Tn6000). The number of Tn records is represented by the bubble size and also indicated on the right of each bubble. * year of discovery as approval never received
Fig. 3.
Number of Transposon records displayed by host at genera level. The range within the 5 columns are representative of the previously published lists [3–5] and before and after allocation by the Tn Registry (starting at Tn6000)
Future perspectives and conclusions
As research into TE biology continues its migration from primarily experimental biology, where mobility is proven, to a more comparative genomics approach it is likely that the available bioinformatic tools which are used to identify TEs (reviewed in [7] and references therein) will be improved, both in terms of their application for interrogating different host species, and delineating different classes of TE. The complexity of TEs; their variety and biology, continue to be better understood and the nomenclature will need to catch up to cope with this increasing knowledge [7]. It is worth reiterating here, the call for a formal discussion within the international community to fully address the problematic issues of TE nomenclature and come up with an agreeable system which can accommodate all classes of TE we see [7, 8] and be future proof to accommodate those that we as yet can only imagine. Until that time, we will continue to operate and update The Transposon Registry with any missing or inaccurate records highlighted by the community. We welcome inclusion of TEs named using other systems to be included within The Transposon Registry and call on the community to provide these details if they wish for them to be included.
Supplementary information
Additional file 1: Table S1. Details of transposon records available in The Transposon Registry (DOCX 529 kb)
Additional file 2: Table S2. List of transposons associated with antibiotic resistance genes (DOCX 24 kb)
Acknowledgements
The authors would like to Mr. Diderik van Halsema at the Liverpool School of Tropical Medicine for his vision of utility for the scientific community. We would also like to thank all those involved in the project at Manta Ray Media Ltd., London, UK for their development expertise and continued platform support.
Authors’ contributions
APR conceptualised and implemented The Transposon Registry. ST and APR wrote the manuscript. ST and MAR compiled data. ST generated the figures and tables. All authors read and approved the final manuscript.
Funding
Not applicable
Availability of data and materials
The dataset used to generate Figs. 2 and 3 and all tables is available in the Additional file 1: Table S1 and Additional file 2: Table S2.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The author declares that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13100-019-0182-3.
References
- 1.Roberts AP, Chandler M, Courvalin P, Guédon G, Mullany P, Pembroke T, et al. Revised nomenclature for transposable genetic elements. Plasmid. 2008;60(3):167–173. doi: 10.1016/j.plasmid.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burrus V, Pavlovic G, Decaris B, Guedon G. Conjugative transposons: the tip of the iceberg. Mol Microbiol. 2002;46(3):601–610. doi: 10.1046/j.1365-2958.2002.03191.x. [DOI] [PubMed] [Google Scholar]
- 3.Campbell A, Berg DE, Botstein D, Lederberg EM, Novick RP, Starlinger P, et al. Nomenclature of transposable elements in prokaryotes. Gene. 1979;5(3):197–206. doi: 10.1016/0378-1119(79)90078-7. [DOI] [PubMed] [Google Scholar]
- 4.Campbell A, Starlinger P, Berg DE, Botstein D, Lederberg EM, Novick RP, et al. Nomenclature of transposable elements in prokaryotes. Plasmid. 1979;2(3):466–473. doi: 10.1016/0147-619X(79)90030-1. [DOI] [PubMed] [Google Scholar]
- 5.Lederberg EM. Plasmid reference center registry of transposon (Tn) allocations through July 1981. Gene. 1981;16(1–3):59–61. doi: 10.1016/0378-1119(81)90060-3. [DOI] [PubMed] [Google Scholar]
- 6.Lederberg EM. Plasmid reference center registry of transposon (Tn) and insertion sequence (IS) allocations through December 1986. Gene. 1987;51(2–3):115–118. doi: 10.1016/0378-1119(87)90299-X. [DOI] [PubMed] [Google Scholar]
- 7.Arkhipova IR. Using bioinformatic and phylogenetic approaches to classify transposable elements and understand their complex evolutionary histories. Mob DNA. 2017;8(1):19. doi: 10.1186/s13100-017-0103-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arensburger P, Piégu B, Bigot Y. The future of transposable element annotation and their classification in the light of functional genomics - what we can learn from the fables of Jean de la Fontaine? Mob Genet Elem. 2016;6(6):e1256852-e. doi: 10.1080/2159256X.2016.1256852. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Details of transposon records available in The Transposon Registry (DOCX 529 kb)
Additional file 2: Table S2. List of transposons associated with antibiotic resistance genes (DOCX 24 kb)
Data Availability Statement
The dataset used to generate Figs. 2 and 3 and all tables is available in the Additional file 1: Table S1 and Additional file 2: Table S2.