Abstract
A heterotrimeric complex containing Lin-10/X11α, Lin-2/CASK, and Lin-7 is evolutionarily conserved from worms to mammals. In Caenorhabditis elegans, it localizes Let-23, a receptor tyrosine kinase, to the basolateral side of vulval epithelium, a step crucial for proper vulva development. In mammals, the complex may also participate in receptor targeting in neurons. Accordingly, phosphotyrosine binding (PTB) and postsynaptic density-95/Discs large/Zona Occludens-1 domains found in X11α and mLin-2/CASK bind to cell-surface proteins, including amyloid precursor protein, neurexins, and syndecans. In this paper, we have further analyzed the X11α–mLin-2/CASK association that is mediated by a novel protein–protein interaction. We show that the mLin-2/CASK calmodulin kinase II (CKII) domain directly binds to a 63 amino acids peptide located between the Munc-18-1 binding site and the PTB domain in X11α. Ca2+/calmodulin association with mLin-2/CASK does not modify the X11α–mLin-2 interaction. A region containing the mLin-2/CASK guanylate kinase domain also interacts with X11α but with a lower affinity than the CKII domain. Immunostaining of X11α in the brain shows that the protein is expressed in areas shown previously to be positive for mLin-2/CASK staining. Together, our data demonstrate that the X11α–mLin-2 complex contacts many partners, creating a macrocomplex suitable for receptor targeting at the neuronal plasma membrane.
Keywords: PDZ, PTB, X11, mLin-2/CASK, CaM kinase, receptor localization
Protein–protein interactions govern signal transduction networks and localization of proteins in the cells. They are mediated by a growing number of protein modules that bind peptide and lipid targets (Pawson and Scott, 1997). Phosphotyrosine binding (PTB) domains found in Shc and insulin receptor substrate-1 bind to tyrosine phosphorylated peptides and play an important role in tyrosine kinase signaling, whereas analogous domains identified in X11, Numb, and Fe65 prefer unmodified peptides (Borg and Margolis, 1998). Postsynaptic density-95/Discs large/Zona Occludens-1 (PDZ) domains bind C-terminal peptides and participate in receptor localization (Fanning and Anderson, 1997). For example, the well characterized postsynaptic density-95 (PSD-95) protein engages interaction with NMDA receptor and K+ channels, a step important for their clustering at the plasma membrane (Kim et al., 1995; Kornau et al., 1995).
We recently described the X11 protein family containing one PTB and two PDZ domains. The X11 PTB domain interacts with a tyrosine-based peptide found in amyloid precursor protein (APP), a protein important for Alzheimer’s disease pathogenesis (Borg et al., 1996; McLoughlin and Miller, 1996; Zhang et al., 1997). This interaction allows X11 proteins to slow the processing of APP to Aβ, a peptide found in the plaques of patients with Alzheimer’s disease (Borg et al., 1998b). The X11 family comprises three members: X11α, X11β, and X11γ, also called Munc-interacting proteins (Mints) (Okamoto and Sudhof, 1997; Borg et al., 1998a). Although X11α and X11β are strictly neuronal, X11γ is expressed in all tissues (Duclos et al., 1993; Borg et al., 1998b). X11α participates in a brain-specific heterotrimeric complex containing two other PDZ domain proteins, mLin-2/CASK and mLin-7 (Borg et al., 1998a; Butz et al., 1998). The mLin-2/CASK protein also contains a calmodulin kinase II (CKII), an SH3, and a guanylate kinase (GK) domain and is related to other membrane associated guanylate kinase (MAGUK) proteins (Hata et al., 1996; Hoskins et al., 1996). InC. elegans, mutations of lin-10,lin-2, and lin-7 genes, representing the respective homologs of X11α, mLin-2/CASK, andmlin-7 in mammals, abrogate the basolateral localization of Let-23 in vulval epithelium and preclude subsequent activation by its ligand (Hoskins et al., 1996; Simske et al., 1996; Kaech et al., 1998). Mislocalization of Let-23 leads to a vulvaless phenotype equivalent to mutations affecting receptor activation.
We and others have demonstrated that the N terminus of X11α binds to mLin-2/CASK and that this interaction is required for thein vivo X11α–mLin-2 interaction (Borg et al., 1998a; Butz et al., 1998; Kaech et al., 1998). Here, we further delineate this binding site to a 63 amino acids peptide in X11α. The mLin-2/CASK CKII domain strongly interacts with this peptide in precipitation and overlay assays. Deletions of the CKII domain indicate that the integrity of the domain is required for this interaction. The calmodulin binding site located C-terminal to the mLin-2/CASK CKII domain is not required for binding to X11α, and binding of calmodulin does not interfere with the X11α–mLin-2/CASK interaction. We also demonstrate that the mLin-2/CASK GK domain and its flanking sequences interact with X11α in vitro. In situhybridization and immunostaining show that X11α has a somatodendritic pattern in neurons and is particularly expressed in olfactory bulb, cerebellum, cortex, and several brainstem nuclei. Cell-surface proteins, such as neurexins, syndecans, and APP, interact with the X11α–mLin-2/CASK complex through PTB and PDZ domain interactions (Borg et al., 1996; Hata et al., 1996; Cohen et al., 1998; Hsueh et al., 1998). Additional binding of mLin-7, Munc-18-1–Syntaxin, and calmodulin generates a neuronal multiprotein complex, which we predict will be involved in receptor localization.
MATERIALS AND METHODS
Antibodies. Anti-Myc 9E10 (Oncogene Research Products, Cambridge, MA) monoclonal antibody was used for immunoprecipitation and immunoblotting. Polyclonal anti-mLin-2/CASK and anti-X11 antibodies were described previously (Borg et al., 1998a). Anti-PSD-95 and anti-calmodulin monoclonal antibodies were from Upstate Biotechnology (Lake Placid, NY). Anti-T7 monoclonal antibody was from Novagen (Madison, WI). Anti-Syntaxin monoclonal antibody was from Sigma (St. Louis, MO). Anti-Munc-18-1 and anti-mLin-2/CASK monoclonal antibodies were from Transduction Laboratories (Lexington, KY). Avidin–biotin blocking kit, biotinylated goat anti-rabbit IgG, and streptavidin biotinylated horseradish peroxidase complex were purchased from Vector Laboratories (Burlingame, CA).
DNA constructs. Full-length X11α,X11β, and X11γ cDNAs have been described elsewhere (Borg et al., 1998a,b). Human lin-2 cDNA assembled from expressed sequence tags (identical to GenBank accession number AF035582) (Borg et al., 1998a) was used as a template to create different constructs, allowing expression of glutathioneS-transferase (GST) fusion proteins. The RK5-myc vector was used to express X11α and mLin-2/CASK fused to the myc epitope (Borg et al., 1996). All constructs were sequenced using Sequenase version 2.0 (Amersham, Cleveland, OH).
Cell culture. Human embryonic kidney 293 and A-172 cells were grown in DMEM (Life Technologies, Grand Island, NY) containing 100 U/ml penicillin and 100 μg/ml streptomycin sulfate, supplemented with 10% fetal calf serum (FCS). NT2 cells were maintained in DMEM–F-12 medium containing 100 U/ml penicillin and 100 μg/ml streptomycin sulfate, supplemented with 10% FCS.
Immunohistochemistry. Five male Sprague Dawley rats from Charles River Laboratories (Wilmington, MA), weighing 250–325 gm, were used in this study. The rats were anesthetized with sodium pentobarbital (50 mg/kg, i.p.) (Butler, Columbus, OH) and perfused transcardially with 0.1 m phosphate buffer (PB), pH 7.6, containing 15,000 U/l heparin, followed by 4% paraformaldehyde in 0.1 m PB. The brains were removed, post-fixed for 1 hr in the same fixative at 4°C, and then cryoprotected overnight at 4°C in 0.1 m PBS, pH 7.6, containing 20% sucrose. The brains were then frozen in isopentane cooled to −80°C. Brains were then sectioned on a Bright–Hacker cryostat, and the 40-μm-thick coronal sections were kept in antifreeze solution at −20°C until processed for immunohistochemistry.
Immunohistochemistry was performed as described previously, with slight modifications (Lõpez-Figueroa et al., 1996). The free-floating sections were washed three times for 10 min each in PBS and incubated in 0.3% H2O2 in methanol for 10 min at room temperature for quenching of the endogenous peroxidase activity. The sections were then washed three times for 10 min each in PBS. To reduce nonspecific background, sections were incubated in avidin–biotin blocking kit for 10 min, followed by incubation in a solution of 5% normal goat serum, 0.1% bovine serum albumin (BSA), and 0.3% Triton X-100 (TX) in PBS for 20 min. Sections were incubated with the specific antisera overnight at 4°C (polyclonal rabbit anti-X11 antibody used at 1:650) diluted in the previous solution. The sections were then rinsed three times for 10 min each in 0.1% TX in PBS, followed by incubation in biotinylated goat anti-rabbit IgG diluted 1:1000 in 0.1% TX in PBS for 1 hr at room temperature. After washing three times for 10 min each in PBS, the sections were incubated with streptavidin biotinylated horseradish peroxidase complex diluted 1:1000 in PBS for 1 hr at room temperature. The sections were then washed with PBS three times for 10 min each and developed with a solution of 0.05% diaminobenzidine, 0.006% nickel ammonium sulfate, and 0.001% H2O2 in 0.1 m sodium acetate. The sections were then washed twice in PBS, mounted, air dried, and coverslipped by use of Permount as mounting medium. Staining specificity was assessed with incubation of parallel sections in preincubated antiserum with its corresponding antigen. Images were captured using an Axiophot microscope (Zeiss, Oberkochen, Germany) and camera (DXC-970; Sony, Tokyo, Japan). In situhybridization. Four rats were killed by rapid decapitation, and their brains were removed and frozen in isopentane cooled to −80°C. Brains were then sectioned on a cryostat, and the 15-μm-thick coronal sections, mounted onto polylysine-coated slides, were kept at −80°C until processed for in situhybridization. Riboprobes encoding the human X11α N terminus were used in this study. Antisense probes were transcribed from the T3 promoter using T3 RNA polymerase. The probe was labeled with [35S]dUTP and [35S]dCTP as described previously (Lõpez-Figueroa et al., 1998). The linearized plasmid was incubated for 2 hr at 37°C in a solution containing 5× transcription buffer, [35S]dUTP and [35S]dCTP, 150 μm dNTPs, 12.5 mm dithiothreitol, 20 U of RNase inhibitor, and 6 U of the corresponding RNA polymerase. The labeled probe was then separated in a Sephadex G50/50 column.
Tissue sections were then processed for in situhybridization histochemistry. The sections were fixed in 4% paraformaldehyde for 1 hr, followed by three washes in 2× SSC (1× SSC is 150 mm NaCl and 15 mm sodium citrate). The sections were then placed in a solution containing acetic anhydride (0.25%) in triethanolamine (0.1 m, pH 8.0) for 10 min at room temperature, rinsed in distilled water (DW), and dehydrated through graded series of alcohol. After air drying, the sections were hybridized with the corresponding 35S-labeled cRNA probe in a hybridization buffer (containing 50% formamide, 10% dextran sulfate, 3× SSC, 50 mm sodium phosphate buffer, pH 7.4, 1× Denhardt’s solution, 0.1 mg/ml yeast tRNA, and 10 mmdithiothreitol) to yield 106 dpm/35 μl. The sections were coverslipped and placed inside a humidified box overnight at 55°C. After hybridization, the coverslips were removed, and the sections were rinsed and washed twice in 2× SSC for 5 min each and then incubated for 1 hr in RNase (200 μg/ml in Tris buffer containing 0.5 m NaCl, pH 8.0) at 37°C. The sections were washed in increasingly stringent solutions of 2×, 1×, and 0.5× SSC for 5 min each, followed by incubation for 1 hr in 0.1× SSC at 65°C. After rinsing in DW, the sections were dehydrated through graded alcohols, air dried, and exposed to a Kodak XAR film (Eastman Kodak, Rochester, NY) for 5–7 d. Finally, the sections were dipped into photographic emulsion (Kodak NTB-2), exposed for 13–17 d, developed in Kodak D-19 developer (2 min), fixed (3 min), and counter-stained with cresyl violet. Sections pretreated for 1 hr with RNase (200 μg/ml) or treated with sense riboprobes from the same plasmid insert were used as controls.
Immunostaining of NT2 cells. Differentiated NT2 cells were plated on acid-treated coverslips coated with poly-d-lysine (Sigma) and Matrigel (Collaborative Research, Bedford, MA). After fixation with PBS–4% paraformaldehyde, cells were washed with PBS–10 mm glycine and permeabilized with PBS–0.1% TX. After blocking for 1 hr in goat serum, coverslips were incubated with antibodies diluted in PBS–2% goat serum in a humidified chamber for 1 hr (affinity-purified anti-X11 at 1:500, anti-mLin-2/CASK at 1:125, anti-giantin at 1:1000, and 6E10 anti-APP at 1:40). All secondary antibodies coupled to FITC or Cy3 (Sigma) were diluted at 1:200 in PBS–2% goat serum. Confocal analysis of immunostaining was performed on a Noran confocal laser scanning imaging system (Noran Instruments, Middleton, WI) with a Nikon Diaphot 200 inverted microscope at the Morphology and Image Analysis Core of the University of Michigan Diabetes Research and Training Center.
Protein procedures. Cells were washed twice with cold PBS and lysed in lysis buffer (50 mm HEPES, pH 7.5, 10% glycerol, 150 mm NaCl, 1% TX, 1.5 mmMgCl2, and 1 mm EGTA) supplemented with 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, and 10 μg/ml leupeptin. After centrifugation at 16,000 × g for 20 min, lysate protein content was normalized using the Bio-Rad (Hercules, CA) protein assay kit. Mouse brain proteins were extracted after a similar procedure. For immunoprecipitation, lysates were incubated with antibodies overnight at 4°C. Protein A–agarose was added, and immune complexes bound to beads were recovered after 1 hr, washed three times with buffer (containing 50 mm HEPES, pH 7.5, 10% glycerol, 150 mm NaCl, and 0.1% TX), boiled in 1× sample buffer, and separated by SDS-PAGE. Transfer and immunoblotting on nitrocellulose using HRP protein A or HRP anti-mouse antibody chemiluminescence method were performed as described previously (Borg et al., 1996). For overlay assays, the membrane was incubated 2 hr at room temperature with soluble His-tagged or GST fusion proteins at 1 μg/ml in TBS–5% BSA and 1 mm DTT. After rinsing with TBS–0.1% TX and TBS buffers, the membrane was incubated with mouse monoclonal anti-T7 or rabbit polyclonal anti-GST antibodies diluted in TBS–5% BSA for 2 hr. The immune complexes were revealed using HRP goat anti-mouse antibody or HRP protein-A chemiluminescence method. Cell transfection, GST production, and GST binding assays were performed as described previously (Borg et al., 1996).
RESULTS
Mapping the X11α—mLin-2 binding sites
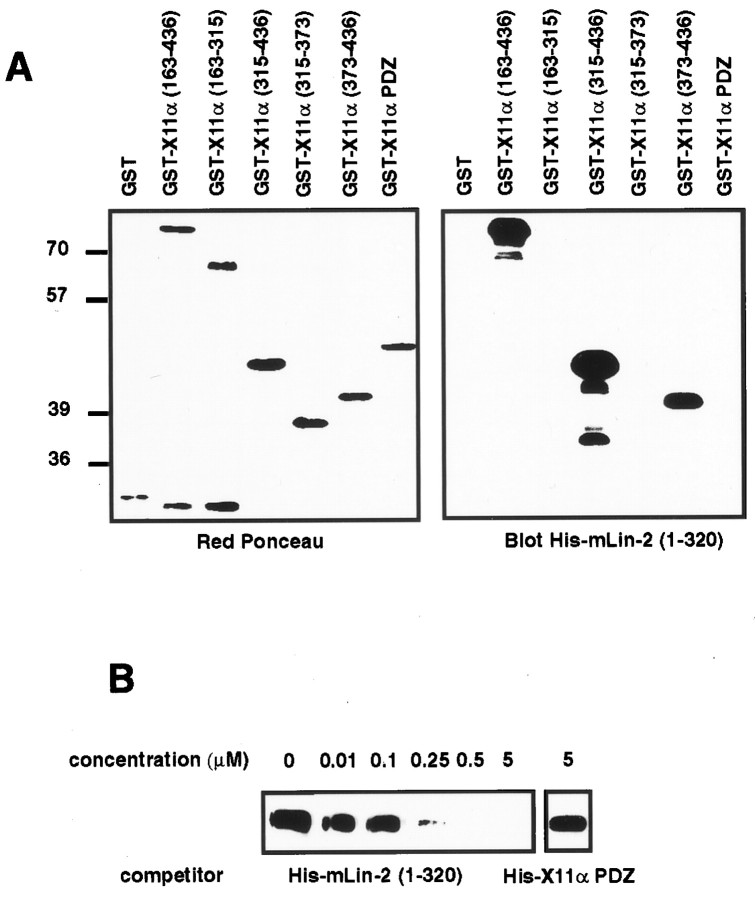
We have further delineated the site of interaction on the X11α N terminus for mLin-2/CASK (Fig.1A). In X11α, the 163–436 amino acids region is responsible for the coimmunoprecipitation between the two proteins (Borg et al., 1998a) and precipitates mLin-2/CASK from mouse brain extracts (Fig.1B). Analogous regions in X11β and X11γ do not interact with mLin-2/CASK. As shown previously, X11α and X11β N termini bind to the complex of Munc-18-1 and Syntaxin (Okamoto and Sudhof, 1997). In contrast, we found no interaction between Munc-18-1 or Syntaxin and the X11γ N terminus. PSD-95 did not bind to X11 fusion proteins (Fig. 1B). We have generated GST fusion proteins representing smaller peptides of X11α and examined the binding to mLin-2/CASK by GST precipitation (Fig. 1C; Table 1). We demonstrate that a region encompassing residues 373–436 in X11α is sufficient to bind mLin-2/CASK. No binding was detected with the X11α PTB and PDZ domains (Fig. 1C). The binding site was further subdivided in shorter peptides, but none of them could bind to mLin-2/CASK, suggesting that the X11α region 373–436 represents the minimal site of interaction (Table 1). As expected, no homologous regions are found in X11β and X11γ. Others have found that the region 226–314 in X11α is sufficient to bind Munc-18-1 (Okamoto and Sudhof, 1997). Accordingly, GST X11α (region 163–315) binds efficiently to the Munc-18-1–Syntaxin complex (Fig. 1C). Together, these data show that mLin-2/CASK and Munc-18-1 proteins bind to the X11α N terminus on two different sites.
Fig. 1.
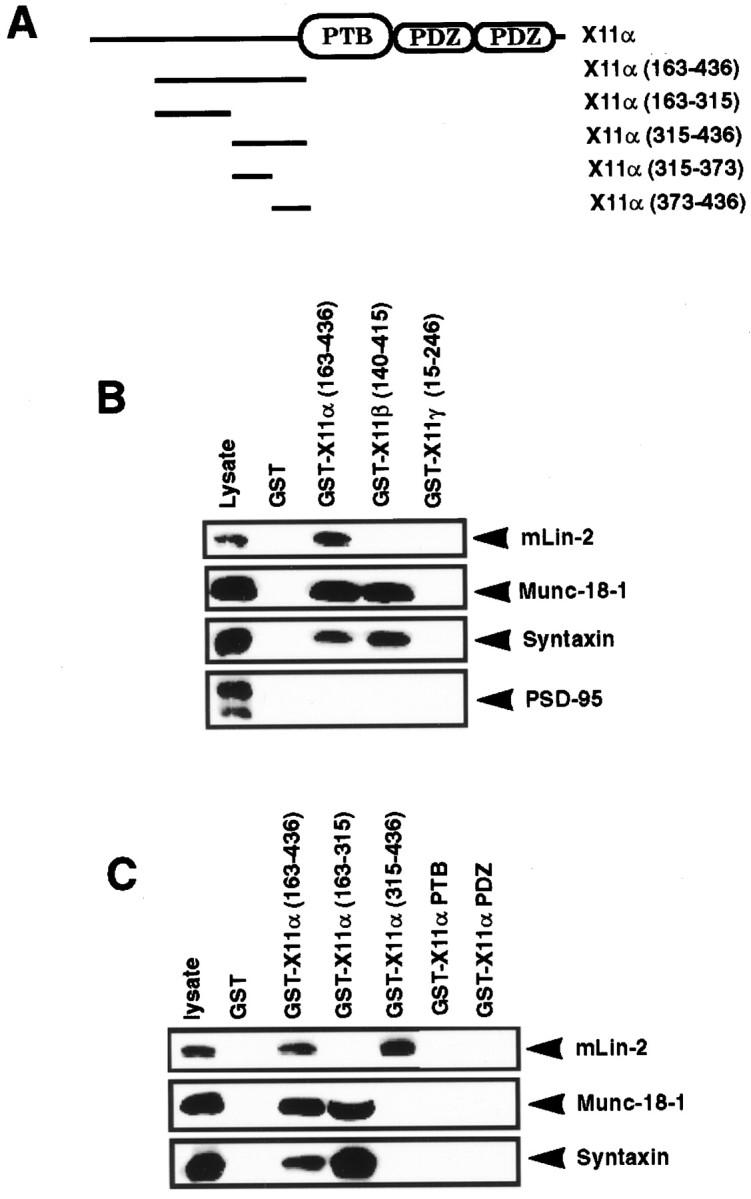
Delineation of the mLin-2/CASK binding site in X11α protein. A, Schematic representation of the X11α protein. The X11α protein contains a central PTB domain, followed at its C-terminal end by two PDZ domains. N-terminal fragments were produced as GST fusion proteins. B, Proteins extracted from mouse brain were precipitated with GST, GST X11α (region 163–436), GST X11β (region 140–415), or GST X11γ (region 15–246) coupled to glutathione beads. These fusion proteins incorporate peptides from the N terminus of these X11 isoforms. After washing, proteins were separated on 10% SDS-PAGE and transferred to nitrocellulose. The membrane was probed with polyclonal anti-mLin-2/CASK and monoclonal anti-Munc-18-1, anti-Syntaxin, and anti-PSD-95 antibodies. One-tenth of the lysate used for precipitation was run as control (lysate). Detection was performed by chemiluminescence. C, Same as B, with additional X11α fusion proteins. The GST X11α PTB and PDZ proteins comprise the PTB and the two PDZ domains of the protein, respectively.
Table 1.
X11α (region 373–436) is the minimal binding site for mLin-2
| X11α peptides | mLin-2 | Munc-18-1 |
|---|---|---|
| 163–436 | +++ | +++ |
| 163–315 | − | +++ |
| 315–436 | +++ | − |
| 315–373 | − | − |
| 373–436 | ++ | − |
| 373–392 | − | NT |
| 373–402 | − | NT |
| 390–408 | − | NT |
| 390–425 | − | NT |
X11α peptides fused to the GST protein were used to pull down mLin-2 from a 293 cells lysate or mouse brain extract. Bound protein was detected by Western blot or overlay assays. Binding to X11α (region 373–436) was reproducibly weaker than to larger peptides. NT, Non-tested.
The first 320 amino acids of mLin-2/CASK encompassing the CKII domain directly binds to full-length X11α by overlay assay (Borg et al., 1998a). We used this assay to show that this region interacts with the X11α (region 373–436) peptide (Fig.2A). Binding of GST mLin-2/CASK (region 1–320) to X11α was inhibited at a concentration of 250 nm soluble His-mLin-2/CASK (region 1–320), whereas 5 μm control protein did not affect the binding (Fig.2B). These data allow us to conclude that mLin-2/CASK (region 1–320) binds tightly to the 63 amino acid peptide found in the X11α N terminus.
Fig. 2.
The mLin-2/CASK CKII domain directly binds to an X11α (region 373–436) peptide. A, GST fusion proteins described in Figure 1C were subjected to SDS-PAGE and transferred to nitrocellulose. Equivalent amounts of proteins were revealed with Ponceau red stain (left). The membrane was probed with soluble His-mLin-2/CASK (region 1–320) protein, and bound proteins were revealed with anti-T7 antibody, followed by HRP goat anti-mouse and chemiluminescence detection (right).B, The same procedure was performed to detect X11α in 293 cell lysate, except that soluble GST mLin-2/CASK (region 1–320) protein was used as primary reagent and anti-GST antibody–HRP protein A as secondary reagents. Increasing concentrations of soluble His-mLin-2/CASK (region 1–320) were mixed with a fixed concentration of soluble GST mLin-2/CASK (region 1–320) protein to compete for binding to X11α. A soluble His-X11α PDZ containing the two X11α PDZ domains was used as a negative control.
The mLin-2/CASK region 1–320 contains a CKII domain, followed by a calmodulin binding site (residues 294–320). We asked whether the peptide (region 294–320) was involved in the binding with X11α. Various mLin-2/CASK GST fusion proteins were used to precipitate myc-tagged X11α expressed in 293 cells (Fig.3A). Bound proteins were resolved by SDS-PAGE, transferred to nitrocellulose, and revealed with anti-myc antibody. GST mLin-2/CASK (region 1–294) does not contain the calmodulin binding site but still binds very well to X11α (Fig.3B). We also introduced deletions within the CKII domain of mLin-2/CASK; truncated CKII protein does not bind to X11α, suggesting that residues 1–294 are required for the proper folding and/or function of the domain (Fig. 3C).
Fig. 3.
An integral mLin-2/CASK CKII domain is required for binding to X11α. A, Schematic representation of mLin-2/CASK and GST fusion proteins used in this study.B, Myc-tagged X11α expressed in 293 cells was precipitated by mLin-2/CASK GST coupled to glutathione beads, and bound proteins were revealed by Western blot with anti-myc antibody.C, Same as B, with different mLin-2/CASK constructs.
Calmodulin binds to mLin-2/CASK and does not affect X11α–mLin-2/CASK interaction
Our next experiments aimed to determine the role of calmodulin in X11α–mLin-2/CASK association. Calmodulin-dependent kinases require the binding of Ca2+/calmodulin to activate their catalytic activity (Goldberg et al., 1996). It has been suggested that Ca2+/calmodulin binds to a GST mLin-2/CASK fusion protein (Hata et al., 1996). We used an in vitro binding assay to detect an interaction between the mLin-2/CASK (region 294–320) peptide and purified calmodulin. Increasing amounts of calmodulin were incubated with immobilized GST mLin-2/CASK (regions 1–320 or 1–294) fusion proteins. Binding assays were performed in the presence of 0.1 mm CaCl2 or 1 mm EGTA. After incubation, beads were washed, and bound calmodulin was revealed by Western blot with anti-calmodulin antibody. Figure 4A shows that calmodulin binds to the peptide (region 294–320) in a Ca2+-dependent manner. No binding was evident when EGTA was added. Furthermore, we could coimmunoprecipitate calmodulin and mLin-2/CASK from A-172 cell lysate (Fig. 4B). Binding of Ca2+/calmodulin to GST mLin-2/CASK (region 1–320) does not affect the binding of X11α to the CKII domain (Fig. 4C), suggesting that the X11α–mLin-2/CASK interaction is not regulated by calmodulin.
Fig. 4.
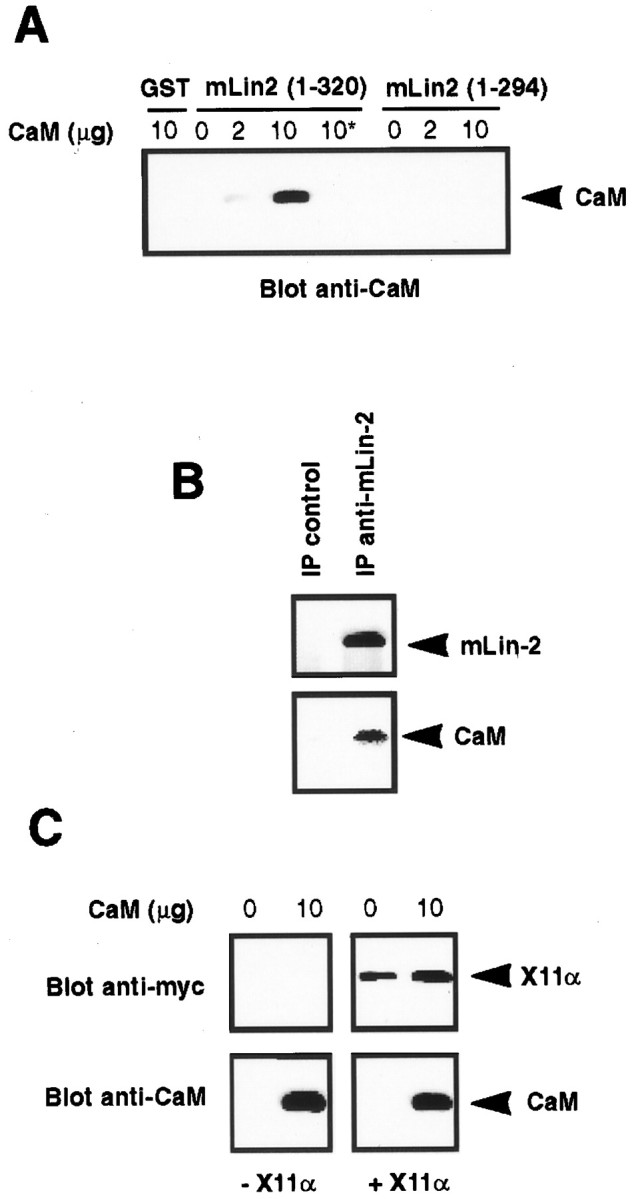
Calmodulin binds to mLin-2/CASK but does not affect X11α–mLin-2 interaction. A, GST and GST mLin-2/CASK (regions 1–320 and 1–294) were incubated with 0, 2, or 10 μg of calmodulin in the presence of 0.1 mmCa2+ or 1 mm EGTA (asterisk). After washing, bound calmodulin was resolved on SDS-PAGE, transferred to nitrocellulose, and detected with anti-calmodulin antibody. B, Lysates from untransfected A-172 cells were immunoprecipitated with preimmune or immune anti-mLin-2/CASK antibodies, and bound proteins were resolved on SDS-PAGE and transferred to nitrocellulose. Proteins were successively revealed with anti-mLin-2/CASK (top) and anti-calmodulin (bottom) antibodies. C, GST mLin-2/CASK (region 1–320) fusion protein immobilized on glutathione beads was incubated with Ca2+/calmodulin, and then lysate with (+X11α) or without (−X11α) myc-tagged X11α was added. Bound calmodulin and X11α was then assessed using immunoblotting.
A region of mLin-2/CASK encompassing the GK domain binds to X11α
We found that the mLin-2/CASK CKII domain is crucial for in vivo interaction with X11α. Indeed, a mLin-2/CASK protein containing only the CKII and PDZ domains (region 1–612) coimmunoprecipitates with X11α, and this binding is conferred by the CKII domain (Borg et al., 1998a). However, in vitro binding assays led us to consider also an interaction between X11α and the second half of mLin-2/CASK containing an SH3 and GK domain (region 578–897). In Figure5A, we show that the GST mLin-2/CASK (regions 1–612 and 578–897) fusion proteins bind to X11α in a specific manner because neither X11β nor X11γ can bind to these fusion proteins (Fig. 5A; data not shown). Binding of X11α to mLin-2/CASK (region 578–897) was reproducibly weaker than binding to mLin-2/CASK (region 1–612). This interaction is direct because soluble GST mLin-2/CASK (region 578–897) binds to X11α in an overlay assay (Fig. 5B). Whereas GST mLin-2/CASK (region 1–320) detects X11α in brain lysate, no signal is obtained with GST mLin-2/CASK (region 578–897), suggesting again a low-affinity interaction. The SH3 domain of mLin-2/CASK does not participate in this interaction because GST mLin-2/CASK (region 658–897) is sufficient for binding to X11α (Fig. 5C). We were also able to exclude a role for the lysine-rich band 4.1 binding regions (Cohen et al., 1998) because an mLin-2/CASK GST construct containing amino acids 700–897 was also able to bind X11α (results not shown). Thus, both the CKII and the GK domain containing regions in mLin-2/CASK bind to X11α. This dual contact probably increases the interaction between the two partners.
Fig. 5.
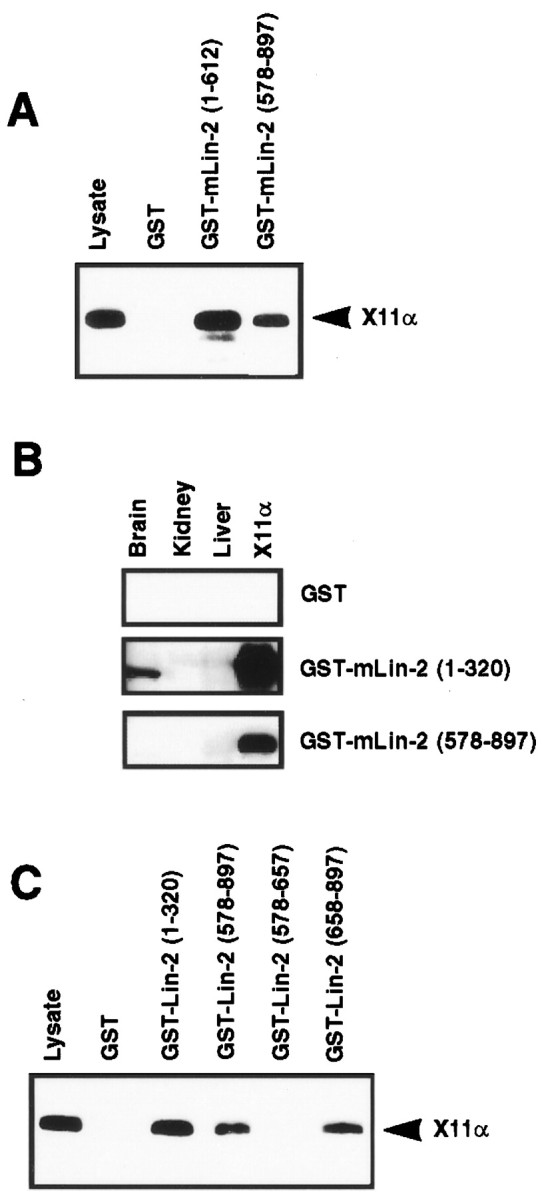
A region of mLin-2/CASK encompassing the GK domain binds to X11α. A, C, Myc-tagged X11α protein expressed in 293 cells was precipitated with different mLin-2/CASK GST fusion proteins and revealed with anti-myc antibody after Western blot. We have also detected binding to nonmyc-tagged constructs (results not shown). B, Brain, liver, and kidney extracts were run on a gel, and proteins were transferred to nitrocellulose. Myc-tagged X11α protein expressed in 293 cells was used as a positive control (X11α). An overlay assay was performed with soluble GST fusion proteins. Bound GST proteins were revealed using anti-GST antibody–HRP protein A chemiluminescence method.
Localization of X11α in the brain
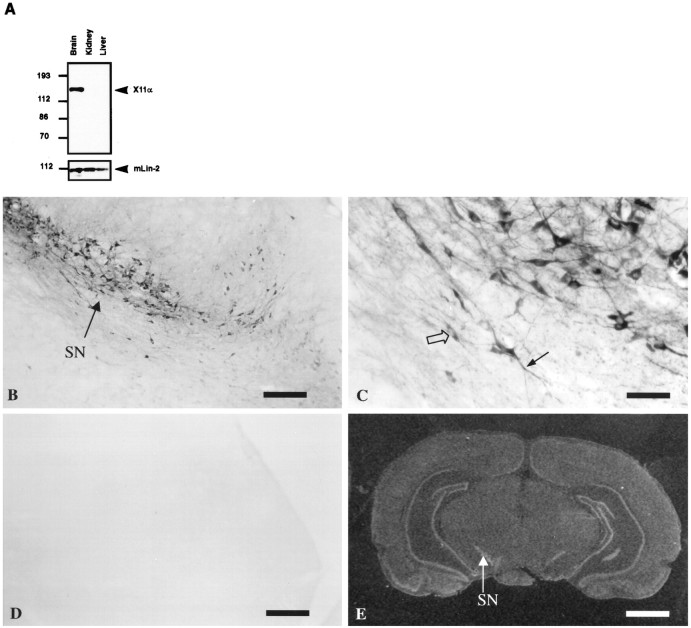
To examine the expression pattern of X11α, protein extracts from various organs were run on SDS-PAGE, transferred to nitrocellulose, and revealed with anti-X11 antibody. X11α is only expressed in the brain, whereas mLin-2/CASK is present in all tissues (Fig.6A). Previous analyses have shown that X11α is detected in the cerebellum and hippocampus in mouse brain by in situ hybridization analysis (Duclos et al., 1993). We obtained similar results with a humanX11α probe in rat brains (Fig. 6E). In addition, strong labeling was observed in the olfactory system, the piriform and enthorinal cortex, the supraoptic nucleus of the hypothalamus, the substantia nigra, and other mesencephalic areas. In control experiments using a sense RNA probe, no signal was observed (data not shown). We performed immunostaining with anti-X11α antibody on rat brain sections to document the localization of the protein. No signal was detected when antibody was preincubated with the immunogen (Fig. 6, compare B, D). The substantia nigra is strongly stained by anti-X11α antibody, and this expression correlates with a positive signal by in situ hybridization (Fig. 6E). High magnification of neurons in substantia nigra shows a diffuse staining in intracellular compartments, with exclusion of nuclei (Fig. 6C).
Fig. 6.
X11α is a brain-specific protein.A, Total proteins extracted from mouse brain, kidney, and liver were resolved by 8% SDS-PAGE and transferred to nitrocellulose. Immunoblot with anti-X11α antibody detects X11α only in the brain. mLin-2/CASK is present in all tissues, as observed previously (Hata et al., 1996; Cohen et al., 1998). B, X11α-positive immunostaining in substantia nigra (SN). Scale bar, 300 μm. C, High magnification of the substantia nigra showing neuronal somata, with exclusion of the nucleus (open arrow) and positive dendrites (filled arrow). Scale bar, 50 μm.D, Same as B, but the anti-X11α antibody was preincubated with X11α peptide. E,In situ hybridization of rat brain with theX11α probe. The arrow shows the substantia nigra labeling. Scale bar, 0.2 cm.
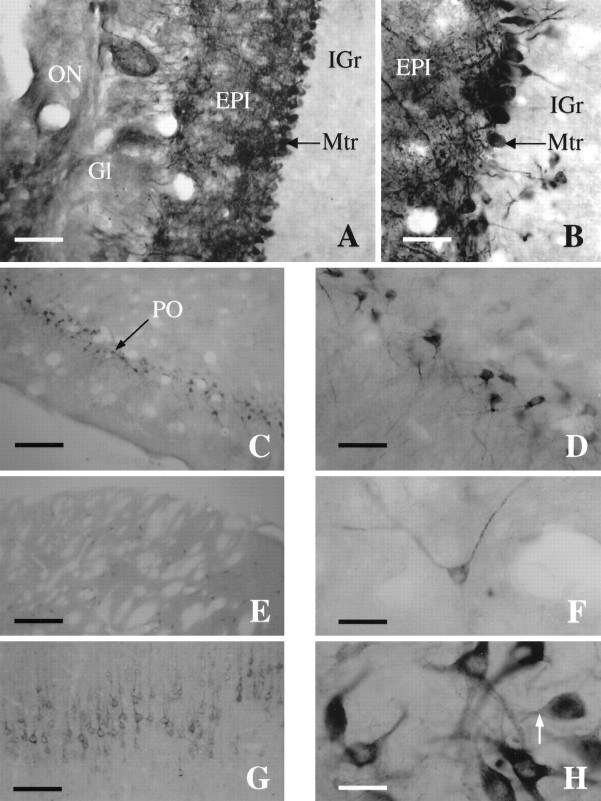
Several rat brain sections were stained to study X11α expression in more detail. X11α-positive immunostaining was observed in several nuclei throughout the rat brain. Within the telencefalon, the main olfactory bulb exhibited heavy staining, especially in the mitral and external plexiform layers. In addition, some staining was observed in the glomerular layer (Fig.7A,B). The internal granule cell layer was basically unstained. The piriform cortex exhibited a large number of intensely stained neurons, which were continuous through the entorhinal cortex (Fig.7C,D). Throughout the cortex, layer V was stained with X11α neurons (Fig. 7G). In addition, layers II–IV also exhibited weak staining, whereas layer I was devoid of staining (data not shown). Within the striatum, X11α-immunopositive medium-sized neurons were scattered in distribution (Fig.7E). The striatal neurons exhibited a characteristic punctuate labeling (Fig. 7F). Furthermore, scattered stained neurons were observed in the septum, nucleus accumbens, substantia innominata, and olfactory tubercle. In the nucleus of the diagonal band and the medial preoptic nucleus, numerous positive cells of various sizes and intensity of staining were observed. Few intensely stained and scattered neurons were observed in the hippocampus. In addition, the dentate gyrus exhibited a denser staining. Several amygdaloid nuclei contained large and intensely X11α-stained neurons. Caudally in the diencephalon, many hypothalamic and thalamic nuclei exhibited positive-stained neurons of various sizes and intensity. For instance, a dense group of heavily stained neurons was present in the supraoptic nucleus and dorsally in the habenula. Intense staining was also present in the median eminence.
Fig. 7.
X11α immunostaining in the rat brain.A, Photomicrograph of a coronal section through the main olfactory bulb (MOB) immunostained for X11α. Scale bar, 15 μm. B, High-magnification photomicrograph of the main olfactory bulb showing dense staining of the mitral cell layer. Note the intensely labeled dendrites extending to the glomerular layer. Scale bar, 40 μm. C, Coronal section through the piriform cortex (PO). Scale bar, 230 μm.D, High-magnification photomicrograph of the intensely stained pyramidal cells of layer 2 of the piriform. Scale bar, 40 μm.E, X11α-immunostained section exhibiting scattered cells in the striatum. Scale bar, 230 μm. F, Detail of a X11α-positive neuron in the striatum. Note the punctuate labeling along the axon. Scale bar, 40 μm. G, Coronal section through the cortex stained with X11α antibody. The staining is most prominent in layer V. Scale bar, 100 μm. H, High-magnification photomicrograph at the level of the substantia nigra with cells strongly stained for X11α. Note the lack of nuclear staining and the punctuate labeling. Scale bar, 40 μm.
Within the mesencephalon, scattered positive neurons were observed in the superior and inferior colliculus. A large number of heavily stained neurons, together with a dense fiber network, was present in the substantia nigra, gigantocellular reticular nucleus, and red nucleus (Fig. 7H). There was also a moderate staining in neurons of the dorsal raphe, as well as in the dorsal cochlear nucleus. Many nuclei within the caudal-most region of the brain corresponding to pons and medulla exhibited a large number of intensely stained neurons and fibers. Cells within these region exhibited a characteristic punctuate staining of the soma, axons, and dendrites, whereas in most cases, the nucleus was devoid of staining. The cerebellum exhibited a moderate staining, with intense staining of the Purkinje cells (data not shown).
Localization of X11α–mLin-2 in NT2 neurons
In our next series of studies, we wanted to determine where these proteins might interact within cells. To examine the localization of these proteins within neurons, we performed immunostaining of human NT2 neurons. NT2 teratocarcinoma cells form neurons when differentiated with retinoic acid (Pleasure and Lee, 1993). X11α is expressed in the differentiated NT2 cells but not in the stem cells (Fig.8A). Staining of cells with anti-X11α antibodies indicates that the endogenous protein is localized in the cytosol and in a perinuclear region (Figs.8A,B). A similar localization was seen when a myc-tagged X11α protein was expressed in PC12 cells (data not shown). Lin-10 has also been localized to the perinuclear region inC. elegans neurons (Rongo et al., 1998). mLin-2/CASK localized to the same regions in cells (Fig. 8B). The perinuclear region represents a component of the wheat germ ag-stained regions (results not shown). Furthermore, X11α colocalizes with giantin, suggesting that X11α and mLin-2/CASK are in a fraction of the golgi apparatus (Linstedt and Hausi, 1993). Interestingly, APP, an X11α partner, is also predominately located in the golgi and trans-golgi network in neurons (Caporaso et al., 1994).
Fig. 8.
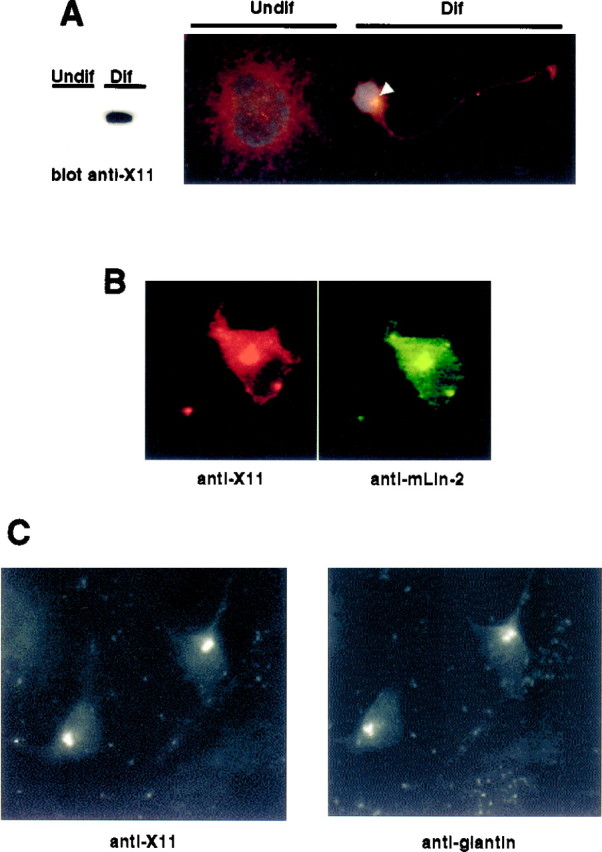
Colocalization of X11α and mLin-2/CASK in differentiated NT2 cells. A, Undifferentiated (Undif) or differentiated (Dif) NT2 cells treated with retinoic acid were lysed, and equal amounts of proteins were subjected to Western blot. The membrane was revealed with anti-X11 antibody (left). Cells were also stained with anti-X11 antibody and the nuclear stain 4-6-diamidino-2-phenylindole. White arrowindicates the restricted localization of X11α in a differentiated (Dif) neuron. The cell at the leftrepresents an undifferentiated (Undif) NT2 cell.B, Immunostaining of a differentiated NT2 cell with anti-X11 (left, Cy3-coupled secondary antibody) and anti-mLin-2/CASK (right, FITC-coupled secondary antibody). C, Immunostaining of a differentiated neuron with anti-X11 (left, FITC-coupled secondary antibody) and anti-giantin (right, Cy3-coupled secondary antibody) antibodies.
DISCUSSION
This study further delineates the interaction between X11α and mLin-2/CASK, two proteins involved in receptor localization in neurons (Borg et al., 1998a; Butz et al., 1998; Kaech et al., 1998; Rongo et al., 1998). We show that a 63 amino acid peptide found in X11α interacts with the mLin-2/CASK CKII domain. Munc-18-1, another X11α partner, binds to a different location in the X11α N terminus. Genetic analyses have suggested that the Lin-2 CKII domain acts like a protein–protein interaction domain rather than a kinase (Hoskins et al., 1996). A similar conclusion is drawn from our biochemical data with the mLin-2/CASK CKII domain. Calmodulin is a cellular calcium sensor for many enzymes and regulates ion channels, cell cycle, and cytoskeletal organization (James et al., 1995). Furthermore, previous studies have demonstrated a role for calmodulin as a negative regulator of protein–protein interactions (Wyszynski et al., 1997). In contrast, we show that calmodulin binding does not affect X11α–mLin-2 interaction. Deletion analysis of the mLin-2/CASK CKII domain has demonstrated that the integrity of the CKII domain is required for proper binding to X11α. We also describe an interaction between the mLin-2/CASK C terminus containing a GK domain and X11α. This interaction is weaker than the interaction between the CKII domain of mLin-2/CASK and X11α. Accordingly, we feel that this interaction involving the GK domain may increase the avidity of X11 with mLin-2/CASK but cannot be solely responsible for the interaction. InC. elegans, a lin-2 transgene, mutated to produce a protein with a catalytically inactive GK domain, is able to rescue the vulvaless phenotype caused by lin-2 mutations (Hoskins et al., 1996). Furthermore, the GK domain of PSD-95 and synaptic scaffolding molecule interacts with proteins found in postsynaptic densities (Kim et al., 1997; Hirao et al., 1998). Together, these data argue that the GK domain of MAGUK proteins acts as a protein–protein interaction domain.
APP, neurexins, and syndecans bind to the PTB and PDZ domains found in the X11α–mLin-2 complex (Borg et al., 1996, 1998a; Hata et al., 1996; Cohen et al., 1998; Hsueh et al., 1998). Additionally, it has been shown that mLin-7 binds tightly to mLin-2/CASK (Borg et al., 1998a; Butz et al., 1998; Kaech et al., 1998). Considering that the PDZ domains of mLin-7 and X11α and the SH3 domain of mLin-2/CASK are also available for interactions, further studies will certainly increase the number of interactors found in this complex. Such complexity is common for PDZ domain proteins. For example, in Drosophila, inactivation no afterpotential D (INAD), a PDZ protein, contacts multiple partners (phospholipase C, calmodulin, rhodopsin, and TRP ion channel) important in phototransduction (Xu et al., 1998). In worms, the Lin-10–Lin-2–Lin-7 heterotrimeric complex targets Let-23 to the basolateral epithelium in vulva (Kaech et al., 1998). In mammals, no precise role has been assigned to the complex, but a role in receptor localization is probable. Munc-18-1–Syntaxin is a brain-specific plasma membrane complex involved in the docking of vesicles during exocytosis (Hata et al., 1993). A possible scenario is that the X11α–mLin-2–mLin-7 complex binds to Munc-18-1–Syntaxin at the plasma membrane to deliver receptors such as APP, neurexins, and syndecans. We have tried to detect an in vivo interaction between the heterotrimeric complex and Munc-18-1–yntaxin. Although we could easily coimmunoprecipitate X11α, mLin-2/CASK, and mLin-7 from mouse brain extracts, our attempts to demonstrate a coimmunoprecipitation with Munc-18-1–Syntaxin were unsuccessful. However, other groups have detected a complex containing X11α/Mint-1 and Munc-18-1 (Okamoto and Sudhof, 1997).
In situ hybridization data show that X11α is present in the hippocampus, cerebral cortex, anterior thalamic nuclei, and cerebellum. In addition, positive signal was observed in the olfactory bulb, the piriform cortex, hypothalamus, and other thalamic areas, as well as numerous brainstem areas. Immunohistochemical data demonstrate that, in general, protein and mRNA follow a similar pattern for X11α distribution. However, in some areas, such as the hippocampus, mRNA expression was highly abundant, but protein levels appeared to be relatively low. These differences may be explained by a high level of mRNA expression versus a low translational rate and/or a high proteolytic rate. mLin-2/CASK is ubiquitously expressed, with a predominant expression in the brain (Hata et al., 1996; Cohen et al., 1998) (Fig. 6A). Like X11α, mLin-2/CASK is distributed in a punctate somatodendritic pattern in neurons. Many regions are positive for X11α and mLin-2/CASK expression. For example, cortical layer V pyramidal neurons and their apical dendrites present an overlapping staining. The same punctate nature of staining is seen in pyramidal cell dendrites, which suggests a synaptic localization of the two proteins. X11α and mLin-2/CASK are also found in neurons of the thalamus and in Purkinje cells of the cerebellum (Hsueh et al., 1998). However, mLin-2/CASK has not been described in some other areas, such as the hypothalamus or brainstem nuclei, where X11α is expressed. A more complete study of the distribution of mLin-2/CASK may reveal a wider distribution than previously described. Unfortunately, the anti-mLin-2/CASK that we produced and the commercially available anti-mLin-2/CASK antibodies did not give any specific signal in rat brain immunohistochemistry. We find this complex in neurons, where it is likely to play an important role in the localization of proteins to presynaptic or postsynaptic sites. In human neurons, X11α is found in the cytosol and in a component of the golgi network. Although the significance of this localization is presently unclear, we speculate that X11α in these compartments is required for the proper targeting of receptors. Although we could not localize mLin-2/CASK to specific structures other than the golgi in NT2 neurons, recent studies have also localized the protein to synapses and basolateral membranes of epithelial cells (Cohen et al., 1998; Hsueh et al., 1998).
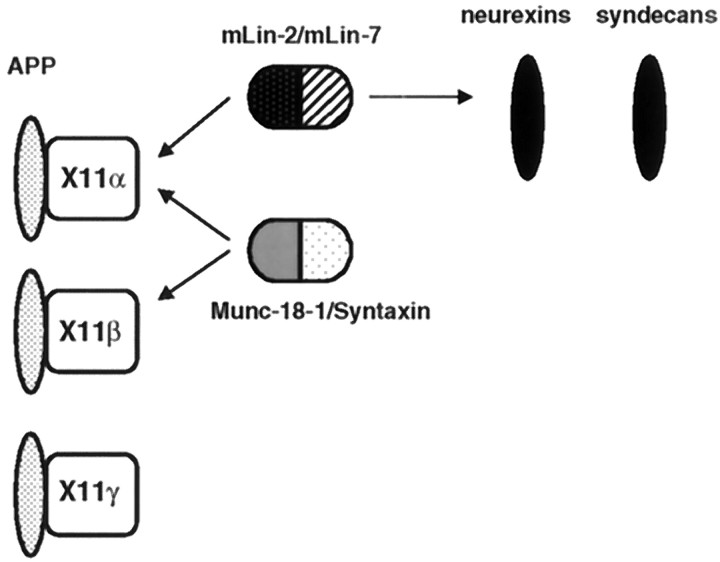
The X11β gene is also expressed in the brain, and the encoded protein is functionally related to X11α because it binds to APP and Munc-18-1–Syntaxin (Okamoto and Sudhof, 1997; Borg et al., 1998b). The lack of binding to mLin-2/CASK probably creates functional differences with X11α. Finally, X11γ does not bind to mLin-2/CASK and Munc-18-1 but still binds to APP. In neurons, APP is associated with at least three different intracellular complexes containing X11 proteins (Fig. 9). An alteration in localization of APP or its retention in a subcellular compartment induced by X11α may explain the effects of X11α on the processing of APP (Borg et al., 1998b). Additionally, members of the Fe65 protein family bind to the cytoplasmic region of APP (Borg et al., 1996; Fiore et al., 1996; Guenette et al., 1996; Trommsdorff et al., 1998). These multiple complexes may play a role in normal and pathological metabolism of APP in neurons.
Fig. 9.
The X11 protein family participates in multiple protein complexes in neurons. Schematic representation of the different proteins interacting with X11 proteins. X11α and X11β are highly expressed in brain, whereas X11γ is ubiquitously expressed. APP binds to all three X11 PTB domains, whereas the mLin-2/CASK–mLin-7 complex only interacts with X11α. The Munc-18-1–Syntaxin neuronal complex interacts with the neuronal X11 species. Neurexins and syndecans binds to the mLin-2/CASK PDZ domain.
Footnotes
This study was supported by National Institute of Mental Health Program Project MH 42251 and the Pritzker Network (M.O.L and S.J.W.). B.M. is an investigator of the Howard Hughes Medical Institute. We thank Dr. Lawrence Mathews for the purified calmodulin and anti-calmodulin antibody. Anti-giantin monoclonal antibody was kindly provided by Dr. Hans-Peter Hausi.
Correspondence should be addressed to Dr. Ben Margolis, Howard Hughes Medical Institute, University of Michigan Medical Center, Room 4570, MSRB II, 1150 West Medical Center Drive, Ann Arbor, MI 48109-0650.
Drs. Borg and Lõpez-Figueroa contributed equally to this work.
REFERENCES
- 1.Borg J-P, Margolis B. Function of PTB domains. Curr Top Microbiol Immunol. 1998;228:23–38. doi: 10.1007/978-3-642-80481-6_2. [DOI] [PubMed] [Google Scholar]
- 2.Borg J-P, Ooi J, Levy E, Margolis B. The phosphotyrosine interaction domains of X11 and FE65 bind to distinct sites on the YENPTY motif of amyloid precursor protein. Mol Cell Biol. 1996;16:6229–6241. doi: 10.1128/mcb.16.11.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borg J-P, Straight SW, Kaech SM, De Taddeo-Borg M, Kroon DE, Karnak D, Turner RS, Kim SK, Margolis B. Identification of an evolutionarily conserved heterotrimeric protein complex involved in protein targeting. J Biol Chem. 1998a;273:31633–31636. doi: 10.1074/jbc.273.48.31633. [DOI] [PubMed] [Google Scholar]
- 4.Borg J-P, Yang Y, De Taddeo-Borg M, Margolis B, Turner RS. The X11α protein slows cellular amyloid precursor protein processing and reduces Aβ40 and Aβ42 secretion. J Biol Chem. 1998b;273:14761–14766. doi: 10.1074/jbc.273.24.14761. [DOI] [PubMed] [Google Scholar]
- 5.Butz S, Okamoto M, Sudhof TC. A tripartite protein complex with the potential to couple synaptic vesicle exocytosis to cell adhesion in brain. Cell. 1998;94:773–782. doi: 10.1016/s0092-8674(00)81736-5. [DOI] [PubMed] [Google Scholar]
- 6.Caporaso GL, Takei K, Gandy SE, Matteoli M, Mundigl O, Greengard P, De Camilli P. Morphologic and biochemical analysis of the intracellular trafficking of the Alzheimer beta/A4 amyloid precursor protein. J Neurosci. 1994;14:3122–3138. doi: 10.1523/JNEUROSCI.14-05-03122.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen AR, Wood DF, Marfatia SM, Walther Z, Chishti AH, Anderson JM. Human CASK/LIN-2 binds syndecan-2 and protein 4.1 and localizes to the basolateral membrane of epithelial cells. J Cell Biol. 1998;142:129–138. doi: 10.1083/jcb.142.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duclos F, Boschert U, Sirugo G, Mandel J-L, Hen R, Koenig M. Gene in the region of the Friedreich ataxia locus encodes a putative transmembrane protein expressed in the nervous system. Proc Natl Acad Sci USA. 1993;90:109–113. doi: 10.1073/pnas.90.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fanning AS, Anderson JM. PDZ domains and the formation of protein networks at the plasma membrane. Curr Top Microbiol Immunol. 1997;228:209–233. doi: 10.1007/978-3-642-80481-6_9. [DOI] [PubMed] [Google Scholar]
- 10.Fiore F, Zambrano N, Minopoli G, Donini V, Duilio A, Russo T. The regions of the Fe65 protein homologous to the phosphotyrosine interaction/phosphotyrosine binding domain of Shc bind the intracellular domain of the Alzheimer’s amyloid precursor protein. J Biol Chem. 1996;270:30853–30856. doi: 10.1074/jbc.270.52.30853. [DOI] [PubMed] [Google Scholar]
- 11.Goldberg J, Nairn AC, Kuriyan J. Structural basis for the autoinhibition of calcium/calmodulin-dependent protein kinase I. Cell. 1996;84:875–887. doi: 10.1016/s0092-8674(00)81066-1. [DOI] [PubMed] [Google Scholar]
- 12.Guenette SY, Chen J, Jondro PD, Tanzi RE. Association of a novel human FE65-like protein with the cytoplasmic domain of the β-amyloid precursor protein. Proc Natl Acad Sci USA. 1996;93:10832–10837. doi: 10.1073/pnas.93.20.10832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hata Y, Slaughter CA, Sudhof TC. Synaptic vesicle fusion complex contains unc-18 homologue bound to syntaxin. Nature. 1993;366:347–351. doi: 10.1038/366347a0. [DOI] [PubMed] [Google Scholar]
- 14.Hata Y, Butz S, Sudhof TC. CASK: a novel dlg/PSD95 homolog with an N-terminal calmodulin-dependent protein kinase domain identified by interaction with neurexins. J Neurosci. 1996;16:2488–2494. doi: 10.1523/JNEUROSCI.16-08-02488.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirao K, Hata Y, Ide N, Takeuchi M, Irie M, Yao I, Deguchi M, Toyoda A, Sudhof TC, Takai Y. A novel multiple PDZ domain-containing molecule interacting with N-methyl-d-aspartate receptors and neuronal cell adhesion proteins. J Biol Chem. 1998;273:21105–21110. doi: 10.1074/jbc.273.33.21105. [DOI] [PubMed] [Google Scholar]
- 16.Hoskins R, Hajnal A, Harp S, Kim S. The C. elegans vulval induction gene lin-2 encodes a member of the MAGUK family of cell junction proteins. Development. 1996;122:97–111. doi: 10.1242/dev.122.1.97. [DOI] [PubMed] [Google Scholar]
- 17.Hsueh Y-P, Yang F-C, Kharazia V, Naisbitt S, Cohen AR, Weinberg RJ, Sheng M. Direct interaction of CASK/LIN-2 and syndecan heparan sulfate proteoglycan and their overlapping distribution in neuronal synapses. J Cell Biol. 1998;142:139–151. doi: 10.1083/jcb.142.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.James P, Vorherr T, Carafoli E. Calmodulin-binding domains: just two-faced or multi-faced? Trends Biochem Sci. 1995;20:38–42. doi: 10.1016/s0968-0004(00)88949-5. [DOI] [PubMed] [Google Scholar]
- 19.Kaech SM, Whitfield CW, Kim SK. The LIN-2/LIN-7/LIN-10 complex mediates basolateral membrane localization of the C. elegans EGF receptor LET-23 in vulval epithelial cells. Cell. 1998;94:761–771. doi: 10.1016/s0092-8674(00)81735-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim E, Niethammer M, Rothschild A, Jan YN, Sheng M. Clus- tering of Shaker-type K+ channels by interaction with a family of membrane-associated guanylate kinases. Nature. 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- 21.Kim E, Naisbitt S, Hsueh Y-P, Rao A, Rothschild A, Craig AM, Sheng M. GKAP, a novel synaptic protein that interacts with the guanylate kinase-like domain of the PSD-95/SAP90 family of channel clustering molecules. J Cell Biol. 1997;136:669–678. doi: 10.1083/jcb.136.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kornau H-C, Schenker LT, Kennedy MB, Seeburg PH. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- 23.Linstedt AD, Hausi H-P. Giantin, a novel conserved golgi membrane protein containing a cytoplasmic domain of at least 350 kDa. Mol Biol Cell. 1993;4:679–693. doi: 10.1091/mbc.4.7.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lõpez-Figueroa MO, Ravault JP, Cozzi B, Moller M. Presence of nitric oxide synthase in the sheep pineal gland: an experimental immunohistochemical study. Neuroendocrinology. 1996;63:384–392. doi: 10.1159/000126979. [DOI] [PubMed] [Google Scholar]
- 25.Lõpez-Figueroa MO, Itoi K, Watson SJ. Regulation of nitric oxide synthase mRNA expression in the rat hippocampus by glucocorticoids. Neuroscience. 1998;87:439–446. doi: 10.1016/s0306-4522(98)00075-x. [DOI] [PubMed] [Google Scholar]
- 26.McLoughlin DM, Miller CCJ. The intracellular cytoplasmic domain of the Alzheimer’s disease amyloid precursor protein interacts with phosphotyrosine-binding domain proteins in the yeast two-hybrid system. FEBS Lett. 1996;397:197–200. doi: 10.1016/s0014-5793(96)01128-3. [DOI] [PubMed] [Google Scholar]
- 27.Okamoto M, Sudhof TC. Mints, Munc18-interacting proteins in synaptic vesicle exocytosis. J Biol Chem. 1997;272:31459–31464. doi: 10.1074/jbc.272.50.31459. [DOI] [PubMed] [Google Scholar]
- 28.Pawson T, Scott JD. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 29.Pleasure SJ, Lee VM. NTera 2 cells: a human cell line which displays characteristics expected of a human committed neuronal progenitor cell. J Neurosci Res. 1993;35:585–602. doi: 10.1002/jnr.490350603. [DOI] [PubMed] [Google Scholar]
- 30.Rongo C, Whitfield CW, Rodal A, Kim SK, Kaplan JM. LIN-10 is a shared component of the polarized protein localization pathways in neurons and epithelia. Cell. 1998;94:751–759. doi: 10.1016/s0092-8674(00)81734-1. [DOI] [PubMed] [Google Scholar]
- 31.Simske JS, Kaech SM, Harp SA, Kim SK. LET-23 receptor localization by the cell junction protein LIN-7 during C. elegans vulval induction. Cell. 1996;85:195–204. doi: 10.1016/s0092-8674(00)81096-x. [DOI] [PubMed] [Google Scholar]
- 32.Trommsdorff M, Borg J, Margolis B, Herz J. Interaction of cytosolic adaptor proteins with neuronal apolipoprotein E receptors and the amyloid precursor protein. J Biol Chem. 1998;273:33556–33565. doi: 10.1074/jbc.273.50.33556. [DOI] [PubMed] [Google Scholar]
- 33.Wyszynski M, Lin J, Rao A, Nigh E, Beggs AH, Craig AM, Sheng M. Competitive binding of α-actinin and calmodulin to the NMDA receptor. Nature. 1997;385:439–442. doi: 10.1038/385439a0. [DOI] [PubMed] [Google Scholar]
- 34.Xu X-Z, Choudhury A, Li X, Montell C. Coordination of an array of signaling proteins through homo- and heteromeric interactions between PDZ domains and target proteins. J Cell Biol. 1998;142:545–555. doi: 10.1083/jcb.142.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Z, Lee CH, Mandiyan V, Borg J-P, Margolis B, Schlessinger J, Kuriyan J. Sequence-specific recognition of the internalization motif of the Alzheimer’s amyloid precursor protein by the X11 PTB domain. EMBO J. 1997;16:6141–6150. doi: 10.1093/emboj/16.20.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]