Abstract
Helicobacter pylori is an infectious agent that colonizes the gastric mucosa of half of the population worldwide. This bacterium has been recognized as belonging to group 1 carcinogen by the World Health Organization for the role in development of gastritis, peptic ulcers, and cancer. Due to the increase in resistance to antibiotics used in the anti-H. pylori therapy, the development of an effective vaccine is an alternative of great interest, which remains a challenge. Therefore, a rational, strategic, and efficient vaccine design against H. pylori is necessary where the use of the most current bioinformatics tools could help achieve it. In this study, immunoinformatics approach was used to design a novel multiepitope oral vaccine against H. pylori. Our multiepitope vaccine is composed of cholera toxin subunit B (CTB) that is used as a mucosal adjuvant to enhance vaccine immunogenicity for oral immunization. CTB fused to 11 epitopes predicted of pathogenic (UreB170–189, VacA459–478, CagA1103–1122, GGT106–126, NapA30–44, and OipA211–230) and colonization (HpaA33–52, FlaA487–506, FecA437–456, BabA129–149, and SabA540–559) proteins from H. pylori. CKS9 peptide (CKSTHPLSC) targets epithelial microfold cells to enhance vaccine uptake from the gut barrier. All sequences were joined to each other by proper linkers. The vaccine was modeled and validated to achieve a high-quality three-dimensional structure. The vaccine design was evaluated as nonallergenic, antigenic, soluble, and with an appropriate molecular weight and isoelectric point. Our results suggest that our newly designed vaccine could serve as a promising anti-H. pylori vaccine candidate.
Keywords: Helicobacter pylori, in silico, multiepitope, oral vaccine, reverse vaccinology
1. Introduction
Helicobacter pylori infection is associated with gastrointestinal tract diseases, including chronic gastritis, peptic ulcer, mucosa-associated lymphoproliferative disorders, and gastric cancer (Marshall and Warren, 1984; IARC, 1994; Wroblewski et al., 2010). Its global prevalence is >50%, which may significantly vary within and among countries, according to geography, ethnicity, age, and socioeconomic factors (Leja et al., 2016; Burucoa and Axon, 2017). The conventional antimicrobial therapy against H. pylori is costly, causes serious side effects and potential reinfections, and eliminates <80% of H. pylori infection, which facilitate drug resistance (Thung et al., 2016). Therefore, considerable efforts have been made to develop a preventive strategy against H. pylori infection, such as recombinant vaccines, with limited effectiveness. Numerous studies of vaccination in animal models have shown that immunization with combinations of antigens confers a better protective immunity against this bacterium; however, the establishment of an immunological response that allows the eradication in gastric mucosa remains a challenge (Rappuoli et al., 2016; Sutton and Boag, 2018).
The technological advances in the fields of genomics, proteomics, human immunology, and structural biology have provided the molecular information for the discovery and prediction by bioinformatics tools of novel antigens, epitopes, and design of vaccines against pathogenic bacteria, such as meningococcus B (Serruto et al., 2012; O'Ryan et al., 2013; Rappuoli et al., 2016). Bioinformatics tools can predict sequences with binding affinity to MHC alleles and epitopes of T and B cells (Rosa et al., 2015). The focus on vaccine design and development has changed to the production of peptides composed of multiple epitopes (multiepitope vaccines), based on linear arrangements, as a novel alternative. Immunological studies have shown that H. pylori infection induces an activation of adaptive B and T cell responses and that a Th1/Th17-polarized immune response is required for its prevention (Müller and Hartung, 2016). Some recent studies have designed multiepitope vaccine of H. pylori using bioinformatics (Li et al., 2014; Meza et al., 2017). In addition, epitope-based vaccines have demonstrated various advantages, including safety, the opportunity to rationally engineer the epitopes for increased potency, breadth, and antigenicity, and the possibility to focus large repertories of immune responses on conserved epitope sequences (Livingston et al., 2002; Oyarzún and Kobe, 2015).
In the present study, a novel oral vaccine against H. pylori was in silico designed.
2. Methods
2.1. Selection and antigenic evaluation of proteins
H. pylori proteins were selected based on the following criteria: (1) reported antigenicity, (2) virulence, and (3) proteins related to the mechanisms of adhesion and colonization.
The entire protein sequences were retrieved from NCBI reference sequence database in FASTA format. For bacterial protein subcellular localization prediction, CELLO v2.5 was used (Yu et al., 2006). The database of Online GEne Essentiality (OGEE) was used to evaluate the essentiality of genes (Chen et al., 2017). PATRIC 3.5.16 database was used for study of the virulence role of proteins (Wattam et al., 2017). Proteins were screened by BLASTp to detect sequence homologs to Homo sapiens. To predict transmembrane (TM) helixes, TMHMM v2.0 was used (Krogh et al., 2001). Compute pI/Mw tool was used to calculate the estimated isoelectric point and molecular weight of all amino acid sequences (Wilkins et al., 1999).
2.2. Conservation, consensus sequence, and alignment
Conservation of selected proteins was analyzed using H. pylori strain 26695 as the reference. For proteins with high variability, a consensus sequence was generated using sequences of world H. pylori representative strains (85 complete genome sequences). The sequence alignment was performed using CLC Main Workbench v7.8 (QIAGEN Bioinformatics), Emboss Cons, and T-Coffee software.
2.3. T cell epitope prediction
To identify MHC-I binding epitopes, NetMHC 4.0 server was used (Andreatta and Nielsen, 2016). Eighty-one human leukocyte antigen (HLA) alleles (HLA-A, -B, -C, and -E) and six murine alleles (H-2) were evaluated. Predictions were calculated for nine-mers epitopes with a threshold for strong binders of 0.5% and a threshold for weak binders of 2%. The second was ProPred-I that implements matrices for 47 MHC Class-I alleles, proteasomal, and immunoproteasomal models. The threshold scoring peptides were 4% (Singh and Raghava, 2003). The third one was IEDB MHC-I Binding Predictions; the best-predicted binders for 27 HLA and 6 H-2 alleles were selected based on a percentile rank of ≤1%.
For MHC-II binding epitopes, ProPred was used; 51 HLA-DR alleles were evaluated using a default threshold of 3% (Singh and Raghava, 2001). The second was IEDB MHC-II Binding Predictions, a reference panel of 27 HLA alleles and 3 H-2-I alleles was tested; the selection was based on a consensus percentile rank of the top 10%. The third was NetMHCII 2.3 server; predictions were obtained for 25 HLA-DR alleles, 20 HLA-DQ, 9 HLA-DP, and 7 mouse H2 class II alleles using a threshold of −99.9, threshold for the strong binder of 2%, and threshold for the weak binder of 10%.
2.4. B cell epitope prediction
Linear B cell epitopes of 20-mers were predicted utilizing ABCpred with a threshold of 0.75. The second one was BcePred. The BCPred server was applied with a specificity threshold of 75%. For BepiPred server, only amino acids with score ≥1.3 were considered for the downstream analysis (Jespersen et al., 2017). DiscoTope 2.0 server was used for identification of discontinuous B cell epitopes with a threshold of −3.7; the sensitivity and specificity were 0.47 and 0.75, respectively (Kringelum et al., 2012). ElliPro server predicts linear and discontinuous epitopes. For prediction, a maximum distance (Å) of six and a minimum score of 0.5 were used (Ponomarenko et al., 2008).
2.5. Selection of epitopes
Epitopes were selected based on the following criteria: (1) 20-mer epitopes, (2) epitopes matching on all algorithms, if possible, and (3) potential to bind with the maximum number of MHC-I and MHC-II alleles. For selection, sequences were aligned and overlapped using Clustal Omega server.
2.6. Vaccine design
The VaccineCAD (EpiVax Inc., RI) and EpiToolKit 2.0 were used to optimize epitope order. They link candidate epitopes into a string-of-beads design, minimizing nonspecific junctional epitopes that may be produced in the linking process (Schubert et al., 2015). In addition, Gly-Pro-Gly-Pro-Gly (GPGPG) and Lys-Lys (KK) spacer sequences were engineered at the interepitope position. Cholera toxin subunit B (CTB) and CKS9 (M cell homing peptide) sequences were added to the final vaccine model at the N-terminal and C-terminal, respectively.
2.7. Protein prediction and validation of secondary and tertiary structures
The secondary structure of the multiepitope antigen was predicted using PSIPRED v3.3 (McGuffin et al., 2000). The three-dimensional (3D) structure modeling was performed using I-TASSER server (Yang and Zhang, 2015). Molecular Operating Environment v2014.0901 and Jmol were used for visualizing 3D structures of proteins. For refinement of 3D model structure, GalaxyRefine and GalaxyLoop were applied (Park et al., 2011).
The best model was validated by the ProSA-web (Wiederstein and Sippl, 2007) and ERRAT (Colovos and Yeates, 1993). The residue-by-residue stereochemical qualities of models were validated by Ramachandran plot obtained from PROCHECK server (Laskowski et al., 2012). The best-refined model was selected.
2.8. Antigenicity, allergenicity, solubility, and physicochemical predictions of the vaccine
For antigenicity prediction, VaxiJen server was used. For allergenicity evaluation, AllerTOP v.2.0 and AlgPred servers were used. For solubility prediction, SOLpro server was used. Finally, ProtParam allowed the computation of various physical and chemical parameters (Wilkins et al., 1999).
2.9. Codon optimization
DNA coding sequence of the oral vaccine was optimized for high-level protein expression in Lactococcus lactis using Codon optimization On-Line server. The tool is based on several design parameters such as individual codon usage, codon pairing, codon adaptation index (CAI), and others (Chin et al., 2014).
3. Results and Discussion
3.1. Protein selection and evaluation
It has been shown that the combination of a long list of antigens can increase the effectiveness of vaccines (Rossi et al., 2004). Malfertheiner et al. (2008) evaluated a multicomponent vaccine in human volunteers; they observed that the vaccine was safe and highly immunogenic, inducing long-lasting humoral and cellular responses to the antigens. Therefore, 11 proteins were selected for our vaccine. UreB has been reported in many studies for antigenicity; VacA, CagA, GGT, NapA, and OipA are the main proteins involved in the pathogenic mechanisms. HpaA, FlaA, FecA, BabA, and SabA were selected for their participation in the mechanisms of adhesion and colonization.
Selected proteins were further screened in the next step for further parameters, including subcellular localization, essentiality, virulence, nonhuman homology, TM helixes, and molecular weight. Our predictions showed that UreB and NapA possessed a cytoplasmic location; CagA, FlaA, VacA, and BabA were extracellular, whereas OipA, SabA, HpaA, and FecA were predicted as outer membrane proteins. In addition, GGT was classified as a periplasmic protein, as shown in Table 1. Surface and extracellular proteins are good targets to develop a vaccine aiming toward prevention of bacterial infections and diseases (Dwivedi et al., 2016).
Table 1.
The Subcellular Localization, Gene Essentiality, Virulence, Human Homology, Transmembrane Helix, Isoelectric Point, and Molecular Weight Predictions of Helicobacter pylori Selected Proteins
| Protein | Accession No. | Locus tag | Subcellular localization | Gene essentiality | Virulent protein | Human homology | TM helix | pI/MW (kDa) |
|---|---|---|---|---|---|---|---|---|
| UreB | NP_206872.1 | HP0072 | C | N-ES | V | N-H | 0 | 5.64/61.68 |
| NapA | NP_207041.1 | HP0243 | C | N-ES | V | N-H | 0 | 5.59/16.93 |
| CagA | NP_207343.1 | HP0547 | E | N-ES | V | N-H | 0 | 8.82/132.4 |
| FlaA | NP_207396.1 | HP0601 | E | N-ES | V | N-H | 0 | 6.03/53.3 |
| OipA | NP_207432.1 | HP0638 | OM | N-ES | V | N-H | 0 | 9.78/34.2 |
| SabA | NP_207519.1 | HP0725 | OM | N-ES | V | N-H | 0 | 9.12/69.4 |
| HpaA | NP_207590.1 | HP0797 | OM | N-ES | V | N-H | 0 | 8.60/29.0 |
| FecA | NP_207600.1 | HP0807 | OM | ES | N-V | N-H | 0 | 9.66/88.9 |
| VacA | NP_207680.1 | HP0887 | E | N-ES | V | N-H | 1 (35–57 aa) | 9.02/139.3 |
| GGT | NP_207909.1 | HP1118 | P | ES | V | N-H | 1 (5–27 aa) | 9.27/61.2 |
| BabA | NP_208035.1 | HP1243 | E | N-ES | V | N-H | 0 | 8.95/79.1 |
aa, amino acid; C, cytoplasmic; E, essential; ES, extracellular; N-ES, nonessential; N-H, nonhomology; N-V, nonvirulent; OM, outer membrane; P, periplasmic; TM, transmembrane; V, virulent.
In the analysis of essential genes by OGEE, HP1118 (GGT) and HP0807 (FecA) genes were identified. Essential genes are those genes of an organism that are critical for its survival; essential genes are of particular importance because of their theoretical and practical applications such as studying the robustness of a biological system, defining a minimal genome/organism and identifying effective therapeutic targets in pathogens (Chen et al., 2017). Furthermore, 9 of the 11 proteins were found in the database of virulence factors of PATRIC; FecA and GGT were not identified in this category. Homology analysis of the 11 prioritized proteins using BLASTp revealed ≤30% identity, which was significant to declare the sequences as nonhuman homologs; Vaccine targets should not be human homologues to avoid autoimmunity. The prediction of the topology of proteins by TMHMM showed that VacA and GGT had one TM helix, whereas other proteins do not unveil the presence of any such topology. VacA and GGT showed the presence of a TM helix located at 35–57 and at 5–27 amino acid positions, respectively. Finally, molecular weights calculated by Compute pI/Mw tool of nine proteins resulted to weigh <110 kDa, whereas VacA and CagA have molecular weights of 139.3 and 132.4 kDa, respectively (Table 1).
Good vaccine candidates are considered as those that do not present homology with human proteins to avoid the generation of a potential autoimmune response; these candidates must also lack TM regions, to facilitate their expression. Another characteristic a good vaccine candidate should have is to possess good antigenic properties, which are important for the pathogenesis of the microorganism and for protection against the disease (Monterrubio-López et al., 2015). Some authors have used these approaches in the selection of candidate proteins for the in silico design of H. pylori vaccines (Naz et al., 2015; Nezafat et al., 2017).
3.2. Consensus antigen
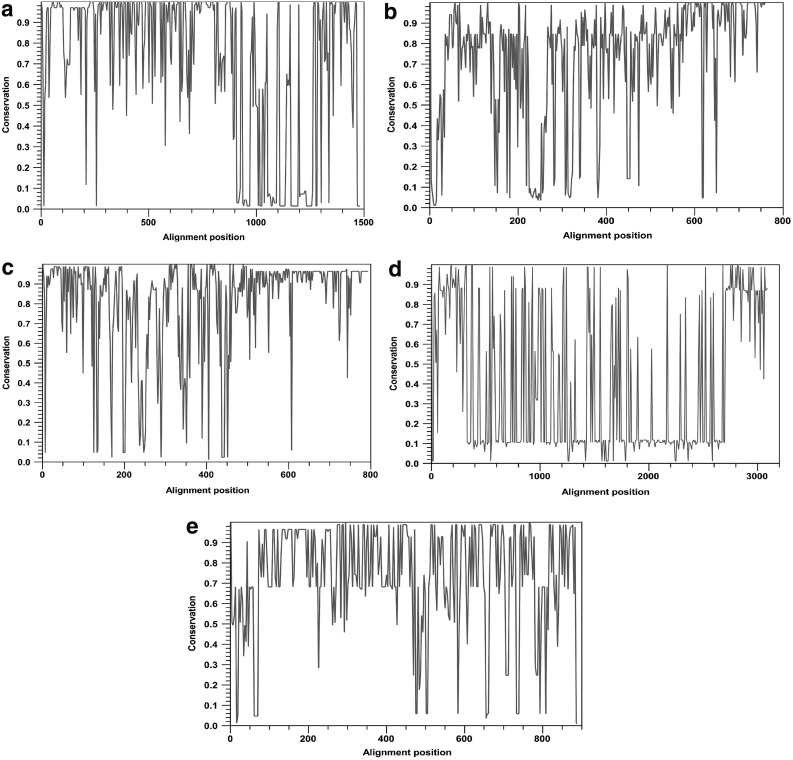
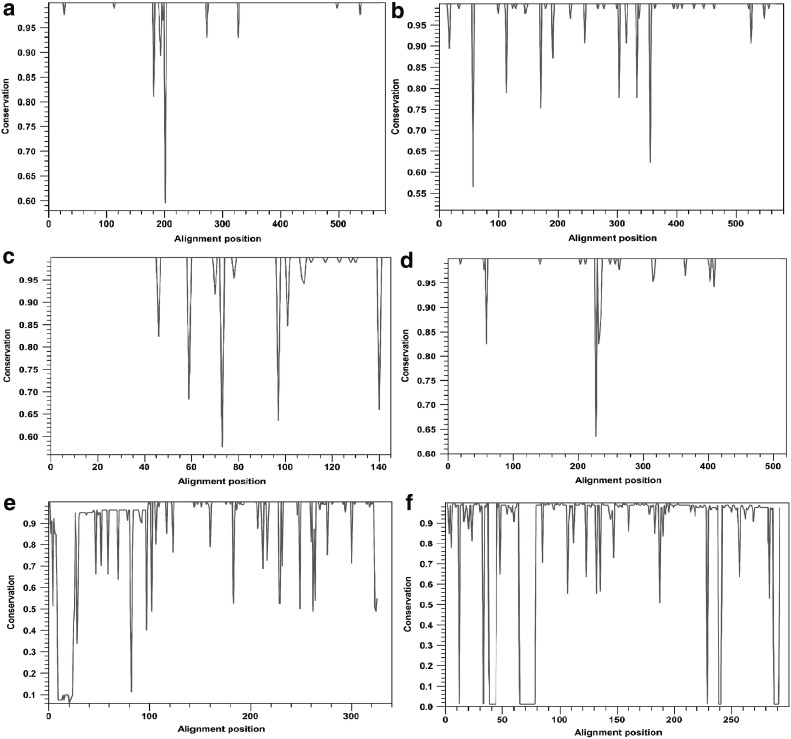
To develop a universal H. pylori vaccine, a conservation analysis of the selected antigens was performed. The amino acid sequences of each protein were compared and aligned with 85 sequences corresponding to fully annotated genomes of H. pylori from different geographical regions at the time of the study. A high variability in the proteins CagA, VacA, FecA, BabA, and SabA was observed, as shown in Figure 1. Consequently, to reduce the variability between H. pylori strains, consensus sequences were obtained. This approach has been used against infectious agents with a high variability rate such as HIV (Thomson et al., 2005; Grimm and Ackerman, 2013). In contrast, the proteins with the highest percentage (≥95%) of conservation were UreB, NapA, GGT, FlaA, HpaA, and OipA (Fig. 2).
FIG. 1.
Analysis of conservation of selected proteins. Entire protein sequences among the 85 annotated genomes of Helicobacter pylori were aligned using CLC Main workbench, and conservation scores were represented in a 2D line chart. (a) CagA, (b) SabA, (c) BabA, (d) VacA, and (e) FecA. 2D, two-dimensional.
FIG. 2.
Analysis of conservation of selected proteins. Entire protein sequences among the 85 annotated genomes of Helicobacter pylori were aligned using CLC Main workbench, and conservation scores were represented in a 2D line chart. (a) UreB, (b) GGT, (c) NapA, (d) FlaA, (e) OipA, and (f) HpaA.
3.3. T and B cell epitopes
The prediction analysis by different bioinformatics servers for T and B cells (using MHC-I/-II alleles for human and mouse BALB/c) allowed the selection of 11 epitopes based on their score, number of alleles, and agreement between the servers used. Epitopes obtained were NapA30–49, CagA1103–1122, FlaA487–506, OipA211–230, GGT106–126, HpaA33–52, BabA129–149, FecA437–456, SabA540–559, VacA459–478, and UreB170–189 (Table 2). HpaA33–52 epitope has been previously reported by Hu et al. (2016); they evaluated the response of antigen-specific CD4+ T lymphocytes stimulated with recombinant HpaA from 101 H. pylori-infected subjects, a significant increase of the interferon-γ producing cells was observed. In addition, nine residues of UreB170–189 epitope predicted in this study match with UreB181–195 epitope reported by Qiu et al.; they observed the positive reactivity with sera from H. pylori-infected patients by enzyme-linked immunosorbent assay; mice immunized with glutathione S-transferase-UreB181–195 fusion peptide showed that epitope-specific antibodies were capable of inhibiting urease enzymatic activity. In another study, Guo et al. (2017) designed and evaluated a multivalent epitope-based vaccine (named CWAE) that includes UreB181–195 epitope. Oral therapeutic immunization with CWAE significantly reduced the number of H. pylori colonies in the stomach of Mongolian gerbils. The protection of CWAE was associated with higher levels of mixed CD4+ T cell response, immunoglobulin G, and secretory immunoglobulin A (Guo et al., 2017).
Table 2.
Potential Antigenic Epitopes Predicted by Different Servers
| T cell epitopes | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B cell epitopes | IEDB | ProPred | NetMHC | ||||||||||||
| Order | Protein | Position | Sequence | BepiPred | BCPRED | ABCpred | DiscoTope | ElliPro | BcePred | I | II | I | II | I | II |
| 1 | NapA | 30–49 | GTDFFNVHKATEEIYEEFAD | 0.520 | 0.573 | 0.31 | −7.806 | 0.511 | 0.826 | 6H 2M | 5H 0M | 17H 3M | 10H | 31H 1M | 12H 0M |
| 2 | CagA | 1103–1122 | IVAHNVGSVPLSEYDKIGFN | 0.087 | 0.45 | 0.86 | −0.852 | 0.928 | 0.953 | 8H 3M | 6H 0M | 24H 6M | 16H | 34H 1M | 7H 0M |
| 3 | FlaA | 487–506 | LAQSGSYAMSQANTVQQNIL | 0.136 | 0.363 | 0.82 | −4.984 | 0.916 | 0.935 | 10H 3M | 5H 1M | 19H 5M | 10H | 46H 2M | 13H 1M |
| 4 | OipA | 211–230 | ASTWFASNNLTPFNQVKSHT | 0.557 | 0.892 | 0.35 | −2.603 | 0.564 | 0.714 | 10H 2M | 2H 0M | 27H 3M | 3H | 46H 2M | 11H 0M |
| 5 | GGT | 106–126 | APLKATKNMFLDKQGNVVPKL | 0.553 | 0.697 | 0.42 | −5.047 | 0.705 | 0.925 | 8H 2M | 4H 1M | 29H 6M | 11H | 43H 2M | 14H 0M |
| 6 | HpaA | 33–52 | IETNEVALKLNYHPASEKVQ | 0.627 | 0.21 | 0.8 | −5.593 | 0.401 | 0.812 | 9H 1M | 7H 0M | 23H 2M | 21H | 37H 3M | 7H 0M |
| 7 | BabA | 129–149 | NANGQKSTSSTTIFNNEPGYR | 1.243 | 0.969 | 0.67 | −11.203 | 0.678 | 0.812 | 13H 2M | 3H 0M | 16H 4M | 0H | 35H 2M | 5H 0M |
| 8 | FecA | 437–456 | NLVVNTGKVKQTFNMGMRFL | −0.398 | 0.617 | 0.85 | −3.091 | 0.681 | 0.842 | 9H 2M | 8H 0M | 26H 5M | 34H | 32H 2M | 15H 1M |
| 9 | SabA | 540–559 | WLNSQYVNLTAFNNPYSAKV | 0.546 | 0.621 | 0.76 | −4.667 | 0.907 | 0.885 | 14H 3M | 7H 0M | 26H 5M | 28H | 67H 3M | 16H 0M |
| 10 | VacA | 459–478 | NQVGGYALAGSSANFEFKAG | 0.563 | 0.558 | 0.85 | −13.843 | 0.261 | 0.893 | 6H 2M | 5H 2M | 15H 4M | 11H | 38H 2M | 11H 1M |
| 11 | UreB | 170–189 | TTITPGRRNLKWMLRAAEEY | 0.649 | 0.845 | 0.87 | −3.764 | 0.584 | 0.790 | 9H 1M | 6H 1M | 21H 3M | 15H | 39H 2M | 13H 0M |
H, human; I, MHC class I; II, MHC class II; M, mouse.
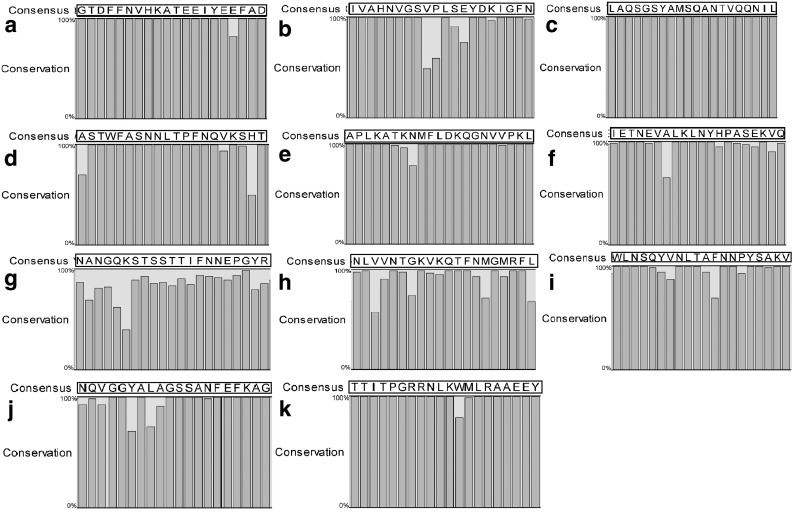
In addition, the residue-to-residue conservation of the selected epitopes was evaluated; H. pylori 26695 was used as the reference. CagA1103–1122, BabA129–149, and FecA437–456 showed one amino acid change; OipaA211–230 showed two changes (Fig. 3).
FIG. 3.
Conservation of selected epitopes among strains of Helicobacter pylori. Individual proteins among the 85 annotated genomes were aligned using CLC Main workbench, and consensus sequence for each epitope was obtained and demonstrated in the black boxes. The percentage of conservation for each residue is shown in a bar chart. (a) NapA30–49, (b) CagA1103–1122, (c) FlaA487–506, (d) OipA211–230, (e) GGT106–126, (f) HpaA33–52, (g) BabA129–149, (h) FecA437–456, (i) SabA540–559, (j) VacA459–478, and (k) UreB170–189.
3.4. Multiepitope antigen design
A step of the design pipeline is concerned with the delivery of the selected epitopes. A common strategy concatenates the epitopes into a so-called string-of-beads polypeptide. The epitope order within a string-of-beads plays a crucial role especially in degradation (Schubert et al., 2015). Therefore it was necessary to optimize the ordering such that the recovery probability of the epitopes was maximal, as shown in Figure 4.
FIG. 4.
Schematic diagram of vaccine construct consists of adjuvant in N-terminal joined to 11 epitopes fused together by proper linkers (GPGPG or KK) and CKS9 M cell homing peptide in C-terminal. CTB, cholera toxin subunit B; GPGPG, Gly-Pro-Gly-Pro-Gly; KK, Lys-Lys.
It has been reported that by coupling the antigen to CTB, a stronger response is achieved through the oral administration route. Moreover, the adjuvant potential of CTB has also been shown in large animal models, indicating that the adjuvant potential would be scalable to higher species (Baldauf et al., 2015). To increase the immunogenic capacity of our vaccine design, the CTB was added in N-terminal.
Oral immunization has ample advantages compared to other administration routes, such as safe and easy administration, high patient compliance without pain, no need of trained medical personnel, easy soak up, and immune responses in systemic and mucosal sites. In the intestine, M cells are antigen-collecting portals located on the follicle-associated epithelium of Peyer's patches, which facilitate to transport antigens from the gut lumen to underneath mucosal immune system (Lelouard et al., 2012). Thus, an M cell targeting peptide ligand (CKS9), consisting of nine amino acids with cyclic conformation, was introduced in C-terminal. Yoo et al. (2010) demonstrated that CKS9 is an efficient target coupled with chitosan nanoparticles by in vitro and in vivo assays.
The final vaccine consists of the adjuvant CTB (CAA53976.1) in the N-terminal fused to 11 epitopes of H. pylori separated by linkers (DPRVPSS, GPGPG, and KK) and fused with CKS9 peptide in the C-terminal. Schematic diagram of the vaccine is displayed in Figure 4.
3.5. Protein structure prediction and validation
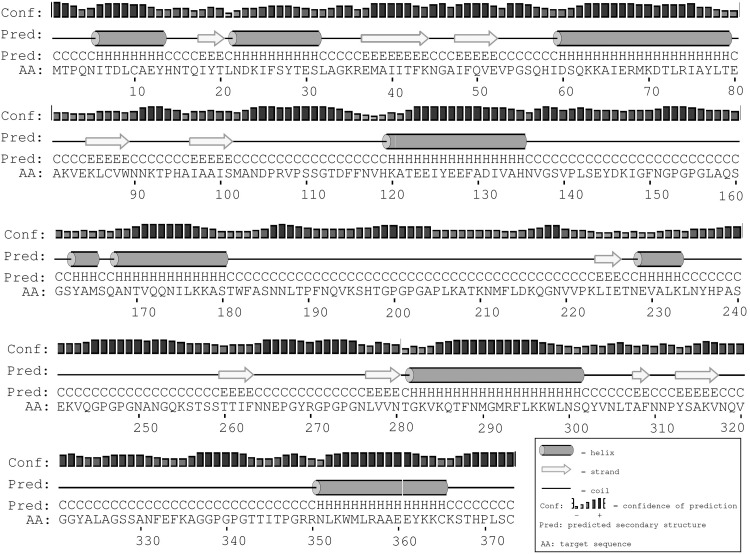
The vaccine is composed of 373 amino acids, and prediction of secondary structure showed that it contains 35% α helixes, 14.0% β sheets, and 49.0% others (random coil and β-turn), as shown in Figure 5.
FIG. 5.
The secondary structure prediction of vaccine by PSIPRED. The protein vaccine consists of 35% α helix (H, cylinder), 14% β strand (E, arrow), and 49% coil (C, line) secondary structural elements. The bar chart represents the percentage of confidence.
Five 3D models of the protein vaccine were generated among which the model with the highest c-score = −0.62 was selected for further refinement; the c-score range is typically from −5 to 2, the higher the value, the higher the confidence.
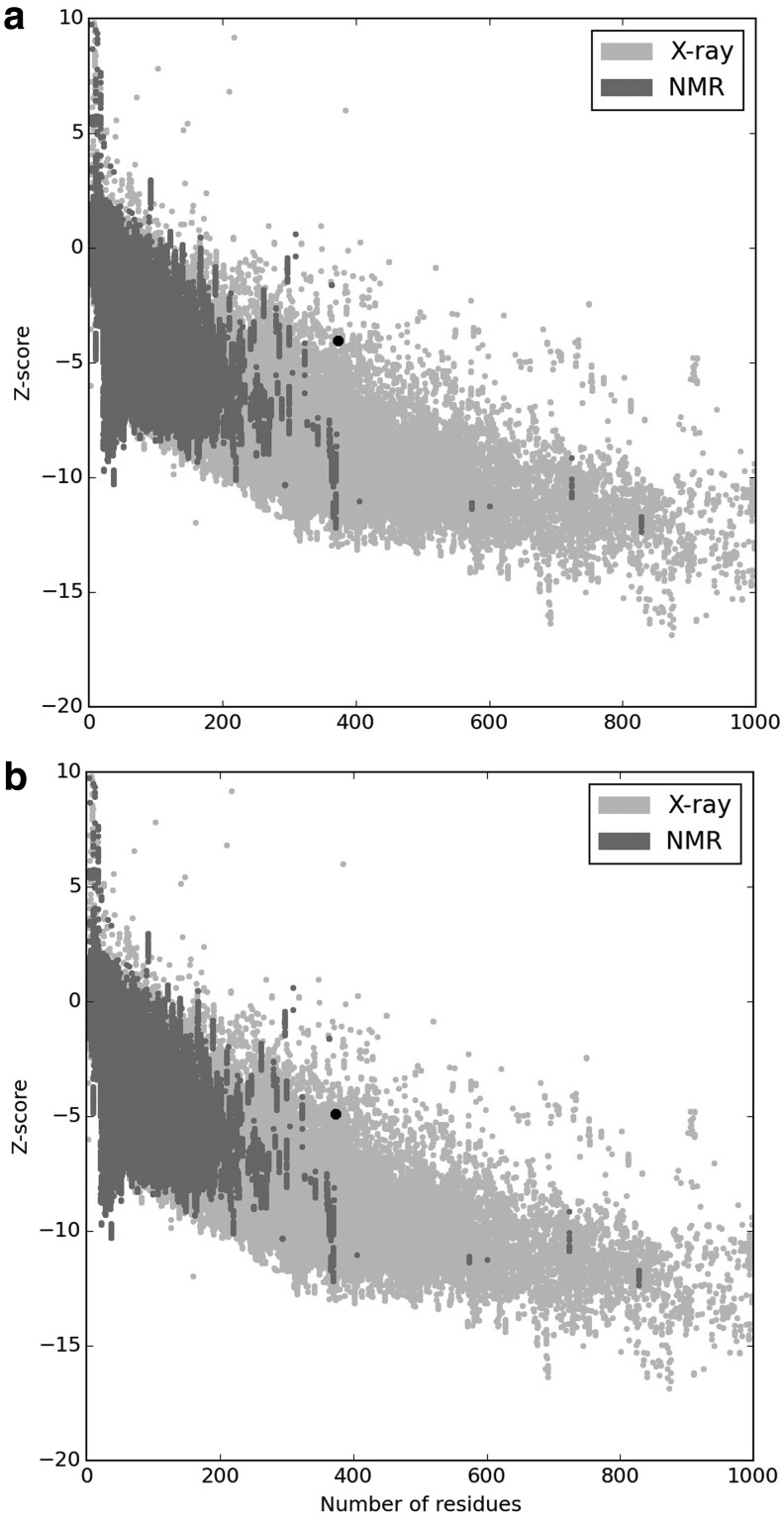
The quality and potential errors in the best model were analyzed. The initial input model z-score was −4.03, which falls within those commonly observed in similar size-native proteins (Fig. 6a). ProSA-web indicated that the preliminary model requires refinement processes. Hence, the crude model was subjected to loop refinement and energy minimization. After all refinement procedures, ERRAT factor was improved from 68.8 to 83.3. The z-score of the final model reached a value of −4.9 (Fig. 6b). The starting and refined models were superimposed (Fig. 7).
FIG. 6.
The z-score plot of unrefined and refined 3D structure of vaccine by ProSA-web. (a) The z-score of the starting model is −4.03, (b) The z-score of model after refinement steps is −4.9. The z-score indicates overall model quality and is depicted as a black spot. The z-score plot contains the z-scores of all experimentally determined protein chains in current protein data bank (PDB) from NMR spectroscopy (charcoal) and X-ray crystallography (silver). 3D, three-dimensional.
FIG. 7.
Comparison of initial and refined 3D protein structure conformations of vaccine by Molecular Operating Environment software. The initial and refined models are colored in silver and charcoal, respectively.
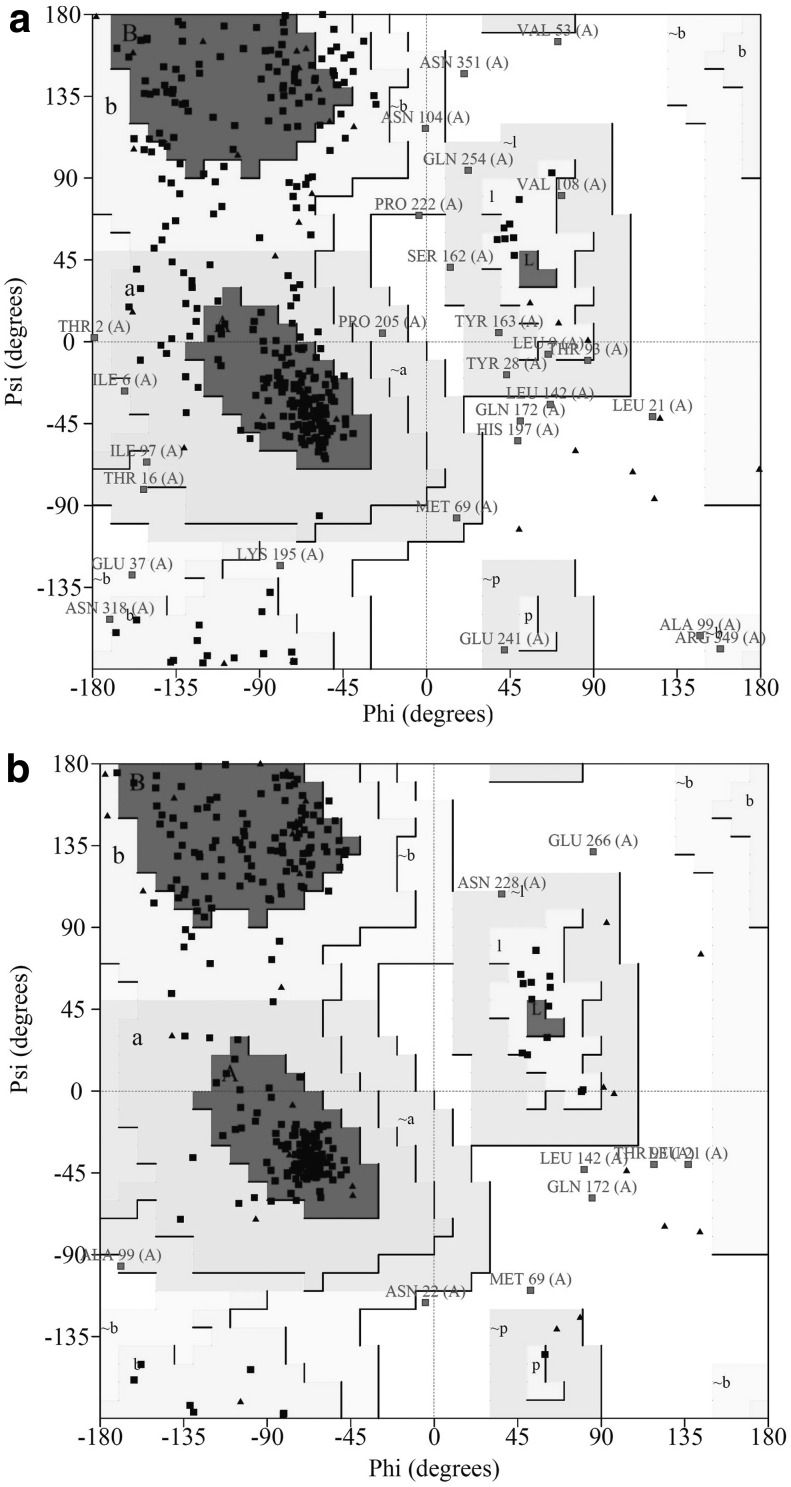
To validate the 3D models, Ramachandran plot analysis was performed before and after refinement processes. The Ramachandran plots of the unrefined model indicated that 63.5% of residues were located in most-favored regions, 28.6% in the additional allowed region, 5.7% in generously allowed regions, and 2.2% in disallowed regions of the plot (Fig. 8a). The refined model showed that 85.1% of residues were located in most-favored regions, 12.1% in additional allowed regions, 0.6% in generously allowed regions, and only 2.2% in disallowed regions (Fig. 8b). Our results indicated that the quality and stability of the final refined model were remarkably improved based on Ramachandran plot predictions.
FIG. 8.
Validation of vaccine 3D model using Ramachandran plot. The Ramachandran plots of (a) the unrefined model and (b) the refined model. The most-favored (A, B, and L) and additional allowed (a, b, l, and p) regions were demonstrated with charcoal and silver gray colors, respectively. The generously allowed regions (∼a, ∼b, ∼l, and ∼p) are indicated in silver, and the disallowed regions are in white color. Glycine residues are shown in black triangles, and other residues of protein are shown in black squares.
3.6. Antigenicity, allergenicity, solubility, and physicochemical parameters of the vaccine
An antigenicity score of 0.5547 was obtained. The allergenicity prediction showed that the vaccine is not allergenic. The molecular weight and theoretical pI of protein were 40.7 kDa and 9.36, respectively. The recombinant protein vaccine solubility upon overexpression in Escherichia coli was 0.821589. Half-life was estimated to be 30 hours in mammalian reticulocytes, >20 hours in yeast and >10 hours in E. coli. The vaccine was founded as stable with instability index of 32.58. GRAVY and aliphatic index were assessed as −0.485 and 67.77, respectively.
3.7. Codon optimization for expression in L. lactis
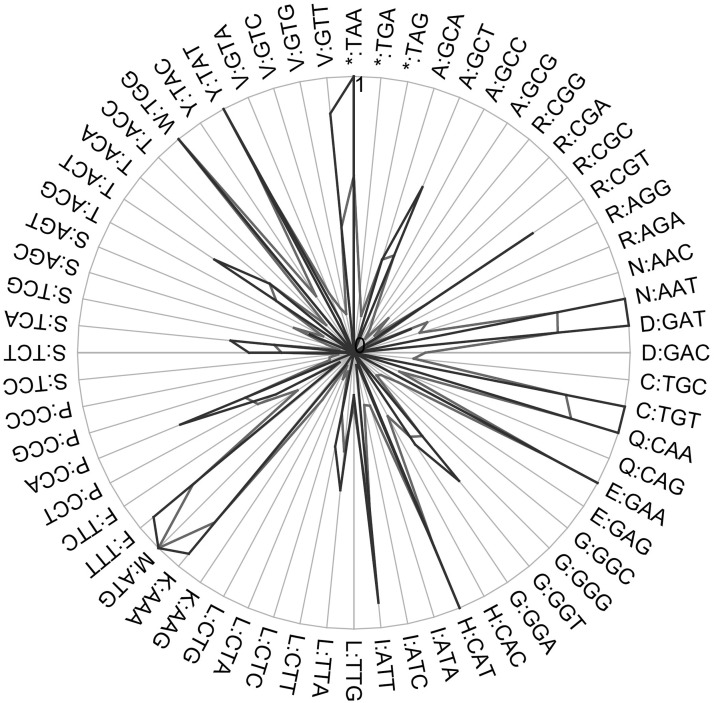
CAI of the gene sequence was 0.9464; a CAI >0.8 is acceptable for high-level expression in the L. lactis host (Fig. 9). The guanine-cytosine (GC) content fitness, individual codon fitness, codon autocorrelation fitness, and 5′ RNA folding instability were −0.0062602, −0.1577, −0.099844, and −1.9, respectively. The overall GC content was 31.63%; the ideal percentage range of GC content ranged from 30% to 70%. Negative CIS elements and repeat sequences were not found in the optimized gene. A total of 78 hidden stop codons were found; Shine-Dalgarno mRNA sequences were excluded. To clone the gene in E. coli-L. lactis pNZ8048 shuttle vector, NcoI and HindIII restriction sites were introduced to 5′ and 3′ ends of DNA sequence, respectively.
FIG. 9.
Comparison of the individual codon usage between host and optimized sequences. The host and optimized sequences are shown in charcoal and black lines, respectively.
4. Conclusion
In this study, we designed a multiepitope oral vaccine against H. pylori based on bioinformatics analysis to produce a robust humoral and cellular immune response for H. pylori prevention. Our novel oral vaccine design could be a good vaccine candidate against H. pylori. However, to validate the prophylactic and therapeutic effects of our oral vaccine design, in vitro and in vivo immunological studies are required.
Acknowledgments
The research and publication of this article were funded, by Consejo Nacional de Ciencia y Tecnología (CONACYT-México) through Programa de estímulos a la investigación, desarrollo tecnológico e innovación 2018 (Grant Nos. 252534 and 251475) and Becas Nacionales 2016 (CVU No. 578753). The authors also thank Unidad de Odontología Integral y Especialidades of Centro de Investigación y Desarrollo en Ciencias de la Salud (UOIE-CIDICS) and Laboratorio de Inmunología y Virología (LIV) of School of Biological Sciences (FCB-UANL) for supporting the development of this study. This article is dedicated to the memory of Nicaraguan students on April 19th, 2018.
Author Disclosure Statement
The authors declare there are no competing financial interests.
References
- Andreatta M., and Nielsen M. 2016. Gapped sequence alignment using artificial neural networks: Application to the MHC class I system. Bioinformatics. 32, 511–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldauf K., Royal J., Hamorsky K., et al. 2015. Cholera toxin B: One subunit with many pharmaceutical applications. Toxins. 7, 974–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burucoa C., and Axon A. 2017. Epidemiology of Helicobacter pylori infection. Helicobacter. 22, e12403. [DOI] [PubMed] [Google Scholar]
- Colovos C., and Yeates T.O. 1993. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 2, 1511–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.-H., Lu G., Chen X., et al. 2017. OGEE v2: An update of the online gene essentiality database with special focus on differentially essential genes in human cancer cell lines. Nucleic Acids Res. 45, 940–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin J.X., Chung B.K.S., and Lee D.Y. 2014. Codon Optimization OnLine (COOL): A web-based multi-objective optimization platform for synthetic gene design. Bioinformatics. 30, 2210–2212 [DOI] [PubMed] [Google Scholar]
- Dwivedi P., Alam S.I., and Tomar R.S. 2016. Secretome, surfome and immunome: Emerging approaches for the discovery of new vaccine candidates against bacterial infections. World J. Microbiol. Biotechnol. 32, 155. [DOI] [PubMed] [Google Scholar]
- Grimm S.K., and Ackerman M.E. 2013. Vaccine design: Emerging concepts and renewed optimism. Curr. Opin. Biotechnol. 24, 1078–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Yang H., Tang F., et al. 2017. Oral immunization with a multivalent epitope-based vaccine, based on NAP, urease, HSP60, and HpaA, provides therapeutic effect on H. pylori infection in Mongolian gerbils. Front. Cell. Infect. Microbiol. 7, 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Chen L., Yang W., et al. 2016. Systematic identification of immunodominant CD4+ T cell responses to HpaA in Helicobacter pylori infected individuals. Oncotarget. 7, 54380–54391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC. 1994. Schistosomes, liver flukes and Helicobacter pylori. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans, Lyon, June 7–14, 1994. IARC Monogr Eval Carcinog Risks Hum. 61, 1–241 [PMC free article] [PubMed] [Google Scholar]
- Jespersen M.C., Peters B., Nielsen M., et al. 2017. BepiPred-2.0: Improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 45, 24–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kringelum J.V., Lundegaard C., Lund O., et al. 2012. Reliable B cell epitope predictions: Impacts of method development and improved benchmarking. PLoS Comput. Biol. 8, e1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., Larsson B., Von Heijne G., et al. 2001. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur M.W., and Thornton J.M. 2012. PROCHECK: Validation of protein-structure coordinates. Int. Tables Crystallogr. F, 684–687 [Google Scholar]
- Leja M., Axon A., and Brenner H. 2016. Epidemiology of Helicobacter pylori infection. Helicobacter. 21, 3–7 [DOI] [PubMed] [Google Scholar]
- Lelouard H., Fallet M., de Bovis B., et al. 2012. Peyer's patch dendritic cells sample antigens by extending dendrites through M cell-specific transcellular pores. Gastroenterology. 142, 592–601 [DOI] [PubMed] [Google Scholar]
- Li X., Xing Y., Guo L., et al. 2014. Oral immunization with recombinant Lactococcus lactis delivering a multi-epitope antigen CTB-UE attenuates Helicobacter pylori infection in mice. Pathog. Dis. 72, 78–86 [DOI] [PubMed] [Google Scholar]
- Livingston B., Crimi C., Newman M., et al. 2002. A rational strategy to design multiepitope immunogens based on multiple Th lymphocyte epitopes. J. Immunol. 168, 5499–5506 [DOI] [PubMed] [Google Scholar]
- Malfertheiner P., Schultze V., Rosenkranz B., et al. 2008. Safety and immunogenicity of an intramuscular Helicobacter pylori vaccine in noninfected volunteers: A phase I study. Gastroenterology. 135, 787–795 [DOI] [PubMed] [Google Scholar]
- Marshall B.J., and Warren J.R. 1984. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1, 1311–1315 [DOI] [PubMed] [Google Scholar]
- McGuffin L.J., Bryson K., and Jones D.T. 2000. The PSIPRED protein structure prediction server. Bioinformatics. 16, 404–405 [DOI] [PubMed] [Google Scholar]
- Meza B., Ascencio F., Sierra-Beltrán A.P., et al. 2017. A novel design of a multi-antigenic, multistage and multi-epitope vaccine against Helicobacter pylori: An in silico approach. Infect. Genet. Evol. 49, 309–317 [DOI] [PubMed] [Google Scholar]
- Monterrubio-López G.P., González-Y-Merchand J.A., and Ribas-Aparicio R.M. 2015. Identification of novel potential vaccine candidates against tuberculosis based on reverse vaccinology. Biomed. Res. Int. 2015, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller A., and Hartung M.L. 2016. Helicobacter pylori and the host immune response, 299–323. In Backert S., Yamaoka Y., eds. Helicobacter pylori Research: From Bench to Bedside. Springer, Japan, Tokyo [Google Scholar]
- Naz A., Awan F.M., Obaid A., et al. 2015. Identification of putative vaccine candidates against Helicobacter pylori exploiting exoproteome and secretome: A reverse vaccinology based approach. Infect. Genet. Evol. 32, 280–291 [DOI] [PubMed] [Google Scholar]
- Nezafat N., Eslami M., Negahdaripour M., et al. 2017. Designing an efficient multi-epitope oral vaccine against Helicobacter pylori using immunoinformatics and structural vaccinology approaches. Mol. Biosyst. 13, 699–713 [DOI] [PubMed] [Google Scholar]
- O'Ryan M., Stoddard J., Toneatto D., et al. 2013. A multi-component meningococcal serogroup B vaccine (4CMenB): The clinical development program. Drugs. 74, 15–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyarzún P., and Kobe B. 2015. Recombinant and epitope-based vaccines on the road to the market and implications for vaccine design and production. Hum. Vaccin. Immunother. 12, 763–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H., Ko J., Joo K., et al. 2011. Refinement of protein termini in template-based modeling using conformational space annealing. Proteins. 79, 2725–2734 [DOI] [PubMed] [Google Scholar]
- Ponomarenko J., Bui H.H., Li W., et al. 2008. ElliPro: A new structure-based tool for the prediction of antibody epitopes. BMC Bioinformatics. 9, 514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qui Y., Wang Y.C., Tao H.X., et al. 2010. Identification of B-cell epitopes urease B subunit of Helicobacter pylori bound by neutralizing antibodies. Vaccine. 28, 5220–5227 [DOI] [PubMed] [Google Scholar]
- Rappuoli R., Bottomley M.J., D'Oro U., et al. 2016. Reverse vaccinology 2.0: Human immunology instructs vaccine antigen design. J. Exp. Med. 213, 469–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa D.S., Ribeiro S.P., Fonseca S.G., et al. 2015. Multiple approaches for increasing the immunogenicity of an epitope-based anti-HIV vaccine. AIDS Res. Hum. Retroviruses. 31, 1077–1088 [DOI] [PubMed] [Google Scholar]
- Rossi G., Ruggiero P., Peppoloni S., et al. 2004. Therapeutic vaccination against Helicobacter pylori in the Beagle Dog Experimental Model: Safety, immunogenicity, and efficacy. Infect. Immun. 72, 3252–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert B., Brachvogel H.P., Jürges C., et al. 2015. EpiToolKit—A web-based workbench for vaccine design. Bioinformatics. 31, 2211–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serruto D., Bottomley M.J., Ram S., et al. 2012. The new multicomponent vaccine against meningococcal serogroup B, 4CMenB: Immunological, functional and structural characterization of the antigens. Vaccine. 30, 87–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H., and Raghava G.P. 2001. ProPred: Prediction of HLA-DR binding sites. Bioinformatics. 17, 1236–1237 [DOI] [PubMed] [Google Scholar]
- Singh H., and Raghava G.P. 2003. ProPred1: Prediction of promiscuous MHC Class-I binding sites. Bioinformatics. 19, 1009–1014 [DOI] [PubMed] [Google Scholar]
- Sutton P., and Boag J.M. 2018. Status of vaccine research and development for Helicobacter pylori. Vaccine. 10.1016/j.vaccine.2018.01.001 [DOI] [PMC free article] [PubMed]
- Thomson S.A., Jaramillo A.B., Shoobridge M., et al. 2005. Development of a synthetic consensus sequence scrambled antigen HIV-1 vaccine designed for global use. Vaccine. 23, 4647–4657 [DOI] [PubMed] [Google Scholar]
- Thung I., Aramin H., Vavinskaya V., et al. 2016. Review article: The global emergence of Helicobacter pylori antibiotic resistance. Aliment. Pharmacol. Ther. 43, 514–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattam A.R., Davis J.J., Assaf R., et al. 2017. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 45, 535–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederstein M., and Sippl M.J. 2007. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 35, 407–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins M.R., Gasteiger E., Bairoch A., et al. 1999. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552 [DOI] [PubMed] [Google Scholar]
- Wroblewski L.E., Peek R.M., Jr., and Wilson K.T. 2010. Helicobacter pylori and gastric cancer: Factors that modulate disease risk. Clin. Microbiol. Rev. 23, 713–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., and Zhang Y. 2015. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 43, 174–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo M.K., Kang S.K., Choi J.H., et al. 2010. Targeted delivery of chitosan nanoparticles to Peyer's patch using M cell-homing peptide selected by phage display technique. Biomaterials. 31, 7738–7747 [DOI] [PubMed] [Google Scholar]
- Yu C.S., Chen Y.C., Lu C.H., et al. 2006. Prediction of protein subcellular localization. Proteins. 64, 643–651 [DOI] [PubMed] [Google Scholar]