Abstract
Uterine fibroids (UFs) are the most common benign neoplastic threat to women’s health and associated with DNA damage and genomic instability. Hypovitaminosis D is a known risk factor for UFs, especially among African Americans. Vitamin D3 has been shown to effectively inhibit UF phenotype, but its mechanisms remain unclear. We hypothesize that Vitamin D3 ameliorates UFs by recovering the damaged DNA repair system, thus inhibits tumor progression. We compared the DNA damage status and Vitamin D receptor (VDR) expression between normal myometrial and UF primary cells. Unrepaired DNA double-strand breaks (DSBs) accumulated but VDR expression decreased in UFs. The RNA and protein levels of key DNA repair members belonging to DNA DSB sensors (MRE11, NBS1, RAD50), mediators and effectors (CHECK2, BRCA1, RAD51) were downregulated in UFs compared with myometrial cells. VDR KD induced DSB accumulation and DNA damage response (DDR) defects in myometrial cells. Using the DNA damage PCR array, the expression of many additional DNA repair genes was downregulated in VDR KD cells. Treatment of UF cells with Vitamin D3 (100 nM) significantly decreased DNA damage and restored DDR concomitant with VDR induction. Notably, the PCR array demonstrated that among 75 downregulated genes after VDR KD, 67 (89.3%) were upregulated after vitamin D3 treatment. These studies demonstrate a novel link between DNA damage and the vitamin D3/VDR axis in UFs. Vitamin D3 suppresses the UF phenotype through orchestrated targeting at multiple molecules in DNA repair pathways, thus offering novel mechanistic insights into the clinical effectiveness of vitamin D3 on UFs.
Keywords: uterine fibroids, DNA damage, DNA repair, hypovitaminosis D, vitamin D3, vitamin D receptor
Introduction
Uterine fibroids (UFs; AKA: leiomyoma) are the most common benign neoplastic threat to women’s health. Tumors occur in 70%–80% of women overall and are clinically manifested in 25%–50% by 50 years of age [1]. Despite being benign, these tumors are nonetheless associated with significant morbidity; they are the primary indication for hysterectomy, and a major source of gynecologic and reproductive dysfunction, ranging from profuse menstrual bleeding and pelvic pain to infertility, recurrent miscarriage, and pre-term labor [2]. UFs therefore represent a significant public health and financial burden with annual health care costs estimated up to $34.4 billion in the United States only and hundreds of billions of dollars worldwide [3].
UF-caused morbidities negatively impact women of all ethnicities, but they disproportionately affect African American (AA) women, who have 3–4 times higher incidence rate and relative risk of UFs than Caucasian (CC) women [4]. AA women develop UFs 10 years earlier and have more and larger UFs than CC women with more aggressive symptoms [1]. While the basis for this risk disparity is not fully understood, recent studies implicate hypovitaminosis D as a major contributor since AA women have a tenfold increased risk of vitamin D deficiency compared with CC women. In addition, we first reported that the risk of UF is inversely correlated with 25-hydroxy vitamin D serum levels [4]. However, it is not clear whether and how the processes that drive UF formation and racial risk disparity are genetically or biochemically linked.
Recently, our group and others have identified somatic mutations in the transcriptional mediator complex subunit 12 (MED12) as a dominant driver of UFs, accounting for ~85% of tumors [5–9]. Notably, MED12-mutant UFs, but not the adjacent myometrium, have been characterized by significant chromosomal loss and rearrangement, suggesting genomic instability as a driving force in tumor progression [10, 11]. Although the cause of these specific fibroid-causing mutations remains unknown, many tissues with defects in DNA repair, indicative of a compromised DNA damage response (DDR), are more susceptible to somatic tumor-forming mutations [12–15]. Our group has recently demonstrated the downregulation of several DNA repair genes in UFs compared with myometrium, suggesting that the impairment of the DNA repair capacity might be linked to UF genomic integrity with subsequent initiation/propagation of the UF tumor [16, 17].
Many epidemiologic and laboratory studies have shown that vitamin D3 exhibits a chemopreventive effect through activation of its receptor (VDR) in different types of cancers [18], including sex steroid-dependent cancers such as breast and prostate cancer, while its deficiency is associated with an increased risk of tumor development [19, 20]. Moreover, studies have shown that vitamin D3 attenuates the level of DNA damage and modulates DDR via induction of DNA repair genes expression. For example, in human and mouse fibroblasts [21], the vitamin D3/VDR axis has been shown to regulate the expression of double-strand break (DSB) repair proteins, and vitamin D3 deficiency leads to reduced homology-dependent DSB repair [19]. Additionally, in human prostate epithelial cells and B-lymphocytes, vitamin D3/receptor signaling has been shown to both prevent DSB formation and promote repair [20, 21]. However, the role of the vitamin D3/VDR axis in DDR of UFs and its functional relationship with MED12-associated genetic instability is unknown.
Our group has previously shown that vitamin D3 effectively suppresses UF growth in both UF cultured cells and UF animal model, supporting the idea that vitamin D3 and its potent analogs might offer promising options for the prevention and early treatment of UFs [4, 19, 22–28]. Nonetheless, a critical barrier to progress in this area is a lack of understanding concerning the relationship between vitamin D3 deficiency and dysfunctional DNA repair machinery, which contribute to the development of UFs.
DNA damage response involving damage sensors, signal transducers, repair effectors, and arrest or death effectors, leading to the most severe lesions, DNA DSBs, must be repaired via homologous recombination or non-homologous end-joining (NHEJ), requiring a high level of accuracy to maintain genome integrity [29–31]. Herein, we hypothesize that hypovitaminosis D exacerbates DNA damage accumulation and genomic instability arising in UFs, leading to enhanced tumor progression and burden. The objective of this study was to investigate the role and molecular mechanism of the vitamin D3/VDR axis in DNA damage repair, further offering a proof of concept for therapeutic intervention in this particular tumor type.
Materials and methods
Human tissue sample collection and primary cell isolation
Freshly collected human fibroid and adjacent myometrial samples were obtained via the Augusta University Biorepository under an approved IRB protocol (IRB No. 644354-6) from AA women of reproductive age (22–55 years old) undergoing hysterectomy or myomectomy for symptomatic UFs. These patients had not received any hormonal supplements, including vitamin D, for 3 months prior to the day of surgery (i.e., the day of sample collection). The 8-cm3 UF tissue samples were collected. Myometrial tissue samples, of at least 2 cm from adjacent UF, were collected to exclude any mechanical or hormonal effects of the fibroids on the adjacent myometrial tissue. For preparation of the primary cell population, the collected samples were washed with calcium- and magnesium-containing Hanks’ balanced salt solution (HBSS) to remove blood and chopped into small pieces. The tissues were then digested for 3.5 h at 37 °C with shaking in an enzyme buffer of calcium- and magnesium-free HBSS containing 1% antibiotics-antimycotics, 2.5% N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid [HEPES], 660 µg/mL collagenase Type IV (Worthington, NJ, USA), and 4.76 µg/mL DNase I (Sigma-Aldrich, St. Louis, MO, USA). The suspension was filtered through a 100-µm sterile nylon mesh cell strainer to remove undigested tissues and then through a 70-µm cell strainer to obtain a single cell suspension. The remaining undigested tissue was suspended in a fresh enzyme buffer and incubated for 14 h at 37 °C and filtered again to obtain a single cell suspension. Cells were plated out and incubated at 37 °C to allow them to attach to a sterile tissue culture-treated plate containing smooth muscle basal medium (SmBM), as described below.
Cell lines and cultures
The immortalized human UF cell line (HuLM) and immortalized human uterine smooth muscle cells (UTSM) were a generous gift from Dr. Darlene Dixon (National Institute of Environmental Health Sciences, Research Triangle Park, NC, USA). These cells were cultured and maintained in SmBM culture medium with 5% fetal bovine serum, 0.1% insulin, 0.2% recombinant human fibroblast growth factor (hFGF-B), 0.1% gentamicin sulfate and amphotericin B mixture and 0.1% human epidermal growth factor (hEGF) at 37 °C in a humidified atmosphere of 5% CO2/95% air. HuLM cells were passaged two times, and when they were 70% confluent, the medium was replaced with phenol red-free, serum-free DMEM/F-12 for vitamin D3 treatment (100 nM vitamin D3 for 3 d). Absolute ethanol was used to dissolve the drugs. The final concentration of ethanol in the culture medium was <0.01%, and the same concentration of ethanol was used as a vehicle in the control cultures.
Primary cells (5 × 105) isolated from fibroids and myometrium, or HuLM cells in the presence or absence of 100 nM vitamin D3 were trypsinized and washed in PBS and centrifuged at 500 × g for 5 min. The supernatant was removed, and the pellets were snap-frozen in liquid nitrogen and stored at –80 °C until further use.
Generation of stable vitamin D receptor knockdown cells
To determine the role of vitamin D signaling in the DNA damage repair system, the VDR gene was knocked down in UTSM cells using a VDR gene-specific short hairpin RNA (shRNA). These UTSM cells expressed normal levels of endogenous VDR protein, as we have shown previously [25], and therefore, UTSM provided an appropriate model. We used lentivirus plasmid constructs containing human VDR gene-specific shRNA (VDR KD) sequences and nonfunctional scrambled-control shRNA (VDR Scr), which were purchased from Origene Inc. (Rockville, MD, USA). These plasmid constructs contain a puromycin selection marker gene. These lentivirus constructs were transiently transfected into human embryonic kidney cells (293 T) using lipofectamine LTX transfection reagent according to the manufacturer’s instructions (Invitrogen). Fifteen hours after transfection, fresh DMEM was added and incubated for another 48 h. Supernatant medium containing lentiviruses was collected and filtered, and subsequently polybrene solution was added at a concentration of 8 µg/mL. These virus-containing media were used to infect cultured UTSM cells. Fifteen hours after infection, fresh medium was added and then cultured for another 48 h. Infected cells were selected with puromycin to generate stable populations, which were used for molecular characterization.
Reagents and antibodies
Bioactive 1, 25 dihydroxyvitamin D3 (vitamin D3) was purchased from Sigma Biochemicals (St. Louis, MO, USA). SmBM was purchased from Lonza (Walkersville, MD, USA). DMED/F-12 was purchased from Gibco/Thermo Fisher scientific (Waltham, MA, USA). The antibodies used in this study are listed in Table 1.
Table 1.
List of antibodies used for Western blotting and immunofluorescence
| Antibodies | Manufacturer and catalog number | Species raised, monoclonal or polyclonal | Techniques and dilutions used |
|---|---|---|---|
| Anti-phospho-histone H2A.X (γ-H2AX) (Ser139) | Cell signaling technology, #9718 | Rabbit, monoclonal | WB, 1:1000 IF, 1:200 |
| Anti-Vitamin D receptor | Abcam, ab134826 | Rabbit, polyclonal | WB, 1:1000 |
| Anti-RAD50 | Cell signaling technology, # 3427 Abcam, ab89 | Rabbit, polyclonalMouse, monoclonal | WB, 1:1000 IF, 1:120 |
| Anti-NBS1 | Cell signaling technology, #14956 | Rabbit, monoclonal | WB, 1:1000 |
| Anti-MRE11 | Cell signaling technology, #4847 | Rabbit, monoclonal | WB, 1:1000 |
| Anti-Check2 | Cell signaling technology, #3440 | Mouse, monoclonal | WB, 1:1000 |
| Anti-BRCA1 | Cell signaling technology, #9010 | Rabbit, polyclonal | WB, 1:1000 |
| Anti-RAD51 | Abcam, ab133534 | Rabbit, monoclonal | WB, 1:5000 |
| Anti-beta actin | Sigma biochemicals, A5316 | Mouse, monoclonal | WB, 1:5000 |
IF: Immunofluorescence, WB: Western blot
Protein extraction and Western blot analysis
Pellets were lysed in lysis buffer with protease and phosphatase inhibitor cocktail (Thermo Fisher scientific, Waltham, MA, USA), and protein was quantified using the Bradford method (Bio-Rad protein Assay kit, Hercules, CA, USA). Next, 30 µg of the protein lysates was resolved in Gradient (4%–20%) MiniPROTEAN TGX Precast Protein Gels (Bio-Rad, Hercules, CA, USA), transferred onto a polyvinylidene fluoride membrane (Bio-Rad), blocked for 1 h at room temperature in either 5% w/v nonfat dry milk or 5% bovine serum albumin (BSA) in 0.05% Tween-supplemented PBS (0.05% PBST) per the antibody specifications and incubated with the specific primary antibodies listed in Table 1 overnight at 4 °C, followed by a 90-min incubation with the appropriate HRP-conjugated secondary antibodies (Cell Signaling Technology, Danvers, MA, USA). The antigen-antibody complex was detected with the Pierce enhanced chemiluminescence detection kit (Thermo Fisher Scientific, Waltham, MA, USA) or Trident Femto Western HRP substrate (GeneTex, Irvine, CA, USA). Specific protein bands were visualized using the ChemiDoc XRS + molecular imager (Bio-Rad, Hercules, CA, USA). The intensity of each protein band was quantified using Bio-Rad Image Lab software and normalized against the corresponding β-actin. The normalized values were used to create data graphs.
Immunohistochemistry
Collected human fibroid and adjacent myometrial samples from four AA women were fixed in 10% buffered formalin for 15–20 h, embedded with paraffin, and subjected to immunohistochemistry (IHC). Paraffin-embedded tissue sections were deparaffinized and rehydrated by being passed through xylene and graded ethanol solutions as previously described. The DNA repair marker RAD51 was analyzed via staining with immunoreactive anti-RAD51 antibody (monoclonal rabbit, 1:1000) from Abcam (Cambridge, MA, USA) and subsequent staining with peroxidase-conjugated secondary antibody.
For visual evaluation, each sample was scored for staining intensity, and positive cells were semi-quantified. To determine the protein immunostaining score, we used a design proposed elsewhere with some modifications. In brief, the semi-quantitative score was established by considering the percentage of labeled cells (0, negative; 1 < 10% of the cells; 2, 10%–50% of the cells; 3, 50%–75% of the cells; 4 > 75% of the cells) and the intensity of immunostaining (0, no staining; 1, weak; 2, mild; 3, strong staining). Multiplication of both scores resulted in a final quotient ranging from 0 to 12. An independent pathologist performed a blind analysis.
Isolation of cellular RNA and cDNA synthesis
Total cellular RNA was isolated from frozen pellets using the Purelink RNA mini kit (Ambion Life Technologies, Waltham, MA, USA). The concentration of total RNA was determined using a NanoDrop (Thermo Scientific, Waltham, MA, USA). One microgram of total RNA from each sample was reverse-transcribed in a 20-µL reaction volume to complementary DNA (cDNA) using Ecodry premix double-primed (Clontech lab, Takara Bio, Japan). The reaction mixture was incubated for 1 h at 42 °C and stopped by incubation at 70 °C for 10 min.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Quantitative real-time PCR was performed to determine the mRNA expression of several genes listed with their primer sequences in Table 2. Primers were purchased from Integrated DNA technologies (IDT, Coralville, Iowa, USA). Equal amounts of cDNA from each sample were added to the Mastermix containing appropriate primer sets and SYBR green supermix (Bio-Rad) in a 20-µL reaction volume. All samples were analyzed in triplicate. Real-time PCR analyses were performed using a Bio-Rad CFX connect. Cycling conditions consisted of denaturation at 95 °C for 2 min followed by 40 cycles of 95 °C for 5 s and 60 °C for 30 s and then 65 °C for 5 s. Synthesis of a DNA product of the expected size was confirmed by melting curve analysis. The 18 S ribosomal RNA values (internal control) were used to normalize the expression data, and normalized values were used to create data graphs.
Table 2.
Human primer sequences for qRT-PCR
| Gene | Forward primer sequence | Reverse primer sequence | |
|---|---|---|---|
| MRE11 meiotic recombination 11 | MRE11 | 5′-CTCAGCAGCAACCAACAAAG-3′ | 5′GTGTCTCTTCCTCTTTGAGACC-3′ |
| RAD50 homolog | RAD50 | 5′-GAATCTAGGCTTCTCAATCAGG-3′ | 5′-CTCTAGCTCGGATATGTTCTTG-3′ |
| Nibrin | NBS1 | 5′-CCACCTCCAAAGACAACTG-3′ | 5′-AGGACGGCAGGAAAGAA-3′ |
| CHECK1 checkpoint homolog | CHECK1 | 5′-TCTGTTGGATGAAAGGGATAAC-3′ | 5′-TCAACAAACGCTCACGATTA-3′ |
| CHECK2 checkpoint homolog | CHECK2 | 5′-GTGTCCACTCAGGAACTCTATTC-3′ | 5′CATCCTGAAGGGCCCATAATC-3′ |
| Breast cancer 1, early onset | BRCA1 | 5′-CAAGAAGAGCAAAGCATGGA-3′ | 5′-GAGGATAGCCCTGAGCAGTC-3′ |
| Breast cancer 2, early onset | BRCA2 | 5′-TTTGCGTTGAGGAACTTGTG-3′ | 5′-AGGTGGCCCTACCTCAAAAT-3′ |
| RAD51 homolog | RAD51 | 5′-AAAGGAAGAGGGGAAACCAG-3′ | 5′-ATCTCCCACTCCATCTGCAT-3′ |
| RAD17 homolog | RAD17 | 5′-GTGGTTTAAGGAGCGGTCAG-3′ | 5′-TTTGCATCCTGAATTAGTCCTTC-3′ |
| MutS homolog 2 | MSH2 | 5′-AAGAAGTGCTATCTGGAAAGAG-3′ | 5′-ACATTTCAGTAAAGGGCATTTG-3′ |
| Exonuclease 1 | EXO1 | 5′-AGGGAAAGCAACTTCTTCG-3′ | 5′-GCCATGGCATGTGTGATA-3′ |
| Vitamin D receptor | VDR | 5′- ATGCCATCTGCATCGTCTC-3′ | 5′-GCACCGCACAGGCTGTCCTA-3′ |
| 18 S Ribosomal RNA | 18 S rRNA | 5′-CGAACGTCTGCCCTATCAACTT-3′ | 5′-ACCCGTGGTCACCATGGTA-3′ |
DNA damage prime PCR array
cDNA from UTSM cells, VDR KD and VDR Scr, or HuLM cells, untreated control and vitamin D treated, were loaded per gene into separate DNA Damage R96 plates (Bio-Rad Inc, Hercules, CA, USA). The expression levels of total 85 genes involved in the DNA damage signaling pathways were simultaneously detected with SsoAdvanced Universal SYBR Green Supermix on a Bio-Rad CFX96 real-time PCR system. The following thermo-cycling conditions were used: 1 cycle of 95 °C for 10 min; 40 cycles of 95 °C for 30 s, and 60 °C for 1 min. For data analysis, the comparative method (∆∆Ct) was used to calculate relative quantities of nucleic acid sequence with Bio-Rad CFX manager software.
Immunofluorescence and laser confocal microscopy
HuLM cells (8 × 104) were cultured on sterile glass coverslips in 6-well plates and were serum-starved overnight when they reached 60% confluence and treated with vitamin D3 100 nM for 3 d. Ethanol vehicle was added to the control cells. Cells were fixed in a 4% formaldehyde solution at room temperature for 15 min. After washing in PBS 3 times, the cells were permeabilized for 15 min using 0.1% Triton X-100/PBS, and then nonspecific binding was inhibited by blocking for 1 h in blocking/incubation solution containing 1% BSA in 0.1% Triton X-100/PBS. Incubation with either primary anti-γ-H2AX (1:200) or anti-RAD50 antibody (1:120) was performed for 2 h followed by incubation with anti-rabbit Alexa Fluor 555 or anti-mouse Alexa Fluor 488-conjugated secondary antibody (Thermo Fisher, Waltham, MA, USA) (1:1000 each) for 1 h, respectively, at room temperature. Cells were washed for 15 min (three washes of 5 min each) with the above-described incubation solution, air-dried, and mounted onto microscopic slides with one drop of Fluorshield (Sigma) containing 4’,6-diamidino-2-phenylindole (DAPI) for nuclear staining. Fluorescent signals were visualized using a Zeiss 780 upright laser confocal fluorescence microscope and ZEN Black 2012 confocal software. Images were captured at 40 × magnification using a 40 × Plan-Apo (oil) - 1.4 NA lens and exported using Zen Blue 2012 software.
Quantification of γ-H2AX foci and RAD50 fluorescence density
Using ImageJ v.1.48k software (NIH, Bethesda, MD, USA), the integrated density was calculated in all images for each nucleus to determine the relative fluorescence density and intensity, corresponding to either nuclear γ-H2AX or RAD50 expression, after subtracting the background signal of each nucleus. The integrated density is a calculus of the mean stained area times the intensity of staining in each pixel in the area and indicates the total amount of stained material in that area.
Statistical analysis
For quantitative RT-PCR data, gene expression results are presented as the fold change ± standard error of the mean (SEM), and the statistical analysis was performed using Bio-Rad CFX Manager 3.1 software. Experiments were performed in triplicate and repeated twice. For Western blot analysis, the results are presented as the mean ± standard deviation (SD) and were analyzed using the two-tailed unpaired Student's t-test. Experiments were performed in triplicate and repeated twice. For immunofluorescence and immunohistochemistry analysis, intensity measurements as described above are represented as the median ± 95% confidence interval and were analyzed using the nonparametric Mann-Whitney test. Differences were considered statistically significant at P < 0.05. GraphPad 7.0 (La Jolla, CA, USA) was used to generate the graphs.
Results
Unrepaired DNA double-strand breaks accumulated but vitamin D receptor expression decreased in human uterine fibroid primary cells
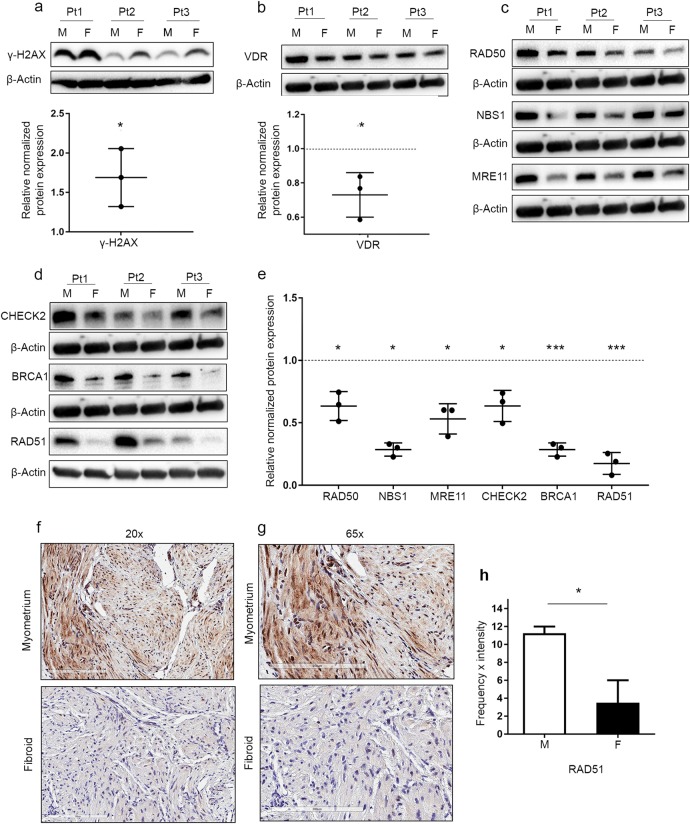
The accumulation of phosphorylated histone H2AX (γ-H2AX) serves as an early event in initiating the DDR, which involves the recruitment of DNA repair proteins, senescence and cell cycle arrest, or in the case of irreparable damage, apoptosis. In this regard, we initially determined the levels of γ-H2AX between primary cells isolated from AA (n = 3) and primary cells isolated from matched adjacent myometrium (n = 3). The expression levels of γ-H2AX were significantly increased in UF primary cells compared to myometrial primary cells (1.69 ± 0.36, P < 0.05), which is indicative of DNA damage accumulation and DSB formation (Fig. 1a). To determine the link between DNA repair damage and the vitamin D/VDR axis, we measured the expression levels of VDR between UFs and matched myometrial primary cells. VDR expression was significantly downregulated in UF primary cells compared with the adjacent myometrial primary cells (0.73 ± 0.13, P < 0.05) (Fig. 1b). To determine whether the DNA repair-related proteins were involved in the accumulation of DNA damage in primary UF cells, Western blot analysis was performed to detect the protein levels of several key DNA damage repair proteins. Our data showed that UF cells exhibited a significant reduction in protein expression levels of DNA DSB-related repair markers compared to myometrial cells. The downregulated DSB-related proteins included the DNA DSB sensors RAD50 (0.63 ± 0.11, P < 0.05), NBS1 (0.28 ± 0.05, P < 0.05) and MRE11 (0.53 ± 0.12, P < 0.05) (Fig. 1c, e). These three DNA repair proteins (MRE11, RAD50 and NBS1) comprise the MRN complex, which is necessary for homologous recombination DDR initiation and serves as a DSB sensor as well as a co-activator for checkpoint signaling. We next determined the protein levels of a key MRN controlling marker, CHECK2, which was significantly downregulated in UF (0.63 ± 0.12, P < 0.05). Similar findings were observed in MRN downstream mediators and effectors of DNA repair proteins such as BRCA1 (0.28 ± 0.05, P < 0.001) and RAD51 (0.17 ± 0.08, P < 0.001) (Fig. 1d, e), which are crucial for DNA DSB binding [26, 27]. The relative protein expression of each DNA repair protein in UF primary cells was normalized to that in the adjacent myometrial primary cells. Since Western blot analysis of RAD51 showed the lowest relative protein expression in UF compared with myometrial primary cells, RAD51 expression was further measured in UF and matched myometrial tissues (n = 4, AA) using IHC analysis. As expected, similar results of significant downregulation were observed in UF (P = 0.0265) (Fig. 1f–h).
Fig. 1.
More DNA damage, an impaired DNA repair capacity, accumulated and less VDR expression in human uterine fibroids compared with the adjacent myometrium from African American patients. Protein lysates were prepared from cultured primary cells (n = 3) isolated from 3 African American (AA) patient (Pt) fibroids (F) and matched myometrial tissues (M). The protein expression of the DNA damage marker γ-H2AX (a) and VDR (b) were determined by Western blot analysis, and relative values were used to generate the corresponding data graph. c The homologous recombination DNA damage response markers RAD50, NBS1 and MRE11 were determined by Western blot analysis. d The protein levels of CHECK2, BRCA1 and RAD51 were determined by Western blot analysis. e The intensity of each band from c and d was quantified and normalized to the corresponding β-actin, and relative values were used to generate the data graph. Normalized values of F relative to M are presented in the graph as the mean ± SD. All experiments were repeated twice. f Immunohistochemical staining of the DNA repair marker RAD51 in matched F and M tissues from AA patients (n = 4, magnification 20 × ). g Immunohistochemical staining of the DNA repair marker RAD51 in matched F and M tissues from AA patients (n = 4, magnification 65 × ). h Semi-quantitative intensity and frequency scores of RAD51 in the evaluated tissues presented as the median ± range. *P < 0.05, ***P < 0.001
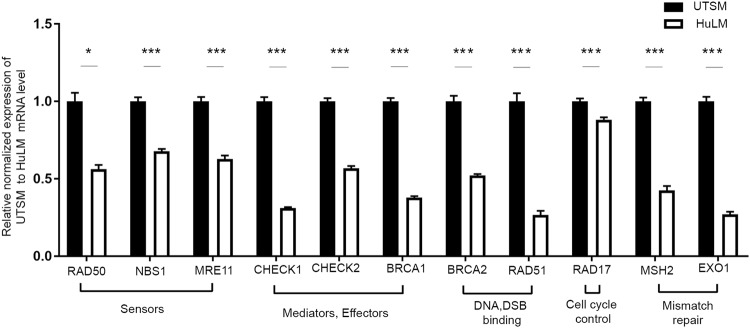
To determine whether transcriptional activity was involved in downregulation of DNA DSB repair-related genes, qRT-PCR was performed in UF and matched myometrial cells. The mRNA expression of several DNA DSB repair-related genes was downregulated in the human UF cell line relative to the myometrial one, including the following (mean fold change ± SEM, P-value): the DSB sensor MRN complex RAD50 (−0.56 ± 0.02, P < 0.05), NBS1 (−0.67 ± 0.01, P < 0.001), and MRE11 (−0.63 ± 0.02, P < 0.001); the DNA repair mediators and effectors CHECK1 (−0.31 ± 0.006, P < 0.001), CHECK2 (–0.56 ± 0.01, P < 0.001) and BRCA1 (−0.37 ± 0.009, P < 0.001); the DNA DSB binding proteins BRCA2 (−0.52 ± 0.008, P < 0.001) and RAD51 (−0.26 ± 0.02, P < 0.001); RAD17 that is involved in cell cycle control (−0.88 ± 0.01, P < 0.001); MSH2 (−0.42 ± 0.02, P < 0.001) and EXO1 (−0.26 ± 0.01, P < 0.001), both of which are involved in mismatch repair and EXO1 is required for DSB end resection [32] (Fig. 2). This DNA repair impairment in human UFs may result in a lack of cell cycle arrest and increased growth of UFs. Our current and previous results show that UFs express lower level of VDR than adjacent normal myometrium [25] and that vitamin D deficiency is associated with UTSM proliferation induction [22]. Next, we determined whether vitamin D deficiency could contribute DNA damage and DNA repair defects.
Fig. 2.
UF cells (HuLM) showed decreased expression of DNA repair-related genes compared with normal UTSM. The mRNA expression of the DNA DSB sensors (RAD50, NBS1, MRE11), mediators and effectors (CHECK1, CHECK2, BRCA1), DSB binding (BRCA2, RAD51), cell cycle check point control (RAD17) and mismatch repair (MSH2, EXO1) were measured in HuLM cells and UTSM cells by qRT-PCR analysis. The mRNA levels were normalized to 18 S rRNA and normalized values were used to generate the graph. Data are presented as the mean ± SEM of triplicate measurement. *P < 0.05, ***P < 0.001. All experiments were repeated twice
Silencing of the vitamin D receptor gene induces DNA damage accumulation and DNA damage response defects in human uterine smooth muscle cells
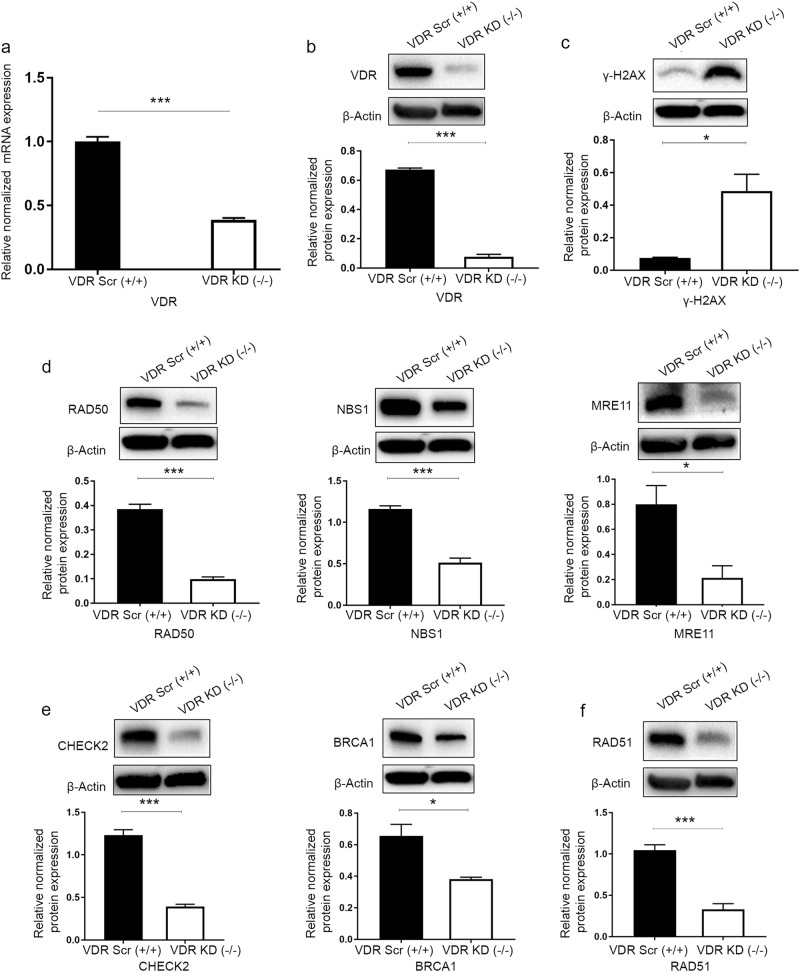
To determine the role of vitamin D signaling in the DNA damage repair system, VDR was depleted in normal myometrial (UTSM) cells using lentiviral-based RNA interference. Cultured UTSM cells were infected with lentiviruses expressing either VDR-specific shRNA or a nonfunctional scramble control. Infected cells were selected with puromycin to generate stable cell populations. VDR KD was confirmed for both RNA and protein expression levels using qRT-PCR and Western blot, respectively. VDR KD cells expressed markedly lower levels of VDR at both RNA and protein levels (P < 0.001) (Fig. 3a, b).
Fig. 3.
Vitamin D receptor (VDR) knockdown induced DNA damage and impaired the DNA damage response in uterine smooth muscle cells (UTSMs). VDR was targeted for knockdown in UTSM cells by infection with lentiviruses expressing VDR-specific shRNA (VDR KD) or scrambled control (VDR Scr). a Real-time PCR analysis was performed to determine the mRNA level of the VDR gene. The mRNA levels were normalized to 18 S RNA, and normalized values were used to generate the graph. Data are presented as the mean ± SEM of triplicate measurements. Cell lysates were analyzed by Western blot analysis using antibodies against (b) VDR, (c) the DNA damage marker γ-H2AX, DNA double-strand break (DSB) repair-related markers, including homologous recombination DNA DSB; (d) sensors (RAD50, NBS1, MRE11); (e) mediators and effectors (CHECK2, BRCA1); and (f) DSB binding (RAD51). The intensity of each protein signal was quantified and normalized to the corresponding β-actin and presented in the graph as the mean ± SD. *P < 0.05, ***P < 0. 001. All experiments were repeated twice
To determine whether VDR KD increased DNA damage in UTSM cells, the level of γ-H2AX protein expression was measured by Western blot analysis, and increased expression levels of γ-H2AX were observed (6.4-fold) in VDR KD UTSM cells in comparison to scrambled control cells (P < 0.05), which is indicative of DNA damage accumulation (Fig. 3c). Next, we determined the three key DNA DSB proteins belonging to the DNA repair sensors. VDR KD in UTSM cells resulted in decreased protein expression as follows: (fold change, P-value) DSB sensors: RAD50 (−3.90, P < 0.001), NBS1 (−2.26, P < 0.001) and MRE11 (−3.75, P < 0.05) (Fig. 3d). Furthermore, the expression levels of downstream signal repair proteins (repair mediators and effectors) were also measured, and three key mediators/effectors, including CHECK2 (−3.14, P < 0.001), BRCA1 (−1.72, P < 0.05), and RAD51 (−3.18, P < 0.001), were significantly downregulated in VDR KD UTSM cells compared with the scrambled control cells (Fig. 3e, f).
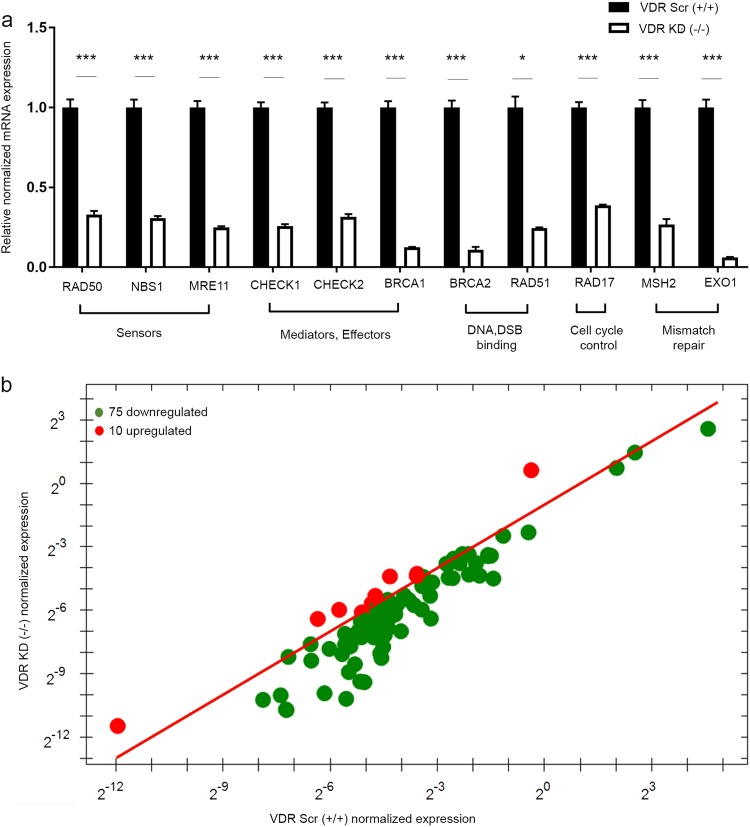
Decreased DNA repair-related protein levels due to depletion of VDR in UTSM occurred concomitantly with a down-regulation of their corresponding transcriptional activities. Using qRT-PCR, a significant decrease in the mRNA expression of several DNA DSB repair-related genes was detected in VDR KD cells compared with the scrambled control, including the following (mean fold change ± SEM, P-value): the DSB sensor MRN complex RAD50 (−0.33 ± 0.02, P < 0.001), NBS1 (−0.3 ± 0.01, P < 0.001), and MRE11 (−0.25 ± 0.006, P < 0.001); the DNA repair mediators and effectors CHECK1 (−0.25 ± 0.01, P < 0.001), CHECK2 (−0.31 ± 0.01, P < 0.001) and BRCA1 (−0.12 ± 0.001, P < 0.001); the DNA DSB binding proteins BRCA2 (−0.10 ± 0.01, P < 0.001) and RAD51 (−0.24 ± 0.005, P < 0.05); RAD17 that is involved in cell cycle control (−0.38 ± 0.004, P < 0.001); and EXO1 (−0.06 ± 0.002, P < 0.001) and MSH2 (−0.26 ± 0.03, P < 0.001) (Fig. 4a).
Fig. 4.
VDR knockdown decreased DNA repair-related gene expression in uterine smooth muscle cells (UTSMs). a Real-time PCR analysis of the mRNA level of DNA DSB sensors (RAD50, NBS1, MRE11), mediators and effectors (CHECK1, CHECK2, BRCA1), DSB binding (BRCA2, RAD51), cell cycle checkpoint control (RAD17) and mismatch repair (MSH2, EXO1) were measured in VDR-specific shRNA (VDR KD) cells compared with the scrambled control (VDR Scr). The mRNA levels were normalized to 18 S RNA and normalized values used to generate the graph. Data are presented as the mean ± SEM of triplicate measurement. *P < 0.05, ***P < 0.001. All experiments were repeated twice. b The Prime PCR DNA damage gene expression array was used to identify additional genes which show differential expression between VDR-specific shRNA (VDR KD) cells and scrambled control cells (VDR Scr). VDR knockdown resulted in the downregulation of 75 DNA repair-related genes, including 11 genes in previous qRT-PCR, while 10 were upregulated
To expand our findings concerning the DDR defect in response to VDR KD and to characterize additional DNA repair-related genes that contribute to the defect in the DNA repair pathway, a gene signature profile of DNA damage was determined using the prime PCR array (Bio-Rad). We identified a total of 75 downregulated versus 10 upregulated genes that were differentially expressed in VDR KD cells compared with VDR scrambled cells (Fig. 4b) (Supplemental Table 1). Our prime PCR array data were consistent with previous qRT-PCR data regarding the differential expression of all measured genes. Moreover, additional key DNA repair genes were identified, which showed reduced expression in response to VDR KD. These downregulated genes in VDR KD cells are involved in nucleotide excision repair and base excision repair (Supplemental Table 1). These data are consistent with previous findings regarding the contribution of vitamin D deficiency in skin cancer pathogenesis via disruption of those DNA repair pathways [33].
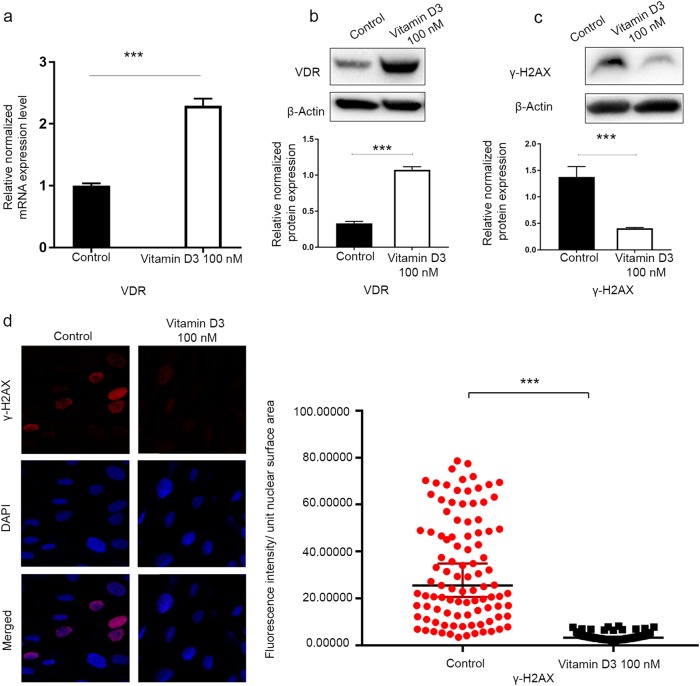
Vitamin D3 treatment suppresses DNA damage and restores the DNA damage response via induction of VDR in human uterine fibroid cells
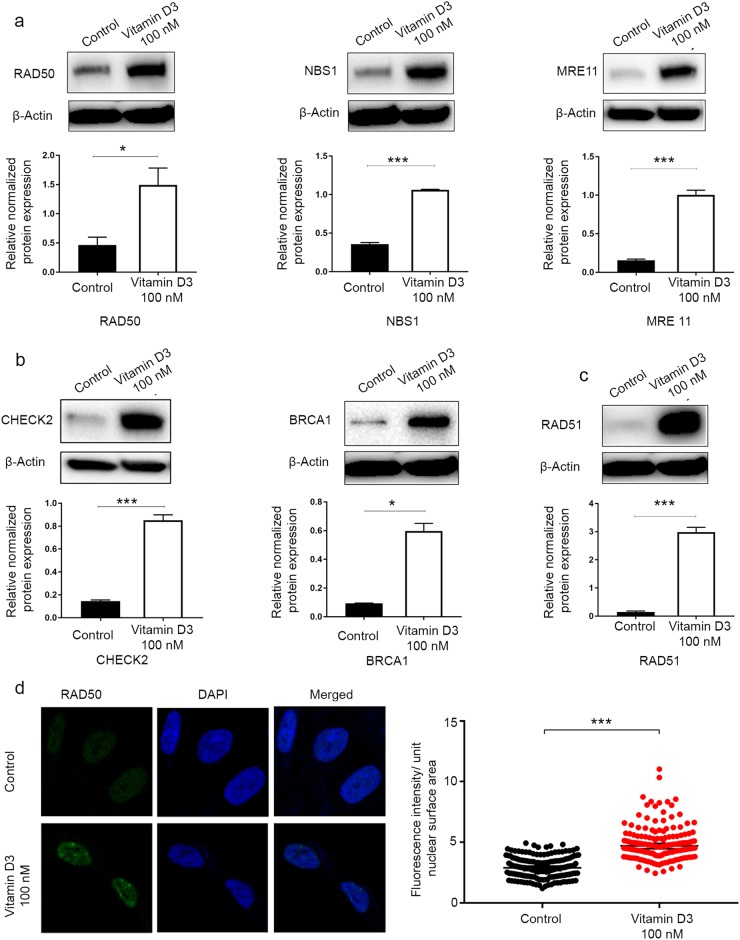
Our previous studies have shown that vitamin D exhibits a potent anti-UF effect both in vitro and in vivo [4]. To determine whether vitamin D3 suppressed the UF phenotype via reparation of the impaired DDR, we treated HuLM cells with 100 nM vitamin D3 and vehicle (ethanol) for 3 d to determine the effect of vitamin D3 on the restoration of DNA repair defects. First, our data showed significant VDR induction in response to vitamin D3 treatment at both transcript and protein levels by qRT-PCR and Western blot analyses, respectively (P < 0.001) (Fig. 5a, b), which was consistent with our previous finding [25]. To investigate the functional relationship between the vitamin D/VDR axis and DNA damage/repair, we investigated the effect of VDR activation in UF-associated DNA damage. Vitamin D3 treatment showed a significant reduction in the protein expression of γ-H2AX in HuLM cells compared with the untreated control by Western blot analysis (P < 0.05) (Fig. 5c), suggesting that vitamin D3 treatment is capable of reducing DNA damage. This finding was further confirmed in vitamin D-treated HuLM cells and vehicle control cells by measuring the fluorescence intensity of γ-H2AX foci. Vitamin D3 treatment significantly reduced DNA damage γ-H2AX fluorescence intensity (P < 0.001) (Fig. 5d) in HuLM cells. These data support the notion that VDR KD contributes to DNA damage accumulation. We further examined whether vitamin D could affect the expression level of DNA DSB repair markers shown to be downregulated in UFs, especially those involved in homologous recombination DDR. Interestingly, vitamin D3 treatment caused a significant upregulation of key DNA repair proteins using Western blot analysis, including the following (fold change, P-value): the DSBs sensors RAD50 ( + 3.23, P < 0.05), NBS1 ( + 2.98, P < 0.001), and MRE11 ( + 6.51, P < 0.001); the repair mediators and effectors CHECK2 ( + 5.85, P < 0.001), BRCA1 ( + 6.37, P < 0.05), and RAD51 (+ 19.9, P < 0.001) (Fig. 6a–c). Furthermore, we confirmed the induction of RAD50 expression in response to vitamin D3 treatment using immunofluorescence staining/confocal laser microscopy. Vitamin D3 treatment significantly increased the fluorescence intensity of RAD50 foci compared with the untreated vehicle control (P < 0.001) (Fig. 6d).
Fig. 5.
The 1, 25 dihyroxyvitamin D3 (vitamin D3) induced VDR expression and a decreased DNA damage load in HuLM cells. HuLM cells were treated with 100 nM of vitamin D3 for 3 d. Ethanol vehicle was added to the control cells. a Real-time PCR analysis of the mRNA level of the VDR gene was performed. The mRNA levels were normalized to 18 S rRNA, and normalized values were used to generate the graph. Data are presented as the mean ± SEM of triplicate measurements. Cell lysates were analyzed by Western blot analysis using (b) anti-VDR antibody and (c) anti-γ-H2AX. The intensity of each protein signal was quantified and normalized to the corresponding β-actin and presented in the graph as the mean ± SD. d HuLM cells (8 × 104) were seeded on sterile glass coverslips in 6-well plates and treated with 100 nM vitamin D3 for 3 d. Ethanol vehicle was added to the control cells. γ-H2AX foci reflecting DNA DSB were assessed using immunofluorescence staining and confocal laser microscopy 40x imaging. Individual data points were the γ-H2AX focus corrected fluorescence intensity per nuclear area, as measured by ImageJ, and lines represent the median ± 95% CI. ***P < 0.001. All experiments were repeated twice
Fig. 6.
The 1, 25 dihyroxyvitamin D3 (vitamin D3) induced DNA repair capacity in HuLM cells. HuLM cells were treated with 100 nM of vitamin D3 for 3 d. Ethanol vehicle was added to the control cells. Cell lysates were analyzed by Western blot analysis to measure the protein expression of DNA double-strand breaks (DSB) repair-related markers, including homologous recombination DNA DSB (a) sensors (RAD50, NBS1, MRE11), (b) mediators and effectors (CHECK2, BRCA1), and (c) DSB binding (RAD51). The intensity of each protein signal was quantified and normalized to the corresponding β-actin and presented in the graph as the mean ± SD. d HuLM cells (8 × 104) were seeded over sterile glass coverslips in six-well plates and treated with 100 nM vitamin D3 for 3 d. Ethanol vehicle was added to the control cells. RAD50 foci were assessed using immunofluorescence staining and confocal laser microscopy 40x imaging. Individual data points were the RAD50 focus-corrected fluorescence intensity per nuclear area, as measured by ImageJ, and lines represent medians ± 95% CI. *P < 0.05, ***P < 0.001. All experiments were repeated twice
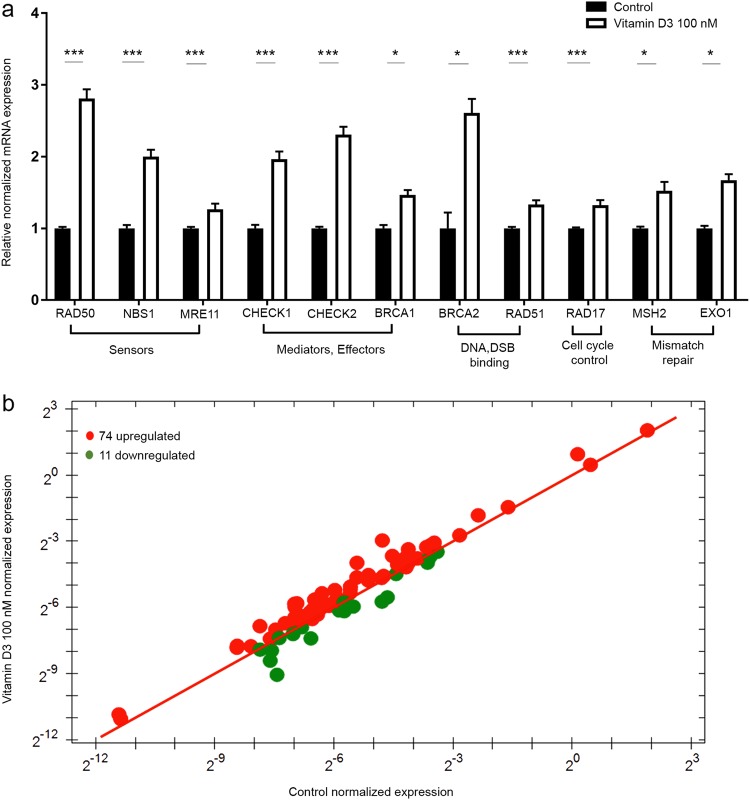
Additionally, vitamin D3 treatment was able to increase the transcriptional levels of the DNA repair-related genes, which have previously been shown to be downregulated in both UFs and VDR-depleted myometrial cells, including the following (mean fold change ± SEM, P-value): the DSB sensor MRN complex RAD50 (2.81 ± 0.12, P < 0.001), NBS1 (2 ± 0.09, P < 0.001), and MRE11 (1.3 ± 0.07, P < 0.001); the DNA repair mediators and effectors CHECK1 (1.97 ± 0.1, P < 0.001), CHECK2 (2.3 ± 0.1, P < 0.001) and BRCA1 (1.5 ± 0.06, P < 0.05); the DNA DSB binding proteins BRCA2 (2.6 ± 0.19, P < 0.05) and RAD51 (1.4 ± 0.06, P < 0.001); RAD17, which is involved in cell cycle control (1.32 ± 0.07, P < 0.001); and MSH2 (1.52 ± 0.12, P < 0.05) and EXO1 (1.7 ± 0.08, P < 0.05). (Fig. 7a).
Fig. 7.
The 1,25 dihyroxyvitamin D3 (vitamin D3) increased DNA repair-related gene expression in HuLM cells. a HuLM cells were treated with 100 nM of vitamin D3 for 3 d. Ethanol was used as vehicle control. Real-time PCR analysis of mRNA levels of DNA double-strand break (DSB) repair-related genes, including homologous recombination DNA DSB sensors (RAD50, NBS1, MRE11), mediators and effectors (CHECK1, CHECK2, BRCA1), DSB binding (BRCA2, RAD51), cell cycle check point control (RAD17) and mismatch repair (MSH2, EXO1) were measured. The mRNA levels were normalized to 18 S rRNA, and normalized values were used to generate the graph. Data are presented as the mean ± SEM of triplicate measurements. *P < 0.05, ***P < 0.001. The experiment was repeated twice. b The prime PCR DNA damage gene expression array was used to identify additional DNA repair genes which show differential expression between vitamin D3 and vehicle-treated cells
To expand these findings that vitamin D3 can restore defective DDR via the induction of DNA repair-related genes and characterize additional DNA repair genes in response to vitamin D3 treatment, we assessed the same gene signature profile of DNA damage by prime PCR array (Bio-Rad). Interestingly, 74 genes were upregulated in response to vitamin D3 treatment, while 11 genes were downregulated in comparison with untreated cells (Fig. 7b) (Supplemental Table 1). Our findings using the DNA damage PCR array confirmed the increased expression of all DNA DSB repair genes in response to vitamin D3 treatment based on the qRT-PCR data. Notably, the expression of 67 downregulated DNA repair genes listed in Prime PCR array after VDR KD were restored in response to vitamin D3 treatment (Supplemental Table 1).
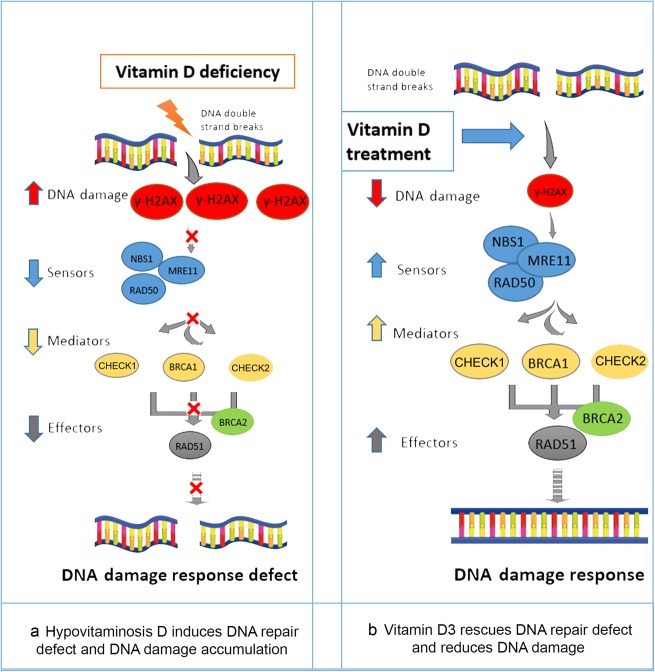
This result might explain the beneficial anti-UF effect of vitamin D3, which we have shown previously both in vitro and in vivo in an animal model [27, 28], through the amelioration of pathogenic DNA damage and restoration of the expression of key DDR genes that are downregulated in VDR-deficient UF cells (Fig. 8).
Fig. 8.
Model of the functional relationship between the vitamin D/VDR axis and DNA damage response. a Hypovitaminosis D induces DNA damage accumulation, as demonstrated by the accumulation of greater amounts of the DNA damage marker γ-H2AX and, consequently, DNA repair defects due to failed recruitment of DNA double-strand break sensors such as RAD50, MRE11 and NBS1, as well as effectors and mediators including CHECK1, CHECK2, BRCA1, BRCA2 and RAD51. This phenomenon results in DNA damage response defects. b Vitamin D3 treatment rescues DNA repair defects and reduces the DNA damage load by reducing the DNA damage marker γ-H2AX and recruiting key DNA repair proteins, including DNA double-strand break sensors such as RAD50, MRE11 and NBS1, as well as effectors and mediators including CHECK1, CHECK2, BRCA1, BRCA2 and RAD51, thus restoring the DNA damage response
Discussion
Uterine fibroids are benign monoclonal neoplasms of the myometrium and represent the most common tumors in women worldwide. UF pathogenesis is not yet clear; however, some genetic drivers were identified and thought to be dominantly responsible for myometrial cell transformation to the tumor. MED12 mutations are by far the most prevalent ones, accounting for almost 70%–80% of UFs. It is notable that MED12-mutant tumors are characterized by significant chromosomal loss and rearrangement, suggesting genomic instability as a driving force in tumor progression. These genomic instabilities and mutations are associated with a defect in DDR. Several studies have shown that tumor or cancer cells have a decreased capacity for DNA repair, which leads to genomic instability and subsequent mutations, bypassing the fate of senescence towards cell division and proliferation [34–36]. The phosphorylated histone H2AX (γ-H2AX) indicates DNA damage and serves as an early event to initiate DDR. Subsequently, the recruitment of DNA repair machineries, including sensors (MRN complex), mediators and effectors, such as γ-H2AX foci, co-localize with most of the DSB-associated DDR factors and are thought to serve as docking sites for recruiting and retaining DNA repair and signaling factors for DSBs. We have previously shown that UFs from AAs show downregulated expression of the DNA repair-related genes RAD51 and BRCA1 compared with the adjacent myometrium [16]. Both genes are essential in the repair of catastrophic DSBs via homologous recombination [37]. In the current study, we performed a comprehensive study to determine defects in the DNA repair system in UFs. We found that UFs accumulate DNA DSBs compared with the matched adjacent myometrium, which was characterized by significant upregulation of H2AX phosphorylated on the 139th serine residue and termed γ-H2AX [38, 39]. γ-H2AX has been investigated in a variety of hormone-dependent cancer types, including breast, cervix, and ovary cancers, highlighting its prognostic value [40]. Moreover, we found that UF cells and tissues exhibited a downregulation of a vast network of DNA repair genes, serving sensor, mediator and effector functions in the DNA repair pathway. For instance, the three markers MRE11, RAD50 and NBS1, which comprise the DSB sensor MRN complex necessary for homologous recombination DDR initiation [41, 42], showed downregulated expression in UF cells in comparison to myometrium. Additionally, homologous recombination mediators and effectors as well as cell cycle checkpoint markers, such as CHECK1, CHECK2, RAD17 and BRCA1 and markers important for DSB binding such as RAD51 and BRCA2, were also diminished. Notably, BRCA2 regulates RAD51 function in DNA repair by recruiting it to sites of DNA breaks [43]. Collectively, these data reveal for the first time that UFs exhibit an accumulation of DSBs along with DDR defects compared with the adjacent myometrium. DDR is crucial for halting proliferation through activation of cell cycle checkpoints and apoptosis induction [44]. Thus, the UF-associated DDR defect could be one of the reasons for the continuous proliferation, impaired apoptosis and further tumorigenesis observed in UFs [45, 46].
Our group has previously shown that VDR depletion in UTSM cells is associated with increased expression of proliferation and fibrosis markers [22]. Additionally, the serum vitamin D3 level correlates inversely with the UF volume in different ethnic groups of patients [47]. African American (AA) women, who have a 10 times higher risk of vitamin D deficiency relative to Caucasian women, have a 3–4 times higher incidence rate of UFs [1, 4]. However, the UF phenotype has never been identified to be linked to DNA repair dysfunction via vitamin D deficiency [4]. In this study, we showed for the first time that the vitamin D3/VDR axis is linked to the DNA repair pathway. The DNA damage in UFs is tightly associated with decreased expression of VDR. The loss of function study demonstrates that VDR depletion is associated with DNA damage accumulation and DNA repair defects in human myometrial cells. In contrast, vitamin D3 treatment is able to reverse the UF-associated DNA damage via the induction of several key DNA repair genes. Our findings for steroid hormone-dependent UFs are consistent with studies in hormone-dependent cancers such as breast and prostate, which have shown a significant impact of vitamin D3 on carcinogenesis [18, 48, 49]. The anti-cancer effect of vitamin D3 treatment is shown through the regulation of the DNA damage process and induction of several DNA repair-related genes in breast and prostate cancers [19–21, 50].
Another important finding of our study is that defects in the vitamin D3/VDR axis alter many DNA repair members, including the DSB sensor MRN complex, mediators and effectors. In this regard, it is quite challenging to use single DNA repair gene therapy to rescue the DNA repair system with patients who have a vitamin D3/VDR defect. In fact, our data show that the expression levels of many DNA repair genes are downregulated in UFs. Interestingly, most UF-associated DNA repair genes exhibit increased expression concomitant with decreased DNA damage in response to vitamin D3 treatment, suggesting that vitamin D3 treatment indeed rescues the DNA repair system by increasing the expression levels of DNA repair genes in UF. Vitamin D3 has been shown to exert its biological function by interacting with and subsequently inducing/activating VDR [51]. Previous studies by our group and others have shown that lower serum vitamin D3 levels and downregulated VDR expression are observed in UFs [23, 25, 47]. In addition, we previously used the Eker rat model of UFs via the administration of vitamin D3, and we observed both increased vitamin D serum levels and VDR expression in UF tumors, suggesting a clear link between vitamin D3 and VDR. In this study, VDR expression in HuLM cells was elevated in response to vitamin D3 treatment, which is consistent with our previous findings [23, 25]. However, the detailed mechanism regarding vitamin D3 induction of VDR upregulation has not been fully identified [52]. A loss-of-function study demonstrated that VDR KD in human fibroblasts results in downregulation of the expression of the homologous recombination-related repair genes RAD51 and BRCA1 and upregulation of γ-H2AX [19]. Another study using a vitamin D-deficient diet in a rat model showed an increase in DNA damage [53]. Our results demonstrate a novel regulation of key DDR genes and DNA repair machinery via the VDR axis in human myometrial cells. VDR depletion with subsequent increases in DNA damage and defects in the consequent response might explain the increased cell proliferation and proliferation marker expression in our and other previous publications [22, 54]. This phenomenon reflects the failure of cell cycle arrest and subsequent apoptosis, which was reversed in response to vitamin D3 treatment in favor of apoptosis induction both in vitro and in an in vivo animal model [27, 28]. Our studies highlight an important role of the vitamin D3/VDR axis in the development of UFs via the DNA repair pathway [33, 55, 56].
Previous studies have shown a positive feedback signaling loop between DNA repair genes and VDR, which explained the cancer chemoprevention effect of vitamin D [20]. A similar finding was also observed in breast cancer cells [19]. However, the mechanism underlying increasing expression levels of DNA repair genes via VitD3/VDR is still largely unknown and necessitates further investigation.
In conclusion, our studies demonstrate that UF cells accumulate unrepaired DSB concomitant with decreased expression of a vast network of DNA repair proteins. Importantly, our gain- and loss-of-function analyses demonstrate for the first time that the vitamin D3/VDR axis is functionally linked to DNA damage and instability status in UF cells. Finally, our data suggest that vitamin D3 treatment could be useful to inhibit UF formation through the amelioration of pathogenic DNA damage.
Electronic supplementary material
Acknowledgements
We would like to thank Dr. Lauren Prusinski and Ms. Archana Laknaur for their help in collecting human tissues and primary cell isolation. This study was supported in part by the National Institutes of Health grants: R01 HD094378-01 (AA) and R01 ES 028615-01(AA) as well as the Augusta University Intramural Grants Program (QY) and Egyptian governmental support (NS).
Author contributions
AA & QY conceived the studies, MA and QY designed the experiments, MA performed all the experiments, AA provided the study materials, MA, NS, SS and QY analyzed the data, MA wrote the paper, QY revised the manuscript, and AA, NS and QY provided the financial support. All authors approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
The online version of this article (10.1038/s41401-018-0184-6) contains supplementary material, which is available to authorized users.
References
- 1.Stewart EA, Laughlin-Tommaso SK, Catherino WH, Lalitkumar S, Gupta D, Vollenhoven B. Uterine fibroids. Nat Rev Dis Prim. 2016;2:16043. doi: 10.1038/nrdp.2016.43. [DOI] [PubMed] [Google Scholar]
- 2.Ali M, Al-Hendy A. Selective progesterone receptor modulators for fertility preservation in women with symptomatic uterine fibroids. Biol Reprod. 2017;97:337–52. doi: 10.1093/biolre/iox094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cardozo ER, Clark AD, Banks NK, Henne MB, Stegmann BJ, Segars JH. The estimated annual cost of uterine leiomyomata in the United States. Am J Obstet Gynecol. 2012;206:e1–9. doi: 10.1016/j.ajog.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brakta S, Diamond JS, Al-Hendy A, Diamond MP, Halder SK. Role of vitamin D in uterine fibroid biology. Fertil Steril. 2015;104:698–706. doi: 10.1016/j.fertnstert.2015.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halder SK, Laknaur A, Miller J, Layman LC, Diamond M, Al-Hendy A. Novel MED12 gene somatic mutations in women from the Southern United States with symptomatic uterine fibroids. Mol Genet Genom. 2015;290:505–11. doi: 10.1007/s00438-014-0938-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al-Hendy A, Laknaur A, Diamond MP, Ismail N, Boyer TG, Halder SK. Silencing Med12 gene reduces proliferation of human leiomyoma cells mediated via Wnt/beta-catenin signaling pathway. Endocrinology. 2017;158:592–603. doi: 10.1210/en.2016-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makinen N, Heinonen HR, Moore S, Tomlinson IP, van der Spuy ZM, Aaltonen LA. MED12 exon 2 mutations are common in uterine leiomyomas from South African patients. Oncotarget. 2011;2:966–9. doi: 10.18632/oncotarget.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mehine M, Kaasinen E, Makinen N, Katainen R, Kampjarvi K, Pitkanen E, et al. Characterization of uterine leiomyomas by whole-genome sequencing. N Engl J Med. 2013;369:43–53. doi: 10.1056/NEJMoa1302736. [DOI] [PubMed] [Google Scholar]
- 9.McGuire MM, Yatsenko A, Hoffner L, Jones M, Surti U, Rajkovic A. Whole exome sequencing in a random sample of North American women with leiomyomas identifies MED12 mutations in majority of uterine leiomyomas. PLoS ONE. 2012;7:e33251. doi: 10.1371/journal.pone.0033251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morton CC. Genetic approaches to the study of uterine leiomyomata. Environ Health Perspect. 2000;108(Suppl 5):775–8. doi: 10.1289/ehp.00108s5775. [DOI] [PubMed] [Google Scholar]
- 11.Mittal P, Shin YH, Yatsenko SA, Castro CA, Surti U, Rajkovic A. Med12 gain-of-function mutation causes leiomyomas and genomic instability. J Clin Invest. 2015;125:3280–4. doi: 10.1172/JCI81534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, et al. GammaH2AX and cancer. Nat Rev Cancer. 2008;8:957–67. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dietlein F, Thelen L, Reinhardt HC. Cancer-specific defects in DNA repair pathways as targets for personalized therapeutic approaches. Trends Genet. 2014;30:326–39. doi: 10.1016/j.tig.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 14.Venkitaraman AR. Tumour suppressor mechanisms in the control of chromosome stability: insights from BRCA2. Mol Cells. 2014;37:95–9. doi: 10.14348/molcells.2014.2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cassidy LD, Liau SS, Venkitaraman AR. Chromosome instability and carcinogenesis: insights from murine models of human pancreatic cancer associated with BRCA2 inactivation. Mol Oncol. 2014;8:161–8. doi: 10.1016/j.molonc.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Q, Nair S, Laknaur A, Ismail N, Diamond MP, Al-Hendy A. The polycomb group protein EZH2 impairs DNA damage repair gene expression in human uterine fibroids. Biol Reprod. 2016;94:69. doi: 10.1095/biolreprod.115.134924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Q, Laknaur A, Elam L, Ismail N, Gavrilova-Jordan L, Lue J, et al. Identification of polycomb group protein EZH2-mediated DNA mismatch repair gene MSH2 in human uterine fibroids. Reprod Sci. 2016;23:1314–25. doi: 10.1177/1933719116638186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu X, Zhou T, Cao N, Ni J, Wang X. Role of vitamin D metabolism and activity on carcinogenesis. Oncol Res. 2014;22:129–37. doi: 10.3727/096504015X14267282610894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graziano S, Johnston R, Deng O, Zhang J, Gonzalo S. Vitamin D/vitamin D receptor axis regulates DNA repair during oncogene-induced senescence. Oncogene. 2016;35:5362–76. doi: 10.1038/onc.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ting HJ, Yasmin-Karim S, Yan SJ, Hsu JW, Lin TH, Zeng W, et al. A positive feedback signaling loop between ATM and the vitamin D receptor is critical for cancer chemoprevention by vitamin D. Cancer Res. 2012;72:958–68. doi: 10.1158/0008-5472.CAN-11-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Halicka HD, Zhao H, Li J, Traganos F, Studzinski GP, Darzynkiewicz Z. Attenuation of constitutive DNA damage signaling by 1,25-dihydroxyvitamin D3. Aging. 2012;4:270–8. doi: 10.18632/aging.100450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Hendy A, Diamond MP, Boyer TG, Halder SK. Vitamin D3 inhibits Wnt/beta-catenin and mTOR signaling pathways in human uterine fibroid cells. J Clin Endocrinol Metab. 2016;101:1542–51. doi: 10.1210/jc.2015-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Hendy A, Diamond MP, El-Sohemy A, Halder SK. 1,25-dihydroxyvitamin D3 regulates expression of sex steroid receptors in human uterine fibroid cells. J Clin Endocrinol Metab. 2015;100:E572–82. doi: 10.1210/jc.2014-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halder SK, Goodwin JS, Al-Hendy A. 1,25-Dihydroxyvitamin D3 reduces TGF-beta3-induced fibrosis-related gene expression in human uterine leiomyoma cells. J Clin Endocrinol Metab. 2011;96:E754–62. doi: 10.1210/jc.2010-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halder SK, Osteen KG, Al-Hendy A. 1,25-dihydroxyvitamin d3 reduces extracellular matrix-associated protein expression in human uterine fibroid cells. Biol Reprod. 2013;89:150. doi: 10.1095/biolreprod.113.107714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Halder SK, Osteen KG, Al-Hendy A. Vitamin D3 inhibits expression and activities of matrix metalloproteinase-2 and -9 in human uterine fibroid cells. Hum Reprod. 2013;28:2407–16. doi: 10.1093/humrep/det265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halder SK, Sharan C, Al-Hendy A. 1,25-dihydroxyvitamin D3 treatment shrinks uterine leiomyoma tumors in the Eker rat model. Biol Reprod. 2012;86:116. doi: 10.1095/biolreprod.111.098145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharan C, Halder SK, Thota C, Jaleel T, Nair S, Al-Hendy A. Vitamin D inhibits proliferation of human uterine leiomyoma cells via catechol-O-methyltransferase. Fertil Steril. 2011;95:247–53. doi: 10.1016/j.fertnstert.2010.07.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guirouilh-Barbat J, Lambert S, Bertrand P, Lopez BS. Is homologous recombination really an error-free process? Front Genet. 2014;5:175. doi: 10.3389/fgene.2014.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rass E, Grabarz A, Bertrand P, Lopez BS. [Double strand break repair, one mechanism can hide another: alternative non-homologous end joining] Cancer Radiother. 2012;16:1–10. doi: 10.1016/j.canrad.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 31.Cannan WJ, Pederson DS. Mechanisms and consequences of double-strand DNA break formation in chromatin. J Cell Physiol. 2016;231:3–14. doi: 10.1002/jcp.25048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Z, Pearlman AH, Hsieh P. DNA mismatch repair and the DNA damage response. DNA Repair (Amst) 2016;38:94–101. doi: 10.1016/j.dnarep.2015.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pawlowska E, Wysokinski D, Blasiak J. Nucleotide excision repair and vitamin D--relevance for skin cancer therapy. Int J Mol Sci. 2016;17:372. doi: 10.3390/ijms17040372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.d’Adda di Fagagna F. Living on a break: cellular senescence as a DNA-damage response. Nat Rev Cancer. 2008;8:512–22. doi: 10.1038/nrc2440. [DOI] [PubMed] [Google Scholar]
- 35.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 36.Blow JJ, Gillespie PJ. Replication licensing and cancer--a fatal entanglement? Nat Rev Cancer. 2008;8:799–806. doi: 10.1038/nrc2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chappell WH, Gautam D, Ok ST, Johnson BA, Anacker DC, Moody CA. Homologous recombination repair factors Rad51 and BRCA1 are necessary for productive replication of human papillomavirus 31. J Virol. 2015;90:2639–52. doi: 10.1128/JVI.02495-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valdiglesias V, Giunta S, Fenech M, Neri M, Bonassi S. GammaH2AX as a marker of DNA double strand breaks and genomic instability in human population studies. Mutat Res. 2013;753:24–40. doi: 10.1016/j.mrrev.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 39.Kuo LJ, Yang LX. Gamma-H2AX - a novel biomarker for DNA double-strand breaks. Vivo. 2008;22:305–9. [PubMed] [Google Scholar]
- 40.Palla VV, Karaolanis G, Katafigiotis I, Anastasiou I, Patapis P, Dimitroulis D, et al. Gamma-H2AX: can it be established as a classical cancer prognostic factor? Tumour Biol. 2017;39:1010428317695931. doi: 10.1177/1010428317695931. [DOI] [PubMed] [Google Scholar]
- 41.Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nat Rev Mol Cell Biol. 2011;12:90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams GJ, Lees-Miller SP, Tainer JA. Mre11-Rad50-Nbs1 conformations and the control of sensing, signaling, and effector responses at DNA double-strand breaks. DNA Repair (Amst) 2010;9:1299–306. doi: 10.1016/j.dnarep.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carreira A, Kowalczykowski SC. Two classes of BRC repeats in BRCA2 promote RAD51 nucleoprotein filament function by distinct mechanisms. Proc Natl Acad Sci USA. 2011;108:10448–53. doi: 10.1073/pnas.1106971108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caldon CE. Estrogen signaling and the DNA damage response in hormone dependent breast cancers. Front Oncol. 2014;4:106. doi: 10.3389/fonc.2014.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dixon D, Flake GP, Moore AB, He H, Haseman JK, Risinger JI, et al. Cell proliferation and apoptosis in human uterine leiomyomas and myometria. Virchows Arch. 2002;441:53–62. doi: 10.1007/s00428-001-0568-7. [DOI] [PubMed] [Google Scholar]
- 46.Wu X, Blanck A, Olovsson M, Henriksen R, Lindblom B. Expression of Bcl-2, Bcl-x, Mcl-1, Bax and Bak in human uterine leiomyomas and myometrium during the menstrual cycle and after menopause. J Steroid Biochem Mol Biol. 2002;80:77–83. doi: 10.1016/S0960-0760(01)00177-7. [DOI] [PubMed] [Google Scholar]
- 47.Sabry M, Halder SK, Allah AS, Roshdy E, Rajaratnam V, Al-Hendy A. Serum vitamin D3 level inversely correlates with uterine fibroid volume in different ethnic groups: a cross-sectional observational study. Int J Women’s Health. 2013;5:93–100. doi: 10.2147/IJWH.S38800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J, Dong Y, Lu C, Wang Y, Peng L, Jiang M, et al. Meta-analysis of the correlation between vitamin D and lung cancer risk and outcomes. Oncotarget. 2017;8:81040–51. doi: 10.18632/oncotarget.18766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fleet JC, DeSmet M, Johnson R, Li Y. Vitamin D and cancer: a review of molecular mechanisms. Biochem J. 2012;441:61–76. doi: 10.1042/BJ20110744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gonzalo S. Novel roles of 1alpha,25(OH)2D3 on DNA repair provide new strategies for breast cancer treatment. J Steroid Biochem Mol Biol. 2014;144(Pt A):59–64. doi: 10.1016/j.jsbmb.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holick MF. Vitamin D: a millenium perspective. J Cell Biochem. 2003;88:296–307. doi: 10.1002/jcb.10338. [DOI] [PubMed] [Google Scholar]
- 52.Pike JW, Meyer MB. The vitamin D receptor: new paradigms for the regulation of gene expression by 1,25-dihydroxyvitamin D(3) Endocrinol Metab Clin North Am. 2010;39:255–69. doi: 10.1016/j.ecl.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Machado Cda S, Venancio VP, Aissa AF, Hernandes LC, de Mello MB, Del Lama JE, et al. Vitamin D3 deficiency increases DNA damage and the oxidative burst of neutrophils in a hypertensive rat model. Mutat Res Genet Toxicol Environ Mutagen. 2016;798-799:19–26. doi: 10.1016/j.mrgentox.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 54.Chatterjee M. Vitamin D and genomic stability. Mutat Res. 2001;475:69–87. doi: 10.1016/S0027-5107(01)00080-X. [DOI] [PubMed] [Google Scholar]
- 55.Reichrath J, Rass K. Ultraviolet damage, DNA repair and vitamin D in nonmelanoma skin cancer and in malignant melanoma: an update. Adv Exp Med Biol. 2014;810:208–33. doi: 10.1007/978-1-4939-0437-2_12. [DOI] [PubMed] [Google Scholar]
- 56.Bikle DD. The vitamin D receptor: a tumor suppressor in skin. Adv Exp Med Biol. 2014;810:282–302. doi: 10.21236/ada614241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.