Abstract
Lysosomes are the terminal organelles of the autophagic-endocytic pathway and play a key role in the degradation of autophagic contents. We previously reported that a natural compound oblongifolin C (OC) increased the number of autophagosomes and impaired the degradation of P62, most likely via suppression of lysosomal function and blockage of autophagosome-lysosome fusion. However, the precise mechanisms of how OC inhibits the lysosome-autophagy pathway remain unclear. In the present study, we investigated the effect of OC on transcription factor EB (TFEB), a master regulator of lysosomal biogenesis, lysosomal function and autophagy. We showed that treatment with OC (15 μM) markedly enhanced the nuclear translocation of TFEB in HeLa cells, concomitantly reduced the interaction of TFEB with 14-3-3 proteins. We further demonstrated that OC caused significant inhibition of mTORC1 along with TFEB nuclear translocation, and OC-mediated TFEB nuclear translocation was dependent on mTORC1 suppression. Intriguingly, this increased nuclear TFEB was accompanied by reduced TFEB luciferase activity, increased lysosomal pH and impaired cathepsin enzyme activities. In HeLa cells, treatment with OC (7.5 μM) resulted in about 30% of cell death, whereas treatment with hydroxycitrate, a caloric restriction mimetic (20 μM) did not affect the cell viability. However, cotreatment with OC and hydroxycitrate caused significantly great cytotoxicity (>50%). Taken together, these results demonstrate that inhibition of lysosome function is mediated by OC, despite evident TFEB nuclear translocation.
Keywords: oblongifolin C, autophagy, TFEB, mTORC1, lysosome
Introduction
Autophagy occurs in eukaryotic cells in a wide variety of physiological and pathological processes; it can be induced by normal developmental processes and by external stresses [1]. Autophagy has a dual role in tumor initiation and development. On the one hand, autophagy can protect normal cells from malignant transformation by removing toxic substances in cells and maintaining genome stability. On the other hand, autophagy can increase tumor cell viability, especially in chemotherapy and radiotherapy, during which autophagy protects tumor cells from the cytotoxicity of chemotherapy drugs. It is generally believed that at the early stage of cancer, autophagy inhibits the initiation of tumorigenesis, whereas during cancer development, autophagy protects tumor cells against environmental stress (such as nutrient starvation before angiogenesis, hypoxia and chemotherapy) and thereby promotes the survival of cancer cells [2].
Lysosomes are the terminal organelles of the endocytic pathway responsible for the degradation of autophagic materials. Over the past few decades, autophagy studies have focused on the molecular mechanisms of autophagosome formation. Recently, researchers have made significant progress in understanding the role of lysosomes in autophagy [3]. Transcription factor EB (TFEB), a master regulator of lysosomal biogenesis as it drives the expression of related genes, has received extensive attention. Some studies show that TFEB is regulated by the lysosome via the mTORC1 pathway [3, 4]. It has been reported that mTORC1 drives cellular growth by controlling numerous processes that regulate protein synthesis and degradation. Under normal conditions, mTORC1 phosphorylates TFEB at serines 221 and 142, thereby retaining TFEB on the lysosomal surface [5, 6]. During nutrient deprivation, TFEB is dephosphorylated and subsequently translocates to the nucleus to drive the expression of transcriptional target genes.
Oblongifolin C (OC), a natural compound found in Garcinia oblongifolia and later isolated from the pericarp of Garcinia yunnanensis, has been found to exhibit antitumor activity in many cancer cell lines [7, 8]. Our previous studies have shown that OC could induce caspase-dependent cancer cell death [9] and inhibit tumor metastasis by inducing expression of keratin 18 and tubulin in cancer cells [10]. OC not only triggered the DNA damage response but also inhibited DNA damage repair [11]. Moreover, a recent investigation has revealed that OC could inhibit autophagic flux, possibly by blocking autophagosome–lysosome fusion and attenuating lysosomal proteolytic activity [12]. However, it remains unclear whether this inhibition of autophagy is associated with its antitumor activity, and the detailed mechanisms of OC-mediated inhibition of autophagy are still unknown.
In this study, we investigated the influence of OC on lysosome activity. We observed that OC treatment not only inhibits the autophagosome–lysosome fusion but also suppresses lysosome biogenesis via regulation of TFEB translocation. Importantly, OC enhances the sensitivity of HeLa cells to hydroxycitrate, a caloric restriction mimetic (CRM) that can induce autophagy and improve anticancer chemotherapy. Our study thus reveals the mechanism by which OC regulates autophagy and the potential for OC combination with CRMs as a novel strategy for anticancer therapy.
Materials and methods
Reagents and antibodies
OC was isolated from fruit extracts of Garcinia yunnanensis Hu. Torin1, PP242 and Anti-FLAG® M2 Affinity Gel were purchased from Sigma (MO, USA). TFEB-luciferase and pCMV-3× Flag-TFEB plasmids were gifts from Dr. A Ballabio (Baylor College of Medicine).
The following antibodies were used in our experiments (used at a 1:1000 dilution for Western blot): TFEB (A303–673A) was purchased from Bethyl Laboratories (TX, USA); TSC2 (4308), phospho-S6 (S235/236) (2211), S6 (2217), phospho-P70S6K (9205), P70S6K (9202) and 14-3-3 (8312) were purchased from Cell Signaling Technology (MA, USA); β-actin (A5441), FLAG (F1804), STX17 (HPA001204) and LaminA + C (L1293) were purchased from Sigma (MO, USA); and GAPDH (ab128915, 1:10,000) was purchased from Abcam (Cambridge, UK).
Cell culture
HeLa cells and MEF cells were obtained from the American Type Culture Collection (ATCC). TSC2-wild-type (TSC2+/+) and TSC2-knockout (TSC2-/-) mouse fibroblasts (MEFs) were obtained from Dr. DJ Kwiatkowski (Brigham and Women’s Hospital, Harvard University). The cells were maintained in DMEM (HyClone, PA, USA) supplemented with 10% fetal bovine serum (HyClone, PA, USA) and 10 U/mL penicillin-streptomycin (Gibco/Invitrogen, NY, USA) in a humidified atmosphere containing 5% CO2 at 37 °C.
Immunoprecipitation and western blotting
For immunoprecipitation (IP) of TFEB protein, 293T cells were transiently transfected with pCMV-3× Flag-TFEB for 24 h, followed by the designated treatment. The cells were lysed using IP lysis buffer (10 mM Tris-HCl, pH 7.4, 100 mM NaCl, 2.5 mM MgCl2, 0.05% Triton-100, protease inhibitors) and sonicated four times. Whole cell lysates obtained by centrifugation were incubated with 10 μL Flag beads overnight at 4 °C with rotation. The protein/bead complexes were washed with IP lysis buffer three times and eluted by boiling with 2× sample-loading buffer. The samples were resolved by SDS-PAGE and transferred onto PVDF membranes. After blocking with 5% nonfat milk, the membrane was probed with the designated primary and secondary antibodies, developed with the enhanced chemiluminescence method and visualized with an ImageQuant LAS 4000 (GE Healthcare, PA, USA).
Immunofluorescence and confocal microscopy
For the immunofluorescence assay, cells were grown on coverslips, rinsed in PBS and fixed with 4% PFA for 30 min at room temperature. Cells were washed three times with PBS and then blocked and permeabilized in PBS containing 3% BSA (Sigma, MO, USA) and 0.1% Tween-20 for 30 min at room temperature. The blocking solution was rinsed off with PBS, and the corresponding antibodies diluted in 1% BSA in PBS were added. Fluorescence images were captured using laser scanning confocal microscopy (OLYMPUS, Japan).
Biochemical fractionation
The nuclear and cytosolic extracts were prepared with NE-PER® nuclear and cytoplasmic extraction reagents (Thermo Fisher, MA, USA) according to the manufacturer’s protocol. Briefly, cells were lysed in cytoplasmic extraction reagent I on ice for 10 min, and then, ice-cold cytoplasmic extraction reagent II was added. The tubes were then centrifuged, and the supernatant (cytoplasmic extract) was recovered. The cell pellet was suspended in ice-cold nuclear extraction reagent and centrifuged. The supernatant containing the nuclear extract was saved.
Quantitative real-time PCR (qRT-PCR)
RNA was extracted with RNeasy (Qiagen, Germany). A reverse transcription reaction was performed using 1 μg of total RNA with a High Capacity cDNA Reverse Transcription kit (Thermo Fisher, MA, USA). The mRNA expression levels were determined by a CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad, CA, USA). TATA-Box Binding Protein (TBP) was used as an internal control of RNA integrity, and the assay was performed in triplicate. The primers used were based on previous reports [4, 6]. The following primer sequences were used in our experiments:
ATP6V1H: 5′-CAGAAGTTCGTGCAAACAAAGC-3′, 5′-TCAGGGCTTCGTTTCATTTCAA-3′; LAMP1: 5′-ACGTTACAGCGTCCAGCTCAT-3′, 5′-TCTTTGGAGCTCGCATTGG-3′; CTSD: 5′-TGCTCAAGAACTACATGGACGC-3′, 5′-CGAAGACGACTGTGAAGCACT-3′; VAMP8: 5′-TGTGCGGAACCTGCAAAGT-3′, 5′-CTTCTGCGATGTCGTCTTGAA-3′; CTSB: 5′-ACAACGTGGACATGAGCTACT-3′, 5′-TCGGTAAACATAACTCTCTGGGG-3′; MCOLN1: 5′-TTGCTCTCTGCCAGCGGTACTA-3′, 5′-GCAGTCAGTAACCACCATCGGA-3′; TBP: 5′-CCACTCACAGACTCTCACAAC-3′, 5′-CTGCGGTACAATCCCAGAACT-3′; LC3: 5′-ACCATGCCGTCGGAGAAG-3′; 5′-ATCGTTCTATTATCACCGGGATTTT-3′; and P62: 5′-AGAGACAGCTCAGCAGCTCCT-3′, 5′-GCCTTGTCAGCCTCCATCAG-3′.
Transient transfection and TFEB luciferase assay
293T cells were transiently cotransfected with pCMV-3× Flag-TFEB and Renilla plasmids using Lipofectamine 2000 (Invitrogen, NY, USA) for 24 h, followed by the designated treatment. The luciferase activity was measured via a Dual-Luciferase Reporter Assay System (Promega, WI, USA) according to the manufacturer’s protocol.
Lysotracker staining
The lysosomal acidity was estimated by LysoTracker. Cells cultured in cover glass slide chambers were first treated as indicated and then incubated for 30 min with 50 nM LysoTracker® Red DND-99 (Invitrogen, NY, USA). Labeled cells were harvested and measured by flow cytometry (BD Biosciences, NY, USA).
Cathepsin activity assay
Enzymatic activity of lysosomal cathepsin B or cathepsin L was detected by Magic Red® reagent using a method described previously with modifications. Cells in 24-well plates with designated treatments were incubated with Magic Red® cathepsin B/L reagent for 15 min. The fluorescence intensities of 10,000 cells per sample were quantified using a BD FACS cytometer (BD Biosciences, NY, USA).
Statistics
Data were expressed as the mean ± SEM. Statistical analyses were performed using GraphPad Prism 5 (GraphPad Software, CA, USA). Western blot and image data were presented as at least three independent experiments. A P-value < 0.05 was considered to indicate significance.
Results
OC blocks autophagosome–lysosome fusion by engaging STX17
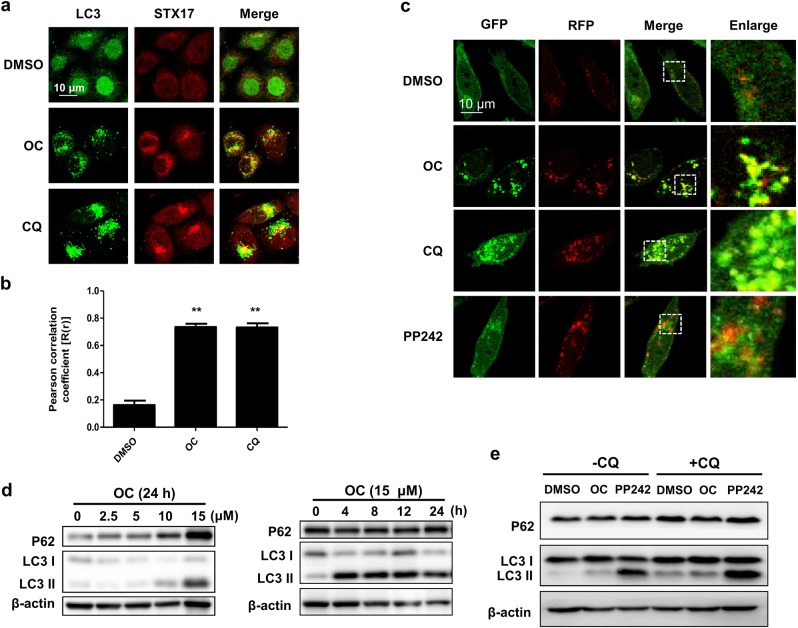
It has been shown that the SNARE protein syntaxin 17 (STX17) is localized at the outer membrane of the completed autophagosome and is required for autolysosome formation [13, 14]. In the course of autophagosome formation, LC3 undergoes lipidation, whereby LC3-I converts to LC3-II [15]. Lipidated LC3 is localized to both the inner and outer membrane of the mature autophagosome and has been used as an autophagosome marker [16]. Our previous data suggested that OC is an autophagy flux inhibitor [12], but whether OC is involved in autophagosome maturation by engaging STX17 is yet unknown. To address this question, we tested the colocalization of LC3 with STX17 in HeLa cells stably expressing GFP-LC3. Confocal images suggested that OC-treated cells exhibited markedly increased colocalization of LC3 with STX17, similar to the pattern observed in chloroquine (CQ)-treated cells (Fig. 1a, b), suggesting that OC promotes autophagosome maturation by engaging STX17. To test whether OC affects the late stage of autophagy when the autophagosome fuses with the lysosome, we used L929 cells stably transfected with mRFP-GFP-LC3 (tfLC3B-L929 cells). GFP signals are quenched in an acidic environment and thus are unstable in the lysosome; mRFP-GFP-LC3, therefore, can only exhibit red signals in the acidic lysosome. As shown in Fig. 1c, in response to PP242 starvation, both the number of GFP and RFP puncta increased, but only very few puncta colocalized with each other. In contrast, CQ could inhibit the acidification of lysosomes and dramatically enhance the colocalization of GFP and RFP puncta. Interestingly, OC treatment led to a marked colocalization of GFP and RFP puncta (Fig. 1c), suggesting that lipidated LC3 was absent in the lysosome following treatment with OC. Consistent with this result, treating HeLa cells with OC dramatically increased the levels of P62 in a dose-dependent and time-dependent manner. This elevation of P62 by OC was not further enhanced by CQ treatment (Fig. 1e), further confirming the inhibition of autophagy at the late stage by OC. Moreover, immunofluorescence images indicated that exposure to OC obviously inhibited the colocalization of LC3 with LysoTracker Red staining, a lysosome dye, suggesting that OC inhibited fusion of the autophagosome and lysosome (Figure S1). Taken together, these data demonstrate that OC enhances autophagosome maturation by engaging STX17 but blocks autophagosome–lysosome fusion.
Figure 1.
OC mediates autophagosome maturation by engaging STX17 and blocks autophagosome-lysosome fusion. a OC mediated autophagosome maturation by engaging STX17. HeLa cells stably expressing GFP-LC3 were treated with OC (15 μM) or CQ (50 μM) for 8 h and then stained with anti-STX17 antibody. The cells were imaged by confocal microscopy. b Quantification of colocalization of LC3 with STX17 using Pearson’s correlation coefficient (Student t-test, **P < 0.01 compared to DMSO). c OC blocked autophagosome-lysosome fusion. The tfLC3B-L929 cells were treated with OC (15 μM), CQ (50 μM), or PP242 (1 μM) for 8 h. The cells were imaged by confocal microscopy. d OC induced the accumulation of P62 and LC3 II in HeLa cells. HeLa cells were treated with OC at the indicated concentration and time. The cell lysates were analyzed by Western blotting using antibodies targeting P62, LC3 and β-actin. e OC inhibited autophagic flux. HeLa cells were treated with OC (15 μM) or PP242 (1 μM) for 24 h in the presence or absence of CQ (50 μM). The cell lysates were analyzed by Western blotting using antibodies targeting P62, LC3 and β-actin
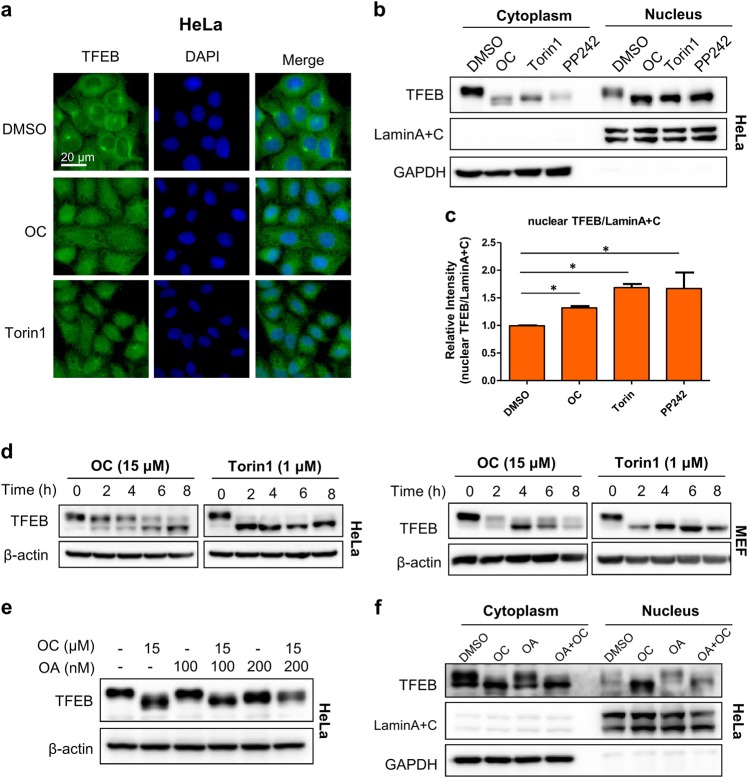
OC induces TFEB nuclear translocation
TFEB plays an important role in the regulation of lysosomal biogenesis and the autophagy process [4, 6]. Because OC has the ability to affect lysosome activity and thereby inhibit autophagy, we assessed the effect of OC on TFEB nuclear translocation by immunostaining. In normal conditions, TFEB was mainly distributed in the cytosol in HeLa cells (Fig. 2a). However, most of the endogenous TFEB translocated to the nucleus after OC treatment, similar to the effects of Torin1 treatment (Fig. 2a). The subcellular fractionation analysis revealed that OC treatment, similar to Torin1 or PP242 treatment, dramatically increased the expression of TFEB in the nucleus (Fig. 2b, c). We then assessed TFEB phosphorylation by evaluating the TFEB mobility-shift. Immunoblotting analysis indicated that treatment with OC significantly promoted the TFEB mobility-shift in HeLa and MEF cells (Fig. 2d), similar to the effect of the mTOR inhibitor Torin1 on TFEB mobility. Furthermore, cotreatment with OC and okadaic acid (OA), a potent inhibitor of specific protein phosphatases, abolished the TFEB band-shift induced by OC (Fig. 2e), indicating that the band-shift caused by OC was due to TFEB dephosphorylation. Furthermore, the subcellular fractionation analysis indicated that while OC treatment induced the translocation of TFEB to the nucleus, a combination of OC and OA treatments could attenuate TFEB dephosphorylation and nuclear translocation (Fig. 2f). Altogether, these results suggest that OC induces TFEB dephosphorylation and subsequent translocation.
Figure 2.
OC induces TFEB nuclear translocation. a OC enhanced TFEB nuclear translocation. HeLa cells were treated with OC (15 μM) or Torin1 (1 μM) for 8 h and then stained with anti-TFEB antibody for immunofluorescence analysis. b, c HeLa cells were treated with OC (15 μM), Torin1 (1 μM) or PP242 (1 μM) for 8 h. Nuclear fractions and cytoplasm lysates were prepared and then analyzed by Western blot using anti-TFEB, anti-LaminA + C and anti-GAPDH antibodies. *P < 0.05, treatment with OC, Torin1 and PP242 compared to DMSO. d OC mediated the TFEB mobility-shift. HeLa or MEF cells were treated with OC (15 μM) or PP242 (1 μM) for 8 h, and endogenous TFEB was analyzed by Western blotting using anti-TFEB and anti-actin antibodies. e Cotreatment with OC and okadaic acid (OA) inhibited TFEB dephosphorylation. HeLa cells were treated with 15 μM OC alone or in combination with OA for 8 h and subjected to Western blot analysis. f The combination of OC and OA inhibited TFEB nuclear translocation. HeLa cells were treated with 15 μM OC alone or in combination with 100 nM OA for 8 h and then subjected to Western blot analysis
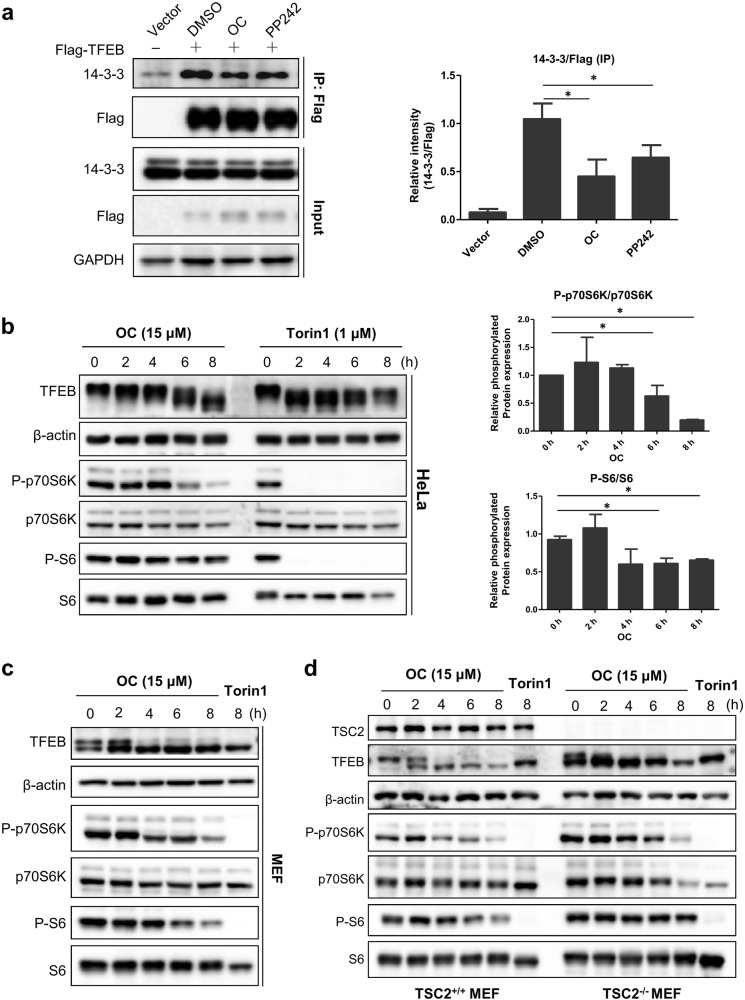
OC-induced TFEB translocation is dependent on mTORC1 activity
TFEB binding to 14-3-3 proteins leads to its cytosolic retention. To investigate whether OC affects the interaction of TFEB and 14-3-3 proteins, we transiently transfected a Flag-TFEB expression plasmid into HEK293T cells and then treated them with either DMSO, OC or PP242. Immunoprecipitation results revealed that both OC and PP242 treatment reduced the binding of 14-3-3 proteins to TFEB (Fig. 3a), suggesting that OC reduced the interaction of TFEB with 14-3-3. This interaction of TFEB with 14-3-3 is dependent on TFEB phosphorylation at Ser142 or Ser211, mediated by mTORC1 [17]. Once mTORC1 activity is inhibited, TFEB undergoes dephosphorylation and translocates to the nucleus [4, 6]. We therefore explored whether OC induces TFEB nuclear translocation by modulating mTORC1 activity. Western blot analysis showed that OC treatment suppressed the phosphorylation of two well-known mTORC1 substrates, namely, p70S6 kinase (P-p70S6K) and ribosomal S6 kinase (P-S6). Torin1, which strongly inhibits mTORC1 activity, was used as a positive control (Fig. 3b, c). TSC2 forms a heterodimeric complex with TSC1 that suppresses mTORC1 activity; a loss of either TSC1 or TSC2 leads to constitutive mTORC1 activation [18, 19]. We next used TSC2-knockout MEF cells (TSC2-/- MEF) to test the effect of OC on mTORC1 activity. Similar to Torin1, OC effectively suppressed mTORC1 activity and induced the TFEB mobility-shift in both TSC2+/+ and TSC2-/- MEF cells (Fig. 3d). A previous study showed that activated protein kinase C (PKC) inactivated GSK3β, leading to reduced phosphorylation, nuclear translocation and activation of TFEB [20]. We observed that OC treatment failed to alter PKC activity in HeLa cells (Figure S2), suggesting that the effect of OC on TFEB was independent of PKC. Collectively, these findings suggest that OC-induced TFEB nuclear translocation is dependent on mTORC1 activity [17].
Figure 3.
OC-induced TFEB nuclear translocation is dependent on mTORC1 activity. a OC attenuated the interaction of TFEB and 14-3-3 proteins. 293 T cells were transfected with a Flag-TFEB plasmid and the next day treated with OC (15 μM) or PP242 (1 μM) for 8 h. Cells were lysed and Flag-TFEB was immunoprecipitated with anti-Flag beads. Cell lysates (input) and immunoprecipitated proteins were immunoblotted with anti-14-3-3, anti-Flag and anti-GAPDH antibodies. Relative 14-3-3/Flag (IP) ratios were measured using ImageJ software. *P < 0.05, treatment with OC and PP242 compared to DMSO. b, c HeLa or MEF cells were treated with OC (15 μM) or Torin1 (1 μM) for a certain time period. TFEB, P-p70S6K and P-S6 were analyzed by Western blotting with the indicated antibodies. *P < 0.05, treatment with OC compared to DMSO. d TSC2+/+ or TSC2-/- MEF cells were treated with OC (15 μM) or PP242 (1 μM) for a certain time period. P-p70S6K and P-S6 were analyzed by Western blotting
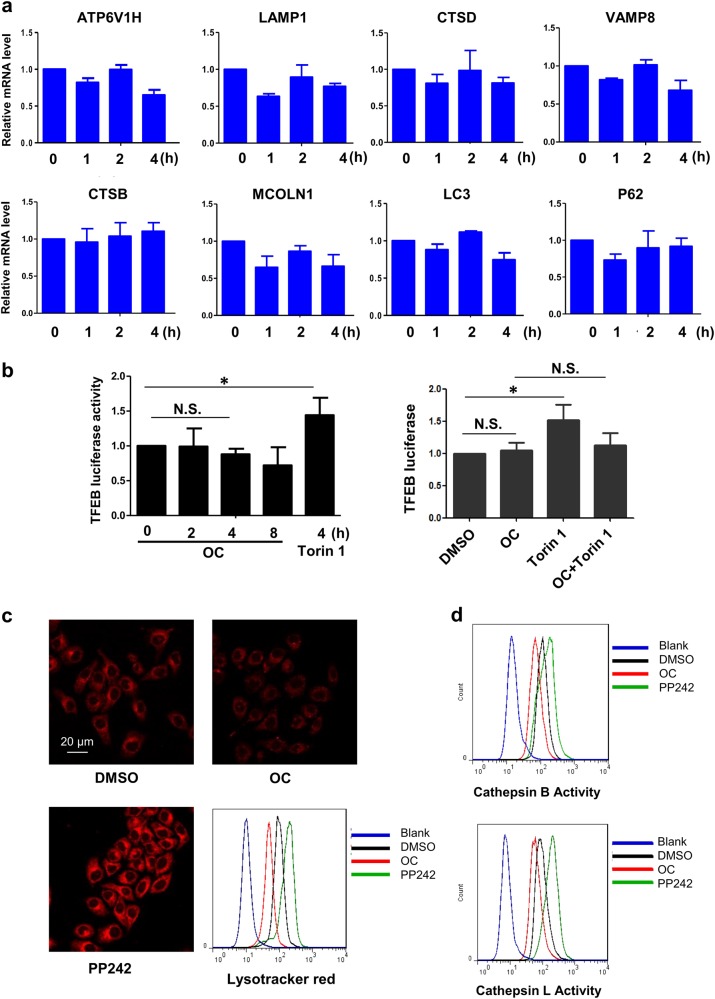
OC fails to increase TFEB-luciferase activity and lysosome function
TFEB nuclear translocation is associated with TFEB transcriptional activity, which is not only related to lysosomal biogenesis and function but also involved in the autophagy process [4]. We then investigated whether OC affects TFEB transcriptional activity. We first tested the expression of TFEB lysosomal and autophagic target genes by qRT-PCR. However, OC did not increase the expression of the TFEB target genes (Fig. 4a). The luciferase assay showed that OC treatment alone did not affect the TFEB-luciferase activity. In contrast, mTOR inhibitor Torin1 clearly enhanced expression of TFEB target genes and luciferase activity (Fig. 4b, left). However, the Torin1-induced elevation in TFEB luciferase activity could be suppressed by OC (Fig. 4b, left). Next, we quantified lysosome biogenesis using LysoTracker, which is a lysosome-specific fluorescent probe. Exposure to OC significantly decreased the intensity of LysoTracker Red staining (Fig. 4c). In addition, we measured the enzyme activities of lysosomal cathepsin B and L, which were elevated by the mTOR inhibitor PP242 (Fig. 4d). Treatment with OC blocked cathepsin B and L activities; this result is consistent with our previous report showing that OC blocks lysosomal function by altering lysosomal pH [12]. In summary, these findings suggest that OC fails to increase TFEB-luciferase activity and lysosome function.
Figure 4.
OC fails to increase TFEB-luciferase activity and lysosome biogenesis. a HeLa cells were treated with OC (15 μM) for a certain time period and analyzed by qRT-PCR for TFEB target gene expression. b 293T cells were transiently transfected with a TFEB-luciferase reporter vector together with a Renilla luciferase vector for 24 h. Cells were then treated with OC (15 µM) for a certain time period or Torin1 (1 μM) for 4 h, and the luciferase activity was evaluated. *P < 0.05, treatment with OC and Torin1 compared to DMSO. (c) HeLa cells were treated with OC (15 μM) or PP242 (1 μM) for 8 h, labeled with LysoTracker Red (50 nM) and analyzed by immunofluorescence and flow cytometry. d HeLa cells were treated with OC (15 μM) or PP242 (1 μM) for 8 h. Cells were labeled with Magic Red Cathepsin B or L reagent for 30 min and measured by flow cytometry
The combination of OC and hydroxycitrate inhibits the growth of HeLa cells
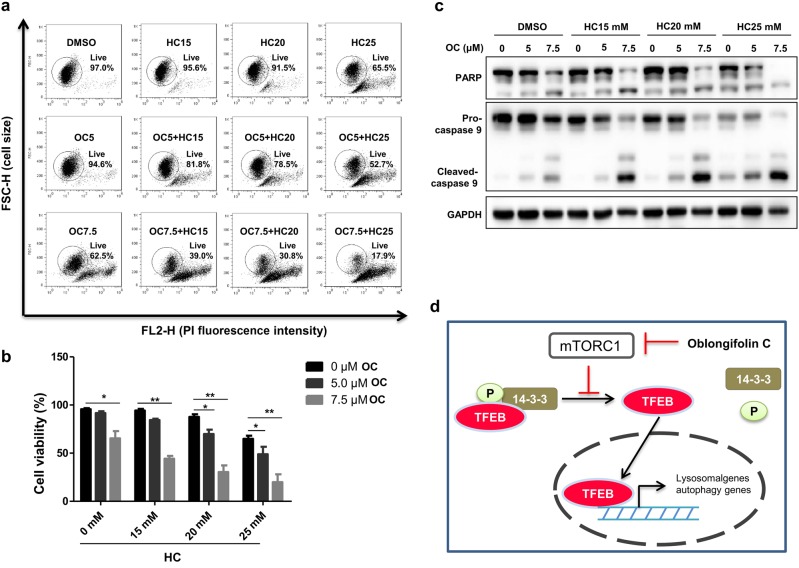
Our previous results showed that a combination of OC treatment with starvation (caloric restriction) significantly reduced tumor size and weight in a xenograft mouse model [12]. A recent study suggested that caloric restriction mimetics (CRMs) induced effects similar to starvation by increasing the autophagic flux. The natural compound hydroxycitrate (HC), a caloric restriction mimetic, is a competitive inhibitor of the ATP citrate lyase that generates cytosolic acetyl-CoA [21]. We therefore investigated whether the combination of OC with HC treatment has the potential to affect cancer cell survival. We examined cell death via a PI-exclusion assay. The flow cytometry analysis suggested that OC treatment alone (7.5 μM) could induce approximately 30% HeLa cell death, and HC treatment alone had almost no effect on HeLa cell death. However, cotreatment with 7.5 μM OC and 20 μM HC resulted in significant cytotoxicity (Fig. 5a, b), and more than 50% of the cells were killed. Consistent with this result, immunoblotting analysis showed that exposure to OC (5 or 7.5 μM) with HC (20 μM) treatment could dramatically enhance PARP and caspase 9 cleavage (Fig. 5c). Thus, the OC-HC combination dramatically inhibited growth of HeLa cells. To determine whether the stimulation of anticancer effects of hydroxycitrate by OC was due to autophagy inhibition, wild-type (ATG5+/+) or autophagy-deficient (ATG5-/-) MEF cells were cotreated with OC and HC for 24 h and then analyzed by flow cytometry. As shown in Figure S3, in ATG5+/+ (-dox) MEF cells, cotreatment with 15 μM OC and 20 mM HC could efficiently induce cell death, while 7.5 μM OC and 20 mM HC produced the same effect in ATG5-/- (+dox) MEF cells, suggesting that the increased anticancer effects of hydroxycitrate by OC was partially due to autophagy inhibition, while OC might regulate other pathways to promote cell death.
Figure 5.
The combination of OC and hydroxycitrate inhibits growth of HeLa cells. a, b HeLa cells were treated with OC (5 or 7.5 μM) alone or in combination with HC (15, 20 or 25 mM) for 24 h and analyzed by flow cytometry. *P < 0.05, **P < 0.01, compared to the related groups linked by lines. c HeLa cells were treated with OC (5 or 7.5 μM) alone or in combination with HC (15, 20, or 25 mM) for 24 h, and the cell lysates were immunoblotted using anti-PARP, anti-caspase 9 and anti-GAPDH antibodies. d The model represents the mechanism by which OC regulates autophagy
Discussion
Autophagy plays different roles in tumor initiation, progression and treatment. In premalignant lesions, it is thought that inducing autophagy can prevent cancer development. In contrast, enhancing autophagy might promote cancer cell growth and survival in advanced cancer [22, 23]. Therefore, strategic utilization of autophagy signaling pathways to develop small molecules against specific targets is a topic of great interest in cancer research. Here, we identified that the natural compound OC can inhibit autophagic flux by blocking lysosome–autophagosome fusion and suppress cell growth in combination with caloric restriction. However, the mechanism by which OC inhibits lysosome–autophagy and lysosome biogenesis remains unknown. Recent studies indicate that many transcription factors regulate autophagy and lysosome genes in mammals [24, 25]. Among these factors, TFEB is a master regulator of lysosomal biogenesis, driving the expression of related genes. During starvation or lysosomal stress, TFEB translocates from the cytoplasm to the nucleus, resulting in the activation of target genes. It has been well known that TFEB translocation is regulated by mTORC1 via phosphorylation. Here, we report that OC can promote TFEB nuclear translocation by inhibiting mTORC1 activity.
Our data demonstrate that although OC promotes TFEB nuclear translocation, lysosome function is still inhibited. Different transcriptional activators and suppressors are involved in the autophagy process. AMPK–SKP2–CARM1 signaling is shown to play vital roles in regulating TFEB. Since CARM1 is a coactivator of TFEB in the regulation of autophagy-related and lysosomal genes during nutrient deprivation [26], we examined whether OC could affect CARM1 protein levels. We found that CARM1 protein levels did not change in the nucleus after OC treatment of HeLa cells, suggesting that OC does not affect the AMPK–SKP2–CARM1 signaling pathway (data not shown). Previous studies indicate that autophagy inhibitors not only block autophagosome–lysosome fusion and lysosome function but also increase TFEB nuclear translation and promote its transcriptional activity. One example is CQ, a weak base that impairs the function of lysosomes by raising their pH, which triggers TFEB accumulation in the nucleus [17]. Another example is fangchinoline, a natural compound that promotes autophagosome accumulation by inhibiting autophagic degradation and inducing TFEB nuclear translocation [27]. Even though OC induces TFEB nuclear translocation (Fig. 5d), its transcriptional activity is still inhibited. We speculate that OC may also affect other pathways, resulting in the inhibition of autophagy and lysosome functions.
Starvation and fasting could improve the efficiency of chemotherapy. Caloric restriction mimetics (CRMs) could be used as an alternative to starvation and fasting. HC is one of the caloric restriction mimetics and can induce autophagy and improve anticancer chemotherapy [21]. A recent report showed that caloric restriction could enhance cisplatin-induced death of melanoma cells [28]. Here, we found that the combination of OC and HC can inhibit the growth of HeLa cells. Our study thus reveals OC treatment as a potential strategy to improve chemotherapy. It has been reported that starvation can enhance anticancer chemotherapy through an immunological mechanism [21]. Further study is needed to understand whether induction of cell death by the combination of HC and OC is related to autophagy and immunosurveillance.
Acknowledgements
We are thankful to Dr. A Ballabio and N Mizushima for providing critical reagents. This work was supported by the National Natural Science Foundation of China (No. 81773951 and No. 81602990), Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning and research grants from NMRC (NMRC-CIRG/1430/2015) to HMS.
Author contributions
HXX, HMS, and YZL designed the research; MW and HST performed most of the experiments and analyzed the data; GL, YR, ZQZ and JY performed some of the Western blotting experiments; WWF contributed material support; YL and MW wrote the manuscript; and HXX and HMS revised the manuscript.
Competing interests
The authors declare no competing interests.
Contributor Information
Han-ming Shen, Email: phsshm@nus.edu.sg.
Hong-xi Xu, Phone: +(86) 21-51323089, Email: xuhongxi88@gmail.com.
References
- 1.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–73. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 2.White E, DiPaola RS. The double-edged sword of autophagy modulation in cancer. Clin Cancer Res. 2009;15:5308–16. doi: 10.1158/1078-0432.CCR-07-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–7. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 4.Settembre C, Di Malta C, Polito VA, Arencibia MG, Vetrini F, Erdin S, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–33. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vega-Rubin-de-Celis S, Pena-Llopis S, Konda M, Brugarolas J. Multistep regulation of TFEB by MTORC1. Autophagy. 2017;13:464–72. doi: 10.1080/15548627.2016.1271514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han QB, Tian HL, Yang NY, Qiao CF, Song JZ, Chang DC, et al. Polyprenylated Xanthones from Garcinia lancilimba showing apoptotic effects against HeLa-C3 cells. Chem Biodivers. 2008;5:2710–7. doi: 10.1002/cbdv.200890225. [DOI] [PubMed] [Google Scholar]
- 8.Yang XW, Grossman RB, Xu G. Research progress of polycyclic polyprenylated acylphloroglucinols. Chem Rev. 2018;118:3508–58. doi: 10.1021/acs.chemrev.7b00551. [DOI] [PubMed] [Google Scholar]
- 9.Feng C, Zhou LY, Yu T, Xu G, Tian HL, Xu JJ, et al. A new anticancer compound, oblongifolin C, inhibits tumor growth and promotes apoptosis in HeLa cells through Bax activation. Int J Cancer. 2012;131:1445–54. doi: 10.1002/ijc.27365. [DOI] [PubMed] [Google Scholar]
- 10.Wang XY, Lao YZ, Xu NH, Xi ZC, Wu M, Wang H, et al. Oblongifolin C inhibits metastasis by up-regulating keratin 18 and tubulins. Sci Rep. 2015;5:10293. doi: 10.1038/srep10293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu W, Cheng M, Lao YZ, Wang XY, Wu J, Zhou L, et al. DNA damage and ER stress contribute to oblongifolin C-induced cell killing in Bax/Bak-deficient cells. Biochem Biophys Res Commun. 2015;457:300–6. doi: 10.1016/j.bbrc.2014.12.104. [DOI] [PubMed] [Google Scholar]
- 12.Lao YZ, Wan G, Liu ZY, Wang XY, Ruan P, Xu W, et al. The natural compound oblongifolin C inhibits autophagic flux and enhances antitumor efficacy of nutrient deprivation. Autophagy. 2014;10:736–49. doi: 10.4161/auto.28034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256–69. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 14.Tsuboyama K, Koyama-Honda I, Sakamaki Y, Koike M, Morishita H, Mizushima N. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science. 2016;354:1036–41. doi: 10.1126/science.aaf6136. [DOI] [PubMed] [Google Scholar]
- 15.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5:ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang J, Manning BD. The TSC1–TSC2 complex: a molecular switchboard controlling cell growth. Biochem J. 2008;412:179–90. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang J, Manning BD. A complex interplay between Akt, TSC2 and the two mTOR complexes. Biochem Soc Trans. 2009;37:217–22. doi: 10.1042/BST0370217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Xu M, Ding X, Yan C, Song ZQ, Chen LW, et al. Protein kinase C controls lysosome biogenesis independently of mTORC1. Nat Cell Biol. 2016;18:1065–77. doi: 10.1038/ncb3407. [DOI] [PubMed] [Google Scholar]
- 21.Pietrocola F, Pol J, Vacchelli E, Rao S, Enot DP, Baracco EE, et al. Caloric restriction mimetics enhance anticancer immunosurveillance. Cancer Cell. 2016;30:147–60. doi: 10.1016/j.ccell.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amaravadi R, Kimmelman AC, White E. Recent insights into the function of autophagy in cancer. Genes Dev. 2016;30:1913–30. doi: 10.1101/gad.287524.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17:528–42. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fullgrabe J, Klionsky DJ, Joseph B. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat Rev Mol Cell Biol. 2014;15:65–74. doi: 10.1038/nrm3716. [DOI] [PubMed] [Google Scholar]
- 25.Fullgrabe J, Ghislat G, Cho DH, Rubinsztein DC. Transcriptional regulation of mammalian autophagy at a glance. J Cell Sci. 2016;129:3059–66. doi: 10.1242/jcs.188920. [DOI] [PubMed] [Google Scholar]
- 26.Shin HJR, Kim H, Oh S, Lee JG, Kee M, Ko HJ, et al. AMPK-SKP2-CARM1 signalling cascade in transcriptional regulation of autophagy. Nature. 2016;534:553–7. doi: 10.1038/nature18014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang ZH, Guo X, Cao WX, Chen X, Lu JJ. Fangchinoline accumulates autophagosomes by inhibiting autophagic degradation and promoting TFEB nuclear translocation. RSC Adv. 2017;7:42597–605. doi: 10.1039/C7RA02738A. [DOI] [Google Scholar]
- 28.Antunes F, Pereira GJ, Jasiulionis MG, Bincoletto C, Smaili SS. Nutritional shortage augments cisplatin-effects on murine melanoma cells. Chem-Biol Interact. 2017;281:89–97. doi: 10.1016/j.cbi.2017.12.027. [DOI] [PubMed] [Google Scholar]