Abstract
Comparative genomics of fungal mitochondrial genomes (mitogenomes) have revealed a remarkable pattern of rearrangement between and within major phyla owing to horizontal gene transfer and recombination. The role of recombination was exemplified at a finer evolutionary time scale in basidiomycetes group of fungi as they display a diversity of mitochondrial DNA inheritance patterns. Here, we assembled mitogenomes of six species from the Hymenochaetales order of basidiomycetes and examined 59 mitogenomes from 2 genetic lineages of Phellinus noxius. Gene order is largely collinear, while intergene regions are major determinants of mitogenome size variation. Substantial sequence divergence was found in shared introns consistent with high horizontal gene transfer frequency observed in yeasts, but we also identified a rare case where an intron was retained in five species since speciation. In contrast to the hyperdiversity observed in nuclear genomes of Phellinus noxius, mitogenomes’ intraspecific polymorphisms at protein-coding sequences are extremely low. Phylogeny network based on introns revealed turnover as well as exchange of introns between two lineages. Strikingly, some strains harbor a mosaic origin of introns from both lineages. Analysis of intergenic sequence indicated substantial differences between and within lineages, and an expansion may be ongoing as a result of exchange between distal intergenes. These findings suggest that the evolution in mitochondrial DNAs is usually lineage specific but chimeric mitotypes are frequently observed, thus capturing the possible evolutionary processes shaping mitogenomes in a basidiomycete. The large mitogenome sizes reported in various basidiomycetes appear to be a result of interspecific reshuffling of intergenes.
Keywords: mitochondrial evolution, mitogenome size variation, population mitogenomics, mitogenomics
Introduction
A typical fungal mitochondrial genome (mitogenome) consists of a circular DNA, which encodes 14 mitochondrial-exclusive protein-coding genes, ribosomal RNAs and transfer RNAs usually in clusters (Férandon et al. 2013; Aguileta et al. 2014; Sandor et al. 2018). Depending on groups, these elements are encoded either on the same strand in ascomycetous or on both in basidiomycetous mitogenomes (Aguileta et al. 2014). These genomes are highly diversified in size ranging from 19,431 bp in Schizosaccharomyces pombe (Bullerwell et al. 2003) to 235,849 bp in Rhizoctonia solani (Losada et al. 2014). Variations in mitogenome size can be large among species within a genus (Bullerwell et al. 2003) or among strains of the same species (Wolters et al. 2015). Presence and expansion of introns in mitogenome protein encoding genes as well as sizable noncoding regions (NCRs) are major determinants of fungal mitogenome size variations (Xu and Wang 2015; Sandor et al. 2018). Given that the highly variable nature of fungal mitogenome size, characterizing the dynamics of introns and determining the origin and function of these NCRs is critical for understanding the evolution of fungal mitogenome.
Inheritance of mitochondrial DNA (mtDNA) is predominantly maternal in sexual plants and animals (Xu and Wang 2015; Sandor et al. 2018). In the Fungi kingdom, however, alternative modes of inheritance have been commonly reported during sexual reproduction (Berger and Yaffe 2000; Basse 2010; Wilson and Xu 2012; Xu and Wang 2015). For instance, the model ascomycete yeasts, Saccharomyces cerevisiae and Schizosaccharomyces pombe, are known to inherit mitochondria biparentally (Wolf et al. 1976; Basse 2010; Xu and Wang 2015). The zygote contains mtDNA from both parents and subsequently segregated into homoplasmic progenies through the process of syngamy. Like animals and plants, the mating process in filamentous ascomycetes usually initiated with the fusion of two morphologically different gametes, and the mitochondria are typically inherited from the larger gamete (Sandor et al. 2018). For basidiomycetes, various modes of mitochondrial inheritance may have been involved in generating recombinant mitochondrial genotypes (Xu and He 2015; Xu and Wang 2015; Sandor et al. 2018).
In fungi, tight regulation of mtDNA transmission is part of molecular mechanisms responsible for sex determination (mating). For model basidiomycetes yeast Cryptococcus neoformans, mtDNA is inherited uniparentally from MATa parent but becomes biparental if the sex-specific homeodomain transcription factors sxi1α and sxi2a are disrupted (Xu et al. 2000; Yan and Xu 2003). A closely related species of Cryptococcusneoformans, Cryptococcusgattii exhibited different inheritance patterns depending on strain combinations and environmental factors such as temperature (Voelz et al. 2013; Zhu et al. 2013; Wang et al. 2015). For Ustilagomaydis, which has a tetrapolar mating system similar to the model mushroom Coprinopsis cinerea, the mtDNA inheritance is controlled by lga2 and rga2 of uncertain function encoded by a2 locus (homologous to C. cinerea matB locus) (Fedler et al. 2009).
Strictly selecting mtDNA from only one parent during mating ensures uniparental inheritance. This is the case for model filamentous basidiomycetes such as C. cinerea and Schiozphyllum commune, where the formation of diploid involves nuclear but not mitochondrial migration, resulting in genetically identical dikaryons with different mitochondria from either parent (Sandor et al. 2018). Nuclear migration is controlled by matB locus encoding peptide pheromones and pheromone receptors (Casselton and Kües 2007), again indicating a strong correlation between mating locus and mitochondria inheritance. Alternatively, the button mushroom Agaricus bisporus and many basidiomycetes do not undergo nuclear migration and clamp formation; instead, they undergo direct hyphal fusion during mating process. This means different mitochondrial genotypes can be present within hyphal cells, suggesting the opportunity to exchange DNA via mitochondrial recombination. Indeed, A. bisporus is known to have uniparental and nonparental mitochondrial inheritance patterns (Hintz et al. 1988; Jin and Horgen 1993, 1994; de la Bastide and Horgen 2003) and have displayed signatures of recombination in natural populations (Xu et al. 2013). Hence, the ephemeral diploid phase may be particularly important in mitochondrial inheritance and diversity (de la Bastide and Horgen 2003; Xu and Wang 2015).
Among the estimated 2.2–3.8 million fungal species (Hawksworth and Lücking 2017), many lineages still lack a formal description or analysis of mitogenomes. One interesting case is the order Hymenochaetales (Agaricomycetes, Basidiomycota), which is dominated by wood decay fungi. Most species in this order are saprotrophic, but some are pathogens involved in major forest incidents since 1971 in different parts of the world (Hepting 1971; Sahashi et al. 2012). In particular, Phellinus noxius has a very wide host range, spanning more than 200 broadleaved and coniferous tree species (at least 59 families), and are the causative agents responsible for brown root rot disease in parts of Asia (Akiba et al. 2015; Chung et al. 2015). A recent comparative and population genomics study of six genomes from this order and 60 P. noxius strains from Taiwan and Japan revealed that P. noxius possesses extraordinarily high diversity in its nuclear genomes (Chung et al. 2017).
Phellinus noxius lacks clamp connections (Ann et al. 2002) and displays a bipolar heterothallic mating system (Chung et al. 2017) with a highly expanded A locus spanning a ∼60-kb region. The availability of whole genome sequences of P. noxius populations prompted us to further understand the evolution of mitogenomes in this hypervariable species. In this study, we first assembled and compared the mitogenomes of P. noxius and five Hymenochaetales species. We demonstrated that core genes are largely in syntenic across these species while both introns and NCRs constitute the majority of P. noxius mtDNA. We then examined mitogenome evolution from two genetically independent lineages of P. noxius and discovered frequent rearrangement in NCRs. The main purpose of this study was to elucidate possible mechanisms driving mitogenome enlargement in P. noxius at a population level. We provided evidences of frequent mitochondrial exchange possibly in heteroplasmy stage during hyphal fusion and mating resulting in recombinant mitotype and expanding mitogenome.
Materials and Methods
De Novo Assembly or Reconstruction of Four Hymenochaetales Mitochondrial Genomes
Sequencing data of P.noxius, Phellinus lamaensis, Porodaedalea pini, and Coniferiporia sulphurascens (syn. Phellinus sulphurascens) were described in Chung et al. (2017). Mitochondrial genomes (mitogenomes) were assembled in 1–6 iterations using Organelle_PBA (v1.0.7; Soorni et al. 2017) with PacBio reads until sequence circularity was detected. Partial mtDNA sequence in the published assembly of each species or conserved protein-coding gene was used as initial reference for the first run. Illumina sequencing data of Fomitiporia mediterranea (Floudas et al. 2012) were obtained from Joint Genome Institute (JGI) database (https://genome.jgi.doe.gov/Fomme1/Fomme1.home.html). The F. mediterranea mitogenome was assembled using NOVOplasty (v2.6.7; Dierckxsens et al. 2017) with Po. pini’s mitogenome as the seed sequence. Circularity was assessed either by in-built function in the assemblers or subsequently by “check_circularity.pl” script distributed with sprai de novo assembly tool (v0.9.9.23; http://zombie.cb.k.u-tokyo.ac.jp/sprai). The consensus of assemblies was improved using pilon (v1.22; Walker et al. 2014). The complete mitogenome of Schizopora paradoxa (Min et al. 2015) is available in JGI database (https://genome.jgi.doe.gov/Schpa1/Schpa1.home.html) and was retrieved for further annotation and analysis.
Annotation of Mitogenomes
Core genes in mitogenomes were annotated using MFannot web service (http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl) with genetic code table 4. Exon boundaries were further manually curated based on multiple sequence alignment of homologs from closely related species using MUSCLE (v3.8.31; Edgar 2004) and RNA-seq alignments where available. tRNA genes were predicted by MFannot, tRNAscan-SE (v2.0; Lowe and Chan 2016), and Aragorn (v1.2.38; Laslett and Canbäck 2004). Only tRNA positions supported by at least two programs were retained for further analysis. Putative open reading frames (ORFs) were predicted by NCBI ORFfinder (v0.4.3; Wheeler et al. 2003) against exon-masked mitogenomes, and hypothetical proteins were searched against Pfam (Finn et al. 2016) and NCBI protein nr database (https://www.ncbi.nlm.nih.gov/protein) for evidence of homing endonuclease (HE) and DNA/RNA polymerase (dpo/rpo) genes. Introns were assigned into subgroups using RNAfinder (v1.40; https://github.com/BFL-lab/RNAfinder). Putative HE domains in introns were searched by PfamScan (v31; Finn et al. 2016). Direct and palindromic repeats were searched using Vmatch (v2.3.0; http://www.vmatch.de) with parameters of “-seedlength 30 -l 30 -exdrop 3 -identity 80.” Tandem repeats were identified using Tandem Repeat Finder (v4.09; Benson 1999) with default parameters. Analysis of simple sequence repeats were performed by MISA (v2.0; Thiel et al. 2003) with default parameters.
RNA Sequencing
The mycelia of P. noxius KPN91 were cultured on the PDA agar plates at 25 °C for about 3–5 days. Total RNA was extracted from mycelia homogenized in liquid nitrogen, following with TRIzol Reagent (catalog no. 15596018, Thermo Fisher Scientific, Inc.). Ploy-A enriched RNA (200 ng) was purified from total RNA by using Dynabeads mRNA purification kit (catalog no. 61006, Thermo Fisher Scientific, Inc.) and was used to conduct cDNA library preparation with Direct cDNA Sequencing kit (SQK-DCS108; Oxford Nanopore Technologies). The reads were sequenced with FLO-MIN106 flow cell on a GridION, Oxford Nanopore and then basecalled by dogfish v0.9.3. Illumina RNA-seq reads of four species in this study (P. noxius, P. lamaensis, Po. pini, and Con. sulphurascens) were downloaded and aligned to corresponding assemblies using STAR (v2.6.0a; Dobin et al. 2013). Nanopore cDNA reads (P. noxius) were aligned using minimap2 (v2.6; Li 2018). Only reads aligned to mitogenome were retained for further analysis.
Assembly and Annotation of P. noxius Isolates
The sequencing data of a total of 59 P. noxius isolates were described in a previous study (Chung et al. 2017). All of the mitogenomes were assembled using the same pipeline with F. mediterranea as described above except that the seed sequence was P. noxius KPN91 mitogenome. Annotation of core genes was searched with query of curated genes of KPN91 using GMAP (v2017-11-15; Wu and Watanabe 2005), followed by multiple sequence alignment using MUSCLE (v3.8.31; Edgar 2004) to identify precise exon–intron boundaries. tRNA annotation was processed as described above. The four (three “large” ranging 6.3–35.4 kb and a small 4.6–8.9 kb) regions were defined and characterized by their frequently rearrangements when compared with KPN91 mtDNA reference (region 1: nad4-rnl, region 2: atp8-cox1, region 3: nad5-rns, and region 4s: cox2-cob).
Phylogenetic Analysis
Phylogenetic analysis of six Hymenochaetales species was carried out using concatenated amino acid alignments of 15 mitochondrial protein-coding genes by MUSCLE (v3.8.31; Edgar 2004), and pruned by trimal strict plus mode (v1.4; Capella-Gutierrez et al. 2009). A maximum-likelihood phylogeny was computed using iqtree (v1.6.6; Nguyen et al. 2015) with LG + F + I + G4 model and 100 bootstrap replicates. The same approach was used to produce the intron phylogeny for P. noxius isolates, except that ModelFinder (Kalyaanamoorthy et al. 2017) in iqtree was invoked to automatically determine the best-fit model. Maximum-likelihood distances inferred by iqtree were passed to SplitsTree4 program to generate a Neighbor-Net (NN) phylogenetic network of concatenated introns (Bryant and Moulton 2004). To validate the robustness of mitochondrial protein-coding genes phylogeny, four unpublished Hymenochaetales species available on JGI database (Onnia scaura, Phellinus ferrugineofuscus, Porodaedalea chrysoloma, and Porodaedalea niemelaei) were used to replot the phylogeny.
DNA Preparation and Polymerase Chain Reaction Condition
Genomic DNA of three isolates (KPN91, KPN323, and Pn113) for intergenic rearrangement validation were prepared as described by Chung et al. (2017). Mitogenomes between these isolates were aligned using MUMmer (v4.0.0beta2; Kurtz et al. 2004) and primers were designed at the synteny blocks within predefined intergenic regions (1–3) using Primer3Plus web service (Untergasser et al. 2012). The primers were further filtered for nonspecificity by BlastN (v2.6.0; McGinnis and Madden 2004) against three mitogenomes. Each PCR composed of ∼10-ng template DNA, 10 μl KAPA HiFi HotStart ReadyMix (2×) (0.4 U of DNA polymerase, MgCl2 at a final concentration of 2.5 mM, each dNTP at a final concentration of 0.3 mM) (Kapa Biosystems), 0.3 μM of each primer, and sterilized distilled water to make up a total volume of 20 μl. Thermo cycling profile included an initial denaturing temperature of 95 °C for 3 min followed by 35 cycles of 98 °C for 20 s, 60 °C for 15 s (for A1_F/B1_R and A1_F/rnl_R using 65 °C, and for F1_F/C1_R and apt8_F/C1_R using 62 °C), and 72 °C for 1 min per kb. The final extension step was 72 °C for 1 min per kb. The PCR products were then analyzed by electrophoresis on 1% agarose gel at 70 V for 45 min. The primer sequences are listed in supplementary table S4, Supplementary Material online.
Pairwise Identity Calculation
Intergenic sequences of four predefined regions were aligned separately using MAFFT (v7.310; Katoh and Standley 2013) with parameters of “–ep 0 –genafpair” to incorporate large indels. The aligned sequences were then concatenated for identity calculation. Direct identity was referred to matched bases over alignment length, and gap-compressed method counted gaps regardless of length as one difference, therefore reduced the extent of large indels on identity score.
Prediction and Significance Test of Rearrangement Breakpoints
All 58 mitogenomes were aligned against KPN91 reference using MUMmer (v4.0.0beta2; Kurtz et al. 2004) to identify the breakpoints of rearrangement segments. A rearrangement segment was defined according to one of the following rules: 1) rearrange across different intergenic regions, 2) rearrange within a single intergenic region, and 3) inversion (supplementary fig. S8, Supplementary Material online). The breakpoints were the boundaries of rearrangement segments and the isolates with <10 breakpoints were omitted. To test the correlation between breakpoints and repetitive elements, we sampled equal number of positions within predefined intergenic regions and the probability of breakpoints locating on repetitive elements was calculated. One thousand bootstrap replicates were performed to infer adjusted P values. The analyses were conducted with python scripting language.
Results
Assembly and Annotation of Six Hymenochaetales Mitochondrial Genomes
There are two published mitogenomes in Hymenochaetales species, with genome size ranging from 57.5 kb in S.paradoxa (Min et al. 2015) to 163.4 kb in P.noxius (Chung et al. 2017). Such large differences in mitogenome sizes prompted us to assemble four more Hymenochaetales species using available Illumina or Pacbio long sequences: P. lamaensis, Po. pini, Con. sulphurascens, and F. mediterranea. The four Hymenochaetales mitogenomes now range from 45.6 to 145.0 kb (table 1), again showing high variations. GC content are low (23.4–34.6%), consistent with the AT-rich nature of fungal mitogenomes (Hausner 2003). Remapping of Illumina reads against the mitogenomes showed no heterozygosity, suggesting that mitogenomes are homozygous in these species with no evidence of heteroplasmy. All six species were predicted to contain 15 protein-coding genes and 2 ribosomal RNA genes (fig. 1 and supplementary figs. S1 and S2A, Supplementary Material online). Twenty-five tRNAs encoding 20 standard amino acids were shared across 6 species and positioned into 3–5 clusters, with the presence of 1–2 species-specific tRNA. All 17 core genes and tRNA clusters of 6 species are encoded on the same strand with an exception of a histidine tRNA in S. paradoxa (supplementary fig. S1E, Supplementary Material online). Interestingly, in four Hymenochaetales species, nad5 starts (ATG) immediately from the last base of adjacent nad4L stop codon (TAA), which has been observed in various fungal mitochondria (Férandon et al. 2013; Wang et al. 2018).
Table 1.
General Statistics of Six Mitogenomes
| Species | P. noxius | P. lamaensis | Po. pini | F. mediterranea | Con. sulphurascens | S. paradoxa |
|---|---|---|---|---|---|---|
| Coverage (Pacbio) | 2,860 | 6,175 | 2,673 | — | 6,477 | — |
| Coverage (Illumina) | 744 | 3,873 | 555 | 1,116 | 1,021 | — |
| Mitogenome length | 163,449 | 45,604 | 144,970 | 115,036 | 58,959 | 57,732 |
| GC content (%) | 28.68 | 34.55 | 28.26 | 23.39 | 33.17 | 28.22 |
| # tRNAs | 27 | 26 | 26 | 27 | 25 | 26 |
| # Exons of 17 standard genes | 78 | 20 | 38 | 45 | 18 | 25 |
| atp6 | 1 | 1 | 1 | 1 | 1 | 1 |
| atp8 | 1 | 1 | 1 | 1 | 1 | 1 |
| atp9 | 1 | 1 | 1 | 1 | 1 | 1 |
| cob | 9 | 3 | 3 | 6 | 2 | 2 |
| cox1 | 24 | 2 | 12 | 13 | 1 | 7 |
| cox2 | 5 | 1 | 1 | 3 | 1 | 1 |
| cox3 | 3 | 1 | 1 | 2 | 1 | 1 |
| nad1 | 5 | 1 | 1 | 3 | 1 | 1 |
| nad2 | 2 | 1 | 1 | 1 | 1 | 1 |
| nad3 | 1 | 1 | 1 | 1 | 1 | 1 |
| nad4 | 2 | 1 | 1 | 2 | 1 | 1 |
| nad4L | 1 | 1 | 1 | 1 | 1 | 1 |
| nad5 | 7 | 1 | 2 | 3 | 1 | 1 |
| nad6 | 1 | 1 | 1 | 1 | 1 | 1 |
| rps3 | 1 | 1 | 1 | 1 | 1 | 1 |
| rnl | 13 | 1 | 8 | 4 | 1 | 2 |
| rns | 1 | 1 | 1 | 1 | 1 | 1 |
| # Introns | 61 | 3 | 21 | 28 | 1 | 8 |
| # ORFs | 95 | 18 | 100 | 66 | 30 | 24 |
| HEGa | 21 | 2 | 15 | 22 | 0 | 6 |
| dpoBb | 3 | 1 | 11 | 4 | 5 | 1 |
| Length of exons | 21,975 (13.44%) | 21,345 (46.81%) | 24,328 (16.78%) | 22,242 (19.33%) | 23,435 (39.75%) | 21,795 (37.75%) |
| Length of introns | 62,449 (38.21%) | 2,829 (6.2%) | 25,366 (17.5%) | 34,228 (29.75%) | 755 (1.28%) | 8,733 (15.13%) |
| Length of intergene | 79,025 (48.35%) | 21,430 (46.99%) | 95,276 (65.72%) | 58,566 (50.91%) | 34,769 (58.97%) | 27,204 (47.12%) |
| Length of repeats | 12,784 (7.82%) | 1,588 (3.48%) | 12,377 (8.54%) | 3,771 (3.28%) | 2,059 (3.49%) | 1,685 (2.92%) |
Homing endonucleases containing LAGLIDADG or GIY-YIG domain.
Family B DNA polymerase.
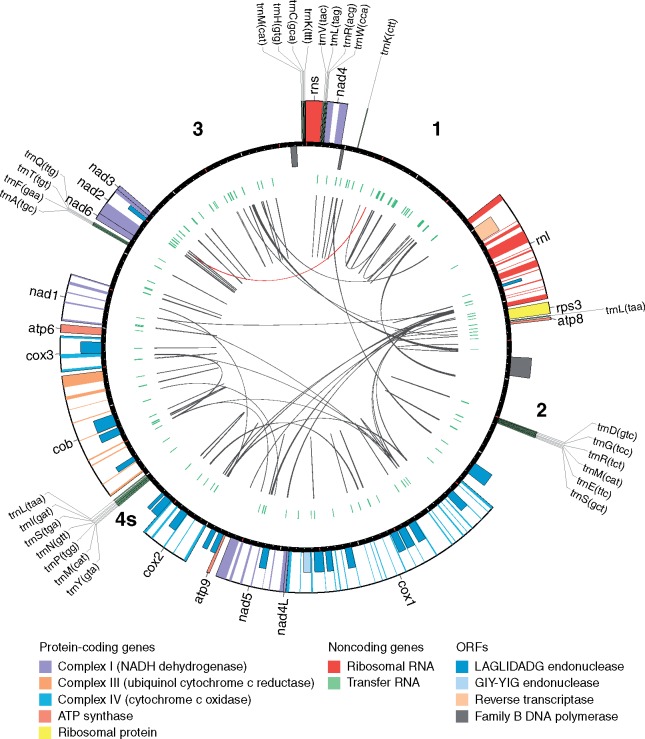
Fig. 1.
—Genetic map of Phellinus noxius KPN91 mitogenome. The first base is defined as the start of rns gene. The ticks on the circle are separated by 1 kb. The boxes adhered to the circle are genes and the half-sized boxes are predicted ORFs. The boxes at outside and inside the circle indicates the genes encoding on positive and negative strand, respectively. Colored regions are exons. In the inner circle, green tiles represent the tandem repeats and the black and red links represent the direct and palindromic repeats, respectively. The bold numbers indicated the four defined intergenic regions (1–3 and 4s). The plot was generated using Circos software (Krzywinski et al. 2009).
Coding contents are relatively conserved across the six Hymenochaetales species (average 22.5 kb), however, huge variations were observed in their intronic and intergenic sequences. Prediction of ORFs in intronic and intergenic regions resulted in 18–100 putative protein-coding sequences (table 1 and supplementary table S1, Supplementary Material online). Majority of the ORFs did not show homology across various databases. The ORFs with confident protein domain profiles assigned were mostly HE (containing LAGLIDADG or GIY-YIG domain) and family B DNA polymerase (dpoB). Phellinus noxius has the largest mitogenome among the six Hymenochaetales species, with 38.2% introns and 48.4% intergene sequences. The mitogenome of Po. pini has an intron content of 17.5% and 65.7% intergene region. In contrast, P. lamaensis has the most reduced mitogenome with highest percentage of coding content (46.8%). We further investigated whether repetitive sequences contributed to the noncoding content of these mitogenomes. The total repeat content accounted for 2.9–8.5% of mitogenomes, with only two large palindromic repeats (>500 bp) observed in Po. pini. These repeats did not contribute to the majority of noncoding content as only 4.7–12.2% of intergenic sequences were classified as repetitive. The invertron-like sequences are plasmids integrated into fungal or plant mtDNA, usually consisting of dpoB and DNA-dependent RNA polymerase between inverted repeats (Hausner 2012; Mardanov et al. 2014). Although intact invertron-like sequences were not observed, we found at least one dpoB gene in each mitogenome suggesting possible ancient integration events.
Gene Synteny
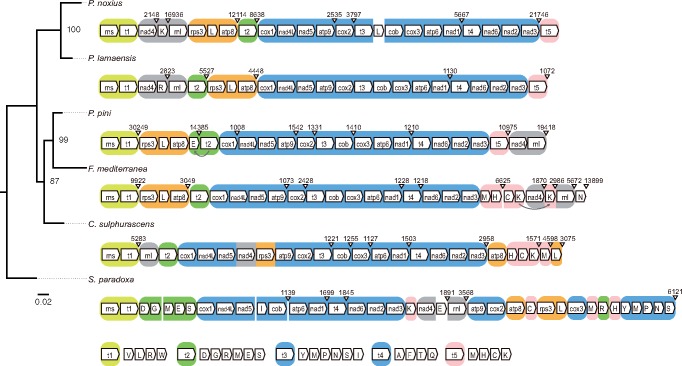
Species phylogeny inferred from 1,127 orthologous single copy genes placed species of similar genome sizes and primary habits together with S. paradoxa being the most diverged clade (Chung et al. 2017). The mitochondrial phylogeny of six Hymenochaetales was inferred from concatenation of 15 protein-coding genes (fig. 2). Different from the nuclear phylogeny, Con. sulphurascens was in the same clade with Po. pini and F. mediterranea. The gene order of 17 core genes and tRNA clusters were considerably conserved across five of the six Hymenochaetales species. For example, the largest synteny block started from cox1 and extended to 22 genes (blue box). Greatest difference was found in S. paradoxa, which agrees with the nuclear phylogeny (Chung et al. 2017). In the clade of P. noxius and P. lamaensis, nad4 and rnl genes were both relocated to downstream of t1 tRNA cluster. Furthermore, the location of t2 tRNA cluster and rps3-atp8 block was swapped in P. lamaensis. Both P.noxius and P. lamaensis harbored a tRNA between nad4 and rnl. The two tRNAs shared 88.9% identity in the 72-bp gene region, suggesting accumulation of mutations in a recently gained gene. Boundaries of intergenic spacers were similar between P. noxius and P. lamaensis but not across other species. Gene duplication events were only observed in t2 in Po. pini and t5 tRNA clusters in F. mediterranea.
Fig. 2.
—Phylogenetic tree of mitochondrial protein-coding genes and gene synteny in six mitogenomes. The colored boxes represent different conserved gene blocks. The intergene length larger than 1 kb is labeled above the gene wedges. The arrows below the gene wedges indicate gene duplication. The bootstrap values were inferred with 100 replicates.
Mitochondrial Intron Dynamics
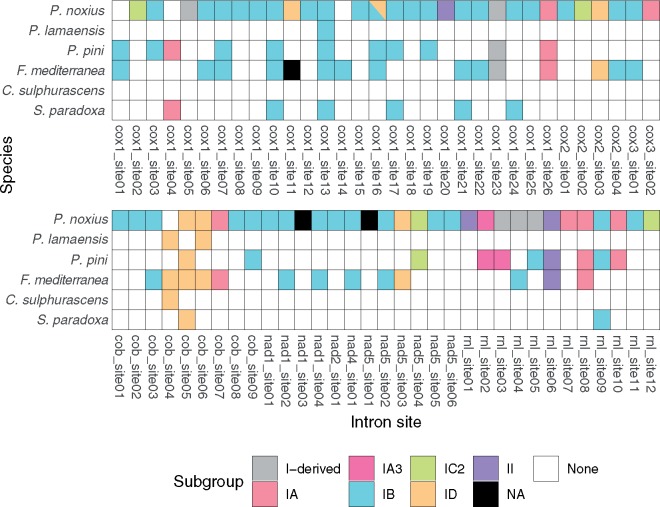
Phellinus noxius mitogenome contains most introns in 6 species, with 61 introns spreading in 9 genes. In contrast, Con. sulphurascens mitogenome contains only one intron in cob gene. The largest reservoir of introns is P. noxius cox1 gene, which retains 23 group I introns and a group II intron (17th intron), accounting for 93.5% of this gene. A total of 122 introns locating in 65 genetic sites were identified in six Hymenochaetales mitogenomes, with 39 sites were shared among species (fig. 3 and supplementary table S2, Supplementary Material online). No introns were found in eight genes (atp6, atp8, atp9, nad3, nad4L, nad6, rns, and rps3), whereas the cob gene contained at least one intron in every species. The majority of introns were of group I (114/122) and were classified into six subgroups (IA, IA3, IB, IC2, ID, and I-derived). Of these, group IB introns accounted for 57.4% of total introns (70/122). Interestingly, the 13th intron of P. noxius cox1 gene (cox1_intron13) was predicted to have two separated complete intron cores (IB and ID) suggesting a twintron: an intron inserted by another intron (Deng et al. 2018).
Fig. 3.
—Shared intron positions and intron subgroups in six Hymenochaetales species. Details of intron properties refer to supplementary table S2, Supplementary Material online. NA: not assigned to any subgroup.
One possible explanation for the prevalence of introns in these species is the presence of HEs, which are highly specific endonucleases playing an important role in intron homing and precise splicing in group I introns (Yan et al. 2018). We searched for protein domains and ORFs, revealing that 65 introns each containing at least one LAGLIDADG or GIY-YIG domain. Nine of these introns did not have any predicted ORF suggesting loss of HE activity. Conversely, 12 intronic ORFs showed no evidence of HE domain. In addition to group I introns, we found 3 out of 5 group II introns comprising a putative LAGLIDADG domain, which has been reported in several filamentous fungi but showed no activity in enhancing splicing itself. LAGLIDADG domain might be an invaded group I HEG in a group II intron (Toor and Zimmerly 2002; Mullineux et al. 2010). Only a group II intron (PNOK_rnl_intron01) comprised an intact ORF with a complete reverse transcriptase domain.
Out of 39 intron sites present in multiple species, 34 comprised introns belonging to the same subgroup suggest a preference of insertion (Swithers et al. 2009). Alternatively, these introns may have already been inserted since their common ancestor; however, this may be less likely as the shared introns showed as low as 74.3% nucleotide identity. This scenario may be more likely in sites where introns are still found in more than three species. There are three such intron sites (cob_site5, cox1_site10, and cox1_site13) and phylogeny indicated two sites (cox1_site10 and cox1_site13) were concordant with the mitochondrial phylogeny, suggesting the intron was present since these species’ common ancestor (supplementary fig. S3, Supplementary Material online).
Population Mitogenomics of P. noxius: General Characteristics
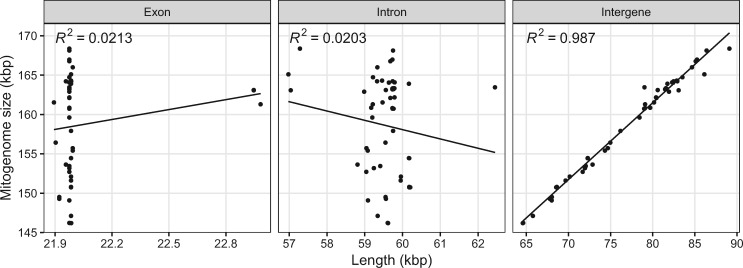
The mitogenomes of additional 58 P. noxius isolates from Japan and Taiwan region were assembled into single circular sequences using NOVOPlasty (supplementary table S3, Supplementary Material online). The mitogenome size can differ by 22 kb ranging from 146,191 bp of Pn102 to 168,365 bp of Pn120. A strong correlation was observed in intergenic region and mitogenome size (R2 = 0.987, P < 0.001; fig. 4) but not exonic and intronic sequences (R2 = 0.0213 and R2 = 0.0203, respectively; fig. 4), suggesting that the expansion of mitochondria was mainly contributed by intergenic regions in P. noxius.
Fig. 4.
—Correlation between mitogenome size and genome properties across 59 Phellinus noxius isolates. Each dot represents an isolate. Details refer to supplementary table S3, Supplementary Material online.
Low Intraspecific Polymorphism in Coding Regions
All 17 core genes were present in all isolates. Gene structures were highly conserved except for two protein-coding genes. Stop codon in the majority of isolates in the cox2 gene was lost in three isolates with two possible changes (minor allele 1: KPN57 and Pn315; minor allele 2: Pn040). The prevalent allele encoded for a 765 bp cox2 gene with five exons, and two minor alleles extended 972 and 1,008 bp in the last exon, respectively. In nad2 gene, a tandem repeat [(AGAAACTAA)3.8–7.8] located in the second exon varied the length of coding regions from 1,695 to 1,731 bp without interrupting the reading frame. Across 4,603 amino acid sites, only 13 and 7 were parsimony-informative and singletons, respectively (supplementary fig. S4, Supplementary Material online). Number of tRNA ranged from 26 to 27. A free-standing tRNA between nad4 and rnl was found to alternatively code for lysine and arginine in these species. Two SNPs were observed in the 72-bp exon region resulting in two different alleles. In five isolates (Pn081, Pn102, Pn329, KPN141, and KPN163), a point mutation in the anticodon period turns the tRNALys(CUU) into an tRNAArg(CCU), accompanying the translocation of surrounding sequences (supplementary fig. S7C, Supplementary Material online). The lysine tRNA was lost in four isolates (Pn015, Pn035, Pn086, and Pn127) due to a large fragment deletion (supplementary fig. S7D, Supplementary Material online), suggesting that it might be a nonessential tRNA and undergoing loss of gene.
Frequent Gain/Loss and Evidences of Rearrangements in Introns
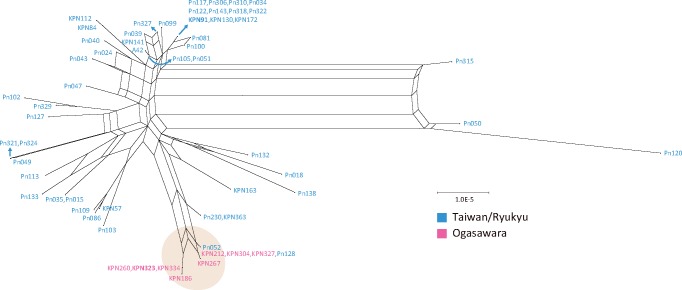
Introns range from 57 to 61 in 9 intron-containing genes in 59 P. noxius isolates (supplementary fig. S5, Supplementary Material online). Phellinus noxius KPN91, which harbored most introns, had an exclusive group II intron of rnl_intron01 with an intact reverse transcriptase domain, suggesting the intron was recently acquired. Completely, loss of introns in some isolates was observed in five intron positions (cox1_intron04, cox1_intron06, cox1_intron07, rnl_intron02, and rnl_intron11). We constructed a phylogeny using the concatenated intronic sequences and showed incongruences to the one constructed based on whole genome variations where two genetical lineages (Taiwan/Ryukyu and Ogasawara) were defined (supplementary fig. S6, Supplementary Material online). To unravel the extent of conflicting signals in the sequences, we constructed a NN phylogenetic network (fig. 5), which is a form of split network where the presences of boxes suggest recombination between connected sequences (Bryant and Moulton 2004). Divergence of two lineages was indeed observed from the network as all eight isolates from Ogasawara island were grouped together. Extensive ambiguous phylogenetic signals were observed within the Taiwanese lineage. Evidence of inter-lineages rearrangements were found as two Taiwanese isolates (Pn052 and Pn128) were clustered together with the Ogasawara isolates and two more (Pn230 and KPN363) were located between the two lineages.
Fig. 5.
—Neighbor-Net phylogenetic network of maximum-likelihood distance inferred from concatenated intronic sequences of 59 Phellinus noxius isolates.
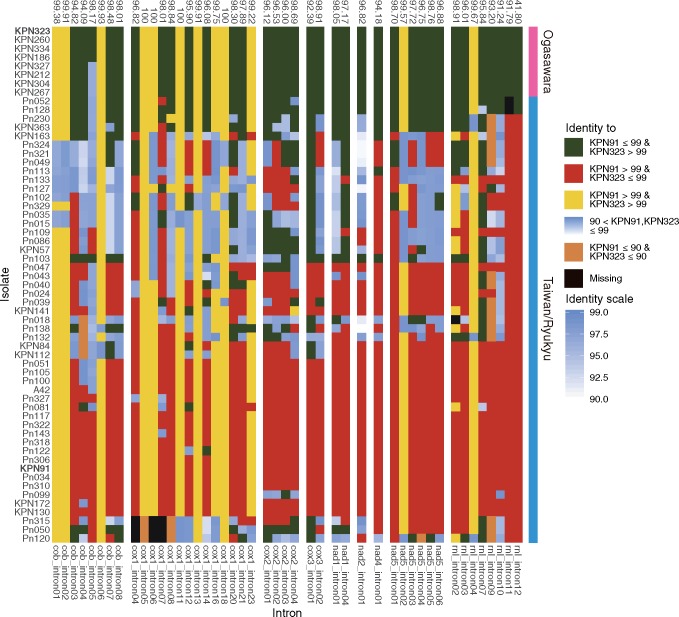
To further visualize the intron dynamics in the Taiwanese/Ryukyu lineage, we first chose one species from Taiwan/Ryukyu (KPN91) and Ogasarawa (KPN323) lineage as a reference and calculated nucleotide identity of all introns against these references. Intron identity of pairwise comparisons between two references is on average 97.1%, with 11 introns identical to each other. The intron with the lowest pairwise identity was rnl_intron12 with 41.8%, and a closer inspection revealed a unique double sized rnl_intron12. We plotted a heatmap of intron identity according to both references and revealed a mosaic pattern of intron distributions. With the double sized rnl_intron12, the introns of the two Taiwanese isolates that were clustered together to the Ogasawara lineage in the intron phylogeny resemble the introns of KPN323 more than that of KPN91 (fig. 6). In contrast, strains belonging to the Taiwan/Ryukyu lineage have revealed striking differences among strains (fig. 6). Three modes were found: 1) strains with introns that completely resemble the Taiwanese/Ryukyu reference, 2) strains with mosaic intron patterns that resemble either Taiwan/Ryukyu or Ogasawara references (i.e., Pn230 and KPN363), and 3) strains with introns that were equally dissimilar to both references. The strains showing the last mode were isolates that have the most ambiguous signals (as represented by boxes) in the NN phylogenetic network. The pattern is likely resulted from extensive recombinations between these isolates. Three Taiwanese isolates (Pn050, Pn120, and Pn315) were particularly distant to the rest of the isolates in the NN phylogenetic network. They showed distinct pattern in introns 4–8 of cox1, with two introns possessing low identity to other strains and absence of intronic sequences at three sites (fig. 6 and supplementary fig. S5, Supplementary Material online). These observations suggest that extensive rearrangement of introns have occurred in the Taiwan/Ryukyu lineage but less so in the Ogasawara lineage.
Fig. 6.
—Heatmap of intron identity against KPN91 and KPN323. Intron positions with all identity >99% against KPN91 are removed. The numbers above the chart indicate the intron identity between KPN91 and KPN323. The tiles on the right of the chart are the lineages defined by intron pattern and intronic phylogeny. The y axis is the order of intronic phylogeny (supplementary fig. S6, Supplementary Material online).
Evidence of Extensive Shuffling between Large Intergene Regions Separated by Gene Clusters
To understand the causes of large intergene that are present in P. noxius, we first defined three large (region 1: nad4–rnl 11.3–25.9 kb, region 2: atp8–cox1 15.3–35.4 kb, and region 3: nad3–rns 6.3–24.9 kb) and a small (region 4s: cox2-cob 4.6–8.9 kb) intergenic regions. A close species P. lamaensis had only 11.3 kb in the regions we defined, comparing to an average of 66.2 kb in P. noxius. MtDNA alignment also showed no similarity with other species in this study, suggesting that these intergenic regions are P. noxius-specific (supplementary fig. S2B, Supplementary Material online).
Frequent intra- and inter-rearrangements between four regions were observed. To validate the existence of rearrangement events, several primers at regions 1–3 were designed (supplementary table S4, Supplementary Material online). PCR for all valid primer combinations were successful and the product sizes were as expected, except two ultra-long PCR products (>13 kb for A1_F/B1_R on Pn113 and >20 kb for F1_F/C1_R on KPN323) due to technical limits (supplementary fig. S7, Supplementary Material online). We first sought if a correlation exists between the intron phylogeny and intergene. However, in the two distinct clades defined by the intron phylogeny, similar synteny was observed in isolates belonging to different clades (supplementary fig. S8A and B, Supplementary Material online). Pairwise identities of the concatenated intergenic sequences were then calculated between all isolates using two different metrics (supplementary fig. S9, Supplementary Material online). Using the gap-compressed identity, that is, not considering indels, isolates were grouped into two lineages where 11 isolates from Taiwan/Ryukyu lineage were also grouped with the Ogasawara isolates (supplementary fig. S9A, Supplementary Material online). In contrast, several isolates form distinct groups if indels were considered (supplementary fig. S9B, Supplementary Material online), suggesting that indels are major contributors to intergenic differences. Interestingly, these isolates were placed in the mosaic group in the intron network (figs. 5 and 6), implying evidences of rearrangement across mitogenomes of these isolates.
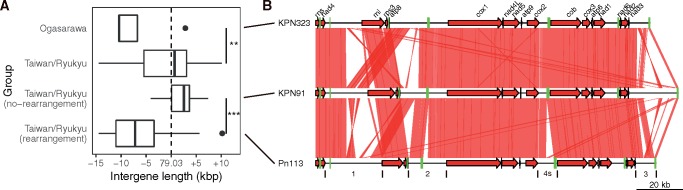
To investigate the effect of rearrangement in mitogenome evolution, we took KPN91 as a reference and sought if any rearrangement event occurred in the four defined intergenic regions (supplementary fig. S10, Supplementary Material online). All eight Ogasawara isolates had similar rearrangement pattern, and the intergene sizes were significantly shorter than that of Taiwan/Ryukyu isolates (P < 0.01; fig. 7). Isolates from the Taiwan/Ryukyu were subsequently classified into two groups based on intergene rearrangement pattern against KPN91 (supplementary fig. S10, Supplementary Material online), which were also significantly different in length (P < 0.001; fig. 7). The length of intergenic spacers in the isolates with rearrangement tended to be smaller than that of the isolates without rearrangement, resulting in a median difference of ∼9.7 kb. On average, 22.17 breakpoints were defined in each isolate when compared with KPN91. To investigate whether these breakpoints were significantly enriched in repetitive regions at intergenes (9.73% in KPN91), we conducted a bootstrapping analysis and revealed observed breakpoints were significantly more commonly found in repeats than random. Out of 28 tested isolates with >10 breakpoints, 26 displayed significance (adjusted P < 0.01; supplementary table S3, Supplementary Material online), suggesting a strong association between rearrangement and repetitive elements at intergenes.
Fig. 7.
—Intergene length distribution and rearrangement events between Ogasawara and Taiwan/Ryukyu lineages. (A) The intergene length between lineages and rearranged or nonrearranged intergene against KPN91 (supplementary fig. S10, Supplementary Material online) are significantly different (**P < 0.01 and ***P < 0.001). The dotted line represents KPN91 intergene length. (B) Rearrangement events in four defined regions between lineages in (A). Red arrows are mitochondrial core genes and green tiles are tRNA genes. Red links between mitogenomes are aligned regions with identity >80%. The intergene boundaries are labeled under Pn113. The plot was generated with genoplotR package in R.
Discussion
In our previous study, we reported four Hymenochaetales genomes and revealed the nature of hyperdiversity of nuclear genomes in two lineages of P. noxius in Taiwan and Japan areas (Chung et al. 2015; Chung et al. 2017). To understand the mitochondrial genomics in Hymenochaetales, we analyzed six mitogenomes in this order. All mitogenomes can be circularized, indicating the high quality of these assemblies. All fungal mitochondrial core genes are present in these species, and the synteny are largely conserved according to their phylogenetic relationships. The overlapping start and stop codons of nad genes have been observed in not only fungi but also animals (Franco et al. 2017; Wang et al. 2019), presumably because of the mRNA transcription of these genes has remained coupled across independent lineages. Only tRNAs were found duplicated and lost, presumably due to the preference of mitochondrial recombination in these regions (Fritsch et al. 2014).
The six mitogenomes varied by at most 3.6-fold in length, from 45.6 kb in P. lamaensis to 163.4 kb in P. noxius. In Organelle Genome Resources NCBI, only six species are larger than P. noxius (Losada et al. 2014; Mardanov et al. 2014; Kanzi et al. 2016; Nowrousian 2016). Gain of introns have been depicted as the main constituents of the enlargement of fungal mitogenomes (Losada et al. 2014; Mardanov et al. 2014; Salavirta et al. 2014; Kanzi et al. 2016). In the case of Hymenochaetales, three out of the six species hold an impressively high number of introns (table 1). In particular, cox1 harbors up to 23 introns in these species which is consistent to previous surveys in fungal mitogenomes as the most intron-containing gene.
Horizontal gene transfer event has been implicated as the main causes and origins of intron gains in fungi (Hausner 2012; Wu et al. 2015). We determined 39 out of 65 shared intron positions, however, only 14 and 3 intron sites were shared in more than 2 or 3 species, respectively. From these sites, we found only two examples of shared intron exhibiting same phylogenetic relationship as the species tree, implying an ancient transposition event. Additional evidence from comparative analysis of multiple species within an order will help reveal whether the retention of such introns has any selective advantage.
Intergenic regions constitute the majority of mitogenomes in all species in this study, consistent with previous findings in yeast mitogenomes where intergenic sequences played a role in genome expansion (Xiao et al. 2017). The initial expansion of intergenic regions may be contributed by the integration of plasmid-like sequences since the few numbers of repetitive elements (Hausner 2003; Férandon et al. 2013). Unlike the introns which need to be spliced precisely for mature mRNA, the integrated plasmid sequences are easily degenerated overtime due to the absence of selection pressure (Hausner 2003; Swithers et al. 2009). Integration of invertron plasmid in mtDNA has been reported in many fungi species including Moniliophthora perniciosa, A.bisporus, and Sclerotinia borealis (Formighieri et al. 2008; Férandon et al. 2013; Mardanov et al. 2014). We found at least one family B DNA polymerase located in intergenic regions in all six species (table 1), but no putative RNA polymerase or invertron-like repetitive elements, suggesting degenerated integrated plasmid.
Our previous investigation on population-scale sequencing of P. noxius nuclear genomes suggests that it is a hypervariable species. Hence, one of the aims of this study was to investigate the evolution history of P. noxius from the mitochondrial perspective. Surprisingly, the analysis of 59 isolates revealed different pace of evolution across coding, intronic, and intergenic regions. The coding regions unusually harbor low variation across the two lineages, suggesting strong purifying selection, that is, arising substitutions were quickly purged from the population. Considering the much higher mutation rate in mitogenome than the nuclear genome (Jung et al. 2012), additional regulatory mechanisms may exist in hypervariable species to reduce potential detrimental effects from the replication of mitogenome. An exception is that a tRNALys(CUU) between nad4 and rnl found deleted in several isolates (supplementary fig. S8C and D, Supplementary Material online). Phellinus noxius mitogenome uses the AAG codon at a very low frequency (<2%) to code for lysine. The lack of mitochondria-encoded tRNA is pretty common across all eukaryotes and may be compensated via different wobble rules, RNA-editing or import from cytoplasm (Salinas-Giegé et al. 2015).
A previous investigation of intraspecific variations on three species of yeast determined introns contributed the highest variance in genome size (Xiao et al. 2017). Our intron analyses of two lineages of P. noxius revealed high similarity but extensive rearrangements of introns among strains of different clades. Our results add up to the expanding list of basidiomycete mitogenomes that have displayed evidences of hybridization and recombination (Xu et al. 2009; Wang et al. 2017). There are two possible scenarios that can account for such observations in a basidiomycete during hyphal fusion between strains of two mitotypes: intron homing or homologous recombination (Haugen et al. 2005; Wu et al. 2015; Leducq et al. 2017). We observed fewer gain and loss of introns at population level so recombination may be a more predominant process, which has been documented in various fungal species (Leducq et al. 2017). Unfortunately, high similarity of intron sequences impedes further recombination-detection based analyses. Future investigations should focus on determination of mitochondrial inheritance mode and the frequency and mechanisms involved in intron rearrangements using artificial hybrids.
In plants (Sloan et al. 2012) and animals (Marlétaz et al. 2017), huge intraspecific variations in intergenic regions have been documented as extremes or unusual atypical to mitogenome evolution. Such large intraspecific variations appear to be more common in fungi which were observed in our investigation, Cryptococcus (Litter et al. 2005; Xu et al. 2009) and other closely related fungal species (Burger et al. 2003; Aguileta et al. 2014; Himmelstrand et al. 2014). We have observed contrasting differences in intergene length between strains of different lineages as well as within a lineage. These findings suggest that, when compared with closely related species, intergene variations are currently driving intraspecific mitogenomic variation in P. noxius and may eventually lead to larger intergene variations. The absolute difference in intergene length led to presence of additional sequences in some strains. Possible origins of these sequences may be a result of horizontal gene transfer (gain), or rearrangement and accumulations of mutations (gain or loss). It is challenging to determine which scenario as these sequences share no homology to publicized genomes. We speculate the later scenario may be more likely as repeats were found associated with rearrangement events identified within the strains of Taiwan/Ryukyu lineage. Intriguingly, these strains were found to possess introns with evidences of rearrangements. Similar extents of rearrangement were reported across the Dikarya and have led to gene order differences (Aguileta et al. 2014; Li et al. 2018). Recombination, break-induced replication (Christensen 2013) coupled with repeats and several nuclear genes (Shedge et al. 2007; Arrieta-Montiel et al. 2009) and accelerated mitochondrial mutation rate have all been proposed to explain the evolution of noncoding sequences in mitogenomes. It remains to be determined how repeats contribute to intergenic rearrangement and expansion without perturbing gene order in mitogenome of P. noxius.
In conclusion, we have revealed frequent rearrangements at intergenic regions play a major role in shaping and extending the mtDNA in Basidiomycetes at inter- and intra-species level. The extreme contrast between nuclear genome and coding region of mitogenome suggests strong purifying selection acting on these genes. Basidiomycetes can be excellent models in understanding mtDNA inheritance. A comprehensive sampling and detailed comparison across mitogenomes will help delineate the relationship between mating and mitochondrial diversity.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Author Contributions
I.J.T. conceived the study. H.-M.K., C.-Y.I.L., T.J.L., and C.-L.C. established the strains collection, carried out experiments, and sequencing. H.-H.L. carried out comparative analysis. I.J.T. and H.-H.L. wrote the manuscript. This work was supported by Academia Sinica of Taiwan (AS-CDA-107-L01 to I.J.T) and Taiwan Ministry of Science and Technology (Grant No. 105-2628-B-001-002-MY3 to IJT). T.J.L was supported by the doctorate fellowship of Taiwan International Graduate Program-Biodiversity, Academia Sinica of Taiwan. C.-L.C was supported by the Office of General Affairs, National Taiwan University.
Supplementary Material
Data deposition: This project has been deposited at NCBI under the accession CM008263 and MK623257–MK623261.
Literature Cited
- Aguileta G, et al. 2014. High variability of mitochondrial gene order among fungi. Genome Biol Evol. 6(2):451–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiba M, et al. 2015. Genetic differentiation and spatial structure of Phellinus noxius, the causal agent of brown root rot of woody plants in Japan. PLoS One 10(10):e0141792.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ann P-J, Chang T-T, Ko W-H.. 2002. Phellinus noxius brown root rot of fruit and ornamental trees in Taiwan. Plant Dis. 86(8):820–826. [DOI] [PubMed] [Google Scholar]
- Arrieta-Montiel MP, Shedge V, Davila J, Christensen AC, Mackenzie SA.. 2009. Diversity of the Arabidopsis mitochondrial genome occurs via nuclear-controlled recombination activity. Genetics 183(4):1261–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basse CW. 2010. Mitochondrial inheritance in fungi. Curr Opin Microbiol. 13(6):712–719. [DOI] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27(2):573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger KH, Yaffe MP.. 2000. Mitochondrial DNA inheritance in Saccharomyces cerevisiae. Trends Microbiol. 8(11):508–513. [DOI] [PubMed] [Google Scholar]
- Bryant D, Moulton V.. 2004. Neighbor-Net: an agglomerative method for the construction of phylogenetic networks. Mol Biol Evol. 21(2):255–265. [DOI] [PubMed] [Google Scholar]
- Bullerwell CE, Leigh J, Forget L, Lang BF.. 2003. A comparison of three fission yeast mitochondrial genomes. Nucleic Acids Res. 31(2):759–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G, Gray MW, Franz Lang B.. 2003. Mitochondrial genomes: anything goes. Trends Genet. 19(12):709–716. [DOI] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T.. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25(15):1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casselton LA, Kües U.. 2007. The origin of multiple mating types in the model mushrooms Coprinopsis cinerea and Schizophyllum commune In: Heitman J, Kronstad JW, Taylor JW, Casselton LA, editors. Sex in fungi: molecular determination and evolutionary implications. Washington: American Society of Microbiology. p. 283–300. [Google Scholar]
- Christensen AC. 2013. Plant mitochondrial genome evolution can be explained by DNA repair mechanisms. Genome Biol Evol. 5(6):1079–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C-L, et al. 2015. The genetic structure of Phellinus noxius and dissemination pattern of brown root rot disease in Taiwan. PLoS One 10(10):e0139445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C-L, et al. 2017. Comparative and population genomic landscape of Phellinus noxius: a hypervariable fungus causing root rot in trees. Mol Ecol. 26(22):6301–6316. [DOI] [PubMed] [Google Scholar]
- de la Bastide PY, Horgen PA.. 2003. Mitochondrial inheritance and the detection of non-parental mitochondrial DNA haplotypes in crosses of Agaricus bisporus homokaryons. Fungal Genet Biol. 38(3):333–342. [DOI] [PubMed] [Google Scholar]
- Deng Y, et al. 2018. Comparison of the mitochondrial genome sequences of six Annulohypoxylon stygium isolates suggests short fragment insertions as a potential factor leading to larger genomic size. Front Microbiol. 9:380.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G.. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, et al. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedler M, Luh K-S, Stelter K, Nieto-Jacobo F, Basse CW.. 2009. The a2 mating-type locus genes lga2 and rga2 direct uniparental mitochondrial DNA (mtDNA) inheritance and constrain mtDNA recombination during sexual development of Ustilago maydis. Genetics 181(3):847–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Férandon C, Xu J, Barroso G.. 2013. The 135 kbp mitochondrial genome of Agaricus bisporus is the largest known eukaryotic reservoir of group I introns and plasmid-related sequences. Fungal Genet Biol. 55:85–91. [DOI] [PubMed] [Google Scholar]
- Finn RD, et al. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44(D1):D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floudas D, et al. 2012. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336(6089):1715–1719. [DOI] [PubMed] [Google Scholar]
- Formighieri EF, et al. 2008. The mitochondrial genome of the phytopathogenic basidiomycete Moniliophthora perniciosa is 109 kb in size and contains a stable integrated plasmid. Mycol Res. 112(Pt 10):1136–1152. [DOI] [PubMed] [Google Scholar]
- Franco MEE, et al. 2017. The mitochondrial genome of the plant-pathogenic fungus Stemphylium lycopersici uncovers a dynamic structure due to repetitive and mobile elements. PLoS One 12(10):e0185545.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch ES, Chabbert CD, Klaus B, Steinmetz LM.. 2014. A genome-wide map of mitochondrial DNA recombination in yeast. Genetics 198(2):755–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugen P, Simon DM, Bhattacharya D.. 2005. The natural history of group I introns. Trends Genet. 21(2):111–119. [DOI] [PubMed] [Google Scholar]
- Hausner G. 2003. Fungal mitochondrial genomes, plasmids and introns In: Arora DK, Khachatourians GG, editors. Fungal Genomics. Amsterdam: Elsevier. p. 101–131. [Google Scholar]
- Hausner G. 2012. Introns, mobile elements, and plasmids. Berlin/Heidelberg (Germany: ): Springer; p. 329–357. [Google Scholar]
- Hawksworth DL, Lücking R.. 2017. Fungal diversity revisited: 2.2 to 3.8 million species. Microbiol Spectr.5(4). [DOI] [PubMed] [Google Scholar]
- Hepting GH. 1971. Diseases of forest and shade trees of the United States. Washington (DC: ): U.S. Department of Agriculture Forest Service. [Google Scholar]
- Himmelstrand K, Olson Å, Durling MB, Karlsson M, Stenlid J.. 2014. Intronic and plasmid-derived regions contribute to the large mitochondrial genome sizes of Agaricomycetes. Curr Genet. 60(4):303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hintz W, Anderson JB, Horgen PA.. 1988. Nuclear migration and mitochondrial inheritance in the mushroom Agaricus bitorquis. Genetics 119(1):35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T, Horgen PA.. 1993. Further characterization of a large inverted repeat in the mitochondrial genomes of Agaricus bisporus (=A. brunnescens) and related species. Curr Genet. 23(3):228–233. [DOI] [PubMed] [Google Scholar]
- Jin T, Horgen PA.. 1994. Uniparental mitochondrial transmission in the cultivated button mushroom, Agaricus bisporus. Appl Environ Microbiol. 60(12):4456–4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung PP, Friedrich A, Reisser C, Hou J, Schacherer J.. 2012. Mitochondrial genome evolution in a single protoploid yeast species. G3 (Bethesda) 2:1103–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzi AM, Wingfield BD, Steenkamp ET, Naidoo S, van der Merwe NA.. 2016. Intron derived size polymorphism in the mitochondrial genomes of closely related Chrysoporthe species. PLoS One 11(6):e0156104.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski MI, et al. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19(9):1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, et al. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5(2):R12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslett D, Canbäck B.. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32(1):11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leducq J-B, et al. 2017. Mitochondrial recombination and introgression during speciation by hybridization. Mol Biol Evol. 34(8):1947–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34(18):3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, et al. 2018. Comparative mitogenomics reveals large-scale gene rearrangements in the mitochondrial genome of two Pleurotus species. Appl Microbiol Biotechnol. 102:6143–6153. [DOI] [PubMed] [Google Scholar]
- Litter J, Keszthelyi A, Hamari Z, Pfeiffer I, Kucsera J.. 2005. Differences in mitochondrial genome organization of Cryptococcus neoformans strains. Antonie Van Leeuwenhoek 88(3-4):249–255. [DOI] [PubMed] [Google Scholar]
- Losada L, et al. 2014. Mobile elements and mitochondrial genome expansion in the soil fungus and potato pathogen Rhizoctonia solani AG‐3. FEMS Microbiol Lett. 352(2):165–173. [DOI] [PubMed] [Google Scholar]
- Lowe TM, Chan PP.. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardanov AV, Beletsky AV, Kadnikov VV, Ignatov AN, Ravin NV.. 2014. The 203 kbp mitochondrial genome of the phytopathogenic fungus Sclerotinia borealis reveals multiple invasions of introns and genomic duplications. PLoS One 9(9):e107536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlétaz F, Le Parco Y, Liu S, Peijnenburg K.. 2017. Extreme mitogenomic variation in natural populations of Chaetognaths. Genome Biol Evol . 9(6):1374–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis S, Madden TL.. 2004. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 32(Web Server issue):W20–W25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min B, et al. 2015. Genome sequence of a white rot fungus Schizopora paradoxa KUC8140 for wood decay and mycoremediation. J Biotechnol. 211:42–43. [DOI] [PubMed] [Google Scholar]
- Mullineux S-T, Costa M, Bassi GS, Michel F, Hausner G.. 2010. A group II intron encodes a functional LAGLIDADG homing endonuclease and self-splices under moderate temperature and ionic conditions. RNA 16(9):1818–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowrousian M. 2016. Complete mitochondrial genome sequence of the Pezizomycete Pyronema confluens. Genome Announc. 4(3):e107536.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahashi N, Akiba M, Ishihara M, Ota Y, Kanzaki N.. 2012. Brown root rot of trees caused by Phellinus noxius in the Ryukyu Islands, subtropical areas of Japan. For Pathol. 42(5):353–361. [Google Scholar]
- Salavirta H, et al. 2014. Mitochondrial genome of Phlebia radiata is the second largest (156 kbp) among fungi and features signs of genome flexibility and recent recombination events. PLoS One 9(5):e97141.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas-Giegé T, Giegé R, Giegé P.. 2015. tRNA biology in mitochondria. Int J Mol Sci. 16(3):4518–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandor S, Zhang Y, Xu J.. 2018. Fungal mitochondrial genomes and genetic polymorphisms. Appl Microbiol Biotechnol. 102(22):9433–9448. [DOI] [PubMed] [Google Scholar]
- Shedge V, Arrieta-Montiel M, Christensen AC, Mackenzie SA.. 2007. Plant mitochondrial recombination surveillance requires unusual RecA and MutS homologs. Plant Cell 19(4):1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Müller K, McCauley DE, Taylor DR, Storchová H.. 2012. Intraspecific variation in mitochondrial genome sequence, structure, and gene content in Silene vulgaris, an angiosperm with pervasive cytoplasmic male sterility. New Phytol. 196(4):1228–1239. [DOI] [PubMed] [Google Scholar]
- Soorni A, Haak D, Zaitlin D, Bombarely A.. 2017. Organelle_PBA, a pipeline for assembling chloroplast and mitochondrial genomes from PacBio DNA sequencing data. BMC Genomics. 18(1):49.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swithers KS, Senejani AG, Fournier GP, Gogarten JP.. 2009. Conservation of intron and intein insertion sites: implications for life histories of parasitic genetic elements. BMC Evol Biol. 9:303.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel T, Michalek W, Varshney R, Graner A.. 2003. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare). Theor Appl Genet. 106(3):411–422. [DOI] [PubMed] [Google Scholar]
- Toor N, Zimmerly S.. 2002. Identification of a family of group II introns encoding LAGLIDADG ORFs typical of group I introns. RNA 8(11):1373–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A, et al. 2012. Primer3—new capabilities and interfaces. Nucleic Acids Res. 40(15):e115.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelz K, et al. 2013. Transmission of hypervirulence traits via sexual reproduction within and between lineages of the human fungal pathogen Cryptococcus gattii. PLoS Genet. 9(9):e1003771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9(11):e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhang S, Li J-H, Zhang Y-J.. 2018. Mitochondrial genome, comparative analysis and evolutionary insights into the entomopathogenic fungus Hirsutella thompsonii. Environ Microbiol. 20(9):3393–3405. [DOI] [PubMed] [Google Scholar]
- Wang P, et al. 2017. Frequent heteroplasmy and recombination in the mitochondrial genomes of the basidiomycete mushroom Thelephora ganbajun. Sci Rep. 7(1):1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wilson A, Xu J.. 2015. Mitochondrial DNA inheritance in the human fungal pathogen Cryptococcus gattii. Fungal Genet Biol. 75:1–10. [DOI] [PubMed] [Google Scholar]
- Wang Z, et al. 2019. The complete mitochondrial genome of Parasesarma pictum (Brachyura: Grapsoidea: Sesarmidae) and comparison with other Brachyuran crabs. Genomics. 111(4):799–807. [DOI] [PubMed] [Google Scholar]
- Wheeler DL, et al. 2003. Database resources of the National Center for Biotechnology. Nucleic Acids Res. 31(1):28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AJ, Xu J.. 2012. Mitochondrial inheritance: diverse patterns and mechanisms with an emphasis on fungi. Mycology 3(2):158–166. [Google Scholar]
- Wolf K, Burger G, Lang B, Kaudewitz F.. 1976. Extrachromosomal inheritance in Schizosaccharomyces pombe. I. Evidence for an extrakaryotically inherited mutation conferring resistance to antimycin. Mol Gen Genet. 144(1):67–73. [PubMed] [Google Scholar]
- Wolters JF, Chiu K, Fiumera HL.. 2015. Population structure of mitochondrial genomes in Saccharomyces cerevisiae. BMC Genomics. 16:451.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Buljic A, Hao W.. 2015. Extensive horizontal transfer and homologous recombination generate highly chimeric mitochondrial genomes in yeast. Mol Biol Evol. 32(10):2559–2570. [DOI] [PubMed] [Google Scholar]
- Wu TD, Watanabe CK.. 2005. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21(9):1859–1875. [DOI] [PubMed] [Google Scholar]
- Xiao S, Nguyen DT, Wu B, Hao W.. 2017. Genetic drift and indel mutation in the evolution of yeast mitochondrial genome size. Genome Biol Evol. 9(11):3088–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, He L.. 2015. Current perspectives on mitochondrial inheritance in fungi. Cell Health and Cytoskelet. 7:143–112. [Google Scholar]
- Xu J, Wang P.. 2015. Mitochondrial inheritance in basidiomycete fungi. Fungal Biol Rev. 29(3-4):209–219. [Google Scholar]
- Xu J, Yan Z, Guo H.. 2009. Divergence, hybridization, and recombination in the mitochondrial genome of the human pathogenic yeast Cryptococcus gattii. Mol Ecol. 18(12):2628–2642. [DOI] [PubMed] [Google Scholar]
- Xu J, Zhang Y, Pun N.. 2013. Mitochondrial recombination in natural populations of the button mushroom Agaricus bisporus. Fungal Genet Biol. 55:92–97. [DOI] [PubMed] [Google Scholar]
- Xu J, et al. 2000. Uniparental mitochondrial transmission in sexual crosses in Cryptococcus neoformans. Curr Microbiol. 40(4):269–273. [DOI] [PubMed] [Google Scholar]
- Yan Z, Xu J.. 2003. Mitochondria are inherited from the MATa parent in crosses of the basidiomycete fungus Cryptococcus neoformans. Genetics 163(4):1315–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, et al. 2018. Deletion of the sex-determining gene SXI1α enhances the spread of mitochondrial introns in Cryptococcus neoformans. Mob DNA 9:24.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Zhai B, Lin X, Idnurm A.. 2013. Congenic strains for genetic analysis of virulence traits in Cryptococcus gattii. Infect Immun. 81(7):2616–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.