Abstract
Robotic high-throughput compound screening (HTS) and, increasingly, DNA-encoded library (DEL) screening are driving bioactive chemical matter discovery in the post-genome era. HTS enables activity-based investigation of highly complex targets using static compound libraries. Conversely, DEL grants efficient access to novel chemical diversity, although screening is limited to affinity-based selections. Here, we describe an integrated droplet-based microfluidic circuit that directly screens solid-phase DELs for activity. An example screen of a 67,100-member library for inhibitors of the phosphodiesterase autotaxin yielded 35 high-priority structures for nanomole-scale synthesis and validation (20 active), guiding candidate selection for synthesis at scale (5/5 compounds with IC50s 4–10 μM). We further compared activity-based hits with those of an analogous affinity-based DEL selection. This miniaturized screening platform paves the way toward applying DELs to more complex targets (signaling pathways, cellular response), and represents a distributable approach to small molecule discovery.
TOC Graphic
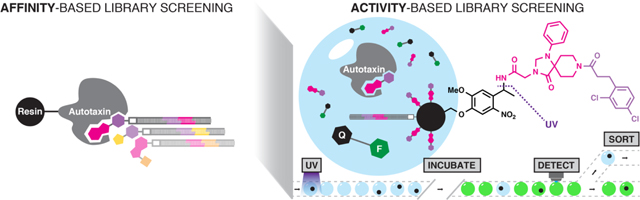
Introduction
High-throughput screening (HTS) — the automated mining of compound libraries for biologically active molecules — has been the primary tool for discovering novel, bioactive chemical matter.1,2 The libraries that fuel these chemical genetics efforts contain compounds of various origins, but many are derivatives of historical drug leads that do not span the appropriate chemical space to investigate increasingly complex target classes, such as protein-protein interactions,3 RNA secondary structures,4 or notoriously difficult targets, such as bacterial growth inhibition.5 Expanding HTS compound libraries by serial synthesis to include such matter is prohibitively expensive and exacerbates the logistics of compound storage.
The search for new and scalable chemical diversity for screening has driven interest in inexpensive, combinatorially-generated chemical libraries, resulting in the now-widespread implementation of DNA-encoded libraries (DELs). DELs, the product of encoded combinatorial synthesis,6 contain millions of distinct small molecules attached to a DNA sequence that encodes the molecule’s synthetic history. Affinity selection and sequencing identifies DEL members that bind a target of interest,7–10 an approach that has yielded ligands for various target classes, including kinases,11 hydrolases,12 cell-surface receptors,13,14 and protein-protein interactions,15 and granted access to macrocyclic peptides and other underrepresented chemical diversity.16–18 However, DEL technology is limited to affinity-based hit identification, making many important target classes (ion channels, receptors, transcription factors, protein complexes, and signal-transduction pathways) refractory to investigation with DELs due to insolubility, instability, or intrinsic disorder.19 These classes still require activity-based biochemical or cellular assays of target response to a single library member in isolation. Such measurements are not possible for DELs because they exist only as complex mixtures.
To exploit the dynamism and chemical diversity of DEL in an activity-based screen, many copies of an individual library member must be allocated its own discrete volume, and this must be parallelized for many library members. “One-bead-one-compound” (OBOC) solid-phase combinatorial synthesis is an efficient strategy for parsing library members into discrete samples using microbeads20–22 where each OBOC library microbead displays 10−15–10−9 moles of an individual library member depending on bead size.23–26 Efficiently distributing these beads into microscopic is readily accomplished using droplet microfluidics,27,28 which achieves rapid and homogenous generation of picoliter volumes, and generates appropriate concentrations for screening from only femtomoles of material.
These scalable strategies for library member parsing and compartmentalization unite here as the foundation of a next-generation approach to drug lead discovery. We demonstrate the synthesis of a 67,100-member DNA-encoded compound library on miniaturized beads29,30 and the analysis of the library using a highly integrated bead-screening microfluidic circuit. The miniaturized HTS platform was used to discover novel inhibitors of a clinically relevant drug target, the phosphodiesterase autotaxin (ATX).31,32 The screen identified 35 high-priority hit compounds comprising multiple chemically distinct scaffolds that validated as active ATX inhibitors (20/35) and suggested 5 structures for scaled synthesis, all 5 of which exhibited IC50 ≤ 10 μM. This new activity-based approach to DEL screening opens the possibility of tackling more complicated biochemical and cellular assays using DEL-derived chemical matter, and represents a platform that could democratize small molecule discovery.
Results and Discussion
The library screening microfluidic circuit architecture (Figure 1) integrates components for water-in-oil flow focusing droplet generation,28 bead loading and compound cleavage,33 content mixing,34 splitting,35 incubation,36 and fluorescence-activated sorting37–39 (Movie S1). The device generated droplets at 60 Hz with 12.5-min assay incubation time, 4% dispersion, and 30,000 beads per hour screening throughput (14% bead occupancy). Droplets of fluorometric ATX activity assay were formed by combining two input streams, one containing the ATX enzyme target and the other containing fluorogenic ATX substrate.36 ATX activity assay performance was statistically satisfactory for HTS (Z′ = 0.71 in droplets) using a positive control ATX inhibitor (Figure S1).40
Figure 1. Integrated microfluidic screening architecture.
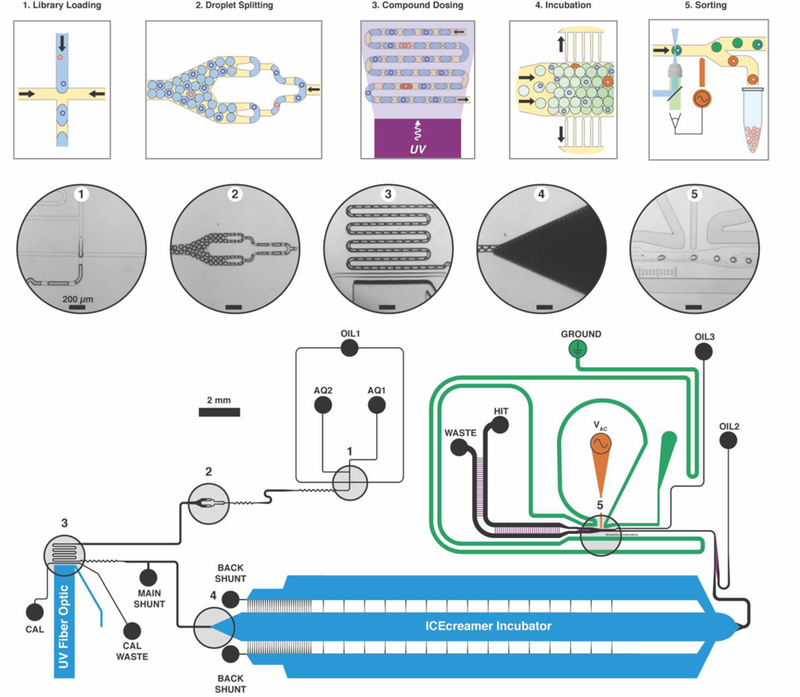
Library beads enter the circuit together with ATX activity-based fluorogenic substrate (AQ1) and mix with enzyme (AQ2) immediately upstream of the water-in-oil (OIL1) flow-focusing junction (1) where droplets form (15 Hz). Flow continues toward the droplet splitter (2), which splits each droplet in half twice (60 Hz). Compound is released from beads into the ATX activity assay droplets as they traverse a serpentine photochemical reactor illuminated with a 365-nm waveguide-coupled LED (3). Oil drains from the droplet flow, packing droplets in the ICEcreamer (4) for assay incubation. At the incubator exit, two oil flows space (OIL2) and focus (OIL3) the droplet stream for analysis. Laser-induced fluorescence assay detection occurs upstream of the droplet sorting junction (5). Hit droplets are statistically significantly less fluorescent than the overall instantaneous droplet population, and trigger a high-voltage AC pulse to the working electrode (VAC) that electrokinetically forces the hit droplet toward the electrode and into the hit collection flow. Channel heights depicted in purple and blue are 30 μm and 250 μm, respectively. All other channel heights are 50 μm.
A solid-phase DNA-encoded library of drug-like small molecules was prepared as input for the screening device via two-cycle split-and-pool combinatorial synthesis.39 The first cycle of chemical diversification contained 110 amino acids (AA) and the second cycle of diversification contained 610 carboxylic acid (CA) caps, yielding 67,100 unique library members. Library bead DNA loading was quantified before and after synthesis (100,000 and 40,000 amplifiable molecules of DNA per bead, respectively). UV-mediated photochemical cleavage of encapsulated library beads during screening released the library members as the primary amides (Figure 2). Computational analysis of library chemical property distributions included median hydrophobicity (1.4), molecular weight (389 Da), hydrogen bond acceptors (7) and donors (2), polar surface area (113 Å2), and number of rotatable bonds (6) (Figure S2).41 In single-bead library quality control (QC) analysis, which compares mass spectral (MS) analysis with DNA sequence-predicted mass,30,42 19/22 QC beads were structure-sequence matched (Table S1).
Figure 2. Library properties.
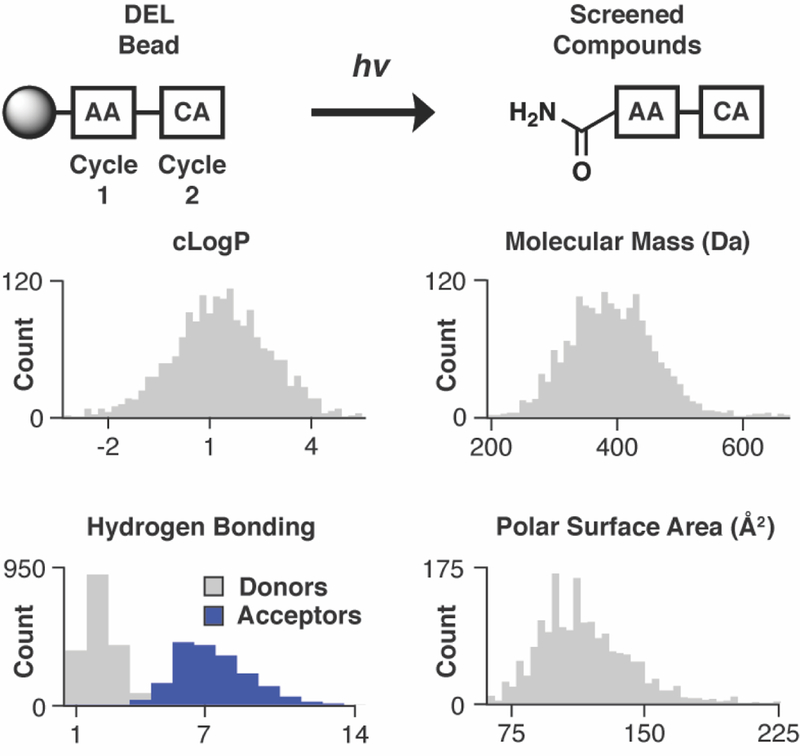
The library was generated with two cycles of acylation chemistry with amino acid (AA) and carboxylic acid (CA) monomers, respectively. Photochemical cleavage releases compound from the library bead as a primary amide. Physicochemical property analysis of photochemically cleaved compounds included hydrophobicity (cLogP), molar mass, hydrogen bond donors/acceptors, and polar surface area. Most compounds analyzed (1,738/1,766) are ‘drug-like’.
One of the major advances of DEL-based combinatorial synthesis is the scope of compounds present in the average library. Unlike the libraries of combinatorial chemistry past, DELs contain ‘drug-like’ diversity by design,43 an observation that also plays out for solid-phase DELs. In the library design of this report, 98% conform to the ‘Rule of 5’44 and 83% fulfill Veber criteria.45 Significantly, the calculated distributions of solid-phase DEL-derived library members directly reflect the properties of the screened molecules as each library member is released from the bead for activity assay. In contrast, solution-phase DEL members are covalently bound to a DNA molecule during affinity selection, complicating library chemical property calculations.
Using the validated solid-phase DEL and droplet-scale ATX assay, we conducted an activity-based screen for compounds that inhibit ATX. During each 3-h screen, the device generated and analyzed 700,000 droplets using 3 μg ATX. For each droplet, in-house generated LabVIEW code calculated the median-smoothed droplet fluorescence values, and performed dynamic statistical hypothesis testing to identify hits by comparing the current droplet’s fluorescence to the mean of the previous 1,000 droplets’ fluorescence. If the current droplet’s fluorescence value fell 4 standard deviations below this population mean, the software identified the droplet as a hit and briefly energized the sorting electrode to collect it (Figure 3A). Droplet generation and hit identification rates were uniform for the duration of the screen, while the bead introduction rate decreased over the course screening (Figure 3B). Assay signal was consistent during screening; 0.7% of droplets fell below the sort threshold and were identified as hits. Most droplets were unoccupied, reflecting the uninhibited ATX activity baseline (Figure 3C). Hit droplet collection images (Figure 3D, Figure S3) verified the sorting efficiency, bead-occupied droplet enrichment, and droplet inhibition. Hit droplet fluorescence was visibly attenuated compared to uninhibited droplets. Most droplets contained a single library bead, though multiply-occupied and unoccupied droplets were also observed. In total, ~7,000 hits were collected from combined screens of ~3 library equivalents (~200,000 beads).
Figure 3. Microfluidic activity-based screening data.
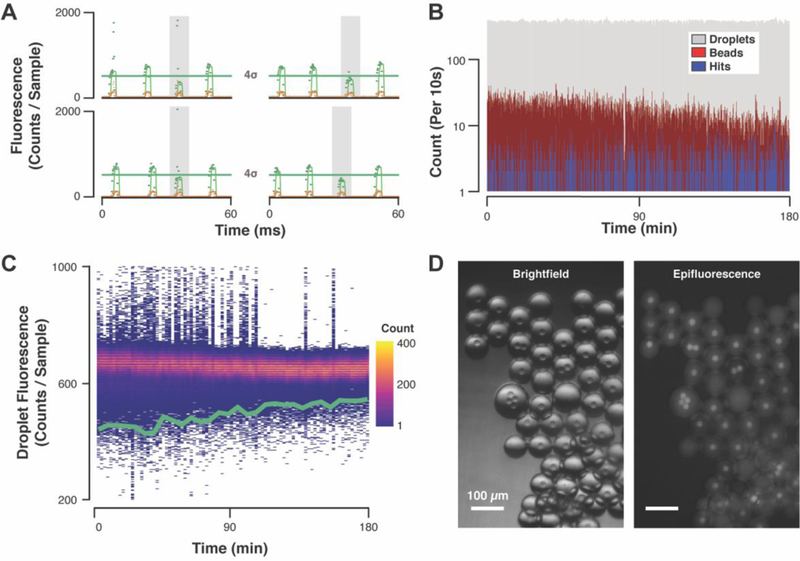
(A) Laser-induced fluorescence traces of hit droplets contain raw data for ATX activity assay signal (520 nm, green points), raw internal standard signal (570 nm, orange points), and median-smoothed data for both channels (green and orange lines). The autofluorescent library beads occasionally pass through the confocal volume, resulting in fluorescence signal spikes (see top and bottom left traces) that are smoothed to identify droplet peak fluorescence, but also used to monitor bead introduction. If a droplet peak fluorescence falls below the sorting threshold (green lines), it is sorted into the hit collection. For each droplet, the mean (μ) and standard deviation (σ) of the previous 1,000 droplet peak fluorescence values is calculated to establish the sorting threshold (μ - 4σ). (B) Droplet generation and hit droplet identification rates were uniform while bead-induced fluorescence signal spikes steadily decreased over the 3 h screen. (C) Hit distribution and negative droplet fluorescence uniformity are visualized in a heat map of binned droplet fluorescence signals (120-s bins) overlaid with sorting threshold (green line). (D) An example hit collection imaged in brightfield and epifluorescence (λex = 470 nm, λem = 525 nm) contains droplets of variably attenuated activity assay signal.
The aggregated screening performance data agreed both with known device operation characteristics and anticipated behavior of a diverse compound library. Droplet generation remained consistent while bead introduction slowed during screening. The bead introduction rate diminishes over time because the beads sediment, but bead introduction rate variability did not impact the assay signal uniformity. Bead-occupied droplets’ fluorescence overlays with the overall population (Figure S4), indicating that the presence of a bead alone was insufficient to inhibit ATX activity. The library, a diverse collection of structures, variably affected ATX activity as reflected in the spread of attenuated-fluorescence droplets. The hit droplet visual inspection is consistent with this observation: hit droplet fluorescence is heterogeneously attenuated. Most hit collection droplets contained only one bead (consistent with ~0.1 bead/droplet mean occupancy), but some droplets were unoccupied. Unoccupied hit droplets could have resulted from sorting errors, but based on the low error rate of sorting39 and the lower fluorescence intensity of the unoccupied droplets in hit collections, it is more likely that hydrophobic, active compounds partitioned between neighboring droplets during incubation.46
DNA from the ~7,000-bead hit collection was amplified, processed for next-generation DNA sequencing, and decoded to yield each bead’s synthesis history. Unique molecular identifiers (UMIs) were used to reject sequencing noise and bead-specific barcodes were used to count replicate hits (beads displaying identical compounds). The data were filtered by UMI complexity and count to yield a final set of 7,100 unique compound-encoding sequences, and then aggregated by monomer usage and replicate class (k class)47 with 35 high-priority hits falling into k classes 3 (33), 4 (1), and 5 (1).39,48 From the bottom-up monomer aggregation analysis, 5 cycle 1 AA monomers are observed in 2 hits, and 3 cycle 2 CA monomers are observed in at least 2 hits (Figure 4A). All 35 hits were prepared via parallel solid-phase synthesis (100 nmol scale); 27/35 compounds yielded the expected product as determined by MS (Table S2). All compounds were evaluated in an ATX inhibition assay using library synthesis and screening conditions, including photocleavage. Most resynthesized compounds (20/35) inhibited ATX activity (< 85 %) in assays of the crude photocleavage reaction supernatants (Figure 4B). Five of the most inhibitory compounds (1–5) were resynthesized at scale, fully characterized, and verified to be active (respective IC50’s in μM: 3.5 ± 0.7, 8 ± 2, 8 ± 1, 10 ± 2, 5 ± 3).
Figure 4. Hit collection deconvolution and validation.
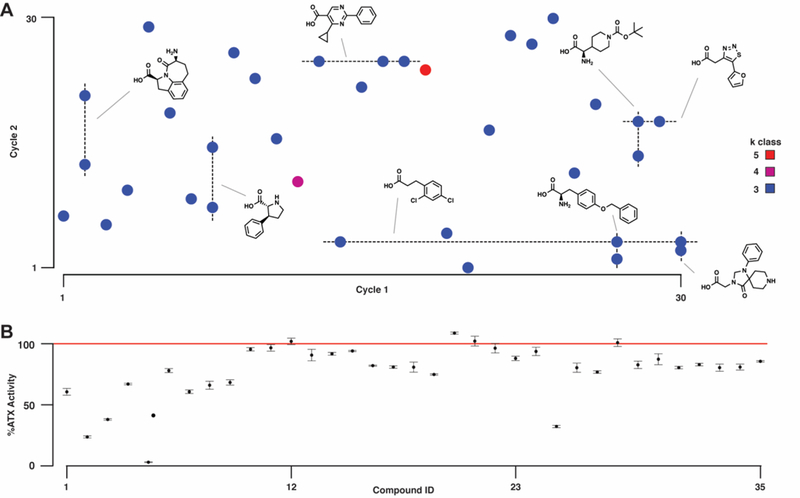
(A) Hit bead sequences were decoded to monomer structures based on their synthesis-encoding region. Replicate hits exhibit a lower false discovery rate (FDR) and thus are statistically more likely to validate as active molecules. Higher numbers of replicates (k class) correlate with exponentially lower FDR. Dashed lines indicate monomer conservation in cycle 1 (vertical) and cycle 2 (horizontal). Monomer conservation in k class ≥ 3 hits is shown. (B) Thirty-five compounds were prepared via parallel solid-phase synthesis, structure-validated by mass spectral analysis, and irradiated (λ = 360 nm) to release compound from the beads. Photocleavage reaction supernatants were then incubated with ATX and the samples subsequently assayed for enzymatic activity using conditions analogous to the droplet-scale ATX assay. Intervals represent standard deviation of the mean.
Analogous to conventional affinity-based DEL screens, the bottom-up analysis of this activity-based screen’s hit collection also indicates monomer conservation. Hit pool monomer conservation may suggest exploitable lead fragments,10 but extensive conservation can also be the result of synthesis truncation.49 Several of the conserved monomers are fragments of known ATX-active compounds. For example, the dichlorobenzene moiety was central to a series of potent ATX inhibitors.50 The triazaspirone appears in inhibitors of related phospholipase D family enzymes,51 and some derivatives have been found to inhibit ATX.43 Conservation among the other indicated fragments may similarly indicate a monomer’s suitability for lead development.
Initial hit compound validation and synthesis was rapid, employing protocols analogous to those of library synthesis and screening. The photochemically cleaved crude product was validated by MS and used for assay with no purification. MS validation suggested that 5/15 inactive compounds were not actually synthesized. The off-bead inhibition results successfully highlighted promising candidates for synthesis at scale (5/5 resynthesized and characterized compounds validated). This semi-quantitative compound validation approach is not conclusive as to whether the remaining 10 compounds are inactive, or if other factors impeded their activity (low synthetic yield, inefficient photocleavage, etc.).52
The hits from the ATX activity-based solid-phase DEL screen were compared to hits identified in a conventional DEL. Affinity selection was performed using a DEL containing 866,250 total compounds,43 13,260 of which were also present in the solid-phase DEL. The 7,423 compounds that enriched during affinity selection were clustered by chemical similarity (1,003 clusters) and ranked by cumulative cluster enrichment. Each ATX hit from the activity-based screen was assigned to one of the 1,003 clusters based on maximum chemical similarity (Figure 5). The solid-phase DEL ATX screening hits spanned a broad range of affinity selection cumulative cluster enrichments. Several hits were highly similar (Tanimoto similarity > 0.7) or identical (1) to members that enriched most during affinity selection. The 5 resynthesized hits’ potency did not deviate greatly (3.5–10 μM), but hit structures exhibited large variation in cluster chemical similarity (0.63 –1) and enrichment (6–1,800).
Figure 5. Activity- and affinity-based screening comparison.
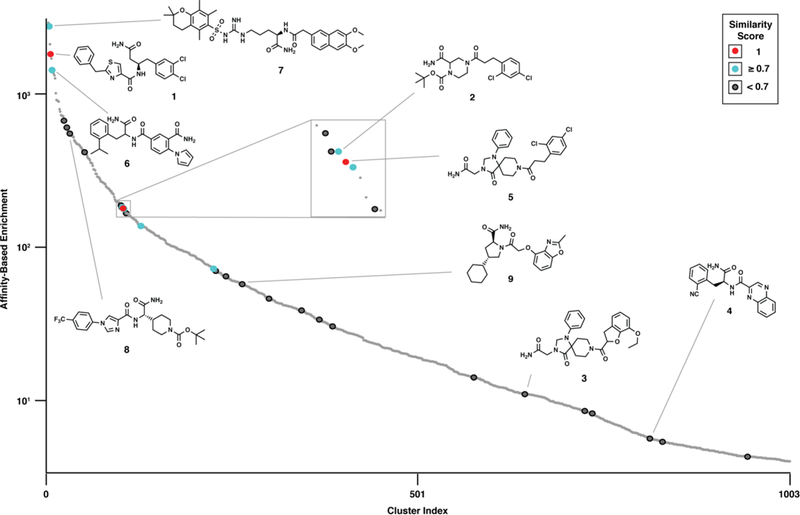
The 35 activity-based screening hits were clustered by chemical similarity with those of an 866,250-member DEL affinity selection. Larger dots indicate that an activity-based hit is included in the cluster. A higher similarity score indicates that the activity-based hit was more similar to the cluster representative of the DEL affinity selection hit. Affinity-based hits discovered as activity-based hits have similarity score = 1. Clusters are ranked by their cumulative enrichment from an affinity-based screen.
Activity-based DEL screening both recapitulates and diverges from affinity-based DEL selection. The two techniques identified the same chemical spaces as ATX ligands (1, 6, 7), suggesting that these molecules are active inhibitors and not merely ATX ligands. The DEL interrogated by affinity selection was much larger and generated many more structural clusters than the solid-phase DEL. Several clusters that strongly enriched during affinity selection were simply not present in the solid-phase library. Three compounds (1, 2, 5) with high similarity to affinity selection hit compounds (Tanimoto similarity = 1.0, 0.9, 1.0, respectively) had effectively identical potency (IC50 = 3.5, 8, and 5 μM, respectively). However, 1 enriched 20-fold more than 2 or 5 during affinity selection. Compounds 3 and 4 were also unremarkable by affinity selection enrichment, but proved to have similar activity against ATX (IC50s = 8 and 10 μM, respectively). Similarly, multiple compounds enriched during affinity selection were not in the activity-based screening hits, perhaps owing to various technique-specific biases (e.g., individual monomer synthetic yields, molecular partitioning, DNA code-specific effects).
The activity-based screening hits reveal non-obvious structure-activity relationships and represent multiple distinct starting points for probe or lead development. Compounds 3 and 5 belong to distinct chemical clusters yet share an AA building block and conjugated rings in the CA building block. The dichlorophenyl of 5 is separated from the amide by an ethylene; compound 3 demonstrates that this spacer/ring can be transformed into a fused, rigid system while retaining activity. Compound 4 is relatively small and hydrophilic compared to most ATX inhibitors and could be a versatile scaffold for medicinal chemistry elaboration. Additionally, 34/35 of the identified hit compounds were chemically distinct (Tanimoto score < 0.7) from any compound within Roche’s 4.2M HTS compound collection (Table S3). These results collectively validate activity-based DEL screening as an effective approach to discovering novel bioactive chemical matter.
HTS is a driving force in drug discovery and has drawn intense interest in miniaturizing the platform as a means of economizing screening. The numerous scaling challenges of HTS, including assay scale and complex robotic automation, have long been ripe targets for microfluidic miniaturization.53–55 Microfluidic instrumentation capable of distributing the screening aspect of HTS ultimately reached the market as one of the first demonstrations of the technology. However, library distribution was and continues to be a significant and separate problem — one where combinatorial library synthesis excels.20–22 The exponential scaling of split-and-pool synthesis and dramatically reduced library footprint are highly attractive, but the unconventional bead format and the eccentricities of mass spectral hit structure deconvolution rendered combinatorial libraries maladapted to standard microplate-based robotic automation. Miniaturization has thus remained a complex problem.
Here, we have presented a potential solution to this long-standing chemistry and engineering challenge. In this first demonstration of activity-based DEL screening, we interrogated a 67,100-member library of drug-like small molecules for inhibitors of a pharmaceutically relevant phosphodiesterase enzyme target. An integrated microfluidic circuit automated and miniaturized the screening process by encapsulating library beads in picoliter-scale droplets of activity assay reagent at a frequency of 60 Hz, and photochemically dosing, incubating, and sorting the droplets containing hit compounds. The library screen required 8 h instrument time and consumed vanishing quantities of activity assay reagents (< 10 μg ATX; < 20 nmol substrate) and library (0.0005 total library). In contrast to affinity selection, the time required to complete an activity-based screen scales linearly with library diversity. Thus, screening an HTS-scale million-compound DEL would require increasing throughput >10-fold, which is within reach given >10 kHz sorting rates of the fastest published droplet sorting rates.56
This work showcases the statistical advantages of the increased sampling inherent t0 microfluidic miniaturization. Sampling is central to the synthesis and analysis of combinatorial libraries, which introduce significant Poisson-derived variances.47 Such variances manifest from stochastic library bead compartmentalization for screening,23–25,57 library member representation,58 bead-to-bead loading capacity,59 and improper hydrophobic compound partitioning between droplets.33,46,60 The increased sampling and dynamic, high-throughput hypothesis testing-based hit identification described here delivers the necessary statistical power to overcome these variances. From the many hit beads collected (7,000), all 5 hits selected for follow-up were found to be active. Of note, the 35 hits are more hydrophobic than the library mean (cLogP = 2.4 and 1.3, respectively), and 6 hits exhibit cLogP > 4. The hits’ high hydrophobicity, consistent with known ATX inhibitor properties,50 could have resulted in active compounds partitioning between droplets46 and concomitantly increased false-positive hit identification. While we did observe evidence for active compounds partitioning between droplets, it did not interfere with hit identification.
Comparing affinity- and activity-based DEL technologies using ATX as a common target illustrates the complementarity of the approaches, despite disparities in library sizes and analysis throughput. Importantly, the solid-phase DEL of these experiments was generated from building block stocks that were used for conventional DEL synthesis. Solid-phase DEL synthesis thus can leverage existing and deep reservoirs of molecular diversity. Solid-phase DELs additionally grant the ability to perform highly parallel hit synthesis after screening; this efficiency is attractive because approximately half of all hits from affinity selections do not validate.43,61 Finally, the droplet-based assay strategy adds options for detecting target engagement. This study used homogeneous fluorescence detection, but other reports have described in-droplet fluorescence anisotropy62,63 or mass spectral detection.55 Such detection modes offer the possibility of using endogenous as opposed to engineered substrates, more closely mimicking the biologically relevant context.
Looking forward, this first disclosure of activity-based DEL screening technology sets the stage for investigating even more complex targets that are beyond the scope of affinity selections. For example, cell surface receptor ligand discovery is often guided by a desire to modulate receptor activity in a particular direction; activity-based screening would reveal such modulation, complementing affinity selection.13,14 Coupled biochemical systems are another interesting opportunity: protein translation is a target for treating genetic and infectious diseases,64–67 and activity-based DEL screens could reveal non-obvious targets for more potently modulating metabolic activity in vitro. Along those lines, phenotypic cell-based screening — a source of many modern first-in-class small molecule drugs68 — is now a distinct possibility given that there are already strong microfluidic cellular analysis foundations in place.69,70 Integrating these HTS-type assay formats with microfluidic miniaturization and the novel chemical matter derived from highly parallel DEL-based molecular diversification collectively constitutes our vision of a distributable and transformative engine for chemical genetics and drug discovery.
Experimental Procedures
Materials Sources.
All reagents were obtained from MilliporeSigma (St. Louis, MO) unless otherwise specified. 1,3-Bis[tris(hydroxymethyl)methylamino]propane (Bis-Tris), trifluoroacetic acid (TFA), triisopropylsilane (TIPS), α-cyano-4-hydroxycinnamic acid (HCCA) (Life Technologies, Carlsbad, CA), N,N’-diisopropylcarbodiimide (DIC, Acrōs Organics, Fair Lawn, NJ), 1-hydroxy-7-azabenzotriazole (HOAt, Accela ChemBio Inc., San Diego, CA), 2,4,6-trimethylpyridine (TMP), ethyl 2-cyano-2-(hydroxyimino)acetate (Oxyma), dimethylformamide (DMF, Thermo Fisher Scientific, Waltham, MA), dichloromethane (DCM, Thermo Fisher Scientific), N,N-diisopropylethylamine (DIEA, Thermo Fisher Scientific), acetonitrile (ACN, Thermo Fisher Scientific), dimethyl sulfoxide (DMSO, AMRESCO Inc., Solon, OH), Poly-(dimethylsiloxane) (PDMS, Dow Corning, Midland, MI), 5-(6)-carboxytetramethylrhodamine (5(6)-TAMRA, Anaspec, Fremont, CA), 4’,6-diamidino-2-phenylindole (DAPI, Anaspec), (4-Fmoc-2-methoxy-5-nitrophenoxy)butanoic acid (Fmoc-PC-OH, Santa Cruz Biotechnology Inc., Dallas, TX), N-α-Fmoc-N-ε−7-methoxycoumarin-4-acetyl-L-lysine (N-α-Fmoc-K(Mca)-OH), Nα-Fmoc-Nω-(2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonyl)-L-arginine (N-α-Fmoc-R(Pbf)-OH, Thermo Fisher Scientific), Fmoc-8-amino-3,6-dioxaoctanoic acid (Fmoc-PEG2, ChemPep, Inc, Wellington, FL), sodium acetate, calcium chloride, Taq DNA polymerase (Taq, New England Biolabs, Ipswich, MA), and 2′-deoxyribonucleotide triphosphate (dNTP, set of dATP, dTTP, dGTP, dCTP, Promega Corp., Milwaukee, WI), were used as provided. Solvents used in solid-phase synthesis were dried over molecular sieves (3 Å, 3.2 mm pellets). Oligonucleotides (Integrated DNA Technologies, Inc. Coralville, IA) were purchased as desalted lyophilate and used without further purification. Oligonucleotide ligation substrates were 5′-phosphorylated (/5Phos/). Amino-modified headpiece DNA (NH2-HDNA, /5Phos/GAGTCA/iSp9//iUniAmM//iSp9/TGACTCCC) was HPLC purified at the manufacturer and used without further purification. Fmoc-amino acid and carboxylic acid building blocks were selected from the Roche Pharma Research and Early Development (pRED) compound collection.
Microfluidic Device Fabrication.
Channel structures were fabricated in PDMS using soft lithography.71 Master channel heights (30 μm, 50 μm, and 250 μm) were measured using a stylus profilometer (DektakXT, Bruker, Billerica, MA). Two-tone PDMS devices were fabricated as previously described.33 Briefly, avobenzone (34 mg) was dissolved in toluene (200 μL) and mixed into PDMS prepolymer (5.5 g, 10:1 elastomer base/curing agent). Degassed avobenzone-PDMS prepolymer was loaded into a syringe (3 mL, BD Medical, Franklin Lakes, NJ), and applied over the ICEcreamer incubator and bead introduction regions of the master. After partial curing (12 min, 80 °C), degassed PDMS prepolymer (44 g, 10:1 base:curing agent) was poured over the master, cured (1 h, 80 °C), cooled (10 min, RT), and peeled from the master. Fluidic ports were punched with a biopsy punch (0.75 mm dia., World Precision Instruments, Inc., Sarasota, FL). PDMS molds and glass slides were cleaned with Safe-Soap (Gold Biotechnologies Inc., St. Louis, MO), then rinsed with DI water and isopropyl alcohol prior to immersion in acid hydrolysis solution (5:1:1 DI H2O/HCl/H2O2, 30 min). Glass slides and PDMS molds were again rinsed with DI water and isopropyl alcohol, and then immediately bonded (16 h, 80 °C).72 Microfluidic devices were fitted with integrated waveguides.33 Waveguide illumination intensity was calibrated by pumping (0.5 μL/min) a solution of DAPI (5 mM, 20 mM Tris-HCl, 150 mM NaOAc, 2.5 mM CaCl2, pH 8.3) through the waveguide calibration channel while varying the LED’s current. Calibrant solution fluorescence emission was measured in the 520 nm channel (100 Hz). Afterward, the calibration channel was rinsed with water, dried with air, and filled with trimethylsiloxy-terminated PDMS (DMS-T22, Gelest, Inc., Tullytown, PA).
Microfluidic droplet based screening assay and device operation.
The droplet-scale autotaxin (ATX) activity assay (Echelon Biosciences Inc., Salt Lake City, UT) was conducted as previously described36 with minor modifications. Briefly, input AQ1 contained fluorogenic ATX substrate FS-373 (FS-3, 10 μM) and internal standard 5(6)-TAMRA (5 μM) in assay buffer. Washed and quantitated library aliquot was combined with AQ1 (~2,000 beads/μL). AQ2 contained ATX (100 nM) in buffer (1/10 diluted kit buffers C and D). OIL1 was squalene (TCI America, Portland, OR) containing KF-6038 (ShinEtsu, Tokyo, Japan, 4% w/w). OIL1 solution was prepared by combining components and mixing with rotation (1 h, 500 rpm). OIL2 and OIL3 were neat squalene.
AQ1 and AQ2, OIL1, OIL2, and OIL3 solutions were loaded into syringes (1/1/1/10/3 mL respectively, BD Medical) fitted with blunt-tip Luer-Lok needles and connected to fluidic inputs via Microbore Tygon tubing. Fluids from syringes were driven through the circuit with displacement syringe pumps (Legato 100, KD Scientific, Holliston, MA). The AQ1-containing syringe barrel was loaded with a PTFE-encapsulated magnet (3 mm dia., V&P Scientific, Inc., San Diego, CA). OIL2 and OIL3 flow (8/3 μL/min, respectively) was initiated while hit and waste tubing was clamped to backfill the circuit. Back shunt outlets were clamped once all tubing was filled with oil. Hit and waste tubing was unclamped once all air was removed from the droplet incubator. AQ1/AQ2/OIL1 flows (400 nL/min) were then initiated and remained constant for the duration of the screen. Droplet generation was allowed to equilibrate (~5 min), and AQ1 solution was homogenized by manually agitating the magnet. The main shunt was subsequently clamped, and droplets began filling the incubator. Data acquisition began after the circuit was equilibrated (25 min).
The confocal LIF detection system and LabVIEW code (National Instruments, Austin, TX) were previously described.39 Briefly, droplet fluorescence emission was detected by photon counting PMTs (H7828, Hamamatsu, Middlesex, NJ) using a custom two-channel (520 and 570 nm) confocal LIF microscope with laser excitation (488 nm, 20 mW, OBIS-488 20LS, Coherent Inc., Santa Clara, CA). LabVIEW code written in house collects signal data and determines droplet sorting decisions. The PMT signals were digitized by a data acquisition board (DAQ, NI USB-6341, National Instruments), and binned into packets of counts (Δt = 0.167 ms). Median-filter smoothing (window width = 5) was applied to the signal in real-time. Droplet signal regions were identified by the 570-nm channel signal (countsn > 60) and 520-nm channel signal maxima (max = countsn−1 when countsn < countsn−1) was returned as the droplet fluorescence value. A dynamic sorting threshold was calculated in real-time to identify ‘hit droplets’ with reduced 520-nm channel fluorescence. The dynamic threshold was calculated as μ - 4σ, where μ and σ were the mean and standard deviation of the last 1,000 droplets’ fluorescence values, respectively. Hit droplets were excluded from the fluorescence values used to calculate population mean and standard deviation. When a hit droplet was detected (droplet fluorescence < μ - 4σ counts), LabVIEW output a TTL pulse from the DAQ board to a waveform generator (Agilent 33210A, Agilent Technologies, Santa Clara, CA), triggering a square wave pulse output (0–7 V, 8.5 kHz, 300 cycles) that was amplified (gain = 100 V/V, TREK Model 2210 high-voltage power amplifier, TREK Inc., Lockport, NY) and conducted through Luer-Lok needle-fitted and Microbore Tygon tubing filled with salt water (4 M NaCl) into a microfabricated electrode channel (VAC).38
Resynthesis linker resin preparation.
Linker and compound synthesis proceeded via iterative cycles of solid-phase peptide synthesis. TentaGel rink amide resin (160 μm, 0.40 mmol/g, 10 mg) was transferred to a fritted spin-column, washed (3 × DCM, 3 × DMF), swelled in DMF (1 h, RT), and washed (3 × DMF). Fmoc was removed (20% piperidine in DMF, 1 × 5 min, 1 × 15 min, 8 rpm, RT), and the resin was washed (3 × DMF, 3 × DCM, 3 × DMF). N-α-Fmoc-K(Mtt)-OH (120 μmol, 0.3 mL) was activated (2 min) with COMU/DIEA (120/240 μmol) in DMF, activated acid was added to resin, and the resin was incubated (30 min, 50 °C, 8 rpm, 2 ×), washed (3 × DMF, 3 × DCM, 3 × DMF), capped (20% acetic anhydride in DMF, 15 min, 50 °C, 8 rpm), and washed (6 × DMF, 3 × DCM, 3 × DMF). Mtt cleavage cocktail (1% TFA, 5% TIPS in DCM) was first used to wash the resin (3 × Mtt cleavage cocktail), then fresh cocktail was added and the resin was incubated (1 × 5 min, 1 × 15 min, 8 rpm, RT). Resin was washed (2 × Mtt cleavage cocktail, 3 × DCM, 3 × DMF, 3 × DCM), and 5(6)TAMRA (200 μmol in DMF, 0.3 mL) was activated (2 min) with COMU/DIEA (200/400 μmol). Activated acid was added to resin and the resin incubated (15 min, 50 °C, 8 rpm, 2 ×). Resin was washed (3 × DMF, 3 × DCM, 3 × DMF), capped (20% acetic anhydride in DMF, 15 min, 50 °C, 8 rpm), and washed (6 × DMF, 3 × DCM, 3 × DMF). Fmoc was removed (20% piperidine in DMF, 1 × 5 min, 1 × 15 min, 8 rpm, RT) and the resin was washed (3 × DMF, 3 × DCM, 3 × DMF). N-α-Fmoc-R(Pbf)-OH (120 μmol in DMF, 0.3 mL) was activated (2 min) with COMU/DIEA (120/240 μmol), added to resin, and the resin was incubated (15 min, 50 °C, 8 rpm) and washed (3 × DMF, 3 × DCM, 3 × DMF). Fmoc was removed (20% piperidine in DMF, 1 × 5 min, 1 × 15 min, 8 rpm, RT) and the resin was washed (3 × DMF, 3 × DCM, 3 × DMF). Fmoc-PEG2 (120 μmol in DMF, 0.3 mL) was activated (2 min) with COMU/DIEA (120/240 μmol). Activated acid was added to resin, and the resin was incubated (15 min, 50 °C, 8 rpm). Fmoc was removed (20% piperidine in DMF, 1 × 5 min, 1 × 15 min, 8 rpm, RT) and the resin was washed (3 × DMF, 3 × DCM, 3 × DMF). Fmoc-PC-OH (200 μmol in DMF, 0.4 mL) was activated with DIC/Oxyma/TMP (300/200/200 μmol), activated acid was added to resin, and the resin was incubated (2 h, 50 °C, 8 rpm) and washed (3 × DMF, 3 × DCM, 3 × DMF).
Five beads were transferred to a new spin column, washed (3 × DCM), and air-dried. Cleavage cocktail (90% TFA, 5% DCM, 5% TIPS, 10 μL) was added to the beads. The beads were incubated (30 min, RT, 8 rpm) and dried in vacuo (60 °C). Compound was resuspended (DMSO, 5 μL), diluted (50% ACN, 0.1% TFA in H2O, 45 μL), and an aliquot (1 μL) co-spotted onto a MALDI-TOF MS target plate with HCCA matrix solution, dried, and analyzed via MALDI-TOF MS.
Hit resynthesis.
Resynthesis linker resin was washed (3 × DMF, 3 × DCM, 3 × DMF), Fmoc was removed (20% piperidine in DMF, 1 × 5 min, 1 × 15 min, 8 rpm, RT), and the resin was washed (3 × DMF, 3 × DCM, 3 × DMF). Resin was transferred (97 nmol/well, 240 μg) to a filtration microplate, washed (3 × DMA), resuspended (DMA), incubated (15 min, RT, 100 rpm), and washed (3 × DMA). The first and second building block couplings consisted of acylation with an N-Fmoc amino acid and a carboxylic acid, respectively. Resin was resuspended (DMA, 45 μL) with building block/HOAt/DIC (1.6/1.6/2.3 μmol, respectively). Plates were covered with adhesive foil and incubated (2 h, 37 °C, 100 rpm). Resin was washed (3 × DMA, 3 × DCM, 3 × DMF), and Fmoc was removed (20% piperidine in DMF, 1 × 5 min, 1 × 15 min, 100 rpm, RT). The resin was washed (3 × DMF, 3 × DCM, 3 × DMA). Resin was resuspended (DMA, 45 μL) with building block/HOAt/DIC (1.6/1.6/2.3 μmol, respectively). Plates were covered with adhesive foil and incubated (2 h, 37 °C, 100 rpm). Resin was washed (3 × DMA, 3 × DCM, 3 × DMF) and transferred to eppendorf tubes for storage (DMF, 0.4 mL, 4°C). Aliquots (40 μl) of each resin were transferred to spin columns, washed (DCM, 3 × 0.3 mL) and acid cleaved/analyzed via MALDI-TOF MS (see above, Table S2).
Bulk resin photocleavage.
Hit compound resin (40 nmol) was transferred to a 96-well white-bottom microplate (Bio-Rad), supernatant was removed, assay buffer (50 μL) was added, incubated (1 h, RT, 600 rpm), and the resin was centrifuged (100 rcf). Three additional buffer exchange/incubations were performed. The compound plate was placed in a 365-nm UV-crosslinker (CL-1000L, Analytik Jena, Jena, Germany), exposed (16 h, 3,000 rpm), and centrifuged (100 rcf). The supernatant was removed and assayed for ATX inhibition.
Hit validation assay.
Hit validation resin photocleavage supernatant (5 μL) was added to ATX (10 nM, 2.5 μL) in 384-well white-bottom microplates (Corning Scientific, Corning, NY) and incubated (30 min, 37 °C). FS-3 was added (20 μM, 2.5 μL) and reaction progress was monitored (Gemini XPS, Molecular Devices, San Jose, CA) immediately (λex = 490 nm, λem = 530 nm, 37 °C). The percent activity was taken as the slope of fluorescence increase over time (5 – 15 min) relative to that of a vehicle control. The assay was conducted identically for IC50 determination, except that all volumes were doubled and a 2× [compound] solution in buffer (20% DMSO in assay buffer, v/v) was incubated with ATX prior to FS-3 addition.
Supplementary Material
Acknowledgments
We gratefully acknowledge Dr. Jianping Cai and Dr. Ann C. Petersen (Roche Pharmaceutical Research and Early Development) for preparing and characterizing compounds 1, 2, and 5, and Dr. Alexander K. Price (Scripps Research) for assistance with data acquisition coding. This work was funded by the National Institute of Health (R01GM120491) and a National Science Foundation CAREER award to B.M.P (1255250). W.G.C. gratefully acknowledges support from a Farris Foundation Graduate Research Fellowship Award.
Abbreviations
- NGS
next-generation sequencing
- HTS
high-throughput screening
- DEL
DNA-encoded library
- OBOC
one bead one compound
- ATX
autotaxin
- AA
amino acid
- CA
carboxylic acid
- QC
quality control
- UMI
unique molecular identifier
- FDR
false discovery rate
- HDNA
headpiece DNA
- OP
oligonucleotide paired stock
- DESPS
DNA-encoded solid-phase synthesis
- TAMRA
tetramethylrhodamine
Footnotes
Associated Content
The Supporting Information is available free of charge on the ACS Publications website.
Experimental details and additional results
The authors declare no competing financial interests.
References
- (1).Schreiber SL; Kotz JD; Li M; Aubé J; Austin CP; Reed JC; Rosen H; White EL; Sklar LA; Lindsley CW; et al. Advancing Biological Understanding and Therapeutics Discovery with Small-Molecule Probes. Cell 2015, 161, 1252–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Brown DG; Boström J Where Do Recent Small Molecule Clinical Development Candidates Come From? J Med Chem 2018, 61, 9442–9468. [DOI] [PubMed] [Google Scholar]
- (3).Scott DE; Bayly AR; Abell C; Skidmore J Small Molecules, Big Targets: Drug Discovery Faces the Protein-Protein Interaction Challenge. Nat Rev Drug Discov 2016, 15, 533–550. [DOI] [PubMed] [Google Scholar]
- (4).Angelbello AJ; Chen JL; Childs-Disney JL; Zhang P; Wang Z-F; Disney MD Using Genome Sequence to Enable the Design of Medicines and Chemical Probes. Chem. Rev. 2018, 118, 1599–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Tommasi R; Brown DG; Walkup GK; Manchester JI; Miller AA ESKAPEing the Labyrinth of Antibacterial Discovery. Nat Rev Drug Discov 2015, 14, 529–542. [DOI] [PubMed] [Google Scholar]
- (6).Brenner S; Lerner RA Encoded Combinatorial Chemistry. Proc Natl Acad Sci U S A 1992, 89, 5381–5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Gartner ZJ; Liu DR The Generality of DNA-Templated Synthesis as a Basis for Evolving Non-Natural Small Molecules. J Am Chem Soc 2001, 123, 6961–6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Melkko S; Scheuermann J; Dumelin C; Neri D Encoded Self-Assembling Chemical Libraries. Nat Biotechnol 2004, 22, 568–574. [DOI] [PubMed] [Google Scholar]
- (9).Mannocci L; Zhang Y; Scheuermann J; Leimbacher M; De Bellis G; Rizzi E; Dumelin C; Melkko S; Neri D High-Throughput Sequencing Allows the Identification of Binding Molecules Isolated From DNA-Encoded Chemical Libraries. Proc Natl Acad Sci U S A 2008, 105, 17670–17675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Clark MA; Acharya RA; Arico-Muendel CC; Belyanskaya SL; Benjamin DR; Carlson NR; Centrella PA; Chiu CH; Creaser SP; Cuozzo JW; et al. Design, Synthesis and Selection of DNA-Encoded Small-Molecule Libraries. Nat Chem Biol 2009, 5, 647–654. [DOI] [PubMed] [Google Scholar]
- (11).Harris PA; Berger SB; Jeong JU; Nagilla R; Bandyopadhyay D; Campobasso N; Capriotti CA; Cox JA; Dare L; Dong X; et al. Discovery of a First-in-Class Receptor Interacting Protein 1 (RIP1) Kinase Specific Clinical Candidate (GSK2982772) for the Treatment of Inflammatory Diseases. J Med Chem 2017, 60, 1247–1261. [DOI] [PubMed] [Google Scholar]
- (12).Litovchick A; Dumelin CE; Habeshian S; Gikunju D; Guie M-A; Centrella P; Zhang Y; Sigel EA; Cuozzo JW; Keefe AD; et al. Encoded Library Synthesis Using Chemical Ligation and the Discovery of sEH Inhibitors From a 334-Million Member Library. Sci Rep 2015, 5, 10916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Wu Z; Graybill TL; Zeng X; Platchek M; Zhang J; Bodmer VQ; Wisnoski DD; Deng J; Coppo FT; Yao G; et al. Cell-Based Selection Expands the Utility of DNA-Encoded Small-Molecule Library Technology to Cell Surface Drug Targets: Identification of Novel Antagonists of the NK3 Tachykinin Receptor. ACS Comb Sci 2015, 17, 722–731. [DOI] [PubMed] [Google Scholar]
- (14).Ahn S; Kahsai AW; Pani B; Wang Q-T; Zhao S; Wall AL; Strachan RT; Staus DP; Wingler LM; Sun LD; et al. Allosteric “Beta-Blocker” Isolated From a DNA-Encoded Small Molecule Library. Proc Natl Acad Sci U S A 2017, 114, 1708–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Kollmann CS; Bai X; Tsai C-H; Yang H; Lind KE; Skinner SR; Zhu Z; Israel DI; Cuozzo JW; Morgan BA; et al. Application of Encoded Library Technology (ELT) to a Protein-Protein Interaction Target: Discovery of a Potent Class of Integrin Lymphocyte Function-Associated Antigen 1 (LFA-1) Antagonists. Bioorg Med Chem 2014, 22, 2353–2365. [DOI] [PubMed] [Google Scholar]
- (16).Wrenn SJ; Weisinger RM; Halpin DR; Harbury PB Synthetic Ligands Discovered by in Vitro Selection. J Am Chem Soc 2007, 129, 13137–13143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Li Y; De Luca R; Cazzamalli S; Pretto F; Bajic D; Scheuermann J; Neri D Versatile Protein Recognition by the Encoded Display of Multiple Chemical Elements on a Constant Macrocyclic Scaffold. Nat Chem 2018, 10, 441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Usanov DL; Chan AI; Maianti JP; Liu DR Second-Generation DNA-Templated Macrocycle Libraries for the Discovery of Bioactive Small Molecules. Nat Chem 2018, 10, 704–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Goodnow RA; Dumelin CE; Keefe AD DNA-Encoded Chemistry: Enabling the Deeper Sampling of Chemical Space. Nat Rev Drug Discov 2017, 16, 131–147. [DOI] [PubMed] [Google Scholar]
- (20).Houghten RA; Pinilla C; Blondelle SE; Appel JR; Dooley CT; Cuervo JH Generation and Use of Synthetic Peptide Combinatorial Libraries for Basic Research and Drug Discovery. Nature 1991, 354, 84–86. [DOI] [PubMed] [Google Scholar]
- (21).Furka A; Sebestyen F; Asgedom M; Dibo G General-Method for Rapid Synthesis of Multicomponent Peptide Mixtures. Int. J. Pept. Protein Res. 1991, 37, 487–493. [DOI] [PubMed] [Google Scholar]
- (22).Lam KS; Salmon SE; Hersh EM; Hruby VJ; Kazmierski WM; Knapp RJ A New Type of Synthetic Peptide Library for Identifying Ligand-Binding Activity. Nature 1991, 354, 82–84. [DOI] [PubMed] [Google Scholar]
- (23).You AJ; Jackman RJ; Whitesides GM; Schreiber SL A Miniaturized Arrayed Assay Format for Detecting Small Molecule-Protein Interactions in Cells. Chem Biol 1997, 4, 969–975. [DOI] [PubMed] [Google Scholar]
- (24).Schullek JR; Butler JH; Ni ZJ; Chen D; Yuan Z A High-Density Screening Format for Encoded Combinatorial Libraries: Assay Miniaturization and Its Application to Enzymatic Reactions. Anal Biochem 1997, 246, 20–29. [DOI] [PubMed] [Google Scholar]
- (25).Borchardt A; Liberles S; Biggar S; Crabtree G; Schreiber S Small Molecule-Dependent Genetic Selection in Stochastic Nanodroplets as a Means of Detecting Protein-Ligand Interactions on a Large Scale. Chem Biol 1997, 4, 961–968. [DOI] [PubMed] [Google Scholar]
- (26).Silen JL; Lu AT; Solas DW; Gore MA; Maclean D; Shah NH; Coffin JM; Bhinderwala NS; Wang YW; Tsutsui KT; et al. Screening for Novel Antimicrobials From Encoded Combinatorial Libraries by Using a Two-Dimensional Agar Format. Antimicrob. Agents Chemother. 1998, 42, 1447–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Thorsen T; Roberts R; Arnold F; Quake S Dynamic Pattern Formation in a Vesicle-Generating Microfluidic Device. Phys Rev Lett 2001, 86, 4163–4166. [DOI] [PubMed] [Google Scholar]
- (28).Anna S; Bontoux N; Stone H Formation of Dispersions Using “Flow Focusing” in Microchannels. Appl Phys Lett 2003, 82, 364–366. [Google Scholar]
- (29).Needels MC; Jones DG; Tate EH; Heinkel GL; Kochersperger LM; Dower WJ; Barrett RW; Gallop MA Generation and Screening of an Oligonucleotide-Encoded Synthetic Peptide Library. Proc Natl Acad Sci U S A 1993, 90, 10700–10704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).MacConnell AB; McEnaney PJ; Cavett VJ; Paegel BM DNA-Encoded Solid-Phase Synthesis: Encoding Language Design and Complex Oligomer Library Synthesis. ACS Comb Sci 2015, 17, 518–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Benesch MGK; Ko YM; McMullen TPW; Brindley DN Autotaxin in the Crosshairs: Taking Aim at Cancer and Other Inflammatory Conditions. FEBS Lett 2014, 588, 2712–2727. [DOI] [PubMed] [Google Scholar]
- (32).Maher TM; van der Aar EM; Van de Steen O; Allamassey L; Desrivot J; Dupont S; Fagard L; Ford P; Fieuw A; Wuyts W Safety, Tolerability, Pharmacokinetics, and Pharmacodynamics of GLPG1690, a Novel Autotaxin Inhibitor, to Treat Idiopathic Pulmonary Fibrosis (FLORA): a Phase 2a Randomised Placebo-Controlled Trial. Lancet Respir Med 2018, 6, 627–635. [DOI] [PubMed] [Google Scholar]
- (33).Price AK; MacConnell AB; Paegel BM hνSABR: Photochemical Dose-Response Bead Screening in Droplets. Anal Chem 2016, 88, 2904–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Song H; Ismagilov RF Millisecond Kinetics on a Microfluidic Chip Using Nanoliters of Reagents. J Am Chem Soc 2003, 125, 14613–14619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Link DR; Anna SL; Weitz DA; Stone HA Geometrically Mediated Breakup of Drops in Microfluidic Devices. Phys Rev Lett 2004, 92, 054503. [DOI] [PubMed] [Google Scholar]
- (36).Cochrane WG; Hackler AL; Cavett VJ; Price AK; Paegel BM Integrated, Continuous Emulsion Creamer. Anal Chem 2017, 89, 13227–13234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Baret J-C; Miller OJ; Taly V; Ryckelynck M; Harrak El, A.; Frenz L; Rick C; Samuels ML; Hutchison JB; Agresti JJ; et al. Fluorescence-Activated Droplet Sorting (FADS): Efficient Microfluidic Cell Sorting Based on Enzymatic Activity. Lab Chip 2009, 9, 1850–1858. [DOI] [PubMed] [Google Scholar]
- (38).Sciambi A; Abate AR Generating Electric Fields in PDMS Microfluidic Devices with Salt Water Electrodes. Lab Chip 2014, 14, 2605–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).MacConnell AB; Price AK; Paegel BM An Integrated Microfluidic Processor for DNA-Encoded Combinatorial Library Functional Screening. ACS Comb Sci 2017, 19, 181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Gierse J; Thorarensen A; Beltey K; Bradshaw-Pierce E; Cortes-Burgos L; Hall T; Johnston A; Murphy M; Nemirovskiy O; Ogawa S; et al. A Novel Autotaxin Inhibitor Reduces Lysophosphatidic Acid Levels in Plasma and the Site of Inflammation. J. Pharmacol. Exp. Ther. 2010, 334, 310–317. [DOI] [PubMed] [Google Scholar]
- (41).Sander T; Sander T; Freyss J; Freyss J; Korff, von M; Korff, von M; Rufener C; Rufener C DataWarrior: an Open-Source Program for Chemistry Aware Data Visualization and Analysis. J Chem Inf Model 2015, 55, 460–473. [DOI] [PubMed] [Google Scholar]
- (42).Dolle RE; Guo J; O’Brien L; Jin Y; Piznik M; Bowman KJ; Li WN; Egan WJ; Cavallaro CL; Roughton AL; et al. A Statistical-Based Approach to Assessing the Fidelity of Combinatorial Libraries Encoded with Electrophoric Molecular Tags. Development and Application of Tag Decode-Assisted Single Bead LC/MS Analysis. J Comb Chem 2000, 2, 716–731. [DOI] [PubMed] [Google Scholar]
- (43).Eidam O; Satz AL Analysis of the Productivity of DNA Encoded Libraries. MedChemComm 2016, 7, 1323–1331. [Google Scholar]
- (44).Lipinski CA; Lombardo F; Dominy BW; Feeney PJ Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv Drug Deliv Rev 1997, 23, 3–25. [DOI] [PubMed] [Google Scholar]
- (45).Veber DF; Johnson SR; Cheng H-Y; Smith BR; Ward KW; Kopple KD Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J Med Chem 2002, 45, 2615–2623. [DOI] [PubMed] [Google Scholar]
- (46).Courtois F; Olguin LF; Whyte G; Theberge AB; Huck WTS; Hollfelder F; Abell C Controlling the Retention of Small Molecules in Emulsion Microdroplets for Use in Cell-Based Assays. Anal Chem 2009, 81, 3008–3016. [DOI] [PubMed] [Google Scholar]
- (47).MacConnell AB; Paegel BM Poisson Statistics of Combinatorial Library Sampling Predict False Discovery Rates of Screening. ACS Comb Sci 2017, 19, 524–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Appell KC; Chung TDY; Ohlmeyer MJH; Sigal NH; Baldwin JJ; Chelsky D Biological Screening of a Large Combinatorial Library. J Biomol Screen 1996, 1, 27–31. [Google Scholar]
- (49).Satz AL Simulated Screens of DNA Encoded Libraries: the Potential Influence of Chemical Synthesis Fidelity on Interpretation of Structure-Activity Relationships. ACS Comb Sci 2016, 18, 415–424. [DOI] [PubMed] [Google Scholar]
- (50).Castagna D; Budd DC; Macdonald SJF; Jamieson C; Watson AJB Development of Autotaxin Inhibitors: an Overview of the Patent and Primary Literature. J Med Chem 2016, 59, 5604–5621. [DOI] [PubMed] [Google Scholar]
- (51).Lavieri R; Scott SA; Lewis JA; Selvy PE; Armstrong MD; Alex Brown H; Lindsley CW Design and Synthesis of Isoform-Selective Phospholipase D (PLD) Inhibitors. Part II. Identification of the 1,3,8-Triazaspiro[4,5]Decan-4-One Privileged Structure That Engenders PLD2 Selectivity. Bioorg. Med. Chem. Lett. 2009, 19, 2240–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Baek HG; Liu R; Lam KS Development of Hydrogel TentaGel Shell-Core Beads for Ultrahigh Throughput Solution-Phase Screening of Encoded OBOC Combinatorial Small Molecule Libraries. J Comb Chem 2009, 11, 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Hadd AG; Raymond DE; Halliwell JW; Jacobson SC; Ramsey JM Microchip Device for Performing Enzyme Assays. Anal Chem 1997, 69, 3407–3412. [DOI] [PubMed] [Google Scholar]
- (54).Thorsen T; Maerkl SJ; Quake SR Microfluidic Large-Scale Integration. 2002, 298, 580–584. [DOI] [PubMed] [Google Scholar]
- (55).Sun S; Slaney TR; Kennedy RT Label Free Screening of Enzyme Inhibitors at Femtomole Scale Using Segmented Flow Electrospray Ionization Mass Spectrometry. Anal Chem 2012, 84, 5794–5800. [DOI] [PubMed] [Google Scholar]
- (56).Sciambi A; Abate AR Accurate Microfluidic Sorting of Droplets at 30 kHz. Lab Chip 2015, 15, 47–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Upert G; Merten CA; Wennemers H Nanoliter Plates-Versatile Tools for the Screening of Split-and-Mix Libraries on-Bead and Off-Bead. Chem Commun 2010, 46, 2209–2211. [DOI] [PubMed] [Google Scholar]
- (58).Burbaum JJ; Ohlmeyer MH; Reader JC; Henderson I; Dillard LW; Li G; Randle TL; Sigal NH; Chelsky D; Baldwin JJ A Paradigm for Drug Discovery Employing Encoded Combinatorial Libraries. Proc Natl Acad Sci U S A 1995, 92, 6027–6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Doran TM; Gao Y; Mendes K; Dean S; Simanski S; Kodadek T Utility of Redundant Combinatorial Libraries in Distinguishing High and Low Quality Screening Hits. ACS Comb Sci 2014, 16, 259–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Gruner P; Riechers B; Orellana LAC; Brosseau Q; Maes F; Beneyton T; Pekin D; Baret J-C Stabilisers for Water-in-Fluorinated-Oil Dispersions: Key Properties for Microfluidic Applications. Current Opinion in Colloid & Interface Science 2015, 20, 183–191. [Google Scholar]
- (61).Goodnow RA A Handbook for DNA-Encoded Chemistry: Theory and Applications for Exploring Chemical Space and Drug Discovery; Wiley, New York, 2014. [Google Scholar]
- (62).Choi J-W; Kang D-K; Park H; deMello AJ; Chang S-I High-Throughput Analysis of Protein-Protein Interactions in Picoliter-Volume Droplets Using Fluorescence Polarization. Anal Chem 2012, 84, 3849–3854. [DOI] [PubMed] [Google Scholar]
- (63).Gielen F; Butz M; Rees EJ; Erdelyi M; Moschetti T; Hyvönen M; Edel JB; Kaminski CF; Hollfelder F Quantitative Affinity Determination by Fluorescence Anisotropy Measurements of Individual Nanoliter Droplets. Anal Chem 2017, 89, 1092–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Pratt SD; David CA; Black-Schaefer C; Dandliker PJ; Xuei XL; Warrior X; Burns DJ; Zhong P; Cao ZS; Saiki A; et al. A Strategy for Discovery of Novel Broad-Spectrum Antibacterials Using a High-Throughput Streptococcus Pneumoniae Transcription/Translation Screen. J Biomol Screen 2004, 9, 3–11. [DOI] [PubMed] [Google Scholar]
- (65).Scheper GC; van der Knaap MS; Proud CG Translation Matters: Protein Synthesis Defects in Inherited Disease. Nat. Rev. Genet. 2007, 8, 711–723. [DOI] [PubMed] [Google Scholar]
- (66).Lowell AN; Santoro N; Swaney SM; McQuade TJ; Schultz PJ; Larsen MJ; Sherman DH Microscale Adaptation of in Vitro Transcription/Translation for High-Throughput Screening of Natural Product Extract Libraries. Chem Biol Drug Des 2015, 86, 1331–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Gao F-B; Richter JD; Cleveland DW Rethinking Unconventional Translation in Neurodegeneration. Cell 2017, 171, 994–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Swinney DC; Anthony J How Were New Medicines Discovered? Nat Rev Drug Discov 2011, 10, 507–519. [DOI] [PubMed] [Google Scholar]
- (69).Brouzes E; Medkova M; Savenelli N; Marran D; Twardowski M; Hutchison JB; Rothberg JM; Link DR; Perrimon N; Samuels ML Droplet Microfluidic Technology for Single-Cell High-Throughput Screening. Proc Natl Acad Sci U S A 2009, 106, 14195–14200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Lan F; Demaree B; Ahmed N; Abate AR Single-Cell Genome Sequencing at Ultra-High-Throughput with Microfluidic Droplet Barcoding. Nat Biotechnol 2017, 35, 640–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Duffy DC; McDonald JC; Schueller OJ; Whitesides GM Rapid Prototyping of Microfluidic Systems in Poly(Dimethylsiloxane). Anal Chem 1998, 70, 4974–4984. [DOI] [PubMed] [Google Scholar]
- (72).Sui G; Wang J; Lee C-C; Lu W; Lee SP; Leyton JV; Wu AM; Tseng H-R Solution-Phase Surface Modification in Intact Poly(Dimethylsiloxane) Microfluidic Channels. Anal Chem 2006, 78, 5543–5551. [DOI] [PubMed] [Google Scholar]
- (73).Ferguson CG; Bigman CS; Richardson RD; van Meeteren LA; Moolenaar WH; Prestwich GD Fluorogenic Phospholipid Substrate to Detect Lysophospholipase D/Autotaxin Activity. Org Lett 2006, 8, 2023–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


