Abstract
To assess karyotypic changes and tumor subpopulations in progression of oligodendroglioma (ODG) we analyzed histologically diagnosed 1p/19q codeleted cases using single nucleotide polymorphism (SNP) microarray data. We separated cases according to grade, which was assigned blind to karyotype information beyond 1p/19q status. The 51 WHO grade II (O2) and 18 WHO grade III (O3) specimens showed frequent chromosomal locations and patterns of change including loss of heterozygosity (LOH), often copy-neutral, on 9p and LOH on 4p and 4q together. Analysis of co-occurrence indicated that most defects were independent but also suggested increased likelihood of defects on 11q, 13q, and 14q in the presence of defects on 18, 4, and 9, respectively. We used the relative degree of change in B-allele frequency as an indicator of an abnormality’s extent, and we present simulated data to clarify how information on subpopulations was thus inferred. Among 9p defects, 89.3% involved the whole tumor, whereas only 47.6% of 4q defects did so. We modeled extent through the tumor as due to a karyotypic change’s likelihood of occurring and the fitness it confers on its subpopulation, and used group data to estimate these values. To assess progression directly, we evaluated specimens from six patients who underwent multiple resections since 1996. Four of these patients had received no chemotherapy or radiation, permitting assessment of the natural history of the tumor karyotype in situ. Defects present throughout a tumor at first resection remained so, whereas among subpopulations, some expanded, some remained constant, and some disappeared. The rate of expansion among subpopulations that did so was not uniform, and estimates of fitness predicted subpopulation composition at recurrence. These results extend prior studies of increased karyotypic abnormality in progression of oligodendroglioma and reveal the complex dynamics of subpopulations in the tumor over time.
Keywords: heterogeneity, subpopulation, progression, microarray, oligodendroglioma, 1p/19q
1. Introduction
The codeletion of chromosome arms 1p and 19q has been recognized to be associated with oligodendroglioma (ODG) for two decades [1], and when strict histological criteria are used for diagnosis, ODG demonstrates 1p/19q codeletion in the great majority of adult cases [2]. 1p/19q codeletion appears to play a role in the pathogenesis of ODG, and gliomas with this karyotype behave more indolently [3]. The importance of this karyotypic change for tumorigenesis and prognosis suggests that improved understanding of other karyotypic abnormalities seen in the tumor could provide insight into tumor development and progression.
The importance of tumor heterogeneity for diagnosis and treatment is increasingly recognized [4, 5]. The more common and lethal glioblastoma, for example, has been shown to have a high degree of intratumor heterogeneity [6, 7]. Understanding of tumor subpopulations is critical for development of maximally effective treatments [8], as chemotherapy may select for resistant clones, or otherwise have undesirable effects on tumor fitness [9], with therapy acting in this sense as a selection pressure [10]. Demonstrating such an unintended consequence, in gliomas with mutations in mismatch repair genes, chemotherapy can disproportionately spare subpopulations with higher mutation rates, thus promoting progression [11]. The importance of subpopulations for outcome is exemplified by medulloblastoma, where genetic analysis of primary tumor and metastases indicates that only a small subset of cells drive dissemination [12]. Heterogeneity has also been identified in ODG [13]. To examine the consequences of heterogeneity for progression, previous reports have examined recurrent ODG by assessing individual genes [14]. Comparative analysis of original tumor and recurrence, a uniquely powerful means of investigating tumor evolution, has been used to identify sequences of molecular alteration during glioma progression [15].
To gain a better understanding of karyotypic changes and heterogeneity in ODG, we examined SNP microarray data for 69 histologically diagnosed adult specimens. We developed new means of visualizing these data, including a method for direct comparison of groups with arbitrary numbers of cases, and here make that software available. Karyotypic defects occur preferentially in a subset of chromosomes, and there is a marked increase in the number of abnormal arms in O3. We utilized spread of B-allele frequency (BAF) as an indicator of proportion of tumor affected by an abnormality, demonstrating karyotypic heterogeneity within tumors and differences among subpopulations in tendency to arise and expand. We formulated a simple means of estimating the relative fitness of a subpopulation bearing a given defect, independent of the defect’s likelihood of occurring. For a case with a histologically identified high grade focus, we separately analyzed tumor regions, demonstrating the averaging of the subpopulation data in results from the pooled sample, and we directly assessed karyotypic changes and subpopulation composition in progression by comparing microarray data from multiple resections from six individual patients.
2. Materials and methods
2.1. Cases and demographics
We searched our clinical database to identify cases of adult O2 or O3 with SNP microarray data from clinical testing since 2011 when the assay entered clinical use, and any earlier resections from the same set of patients. Diagnosis and grade were assigned at the time of signout by the neuropathology service according to WHO criteria [16] and were blind to karyotype information other than 1p/19q status, as these are not reported by the molecular pathology laboratory. For the paired cases retrieved from archives the block that was used at time of signout for immunostaining was cut for microarray analysis and tissue was selected by study pathologists for microdissection. All specimens confirmed to be 1p/19q codeleted, i.e. having complete LOH of a homogeneous character over each of these two arms, are included here as are all paired samples found in the record and retrieved from archives that produced interpretable microarray data; these also all showed the codeletion. We analyzed SNP microarray data from 51 O2 and 18 O3. The O2 group included 29 cases from males and 22 from females. O2 patients ranged in age from 19 to 81, with a mean of 46.5 and a median of 46. O3 included 13 cases from males and five from females. Of the five O3 with the fewest SNP abnormalities, four were from female patients. O3 patients ranged in age from 28 to 71 and were older on average with a mean age of 51.6 and a median of 51. We evaluated BAF and log-R ratio (LRR) for chromosomes 1–22 (q arm only for acrocentric chromosomes 13, 14, 15, 21, and 22) and for the X chromosome for female patients. For three tumor pairs (2_19 and 2_22,2_29 and 2_24, and 2_2 and 3_18) DNA fingerprinting was performed on both samples, confirming for each pair that both cases originated from the same patient. This study was approved by the Johns Hopkins Institutional Review Board.
2.2. SNP microarray and analysis
During neuropathologic assessment, a portion of the tissue containing relatively pure tumor was selected for microdissection. Samples were then run on the HumanCytoSNP-12 BeadChip platform (Illumina) and the results tabulated with BPM and EGT files using GenomeStudio (Illumina). The platform provides information on 298 563 SNPs. We imported the data into Matlab (Mathworks) and wrote custom Matlab functions for analysis. These are available for download (http://pages.jh.edu/~dwnauen/snp_microarray_analysis/). We ignored SNPs with indeterminable location (‘chromosome 0’; 1082 per case), low-confidence BAF (a ‘G-type’ of NC; approximately 20 000 per case) and unassigned BAF (approximately 50 per case). LRR was smoothed in GenomeStudio and additionally smoothed for plotting here with a 30-element boxcar average. In individual case BAF plots, each chromosome was plotted on the full vertical axis, and SNP positions were assigned according to Genetic Distances Build from the Human Genome Project. For plotting of summary BAF data, mapped sites were plotted on the full vertical axis, and values at each SNP site were binned into 13 slots. These values were then binned in groups of 50 successive SNPs along the chromosome. Fixing the number of SNPs per bin ensured that the colorimetric scale has the same values across all chromosomes. The data were smoothed for plotting by a function that weights each element according to the values of contiguous elements, with elements sharing an edge weighted at 0.9 and elements sharing only a vertex weighted at 0.6. Values were normalized to the number of cases in the respective group to provide the same color scale across the plots. SNP positions in the BAF summary data are simply sequential and do not reflect chromosomal distance.
All assessments of BAF and LRR were done manually. Although LRR is needed to assess the cause of observed changes in BAF, in our experience LRR from formalin- fixed, paraffin-embedded (FFPE) tissue can be at times difficult to interpret or misleading (supplementary figure S1 available at stacks.iop.org/CSPO/1/015001/mmedia). Our analysis of SNP data thus always proceeded by evaluation of BAF-only plots to identify abnormalities followed by evaluation of detected abnormalities using BAF-LRR plots to assess karyotypic change. LRR of 1p and 19q almost always showed loss of heterozygosity (LOH) to be due to deletion, but in a few cases, generally older material, LRR on 19q or in one case 1p was indeterminate. The assay showed a high degree of technical replicability (supplementary figure S2 available at stacks.iop.org/CSPO/1/015001/mmedia).
2.3. Assessment of subpopulations
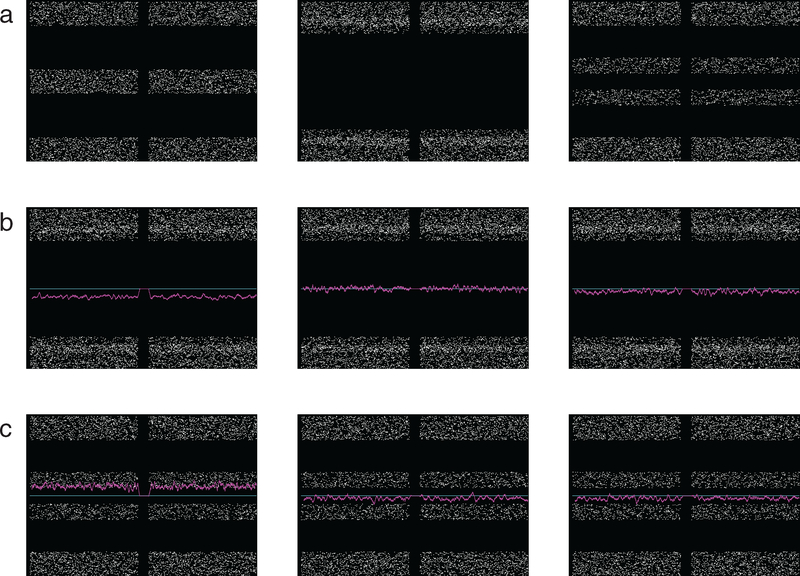
Subpopulation analysis was performed according to the method shown using simulated data in figure 1. LOH leads to spreading or ‘splitting’ of the AB portion of the BAF plot in proportion to the ratio of affected to unaffected cells sampled. We observed varying degrees of BAF spreading (figure 1(a)). 1p/19q is assumed to be an early event and hence present in all tumor cells in ODG. Supporting this, in each case the two arms showed the same degree of spreading, and spreading in these arms was always as wide as or wider than all other tumor-related BAF changes. Thus we normalized for sample purity by assessing spreading for each case relative to the BAF seen on 1p and 19q. We categorized BAF on each arm as normal, broadly split, i.e. as wide as 1p/19q, or thinly split, i.e. definitively split, but less widely so than 1p/19q. Determination of the karyotypic alteration underlying the observed change in BAF requires assessment of the LRR. For broadly split BAF, LRR can indicate single-chromosome loss or copy-neutral change, or it can be indeterminate (figure 1(b)). No homozygous deletions were detected. FFPE tissue suffers from noisy readings, and LRR was more likely to permit inference of underlying karyotypic change for broadly split BAF. Even for thinly split BAF, however, LRR clearly indicated the karyotypic change in some cases. The amount of DNA present at baseline is two-thirds that present following chromosomal gain, whereas loss reduces the amount to half of the original level, so signal change due to gain is expected to be less pronounced, i.e. thinly split (figure 1(c), left). Once gain was excluded, the degree of splitting allowed categorization of each change as affecting all of the tumor (broad splitting) or a subpopulation (thin splitting). Thin splitting as seen when an abnormality is limited to a subpopulation of the tumor is shown with indeterminate LRR in this example (figure 1(c), middle). Microarray results on an individual chromosome from a low-purity sample (figure 1(c), right) can resemble results indicating a subpopulation, in that BAF spreading is limited and LRR may be indeterminate, but in low purity, even 1p and 19q would have limited BAF spread.
Figure 1.
Simulated data to demonstrate intepretation of B-allele frequency (BAF, white dots) and log R ratio (LRR, magenta line). Each plot depicts data for one chromosome with two arms separated by the centromere, p-arm on left. The centromeric region generates no reads. A cyan line is plotted at neutral LRR position for clarity. (a) BAF plots representative of normal data (left) and BAF with wide (middle) and less wide (right) spreading of the values in the middle (‘AB’) portion of the plot. If the spread in these values is as wide as that of 1p/19q in the case, BAF is classified as broady split; if there is spread, but it is less wide, BAF is classified as thinly split. (b) LRR and interpretation in broadly split BAF, with chromosome loss or hemizygosity (left), copy neutral loss of heterozygosity (middle) and indeterminate LRR (right). (c) LRR and interpretation in thinly split BAF, with chromosome gain or trisomy (left) and tumor containing abnormality at this locus in only a subpopulation, with LRR indeterminate as to the chromosomal change (middle). Microarray data from sample containing a large component of non-tumor tissue mixed with tumor (right) can resemble on an individual chromosome the result when a defect is present only in a tumor subpopulation, but in the former case BAF plots even from 1p and 19q would not show wide spreading—a low purity sample would not show wide spread of BAF in any chromosome, whereas a sample only slightly contaminated by normal tissue would show wide spread for those abnormalities that are present through the entire tumor.
Each clone’s size within the tumor is a function both of its likelihood of arising and its relative fitness. To estimate these values for a few common defects, we defined occurrence (O) as the proportion of all assayed cases in which the defect was present. We estimated extent (E) as the proportion of all affected cases in which the defect involved the entire tumor, i.e. those that were broadly split. A defect’s extent is influenced by the growth rate of the affected subpopulation relative to the rest of the tumor, the point in the history of the tumor when the defect arises, and the chance of the defect arising independently elsewhere in the tumor. We assumed the latter two factors to be proportional to occurrence, allowing us to divide extent by occurrence for an estimate of the growth rate or fitness (F), the tendency for a clone bearing the defect to undergo relative expansion: E/O = F.
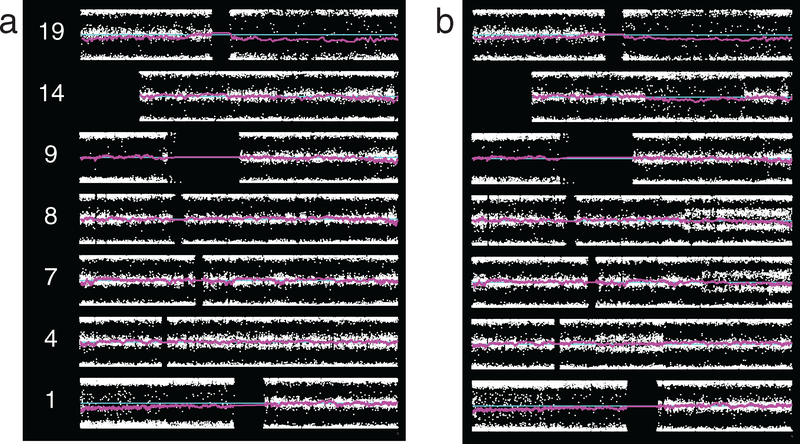
Examples in patient material of changes involving the whole arm or a segment, with broad or thin splitting of BAF, and with LRR indicating either deletion, copy-neutral change, or indeterminate change are shown in figure 2. Deletion of chromosome arms 1p and 19q is seen in both cases. Case 2_40 also demonstrates thinly split BAF along most of 4q, with LRR indeterminate as to the underlying karyotypic change, and broadly split BAF along most of 9p due to copy-neutral change. This pattern of involvement of almost all sites on the arm with sparing of a small centromeric portion was seen commonly on 9p, as was copy-neutral change. Case 3_11 similarly shows copy-neutral change on 9p, although in this instance it involves all SNP sites. This case also shows thin splitting on segments of 4q, 7q, and 8q due to indeterminate underlying karyotypic change, as well as a segment of 14q with broad splitting due to deletion.
Figure 2.
Representative SNP microarray data plots from ODG, two patients. Plots show chromosomes of interest, p-arm on left, with BAF at each SNP location (white dots), smoothed LRR (magenta), and neutral LRR position (cyan). (a) Grade II tumor, case 2_40. Whole arm broadly split BAF is seen on 1p and 19q. LRR shows this to be deletion. In addition to 1p/19q, this case demonstrates thinly split BAF along most of 4q, karyotypic change indeterminate, and broadly split BAF along most of 9p due to copy-neutral change. (b) Grade III tumor, case 3_11. Beyond 1p/19q, this case demonstrates broad splitting on the whole of 9p due to copy-neutral LOH, segmental thin splitting on 4q, 7q, and 8q due to indeterminate karyotypic change, and and broad splitting on a segment of 14q due to deletion.
In some instances the AB portion of the BAF plot was wider than expected for the case and appeared duplicated, but change was not definitive. BAF in these regions was designated ‘suggestive’ of splitting; this change was not counted in totals. In 2 O2, Xp and q (Xpq) showed whole-arm thinly split BAF with indeterminate LRR. In 1 O3, Xpq showed a suggestion of whole arm thinly split BAF. BAF was normal on X in all other female patients. X chromosome changes are not counted in totals to facilitate comparisons among all patients.
2.4. Alternate pattern of BAF
An alternate pattern of BAF was seen on one arm in three cases (2_10, 2_11, and 2_28), and on multiple arms in four cases (2_1 and 2_48 from the same patient, 2_51, and 3_3). This resembled broad splitting, but unusually, it was broader than the BAF alteration on 1p and 19q. Moreover, because even the most carefully selected tumor sample contains non-tumor cells such as endothelia and lymphocytes, the edges of the BAF plot are thicker in areas of broad splitting than in adjacent normal areas, but in areas of this alternate pattern of karyotypic change, the edges of the BAF plot were as thin as those in the normal areas adjacent. This suggests that the alterations are present in every cell. The presence of this defect in two samples from one patient, collected 16 years apart and assessed by microarray more than a year apart, renders artifact extremely unlikely and demonstrates the stability of these abnormalities. We designated this alternate pattern ‘A-type change’ and did not include it in totals.
3. Results
3.1. Group data on chromosomes affected
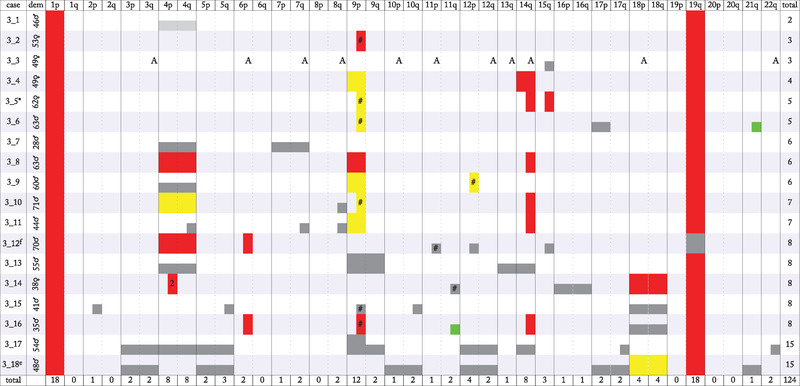
With this framework we analyzed 51 O2 and 18 O3, all 1p/19q codeleted. Results are shown in figures 3 and 4 where cases are ordered by number of arms with abnormalities and chronologically within that. O2 had a mean of 3.7 abnormal arms per case (median 3), with a standard deviation of 1.9. O3 showed on average more abnormalities, with a mean of 6.9 abnormal arms per case (median 6.5) with a standard deviation of 3.5. 17 O2 and only one O3 showed no definitive tumor-related abnormality beyond 1p/19q. 17 of 39 chromosome arms showed no abnormality in any of the 51 O2, whereas only seven arms had no abnormality in any of the 18 O3. Apart from 1p/19q, abnormalities in O2 were concentrated on 9p, 4pq, 18pq, 11q, and acrocentric 13q, 14q, and 15q. Four cases (three from one patient) showed abnormal 1q. Only 15 of 190 abnormalities (7.9% of total) lay on the remaining 27 arms (69.2% of total). The list of arms with most defects was similar in O3 but not identical, highlighting 9p, 4pq, 14q, 18pq, and 12p. Almost all arms with defects in both O2 and O3 were equivalently or more likely to be abnormal in O3, though 13q was only 0.6 fold as likely. In 14 of the 16 O2 with change on 9p this involved all or the large majority of the assayed sites on the arm apart from a small centromeric portion, with the latter pattern seen in seven of the 14. 9p change was copy-neutral in seven of 16 O2. In six O3 the defect on 9p involved all sites on the arm, whereas in the remaining six cases with defect on 9p, almost all sites were affected, with the small centromeric region showing sparing as seen in half of 9p defects in O2. Also as in O2, copy-neutral loss of heterozygosity was common among the 9p abnormalities in O3, accounting for six of the 12 cases. All 10 O2 with 4p defects also had 4q abnormalities, and all five cases with 18p defects also had 18q abnormalities; when both arms of 4 or 18 were involved the defects extended over the entirety of the mapped arms of that chromosome and shared the same degree of splitting and same karyotypic change, either hemizygosity, copy-neutral LOH, or indeterminate. In O3, 4p and 4q alterations were each definitively present in eight cases, seven of those with abnormalities in the whole of both 4p and 4q and sharing the same degree of splitting and same karyotypic change. Defects were present on 18p and 18q in four O3, all of them together and extending over the whole arms, with the same degree of splitting and same karyotypic change. Defects on each arm of 4, 9 and 18 ranged from 1.7 to 2.3 times as frequent in O3 as in O2, whereas 14q abnormality was 5.7 times as frequent in O3, demonstrating abnormal BAF in eight cases, six of them attributable to deletion. Gain was seen in only two O2, both on 11q, and in only two O3, one on 11q and one on 21q.
Figure 3.
Summary of SNP array data for grade II ODG. In addition to 1p/19q, abnormalities were concentrated on 4, 9p, 11q, acrocentric chromosome arms 13q, 14q, and 15q, and 18, along with 1q (3 cases from 1 patient); only 15 of 190 abnormalities (7.9% of total) lay on the remaining 27 arms (69.2% of total). Most abnormalities on 4 and 18 involved both arms. Copy-neutral change was seen frequently on 9p. Identified gain was rare, accounting for 2 abnormalities, both on 11q. Among 9p abnormalities, 14 of 16 were broadly split, whereas only 7 of 13 4q abnormalities and 2 of 5 18p abnormalities were broadly split, indicating differences in the extent of the respective subpopulations.
Figure 4.
Summary of SNP array data for grade III ODG. Data depicted as in figure 3, although here ‘2’ indicates 2 discontiguous abnormalities on one arm, and * indicates that Xp and Xq showed a suggestion of whole-arm thinly split BAF. Gain was identified on only 2 of the 124 arms with abnormalities. 9p showed frequent abnormalities occupying all or nearly all of the arm, with copy-neutral change common. 4 and 14q were the next most common sites of abnormalities, followed by 12p and 18. Defects on 4 and 18 usually involved both arms. 14q defects were generally broadly split BAF attributable to deletion. Among 9p defects, 11 of 12 were broadly split, whereas only three of eight 4q defects, and only two of four 18p defects, showed broad splitting. These differences indicate dissimilarity in the extent of subpopulations containing the defects.
We also investigated the co-occurrence of karyotypic defects by identifying cases containing a defect on a given arm, then considering the proportion of these having defects on each other arm (table 1). Aside from 4p/4q and 18p/18q, defects on most arms did not show evidence of co-occurrence beyond chance levels, but indications were present for a few. Among the eight cases with defects on 11q, 63% also had a defect on 18p and 18q. Five of the six cases with 13q defects also had a defect on 4p and 4q, and 10 of the 12 cases with 14q defects also had a defect on 9p.
Table 1.
Co-occurrence of karyotypic defects. Cases with defects on a given arm (‘primary arm’, rows) were assayed for defects on each other arm (‘secondary arm’, columns).
| Secondary arm | |||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1p | 1q | 2p | 2q | 3p | 3q | 4p | 4q | 5p | 5q | 6p | 6q | 7p | 7q | 8p | 8q | 9p | 9q | 10p | 10q | 11p | 11q | 12p | 12q | 13q | 14q | 15q | 16p | 16q | 17p | 17q | 18p | 18q | 19p | 19q | 20p | 20q | 21q | 22q | Total | ||
| primary arm | 1p | 6 | 1 | 3 | 3 | 26 | 30 | 3 | 4 | 4 | 1 | 1 | 3 | 3 | 41 | 7 | 1 | 3 | 6 | 12 | 6 | 3 | 9 | 17 | 10 | 3 | 6 | 3 | 4 | 13 | 14 | 100 | 4 | 3 | 69 | ||||||
| 1q | 100 | 75 | 100 | 4 | |||||||||||||||||||||||||||||||||||||
| 2p | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 1 | |||||||||||||||||||||||||||||||||
| 2q | 0 | ||||||||||||||||||||||||||||||||||||||||
| 3p | 100 | 100 | 50 | 50 | 100 | 100 | 50 | 50 | 50 | 50 | 100 | 100 | 50 | 50 | 100 | 50 | 50 | 100 | 50 | 50 | 2 | ||||||||||||||||||||
| 3q | 100 | 100 | 50 | 50 | 100 | 100 | 50 | 50 | 50 | 50 | 100 | 100 | 50 | 50 | 100 | 50 | 50 | 100 | 50 | 50 | 2 | ||||||||||||||||||||
| 4p | 100 | 6 | 6 | 94 | 6 | 6 | 11 | 6 | 6 | 6 | 6 | 50 | 22 | 17 | 17 | 17 | 6 | 28 | 28 | 11 | 6 | 6 | 6 | 28 | 28 | 100 | 6 | 18 | |||||||||||||
| 4q | 100 | 5 | 5 | 81 | 5 | 5 | 10 | 5 | 5 | 10 | 10 | 57 | 24 | 14 | 10 | 14 | 5 | 24 | 29 | 10 | 5 | 19 | 19 | 100 | 5 | 21 | |||||||||||||||
| 5p | 100 | 100 | 100 | 50 | 50 | 100 | 50 | 50 | 50 | 50 | 100 | 100 | 50 | 50 | 100 | 50 | 50 | 100 | 50 | 50 | 2 | ||||||||||||||||||||
| 5q | 100 | 33 | 67 | 67 | 33 | 33 | 67 | 67 | 33 | 33 | 67 | 67 | 67 | 33 | 33 | 67 | 67 | 67 | 100 | 33 | 33 | 3 | |||||||||||||||||||
| 6p | 100 | 67 | 67 | 33 | 67 | 33 | 33 | 33 | 33 | 33 | 33 | 33 | 33 | 100 | 3 | ||||||||||||||||||||||||||
| 6q | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 1 | |||||||||||||||||||||||||||||||||
| 7p | 100 | 100 | 100 | 100 | 100 | 1 | |||||||||||||||||||||||||||||||||||
| 7q | 100 | 50 | 100 | 50 | 50 | 50 | 50 | 100 | 2 | ||||||||||||||||||||||||||||||||
| 8p | 0 | ||||||||||||||||||||||||||||||||||||||||
| 8q | 100 | 50 | 100 | 50 | 100 | 100 | 100 | 2 | |||||||||||||||||||||||||||||||||
| 9p | 100 | 11 | 4 | 4 | 4 | 32 | 43 | 4 | 7 | 7 | 4 | 4 | 7 | 14 | 4 | 4 | 11 | 7 | 4 | 14 | 36 | 4 | 4 | 4 | 14 | 14 | 100 | 4 | 4 | 28 | |||||||||||
| 9q | 100 | 20 | 20 | 80 | 100 | 20 | 20 | 80 | 20 | 20 | 40 | 60 | 20 | 20 | 20 | 100 | 20 | 5 | |||||||||||||||||||||||
| 10p | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 1 | ||||||||||||||||||||||||||
| 10q | 100 | 50 | 50 | 50 | 50 | 100 | 50 | 50 | 50 | 50 | 50 | 50 | 100 | 100 | 100 | 50 | 2 | ||||||||||||||||||||||||
| 11p | 100 | 75 | 75 | 25 | 25 | 50 | 25 | 25 | 25 | 50 | 50 | 100 | 25 | 4 | |||||||||||||||||||||||||||
| 11q | 100 | 38 | 25 | 13 | 38 | 25 | 13 | 25 | 13 | 13 | 38 | 63 | 63 | 100 | 8 | ||||||||||||||||||||||||||
| 12p | 100 | 50 | 50 | 75 | 75 | 50 | 50 | 25 | 50 | 25 | 25 | 25 | 25 | 50 | 25 | 25 | 25 | 50 | 25 | 25 | 100 | 25 | 25 | 4 | |||||||||||||||||
| 12q | 100 | 100 | 100 | 50 | 50 | 100 | 100 | 50 | 50 | 50 | 50 | 100 | 50 | 50 | 100 | 50 | 50 | 100 | 50 | 50 | 2 | ||||||||||||||||||||
| 13q | 100 | 83 | 83 | 17 | 17 | 67 | 33 | 17 | 33 | 100 | 6 | ||||||||||||||||||||||||||||||
| 14q | 100 | 8 | 8 | 42 | 50 | 8 | 8 | 8 | 8 | 17 | 83 | 25 | 8 | 17 | 8 | 8 | 17 | 8 | 8 | 17 | 17 | 100 | 17 | 12 | |||||||||||||||||
| 15q | 100 | 29 | 29 | 14 | 14 | 14 | 14 | 14 | 14 | 14 | 14 | 100 | 7 | ||||||||||||||||||||||||||||
| 16p | 100 | 50 | 50 | 100 | 50 | 50 | 100 | 2 | |||||||||||||||||||||||||||||||||
| 16q | 100 | 25 | 75 | 50 | 50 | 50 | 100 | 4 | |||||||||||||||||||||||||||||||||
| 17p | 100 | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 100 | 100 | 2 | |||||||||||||||||||||||||
| 17q | 100 | 67 | 67 | 33 | 33 | 67 | 67 | 33 | 33 | 33 | 33 | 67 | 67 | 33 | 33 | 33 | 33 | 100 | 33 | 33 | 3 | ||||||||||||||||||||
| 18p | 100 | 11 | 11 | 11 | 56 | 44 | 11 | 22 | 11 | 44 | 11 | 11 | 22 | 22 | 56 | 11 | 11 | 22 | 11 | 11 | 22 | 11 | 11 | 100 | 100 | 11 | 9 | ||||||||||||||
| 18q | 100 | 10 | 10 | 10 | 50 | 40 | 10 | 20 | 10 | 40 | 10 | 10 | 20 | 20 | 50 | 10 | 10 | 20 | 10 | 10 | 20 | 10 | 10 | 90 | 100 | 20 | 10 | ||||||||||||||
| 19p | 0 | ||||||||||||||||||||||||||||||||||||||||
| 19q | 100 | 6 | 1 | 3 | 3 | 26 | 30 | 3 | 4 | 4 | 1 | 1 | 3 | 3 | 41 | 7 | 1 | 3 | 6 | 12 | 6 | 3 | 9 | 17 | 10 | 3 | 6 | 3 | 4 | 13 | 14 | 4 | 3 | 69 | |||||||
| 20p | 0 | ||||||||||||||||||||||||||||||||||||||||
| 20q | 0 | ||||||||||||||||||||||||||||||||||||||||
| 21q | 100 | 33 | 33 | 33 | 33 | 33 | 33 | 33 | 33 | 33 | 67 | 33 | 33 | 67 | 100 | 3 | |||||||||||||||||||||||||
| 22q | 100 | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 100 | 50 | 100 | 2 | |||||||||||||||||||||||||
| Mean | 100 | 8 | 16 | 42 | 42 | 54 | 60 | 42 | 50 | 19 | 10 | 13 | 23 | 14 | 55 | 29 | 32 | 41 | 19 | 30 | 43 | 42 | 29 | 39 | 14 | 14 | 28 | 36 | 44 | 45 | 47 | 100 | 39 | 25 | |||||||
Note: The proportion of cases with a defect on the primary arm that also have a defect on the secondary arm is given as a percentage. The bottom row gives the average value for instances in which the secondary arm has defects, and the rightmost column gives the total number of cases with a defect on the primary arm.
3.2. Group data on subpopulations
Assessment of subpopulations showed that 87.5% and 91.7% of O2 and O3 9p defects, respectively, were broadly split, yet only 53.8% and 37.5% of 4q defects, and only 40.0% and 50.0% of 18p defects, were broadly split in O2 and O3. Numerous instances of deletion or copy-neutral change were identified on each of these arms, suggesting that one of these two alterations underlay all observed defects. Hence thinly split change in these arms could be reliably interpreted as involvement limited to a subpopulation, whereas broadly split change demonstrated involvement of the whole tumor. Thus subpopulations with the the defects differed in extent. If we consider O2 and O3 together to estimate the fitness of these common defects (Methods), 9p occurred in 28 of 69 cases (occurrence = 28/69) and was broadly split in 25 of those 28 (extent = 25/28), for a fitness of 2.2. By the same calculation, 4q had a fitness of 1.6, whereas 18p had a fitness of 3.4. This indicates that although 18p defects have a lower likelihood of occurring, subpopulations containing those that do occur have a higher likelihood of becoming predominant.
3.3. Direct comparison of groups
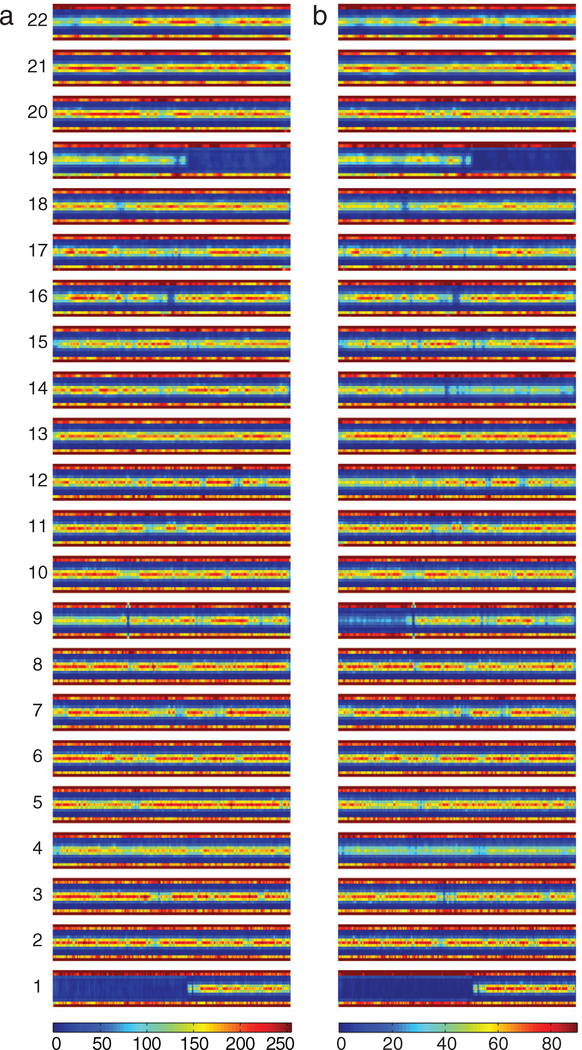
To extend our understanding of differences in SNP microarray readings from O2 and O3 we developed a means of displaying BAF data from multiple cases simultaneously (figure 5; Methods). Although this display avoids the case-by-case subjectivity of the manual assessment involved in producing summary figures, it is affected by the exact position of the defect on the chromosome, as overlapping defects will add whereas adjacent defects will not. This may account for the failure of some of the manually detected changes depicted in figures 3 and 4 to be readily evident here. The apparent reduced signal in both O2 and O3 in areas such as 19p and the centromeric part of 22q may reflect lower densities of SNP sites. This analysis shows that 4pq are abnormal in O2 but more markedly so in O3, as expected for involvement of a larger proportion of cases. 9p follows the same pattern. Relative to O2, in O3 12p is more affected, 14q is prominently altered, and 18pq are also more involved.
Figure 5.
Direct comparison of BAF data from all grade II ODG (O2) and all grade III ODG (O3). (a) O2. (b) O3. Color bars show number of values per plot element; colors are normalized to reflect the same proportion of cases in each group (Methods). All cases show 1p/19q. Altered BAF on 4pq is evident in O2, but more striking in O3. Changes on 9p are seen in O2 but are more marked in O3. 14q is prominently altered in O3.
3.4. Recurrent tumors
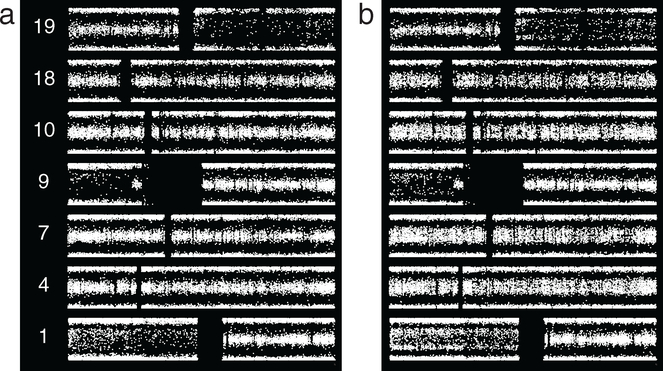
Having established the increased number of SNP microarray abnormalities and common patterns of subpopulation change in ODG progression using group data, we turned to assessment of individual tumors over time, using sequential resections from individual patients. In light of the rarity of microarray data from multiple resections over time, BAF is shown for each case pair. In one instance, three samples resected over a 6 year period from one patient showed no change in BAF data (supplementary figure S2 available at stacks.iop.org/CSPO/1/015001/mmedia), demonstrating both the reproducibility of the assay and the potential for karyotypic stability in ODG. BAF changes were seen in the remainder of the instances where multiple specimens could be examined from one patient. Figure 6 depicts BAF from two specimens from one patient resected approximately 6 years apart. In the earlier case, 2_19, 1p/19q are accompanied by broad splitting on most of 9p. Case 2_22 added to these suggested thin splitting on 4pq, 7pq, 10pq, and possibly 18pq. Defects present only in subpopulations could be obscured by the lower tumor purity of the second sample, which is indicated by BAF values for the broadly split abnormalities. Lower purity may also account for the copy-neutral change on 9p becoming indeterminate. The subpopulation defects likely arose in the interval between procedures.
Figure 6.
Grade II ODG from one patient, resections separated by approximately 6 years. (a) Case 2_19 shows 1p/19q, along with broad splitting of BAF on most of 9p. (b) Case 2_22 shows these abnormalities, with suggested thin splitting on 4pq, 7pq, 10pq, and possibly 18pq. Values for 1p, 19q, and 9p indicate lower purity of the more recent sample. The abnormalities suggested in the second resection likely arose and/or propagated in the interval between cases.
Approximately 16 years separated the procedures producing the cases shown in figure 7. In addition to ‘A-type change’ (Methods), the older case, 2_1, has only 1p/19q. Case 2_48 adds segmental broad splitting on 9p and thin splitting on 13q as well as whole arm thin splitting on 4pq and 6pq. The diagnostic neuropathology report for the more recent specimen noted that it had few mitoses and its Ki-67 proliferation index was not high, but it was ‘more cellular and cytologically more atypical than the 1996 specimen’. 16 years was sufficient in this case for increases in karyotypic and cytologic abnormalities, though mitotic rate and proliferative index remained low.
Figure 7.
ODG, two resections from same patient separated by approximately 16 years, first resection not graded, second grade II. (a) Apart from 1p/19q, case 2_1 shows broadly split BAF on segments of 2p, 3q, 4q, 5q, 10q, 12pq, and 15q. Some of these defects are more readily visualized in the newer specimen. The BAF splitting at the non-1p/19q sites is atypical (see text). (b) Case 2_48 shows the same abnormalities, as well as segmental abnormalities on 9p (broadly split) and 13q (thinly split), and whole-arm thin BAF splitting on 4pq and 6pq. The atypical defects, thought to be unrelated to the neoplasm, were stable over 16 years, while the set of tumor-related abnormalities expanded.
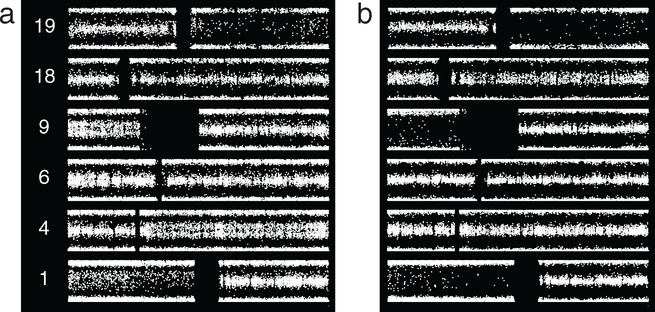
Results shown in figure 8 from resections approximately 7 years apart highlight the complex possibilities for subpopulation change over time. Case 2_29, the older of the two cases, shows 1p/19q and thinly split BAF on most of 4q and all of 9p, as well as suggested thin splitting on a segment of 6p. Case 2_24, the more recent sample from the same patient, shows broadly split BAF on 9p, indicating that abnormality is now present in essentially the whole tumor. In contrast, chromosomes 4 and 6 show normal BAF, indicating that the subpopulation or subpopulations containing abnormalities on 4q and possibly 6p are no longer present. A suggestion of thin splitting is seen on 18pq in the newer sample. Thus, within a given tumor, some subpopulations expand while others disappear.
Figure 8.
Grade II ODG, two resections from one patient separated by 7 years. (a) Apart from 1p/19q, case 2_29 shows thinly split BAF on most of 4q and all of 9p, and a suggestion of thin splitting on a segment of 6p. (b) For the later case, 2_24, the 9p abnormality is now broad splitting, whereas the abnormality on 4 is nearly absent, and no abnormality is seen on 6p. A suggestion of thin splitting is seen on 18pq. Expansion or extinction are possible trajectories over time for a given subpopulation.
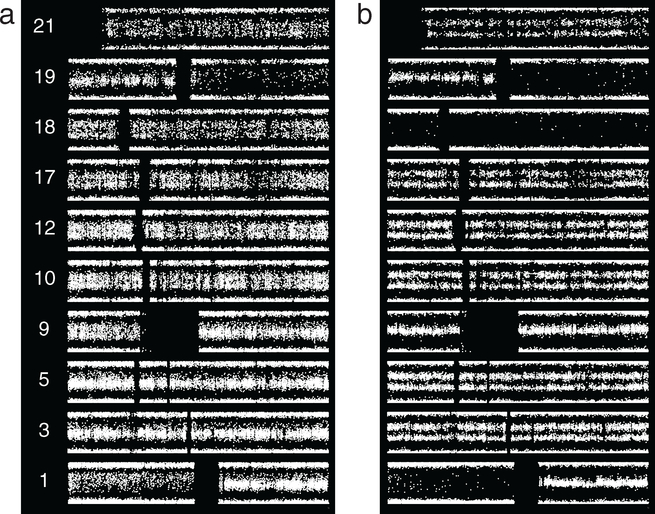
The microarray data shown in figure 9 correspond to O2 that progressed over approximately 7 years to O3. The first case, 2_2, shows suggested thin splitting on 9p, 10pq, 12pq, 17pq, 18pq, and 21q. In case 3_18, 10pq, 12pq, 17pq, and 21q all show definitive thin splitting, but 9p shows no abnormality, and 18pq has become broadly split. 3pq and 5pq also show thin splitting. As with the pair of cases shown in figure 8, these data demonstrate that some subpopulations disappear from the tumor while others expand. The differential change on 18 relative to those on 10, 12, 17 and 21 indicates that these are not all present in the same subpopulations. A subpopulation containing the abnormality on 18 may be outcompeting others, or, alternatively, the defect on 18 may arise independently multiple times. As shown below, fitness estimation favors the former explanation.
Figure 9.
Grade II (left) versus grade III ODG from the same patient, 7 year interval. (a) In addition to 1p/19q, case 2_2 demonstrates a suggestion of thin BAF splitting on 9p, 10pq, 12pq, 17pq, 18pq, and 21q. (b) Case 3_18 demonstrates thin splitting on 10pq, 12pq, 17pq, and 21q, as well as 3pq and 5pq. In the grade III tumor the potential 9p abnormality is absent whereas the chromosome 18 abnormality is broad splitting of BAF, indicating the defect occupies essentially the entire sampled tumor. The differential changes suggest differences in the rate of expansion or the likelihood of repeated occurrence. For the subpopulation with abnormality on 18, fitness estimation (text) favors the former interpretation.
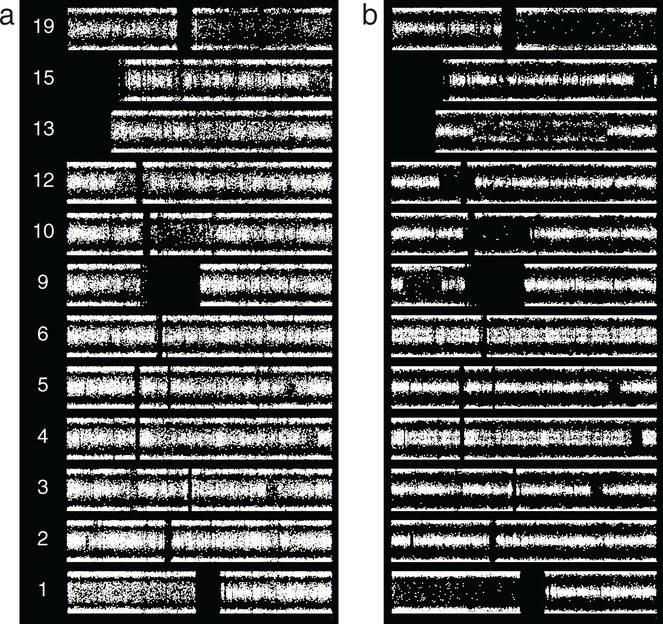
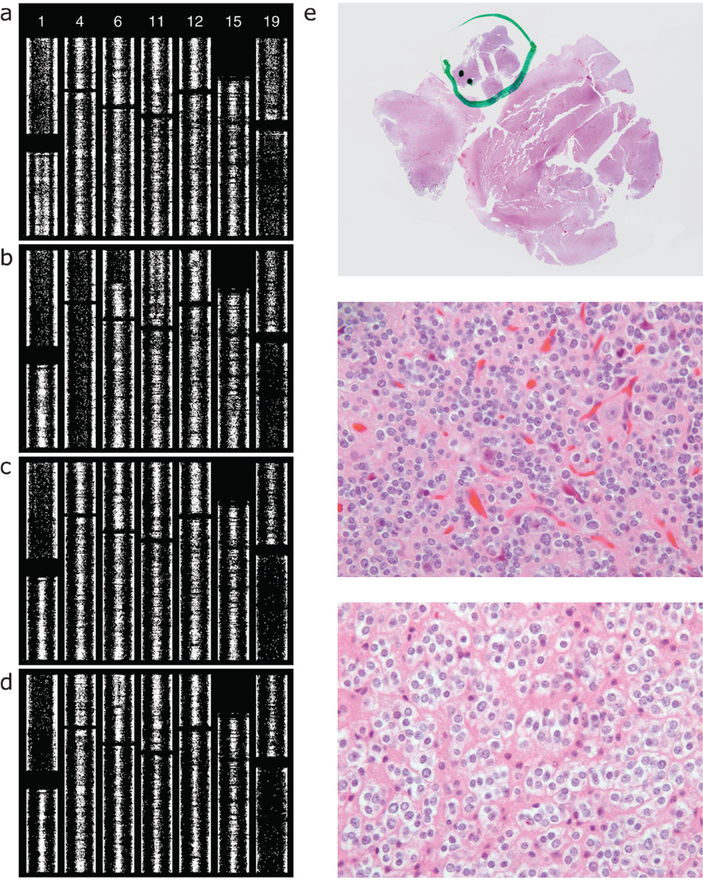
We assessed two resections from one patient separated by an 11 year interval and graded as O2 then O3 (figure 10). Case 2_18 shows thin splitting on all of 1q and a suggestion of thin splitting on all of 19p, with no other abnormalities apart from 1p/19q. The neuropathology report for the more recent specimen, case 3_12, included a comment stating that ‘while most of the lesion appears low grade, and no necrosis or vascular proliferation are identified, in one cellular region more than five mitotic figures are identified in 10 high power fields, and the Ki67 proliferation index is over 30%’. We performed microarray analysis separately on the bulk of the tumor and the portion identified as high-grade at the time of diagnostic assessment. The high grade portion shows increased abnormalities: broadly split BAF on all of 4pq and a segment of 6p as well as thin splitting on segments of 11p, 12p, and 15q (figure 10(b)). It is this high-grade portion of this tumor that is represented as case 3_12 in figure 4 and included with the O3 cases in figure 5. The low-grade portion shows no abnormality beyond 1p/19q (figure 10(c)). Reviewing the slides from which tissue was scraped, we confirmed that SNP assay of the second specimen done at the time of signout included material from all regions in the block. This SNP data shows thin splitting or suggested thin splitting on the whole of 4pq as well as a segment of 6p (figure 10(d)), demonstrating averaging of the signals from the high grade area (figure 10(b)) and the much larger low grade area (figure 10(c)). The portion identified as high grade at the time of signout shows increased cellularity, hyperchromasia, irregular nuclear membranes, and mitoses (figure 10(e)).
Figure 10.
SNP microarray data, two resections from the same patient, separated by an 11 year interval. The first resection was diagnosed as grade II, the second, grade III. The report for the second specimen included a comment attributing the assigned grade to a focal region. For the present study we repeated the microarray analysis of tissue from the second resection, analyzing separately the portion that was assessed as high grade. Here SNP data are shown vertically. (a) From first resection. Apart from 1p/19q, case 2_18 shows thinly split BAF on the whole of 1q and a suggestion of thin splitting on 19p. (b) From second resection, case 3_12, high grade portion. Beyond 1p/19q this focus shows broadly split BAF on the whole of 4pq and a segment of 6p, and thinly split BAF on segments of 11p (nearly whole-arm), 12p (focal), and 15q. The suggested 1q/19p abnormality of the original tumor did not propagate. (c) From second resection, case 3_12, low grade portion. 1p/19q is the only abnormality. (d) SNP array results from the assay on the second resection done at time of signout. Review of the slides used showed that essentially all tissue was sampled. In addition to 1p/19q, thin splitting, or a suggestion thereof, is present on 4pq and on a segment of 6p. Comparison of panels (b)–(d) demonstrates that a broadly split abnormality admixed with other tumor normal in that region results in thin splitting. (e) Hematoxylin and eosin slide from case 3_12, with markings applied at time of signout (top) indicating the high grade area. This focus is shown in the middle panel, with an area of the remaining tumor shown in the bottom panel (original magnification of these two panels 400×). The high grade component shows greater cellularity, nuclear hyperchromasia, abnormal nuclear contours, and mitotic activity. A high quality version of this figure is available as a supplemenatary file (figure S3, available at stacks.iop.org/CSPO/1/015001/mmedia).
For the five tumors where SNP microarray results changed between initial and later resections, subpopulation changes are summarized in table 2.
Table 2.
Summary of subpopulation changes in second biopsies of case pairs.
| Pair | Gap | Treatment | Summary |
|---|---|---|---|
| 2_19 2_22 | 6y | RCHOP 4y after first resection, for lymphoma | 9p defect is present throughout tumor in both cases, and suggested subpopulations with defects on 4pq, 7pq, 10pq, and possibly 18pq arise |
| 2_1 2_48 | 16y | Conservative | Appearance of 9p defect throughout the tumor, and appearance of subpopulation(s) with defects on 4pq, 6pq, and 13q, indicating 9p change is earlier to occur and/or faster to expand |
| 2_29 2_24 | 7y | Conservative | Subpopulation with 9p defect becomes predominant; subpopulation with 4q abnormality becomes nearly undetectable and subpopulation with suggested 6p abnormality disappears; subpopulation with 18pq defect may arise |
| 2_2 3_18 | 7y | Conservative | Initially only suggested subpopulation with 18pq defect becomes predominant relatively quickly; initially only suggested subpopulation(s) with 10pq, 12pq, 17pq, and 21q defects appear(s) to expand, but relatively more slowly; appearance of subpopulation(s) with 3pq and 5pq defects; disappearance of suggested subpopulation with 9p defect, demonstrating that subpopulation with 9p may be outcompeted by other subpopulations |
| 2_18 3_12 | 11y | Radiation (5400 Gy) 4y after first resection | Relatively rapid/early appearance of subpopulation(s) with 4pq and 6p defects; relatively slow/late appearance of subpopulation(s) with 11p, 12p, and 15q defects; disappearance of subpopulation(s) with 1q and suggested 19p defects, suggesting low propagation of such subpopulation(s) |
Note: For simplicity descriptions take a given karyotypic change to be limited to a single subpopulation.
3.5. Evaluating estimates of subpopulation fitness
We then returned to our consideration of fitness, using those case pairs where the initial resection permitted comparison as it contained two of the subpopulations for which fitness was calculated. In the case pair of figure 8, a subpopulation with a 9p abnormality came to predominate within the tumor whereas a subpopulation with 4q, which seemed to occupy a comparable proportion of the tumor initially, disappeared. In the case pair of figure 9, a clone with abnormality on 18pq came to predominate, whereas a clone with 9p abnormality disappeared. Although very few, these direct tests accord with the relative fitness we estimated from all cases for these subpopulations. To remove the possibility of fitness estimates being driven by these case pairs, we excluded them and repeated the estimation. This yielded fitness values for 18p, 9p, and 4q of 3.0, 2.3, and 1.6, respectively, again predicting the outcome of the subpopulation competitions.
4. Discussion
Using single nucleotide polymorphism microarray data, we assessed 69 specimens of 1p/19q codeleted, diagnostically graded ODG. We found an increased number of chromosomal abnormalities in O3, confirming previous work, with no single change associated with progression or higher grade. After 1p/19q, we found the largest number of abnormalities in both O2 and O3 on 9p, and observed that this change was copy-neutral in approximately half of cases. Direct comparison of all cases in the two groups highlighted abnormalities on 4pq and 9p, and in O3 the increased proportion of these with defects, as well as those on 12p, 14q and 18pq. The 9p defect involved all assayed sites on the arm in approximately half of the cases, whereas defects on 4 and 18 frequently involved the entire mapped chromosome. By comparison with the 1p/19q data from the same case, we categorized each abnormality as belonging to a subpopulation or the whole tumor. Among affected specimens, the 9p defect involved the whole tumor in roughly 90% of cases, whereas the next most common defect, 4q, involved the entirety of the tumor in roughly one-half of cases, as did 18p. Using these three abnormalities, we developed a simple framework for distinguishing the fitness with which a subpopulation expands within the tumor from that defect’s likelihood of occurring. Using each karyotypic defect as a marker for a subpopulation or subpopulations, we evaluated specimens from six patients with multiple resections spanning the previous 18 years. We demonstrated the averaging of the microarray signal in a sample that included multiple subpopulations, and we showed that subpopulations exhibit rich dynamics, with some expanding within the tumor, others changing little, and others disappearing. All changes present throughout the tumor at first resection remained so. Our estimates of fitness predicted composition at recurrence for the two tumors containing multiple evaluable subpopulations in the first resection. A 1q/19p translocation has been implicated in the mechanism of the 1p/19q codeletion, and we identified in one case a candidate 1q/19p codeleted subpopulation that was absent at recurrence.
4.1. Chromosomes affected
Several chromosome changes we observed accord with findings of other studies, among them changes on X in a subset of female patients [17] and loss on 13q [2, 18]. Loss on 14q, which we observed at higher frequency in O3, was identified in an early study of ODG karyotype [19]. In a study of ODG recurrence, the second codeleted case showed loss of 12p, 15q, and 18 [17], sites frequently altered among cases we assessed. Several of our findings also differed from earlier results. Although a meta-analysis of CGH data found loss on 15q significantly correlated with higher grade malignancy in 1p/19q codeleted ODG [20], in the cases we studied 15q abnormalities were approximately half as common in O2 as in O3, similar to the ratio of abnormalities in general, and 15q defects in each group appeared scattered through the cases rather than concentrated in cases with greater numbers of defects. Loss on chromosome 4, which we observed in a number of cases, was also reported in multiple studies [2, 17, 18], but a report that subdivided O3 into three groups found the highest-grade subdivision had more chromosomal alterations and greater loss of chromosome 4 [21], whereas we observed change commonly on 4 in O2, and no disproportionate increase in O3.
In the cases we evaluated, 9p was the most common site of non-1p/19q karyotypic defect, occurring in 31% of O2 and 67% of O3. Change on chromosome 9 in ODG was described in numerous earlier studies [2, 18], including the initial report of 1p/19q loss in ODG [1]. 9p abnormalities were found in three codeleted gliomas at recurrence [17, 22]. Another study found that 45.6% of anaplastic ODG showed LOH on 9p [23], whereas we found 9p LOH in 2/3 of O3. 9p deletion was associated with necrosis or microvascular proliferation in 1p/19q codeleted ODG [24], and in the study that subdivided codeleted O3 into three groups based on microvascular proliferation and necrosis, the two higher subdivisions had greater losses of 9p and 9q [21]. A recent study found that the presence of contrast enhancement, likely correlating with higher histologic grade, was associated with greater loss of chromosome 9p [25]. In contrast to these associations with higher grade, we commonly found 9p defects in O2. Moreover, if we assume fewer karyotypic abnormalities to reflect lower grade, in six O2 cases, 9p abnormality was the only defect beyond 1p/19q. Similarly, a study of 12 recurrent codeleted ODG found no link between 9p abnormalities and progression [26]. Among cases we analyzed with 9p alteration, we found it to be copy-neutral in 43.8% of O2 and 50.0% of O3. The high proportion of copy-neutral change among defects on 9p may reflect in part the tendency of the abnormality to involve the entire tumor, making determination of karyotypic change more feasible. Another study also found that copy-neutral LOH was most common on 9p, occurring in 13.2% of O3 [23]. The high proportion of copy-neutral change on 9p we saw in both O2 and O3 argues against association with higher grade, and in six of the seven O2 in which copy-neutral LOH was present on 9p there was at most one additional defect beyond 1p/19q. 9p is a common site of copy-neutral LOH in other tumors [27]. Copy-neutral LOH may promote neoplasia by a number of mechanisms, including unmasking of recessive mutant alleles or alteration of the expression level of imprinted genes [28, 29]. Chromosomal defects affecting most of the cells sampled tended to produce more readily interpretable LRR, but in some cases we did not observe this on 9p, even in specimens of high purity and signal quality. One possibility is that deleted and copy-neutral subpopulations were both present, leading to an intermediate degree of LRR change. This may reflect the loss of one arm in the majority of tumor cells followed by duplication of the remaining arm in a subset [30]. Absence by immunohistochemistry of the protein product of the CDKN2A gene on 9p was seen in one-third of cases with copy-neutral LOH there [23], potentially implicating altered expression in tumor progression. The ramification of the near-whole arm change with sparing of centromeric sites on 9p, which we saw on 7/16 O2 and 6/12 O3 with 9p defect, is unclear.
Among the cases we examined, only four of 2691 arms demonstrated abnormal BAF attributable to gain, with three of the instances occurring on 11q. Although an earlier study found losses commonly on 11q and 13q in histologically-defined ODG [31], gain on 11pq was reported in a 1p/19q codeleted ODG [17], and a more recent study found that 11q was most common gain among 1p/19q codeleted ODG, occurring in 19.1% of cases [23]. The smaller proportion seen in our data could reflect limitations of analysis of FFPE tissue. Reports using frozen tissue found gain in a few ODG on 7pq [22, 23, 25], and one ODG with a colony with extra copy of chromosome 7 was noted in an early report [32]. The suggested or indeterminate changes on 7 we occasionally observed may be due to gain.
We found no evidence for interdependence among defects on most arms, but a surprising frequency of co-occurrence among a few: 18pq defects among cases with defects on 11q, 4pq defects among cases with defects on 13q, and 9p defects among cases with defects on 14q. We referred to the sites of defects as primary and secondary in reference to our method of searching for relations, but the terms do not refer to time of occurrence or to causality. Because the ‘secondary’ arm in each of these relations is a more common site of defects than the ‘primary’ arm, the most straightforward interpretetation is that the presence of defect on the secondary arm increases the likelihood of defect on the primary arm. This would mean that most cases with the primary defect would also have the secondary one, as we observed for defects on these arms. Determination of the true conditional probability will require more cases and normalization for the number of other defects present.
4.2. Subpopulations
The presence of subpopulations in glioma has been appreciated for several decades [32], and a recent study confirmed that ODG is a heterogeneous tumor [13]. Moreover, the surgical resection is on some level a random sampling, although the surgeon will resect as much of the tumor as safely possible. To minimize heterogeneity we limited our consideration to histologically diagnosed ODG in which 1p and 19q showed LOH on all assayed sites. BAF data showed that 1p and 19q abnormalities were present in the great majority of sampled cells in all cases, indicating extent throughout the tumor [33]. When we evaluated recurrent tumor, these defects and all those present throughout the whole tumor were preserved in every case [15]. In contrast, one study found 3/10 gliomas showed LOH on 1p and 19q only in recurrence and 2/10 gliomas showed 1p and 19q LOH only in the initial resection [14], and normal 1p karyotype status has also been reported in recurrent tumor after the primary tumor showed loss of 1p [34], but these may not have been codeleted ODG. By visual assessment of cases reported here the spreading of the AB portion of the BAF plot for 1p was always equal to that for 19q, suggesting that in no case did a subpopulation arise with only one of these defects. This in turn suggests that the contribution to tumorigenesis may involve codeletion per se rather than abnormal genes on one or the other arm revealed by the codeletion. Consideration of cases with ODG histology and no codeletion would be required to assess this, but in practice the association we observe between ODG histology in adult tumors and codeletion is strong, potentially limiting availability of cases for study. The likely LOH on 1q and 19p we observed in the early material from one patient (figure 10) may relate to a role for this translocation in 1p/19q LOH and tumorigenesis [35, 36]. In an earlier report abnormality on 19p was seen in the initial resection, but not the recurrence, of a 1p/19q codeleted ODG [17]. The potential for a portion of tumor hemizygous for 1q/19p to exist as a subpopulation distinct from that hemizygous for 1p/19q may suggest separation of the two abnormalities during an early mitotic event. The normal appearance of 1q and 19p in later material from the same patient indicates that a subpopulation with that abnormality may be prone to disappear as the tumor progresses.
Results from multiple specimens from individual patients over time highlight the potential for subpopulations to arise, expand, remain proportionally stable, or contract. The sequential resections also confirm that the thinly split defects involve different subpopulations, as expected from the abnormalities’ relatively low frequency and their general apparent independence.
If a clone’s extent within the tumor is a function of its fitness and its likelihood of occurring, the fact that defects on 9p often involved the entire tumor could in principle be due to high likelihood of occurrence, which could cause them to appear early, giving time for their subpopulation to comprise a greater proportion of the tumor, and/or to arise independently multiple times. Alternatively the presence of 9p defect throughout the tumor could be due to high fitness permitting rapid growth relative to the rest of the tumor. To assess the relative contributions of fitness and chance of occurrence, using group data we estimated these parameters for a few defects of reasonable frequency. The interaction of tumor subpopulations can be modeled using differential equations [37], but as a simple assessment we assumed no interaction. Our estimates identified the relative fitness of 18p as greater than that of 9p, and that of 9p as greater than that of 4q. We then used the estimate of fitness to predict composition at recurrence in the two recurrent tumors that harbored multiple clones at first resection, finding the higher-fitness subpopulation extended throughout the tumor and the other subpopulation lost in both cases. Based on its frequency among all cases, occurrence, and hence repeat occurrence, is more likely for 9p than for 18p. Without unduly generalizing from a single case, we may infer that the expansion of 18p and loss of 9p in recurrence suggests that fitness is a more important determinant of outcome than is the possibility of repeat occurrence. Indeed, repeat occurrence is expected to be uncommon, given the low overall occurrence rates. This approach suggests that the tendency of 9p to involve the whole tumor is largely attributable to its occurrence being slightly less uncommon than that of other defects, permitting 9p defects to arise early and extend throughout. However, when two subpopulations are present, the uncommonness of defects arising makes fitness more relevant than the possibility of additional occurrence of either defect.
Such an approach can identify karyotypic defects that occur rarely but have wide extent, and hence high fitness. This may have important implications for treatment strategies, as it may be advantageous to guide therapy with an understanding of clonal composition of a given tumor and patterns of subpopulation responses to treatment established from previous cases [38]. Analysis of primary and metastatic tumor samples demonstrates a high degree of karyotypic heterogeneity in other cancers [39], underlining complexities facing truly individualized therapy. Similar mechanisms of increased tumor aggressiveness conferred by subpopulation changes are likely also involved in ODG.
We have framed the interaction among subpopulations as competition, but mutualism, parasitism, and other relationships are also probable. Animal and modeling studies indicate that a subpopulation can create a microenvironment favoring tumor growth [40]. In glioblastoma, subpopulations communicate by paracrine mechanisms in a fashion that supports the growth of the whole tumor [41]. Even when subpopulations of a tumor lack the ability to invade, their joint presence can confer this ability [42]. The tumor cell lineage can be traced from the multiple mutations that occur [43], and it is likely that greater availability of microarray data will lead to increased understanding of the subpopulation dynamics over time and their relation to disease pathogenesis and progression. A larger number of repeated samples from individual patients might permit determination of the range of rates of acquisition of abnormalities, which would in turn allow dating of the neoplasm. In other institutions where 1p/19q status is assessed by microarray, the data from sizable numbers of cases may already have been collected, and could be used for analyses of this type.
In evaluating tumor subpopulations, we observed an alternate pattern of change in BAF in a few cases, characterized by very wide spreading of BAF with no ‘rim’ of values at the edges, suggesting every cell sampled has the abnormality. The relationship of this pattern of change in BAF to the development of ODG is unclear, but it was present in only a few cases, the affected patients were not as a group young, and the multiply sampled tumor with this type of change remained low grade histologically 16 years after the first surgery, all suggesting that a relationship with tumor development or progression is unlikely. Awareness of this type of BAF is important for manual or automated analysis of SNP microarray data.
Use of SNP microarray data for assessment of 1p/19q status in oligodendroglioma has the advantage of providing information on both the karyotypic site of abnormalities and their extent within the tumor. This information can be used to assess subpopulations. Understanding of tumor development and progression will benefit from knowledge of the presence and trajectories of these subpopulations, which are likely to influence responses to therapy.
Supplementary Material
Acknowledgments
The authors would like to acknowledge D Anderson and R Shane of the Molecular Pathology Laboratory as well as S Flowers-Yox, A Jackson, and M Taylor of the Histology Reference Laboratory for expert technical assistance. This project was supported by the National Institute for Neurological Disorders and Stroke R25NS079185 (DN).
Footnotes
Disclosure/conflict of interest
The authors have no disclosures to report.
Supplementary material for this article is available online
References
- [1].Reifenberger J, Reifenberger G, Liu L, James CD, Wechsler W and Collins VP 1994. Molecular genetic analysis of oligodendroglial tumors shows preferential allelic deletions on 19q and 1p Am. J. Pathol 145 1175–90 [PMC free article] [PubMed] [Google Scholar]
- [2].Burger PC, Minn AY, Smith JS, Borell TJ, Jedlicka AE, Huntley BK, Goldthwaite PT, Jenkins RB and Feuerstein BG 2001. Losses of chromosomal arms 1p and 19q in the diagnosis of oligodendroglioma. A study of paraffin-embedded sections Mod. Pathol 14 842–53 [DOI] [PubMed] [Google Scholar]
- [3].van den Bent MJ, Looijenga LHJ, Langenberg K, Dinjens W, Graveland W, Uytdewilligen L, Sillevis Smitt P A, Jenkins RB and Kros JM 2003. Chromosomal anomalies in oligodendroglial tumors are correlated with clinical features Cancer 97 1276–84 [DOI] [PubMed] [Google Scholar]
- [4].Merlo LMF, Pepper JW, Reid BJ and Maley CC 2006. Cancer as an evolutionary and ecological process Nat. Rev. Cancer 6 924–35 [DOI] [PubMed] [Google Scholar]
- [5].Martinez P et al. 2013. Parallel evolution of tumour subclones mimics diversity between tumours J. Pathol 230 356–64 [DOI] [PubMed] [Google Scholar]
- [6].Sottoriva A, Spiteri I, Piccirillo SGM, Touloumis A, Collins VP, Marioni JC, Curtis C, Watts C and Tavaré S 2013. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics Proc. Natl Acad. Sci. USA 110 4009–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Patel AP et al. 2014. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma Science 344 1396–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Nowell PC 1976. The clonal evolution of tumor cell populations Science 194 23–8 [DOI] [PubMed] [Google Scholar]
- [9].Parkin B, Ouillette P, Li Y, Keller J, Lam C, Roulston D, Li C, Shedden K and Malek SN 2013. Clonal evolution and devolution after chemotherapy in adult acute myelogenous leukemia Blood 121 369–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kreso A et al. 2013. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer Science 339 543–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hunter C et al. 2006. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy Cancer Res. 66 3987–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wu X et al. 2012. Clonal selection drives genetic divergence of metastatic medulloblastoma Nature 482 529–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ray M, Goldstein S, Zhou S, Potamousis K, Sarkar D, Newton MA, Esterberg E, Kendziorski C, Bogler O and Schwartz DC 2013. Discovery of structural alterations in solid tumor oligodendroglioma by single molecule analysis BMC Genomics 14 505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lass U. et al. Clonal analysis in recurrent astrocytic, oligoastrocytic and oligodendroglial tumors implicates IDH1-mutation as common tumor initiating event. PLoS One. 2012;7:e41298. doi: 10.1371/journal.pone.0041298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Jeuken JWM, Sijben A, Bleeker FE, Boots-Sprenger SHE, Rijntjes J, Gijtenbeek JMM, Mueller W and Wesseling P 2011. The nature and timing of specific copy number changes in the course of molecular progression in diffuse gliomas: further elucidation of their genetic ‘life story’ Brain Pathol. 21 308–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P 2007. The 2007 WHO classification of tumours of the central nervous system Acta Neuropathol. 114 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jeuken JWM, Sprenger SHE, Vermeer H, Kappelle AC, Boerman RH and Wesseling P 2002. Chromosomal imbalances in primary oligodendroglial tumors and their recurrences: clues about malignant progression detected using comparative genomic hybridization J. Neurosurg 96 559–64 [DOI] [PubMed] [Google Scholar]
- [18].Kitange G, Misra A, Law M, Passe S, Kollmeyer TM, Maurer M, Ballman K, Feuerstein BG and Jenkins RB 2005. Chromosomal imbalances detected by array comparative genomic hybridization in human oligodendrogliomas and mixed oligoastrocytomas Genes Chromosomes Cancer 42 68–77 [DOI] [PubMed] [Google Scholar]
- [19].Cowell JK, Barnett GH and Nowak NJ 2004. Characterization of the 1p/19q chromosomal loss in oligodendrogliomas using comparative genomic hybridization arrays (CGHa) J. Neuropathol. Exp. Neurol 63 151–8 [DOI] [PubMed] [Google Scholar]
- [20].Koschny R, Holland H, Koschny T and Vitzthum H-E 2006. Comparative genomic hybridization pattern of non-anaplastic and anaplastic oligodendrogliomas: a meta-analysis Pathol. Res. Pract 202 23–30 [DOI] [PubMed] [Google Scholar]
- [21].Figarella-Branger D et al. 2014. Mitotic index, microvascular proliferation, and necrosis define 3 groups of 1p/19q codeleted anaplastic oligodendrogliomas associated with different genomic alterations Neuro. Oncol 16 1244–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Martinez R, Schackert H-K, Kirsch M, Paulus W, Joos S and Schackert G 2005. Comparative genetic analysis of metachronous anaplastic oligoastrocytomas with extended recurrence-free interval J. Neurooncol 72 95–102 [DOI] [PubMed] [Google Scholar]
- [23].Idbaih A. et al. SNP array analysis reveals novel genomic abnormalities including copy neutral loss of heterozygosity in anaplastic oligodendrogliomas. PLoS One. 2012;7:e45950. doi: 10.1371/journal.pone.0045950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Godfraind C, Rousseau E, Ruchoux M-M, Scaravilli F and Vikkula M 2003. Tumour necrosis and microvascular proliferation are associated with 9p deletion and CDKN2A alterations in 1p/19q-deleted oligodendrogliomas Neuropathol Appl. Neurobiol 29 462–71 [DOI] [PubMed] [Google Scholar]
- [25].Reyes-Botero G et al. 2014. Contrast enhancement in 1p/19q-codeleted anaplastic oligodendrogliomas is associated with 9p loss, genomic instability, and angiogenic gene expression Neuro Oncol. 16 662–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Campbell BA, Horsman DE, Maguire J, Young S, Curman D, Ma R and Thiessen B 2008. Chromosomal alterations in oligodendroglial tumours over multiple surgeries: is tumour progression associated with change in 1p/19q status? J. Neurooncol 89 37–45 [DOI] [PubMed] [Google Scholar]
- [27].Kralovics R, Guan Y and Prchal JT 2002. Acquired uniparental disomy of chromosome 9p is a frequent stem cell defect in polycythemia vera Exp. Hematol 30 229–36 [DOI] [PubMed] [Google Scholar]
- [28].Lapunzina P and Monk D 2011. The consequences of uniparental disomy and copy number neutral loss-of-heterozygosity during human development and cancer Biol. Cell 103 303–17 [DOI] [PubMed] [Google Scholar]
- [29].Makishima H and Maciejewski JP 2011. Pathogenesis and consequences of uniparental disomy in cancer Clin. Cancer Res. 17 3913–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kawashima-Goto S et al. 2014. Identification of a homozygous JAK3 V674A mutation caused by acquired uniparental disomy in a relapsed early T-cell precursor ALL patient Int. J. Hematol 101 411–6 [DOI] [PubMed] [Google Scholar]
- [31].Rossi MR et al. 2005. Identification of consistent novel submegabase deletions in low-grade oligodendrogliomas using array-based comparative genomic hybridization Genes Chromosomes Cancer 44 85–96 [DOI] [PubMed] [Google Scholar]
- [32].Jenkins RB, Kimmel DW, Moertel CA, Schultz CG, Scheithauer BW, Kelly PJ and Dewald GW 1989. A cytogenetic study of 53 human gliomas Cancer Genet. Cytogenet 39 253–79 [DOI] [PubMed] [Google Scholar]
- [33].Kros JM, van der Weiden M, Zheng P-P, Hop WC, van den Bent MJ and Kouwenhoven MCM 2007. Intratumoral distribution of 1p loss in oligodendroglial tumors J. Neuropathol. Exp. Neurol 66 1118–23 [DOI] [PubMed] [Google Scholar]
- [34].Kuo L-T, Tsai S-Y, Chang C-C, Kuo K-T, Po-Hao Huang A, Tsai J-C, Tseng H-M, Kuo M-F and Tu Y-K 2013. Genetic and epigenetic alterations in primary-progressive paired oligodendroglial tumors PLoS One 8 e67139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Griffin CA, Burger P, Morsberger L, Yonescu R, Swierczynski S, Weingart JD and Murphy KM 2006. Identification of der(1;19) (q10;p10) in five oligodendrogliomas suggests mechanism of concurrent 1p and 19q loss J. Neuropathol. Exp. Neurol 65 988–94 [DOI] [PubMed] [Google Scholar]
- [36].Jenkins RB et al. 2006. A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma Cancer Res. 66 9852–61 [DOI] [PubMed] [Google Scholar]
- [37].Nagy JD 2004. Competition and natural selection in a mathematical model of cancer Bull. Math. Biol 66 663–87 [DOI] [PubMed] [Google Scholar]
- [38].Swanton C 2012. Intratumor heterogeneity: evolution through space and time Cancer Res. 72 4875–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Gerlinger M et al. 2012. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing New Engl. J. Med 366 883–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Marusyk A, Tabassum DP, Altrock PM, Almendro V, Michor F and Polyak K 2014. Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity Nature 514 54–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Inda M-d -M et al. 2010. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma Genes Dev. 24 1731–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wu M, Pastor-Pareja JC and Xu T 2010. Interaction between Ras(V12) and scribbled clones induces tumour growth and invasion Nature 463 545–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Navin NE and Hicks J 2010. Tracing the tumor lineage Mol. Oncol 4 267–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.