Abstract
A long-term hepatocyte culture maintaining liver-specific functions is very essential for both basic research and the development of bioartificial liver devices in clinical application. However, primary hepatocytes rapidly lose their proliferation and hepatic functions over a few days in culture. This work is to establish an ornithine transcarbamylase deficiency (OTCD) patient-derived primary human hepatocyte (OTCD-PHH) culture with hepatic functions for providing an in vitro cell model. Liver tissue from an infant with OTCD was dispersed into single cells. The cells were cultured using conditional reprogramming. To characterize the cells, we assessed activities and mRNA expression of CYP3A4, 1A1, 2C9, as well as albumin and urea secretion. We found that the OTCD-PHH can be subpassaged for more than 15 passages. The cells do not express mRNA of fibroblast-specific maker, whereas they highly express markers of epithelial cells and hepatocytes. In addition, the OTCD-PHH retain native CYP3A4, 1A1, 2C9 activities and albumin secretion function at early passages. The OTCD-PHH at passages 2, 6, 9 and 13 have identical DNA fingerprint as the original tissue. Furthermore, under 3D culture environment, low urea production and hepatocyte marker staining of the OTCD-PHH were detected. The established OTCD-PHH maintain liver-specific functions at early passages and can be long-term cultured in vitro. We believe the established long-term OTCD-PHH culture is highly relevant to study liver diseases, particularly in infants with OTCD.
Keywords: primary human hepatocyte (PHH), long-term culture, CYP450 activity, ornithine transcarbamylase deficiency (OTCD)
Introduction
Liver is a vital organ in humans with various functions in metabolism, protein synthesis, detoxification and bile acid production. Urea cycle, which converts highly toxic ammonia to urea for excretion, is a key metabolic event that is primarily taken place in liver. Ornithine transcarbamylase deficiency (OTCD) is the most common urea cycle disorder caused by complete or partial loss of the ornithine transcarbamylase (OTC) enzymatic activity in liver, which is due to genetic mutations in the OTC gene in patients. However, to our knowledge, there is no cell model to study this disease. The challenges in culturing primary human hepatocyte (PHH) include rapid loss of morphology and liver specific functions, becoming fibroblast-like and weak proliferation ability.1, 2 Several diverse medium formulations have been used to culture PHH, such as modified DMEM,3, 4 modified MCM,2, 5 Williams’ E medium 3 and hepatocyte medium 6. Besides, 3D technique has been used to culture PHH.7, 8 The conditional reprogramming cell technology uses the combination of irradiated mouse fibroblast J2 cells (feeder cells) and the Rho-associated kinase inhibitor (Y-27632) to propagate human epithelial cells.9 It has been used to establish patient-derived cells from diverse human biospecimens, such as breast, skin, prostate and colon, with an indefinite proliferative state without the use of exogenous viral or genetic manipulation.9 Y-27632 is able to increase the viability of human bone marrow-derived mesenchymal stem cells10 and human keratinocyte stem cells11 and induce indefinite cell proliferation.12, 13
Our previous study showed that conditional reprogramming enables long-term in vitro cultivation of primary human hepatocytes, especially when they are derived from young patients.14 In the present study, we established long-term cultured PHH derived from a 4-month-old male patient who had OTCD. We cultured the OTCD patient-derived primary human hepatocyte (OTCD-PHH) for more than 120 days (15 passages) using conditional programming (F medium combined with conditional medium in the presence of Y-27632). Most importantly, we detected that the cells at early passages retain cytochrome P450 enzyme functions and albumin secretion activity. In addition, they express mRNA of CYP family and albumin. Through short tandem repeat (STR) analysis of the cells at passages 2, 6, 9 and 13, we verified that the cells have the same DNA fingerprint as the original tissue. Furthermore, we detected urea production and hepatocyte marker staining of cells in 3D culture environment.
Materials and Methods
In vitro culture of a patient-derived primary hepatocyte
Fresh liver tissue was obtained upon liver transplant of a 4-month-old male patient who had ornithine transcarbamylase deficiency. The experimental protocol used in this study was approved by the Georgetown University Institutional Review Board (IRB). As the tissue was considered a bio-waste, the requirement for informed consent was waived by the IRB. 2cm × 2cm × 1cm liver tissue was dispersed into single cells by enzymatic digestion. Cells were cultured in F medium combined with conditioned medium (CM) in the presence of 10μM Y-27632 with a ratio of 1:3. CM is the medium conditioned by irradiated 3T3-J2 cells.9 Cells were grown in a 37°C incubator with 5% CO2. Medium was changed every other day or every three days. Cells were passaged when 80–90% confluence was reached. The cell concentration and viability were obtained by incubation with trypan blue and then counted by cell counter. HepG2 was obtained from Tissue Culture and Biobanking Shared Resource, Georgetown University. 3T3-J2 and human fibroblast cells were provided by the Center for Cell Reprogramming at Georgetown University.
Short tandem repeat analysis
Genomic DNA was isolated from cells at different passages using DNeasy Blood & Tissue kit (QIAGEN, Hilden, Germany). Short tandem repeat (STR) analysis was performed by Genetica DNA Laboratories (Burlington, USA). We tested 15 STR markers: TH01, D21S11, D18S51, Penta E, D5S818, D13S317, D7S820, D16S539, CSF1PO, Penta D, vWA, D8S1179, TPOX, FGA and Amelogenin. Data analysis and allele size determination were carried out using GeneMapper software.
Cytochromes P450 activity measurement
For characterization of the cells, we performed cytochromes P450 (CYP450) 3A4, 1A1 and 2C9 assays following the protocol of the assay kits (Promega, Fitchburg, USA) when we passaged the cells. Briefly, 20,000 cells were seeded into 96-well plates. On the following day, we replaced culture medium with 50 μL fresh medium containing corresponding luminogenic CYP substrate and incubated cells at 37°C for 1h (for CYP3A4) or 3h (for both CYP1A1 and CYP2C9 ). After incubation, we transferred 25μL of culture medium from each well into a 96-well opaque white luminometer plate, and added 25μL of luciferin detection reagent to initiate a luminescent reaction. After the plate was incubated at room temperature for 20 minutes, luminescence was read using a luminometer (Wallac Victor 1420, Perklin Elmer, Waltham, USA). After luminescence assay, 25μL of medium remained in the wells into which an equal volume (25μL) of the CellTiter-GloR reagent was added to determine cell viability. The contents were mixed for two minutes on an orbital shaker to induce cell lysis. The plate was then incubated at room temperature for 10 minutes to stabilize luminescent signal. We removed 40μL cell lysate from each well to a 96-well opaque white luminometer plate to record luminescence for cell viability. We used HepG2 and human fibroblast cells as a positive control and negative control, respectively.
Albumin secretion assay
Cells were cultured into 96 well plates with a density of 50 000 cells per well with 200 μL F-Y medium for 2, 4, 6 or 24 hours. The culture media was collected and centrifuged at 1000g for 10 minutes at 4°C to remove debris. The collected supernatant was stored at −80°C. We used albumin human ELISA kit (abcam, Cambridge, United Kingdom) to detect human albumin secretion of OTCD-PHH at passage 2, 4 and 5. Following the protocol, 50 μL albumin standard with a series of dilution or samples were added in each well of albumin microplate and incubate for 1 hour. The incubation wells were washed using wash buffer. Afterwards, 50 μL of biotinylated albumin antibody was added into each well and incubated for 30 minutes. Each well was washed. 50 μL SP conjugated was added into each well and incubated for 30 minutes. The microplate was washed. 50 μL of chromogen substrate was added into each well and incubated for 25 minutes when the optimal blue color density formed. We added 50 μL of stop solution into each well. The absorbance of each well was recorded on a microplate reader at 450 nm immediately. The concentration of albumin secretion was determined from the standard curve which is generated by standard albumin samples. F-Y medium was used as blank.
Real-Time PCR
Total RNA was extracted using Direct-zol RNA MiniPrep kit (Zymo Research, Irvine, USA) and treated with DNase (Zymo Research, Irvine, USA) according to the manufacturers’ instructions. First-strand cDNA was synthesized with an equal amount of RNA using High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, USA). Real time PCR was performed in a QuantStudio 12K Flex system (Thermo Fisher, Waltham, USA) using Applied Biosystems™ Power SYBR™ Green PCR Master Mix (Applied Biosystems, Foster City, USA). The relative quantities of mRNA were analyzed using the 2−ΔΔCT method.15 Primer sequences are available upon request.
Three-dimensional culture
Single-cell suspension of hepatocytes was dispersed into Corning Matrigel basement membrane matrix, phenol redfree, LDEV-free (New York, USA) and seeded into 6-well plates with a density of 700 000 cells/well. 2mL phenol red-free F medium combined with CM containing 10μM Y-27632 was added on the top of matrigel and medium was changed every other day.
Urea secretion
Cells were cultured under 2D or 3D conditions in phenol red-free medium. Fresh medium was changed every other day and the culture medium was stored in −80°C for the following experiment. 50 μL culture medium was mixed with 75μL reagent A and B in 96-well plates following the protocol of urea nitrogen (BUN) colorimetric detection kit (Invitrogen, Carlsbad, USA). The plate was incubated for 30min at room temperature and read the absorbance at 450 nm (Wallac Victor 1420, Perklin Elmer, Waltham, USA). The concentration of urea in the culture medium was read from the standard curve which was made by serial dilutions of urea nitrogen standard. Fresh medium was used as blank.
Immunohistochemistry assay
Cells were collected from 2D or 3D culture and washed with PBS three times. After the cells were fixed with 10% formalin overnight at 4 °C, they were embedded, sliced and stained with monoclonal mouse anti-human hepatocyte antibody (Agilent M715801–2, Santa Clara, USA) at a dilution of 1:50 for 1 hour. Then, the slides were washed and incubated with biotin-conjugated goat anti-mouse secondary antibody at a dilution of 1:200 for 30 minutes. Bright field images were taken using Olympus BX61.
Results
Successful establishment of long-term cultured OTCD-PHH
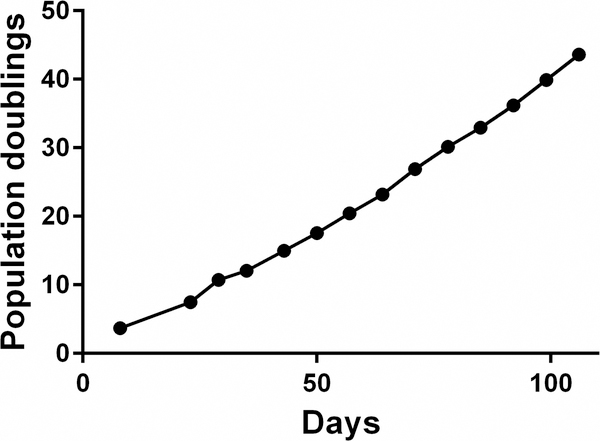
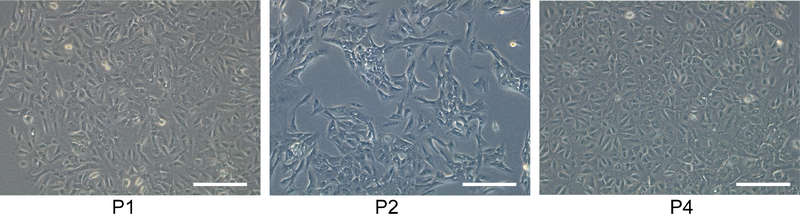
We found that the isolated OTCD-PHH possess highly proliferative ability and can be passaged for more than 15 passages (at least 120 days and 50 population doublings) until the experiment was terminated (Figure 2). Viability of cells is 60–95% at each passage and the cell morphology of p5 is consistent with the early passage (Figure 1). STR analysis of the cells at different passages confirmed genetic stability by comparing them to the original tissue (Table 1).
Figure 2.
OTCD-PHH were passaged for more than 15 passages (120 days). The cell number and viability were recorded at each passage and a plot of population doublings versus time (days) was constructed using Graphpad Prism 6.
Figure 1.
The representative phase contrast images of OTCD-PHH at passages 1, 2, 4. Magnification, 10 ×; bar, 400 μm.
Table 1.
STR analysis of original tissue and cells at passages 2, 6, 9 and 13
| D3S1358 | TH01 | D21S11 | D18S51 | Penta E | D5S818 | D13S317 | D7S820 | D16S539 | CSF1PO | Penta D | vWA | D8S1179 | TPOX | FGA | AMEL | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| tissue | 14, 16 | 7, 9.3 | 28, 29 | 14, 16 | 13, 14 | 11, 13 | 12, 14 | 8, 11 | 8, 11 | 11, 12 | 2.2, 10 | 14, 17 | 13, 14 | 8 | 23 | X, Y |
| P2 | 14, 16 | 7, 9.3 | 28, 29 | 14, 16 | 13, 14 | 11, 13 | 12, 14 | 8, 11 | 8, 11 | 11, 12 | 2.2, 10 | 14, 17 | 13, 14 | 8 | 23 | X, Y |
| P6 | 14, 16 | 7, 9.3 | 28, 29 | 14, 16 | 13, 14 | 11, 13 | 12, 14 | 8, 11 | 8, 11 | 11, 12 | 2.2, 10 | 14, 17 | 13, 14 | 8 | 23 | X, Y |
| P9 | 14, 16 | 7, 9.3 | 28, 29 | 14, 16 | 13, 14 | 11, 13 | 12, 14 | 8, 11 | 8, 11 | 11, 12 | 2.2, 10 | 14, 17 | 13, 14 | 8 | 23 | X, Y |
| P13 | 14, 16 | 7, 9.3 | 28, 29 | 14, 16 | 13, 14 | 11, 13 | 12, 14 | 8, 11 | 8, 11 | 11, 12 | 2.2, 10 | 14, 17 | 13, 14 | 8 | 23 | X, Y |
OTCD-PHH culture is not fibroblast contaminated
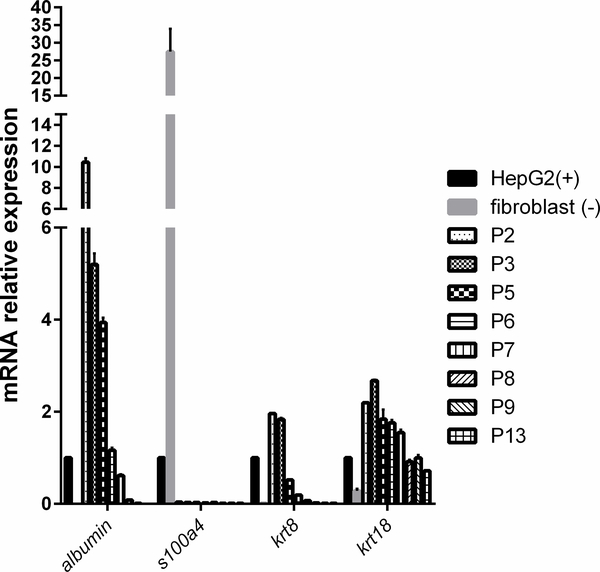
Fibroblast over-growth is the most common phenomenon in long-term culture of primary cells, including hepatocytes. To evaluate the presence of fibroblast, we compared the mRNA expression of biomarkers of epithelial and fibroblast cells in OTCD-PHH culture at different passages versus human fibroblast cells and HepG2. Epithelial-fibroblast transition is one type of epithelial-mesenchymal transition (EMT). S100A4, a fibroblast-specific protein, is a reliable marker to characterize the mesenchymal products caused by EMT that occur during the formation of fibrosis in various organs. Cytokeratin are markers used to identify epithelial cells to fibroblast in kidney, liver and lung. Keratin 8 and keratin18 are two kinds of cytokeratin.16 In contrast to fibroblast cells in which s100a4 is highly expressed, we found that the OTCD-PHH did not express s100a4 mRNA at all passages we assessed. Additionally, consistent with HepG2 cells, OTCD-PHH significantly expressed higher mRNA level of krt18 in all passages and krt8 in early passages, whereas fibroblasts did not (Figure 3). Importantly, the OTCD-PHH maintain high gene expression level of the liver-specific albumin gene up to passage 7. These results clearly indicate that this PHH culture does not undergo fibroblast over-growth in long-term in vitro culture.
Figure 3.
mRNA expression of krt8, krt18, s100a4 and albumin in PHH at different passages, human fibroblast cells (negative control) and HepG2 (positive control). The mRNA levels of corresponding genes relative to gapdh (a house keeping gene) were determined by the quantitative real-time PCR. The results were obtained from three replicate wells.
OTCD-PHH culture possesses cytochromes P450 enzymatic activities
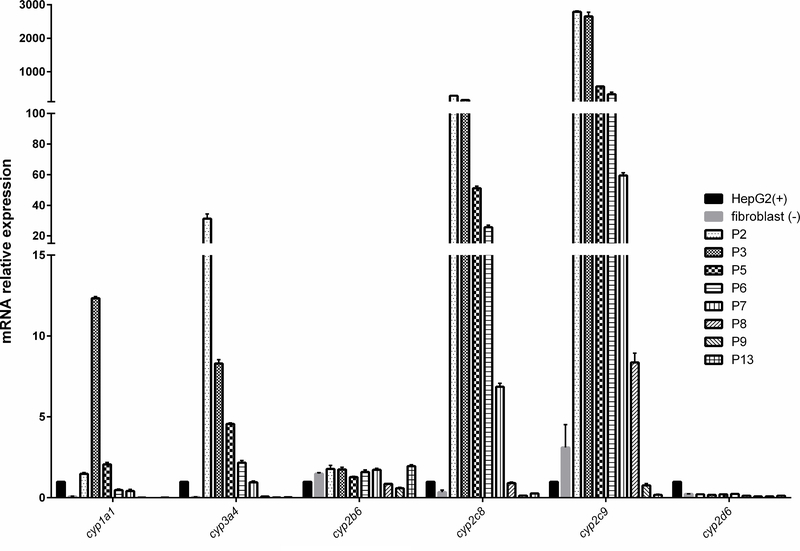
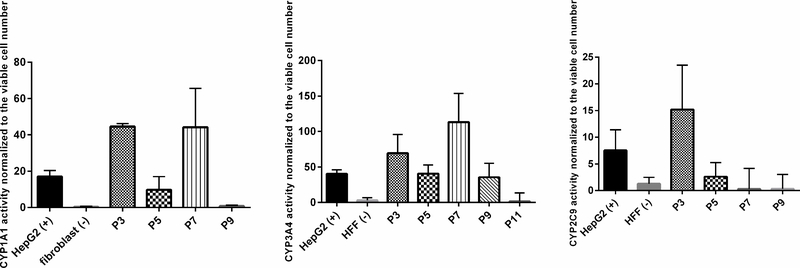
Cytochromes P450 enzymes are responsible for the metabolism of xenobiotic and endogenous compounds metabolism in liver. CYP3A4 and 2C9 are the most important enzymes in liver and contribute to the metabolism of more than 50% of the drugs.17 They are also involved in the metabolic activation of drug-induced toxicity.18 CYP1A1 is involved in steroid hormones biosynthesis.19 To characterize the cells, we measured gene expression and enzymatic activities of several CYP450 family members in the OTCD-PHH. The gene expression analysis showed that the OTCD-PHH express significantly higher cyp1a1, cyp3a4, cyp2c8 and cyp2c9 at early passages than that in HepG2. However, the cyp2b6 and cyp2d6 mRNA expression between PHH, HepG2 and fibroblast did not exhibit differences (Figure 4). Using cytochromes P450 assay, we found that up to passage 7, OTCD-PHH exhibited stronger CYP3A4 activities compared to the positive control, HepG2. Besides, at passage 3 and 7, PHH has higher CYP1A1 activities compared with HepG2. Moreover, OTCD-PHH displayed higher CYP2C9 enzymatic activity at passage 3 compared with HepG2 cells. (Figure 5). These observations suggest that the OTCD-PHH retain native cytochromes P450 enzymes activities in early passages.
Figure 4.
mRNA expression of cyp1a1, 3a4, 2b6, 2c8, 2c9 and 2d6 in PHH at different passages, human fibroblast cells (negative control) and HepG2 (positive control). The mRNA levels of corresponding genes relative to gapdh (a house keeping gene) were determined by the quantitative real-time PCR. The results were obtained from three replicate wells.
Figure 5.
CYP 3A4, 1A1 and 2C9 activities of OTCD-PHH. The assays were carried out following the protocol of P450-Glo CYP3A4, 1A1 and 2C9 assay kits. The enzymatic activities were normalized to cell viability. HepG2 and human fibroblast cell were positive control and negative control, respectively. The results were obtained from three replicate readings.
OTCD-PHH culture preserves albumin secretion functions at early passages
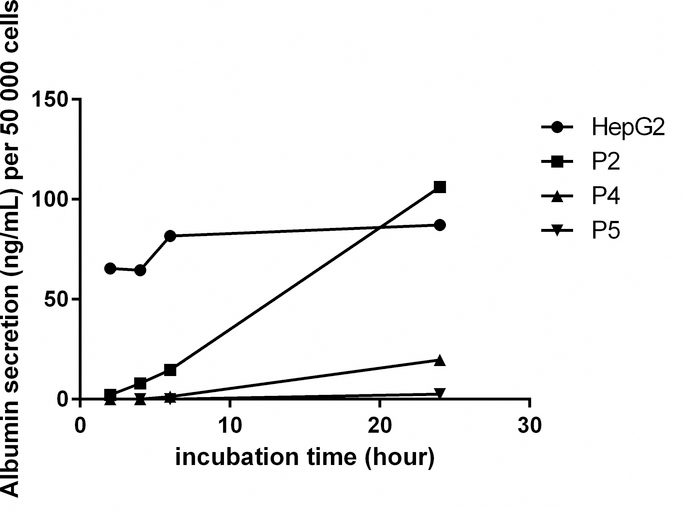
Albumin secretion activity is an important biomarker to characterize hepatocyte. We found that although OTCD-PHH at passage 2 released lower amount of albumin compared to HepG2 after 2, 4, and 6-hour incubation, their albumin production reached over 100 ng/mL, which is higher than HepG2 after 24-hour incubation (Figure 6). The cells at passage 4 and 5 could produce 20 and 3 ng/mL, respectively, after 24-hour incubation (Figure 6). These results suggest that OTCD-PHHs have strong albumin secretion function at early passages and gradually lose this activity over the incubation time.
Figure 6.
Albumin secretion of PHH after 2, 4, 6 or 24 hours incubation. Mean values of each standard and sample were obtained from replicate wells. The standard curve was produced by plotting the concentration of standard on the x-axis and the corresponding 450 nm absorbance on the y-axis. The best-fit line was generated by regression analysis using four-parameter logistic curve-fit. The albumin concentration of samples was calculated from the standard curve.
OTCD-PHH culture secretes low urea under 3D differentiation culture
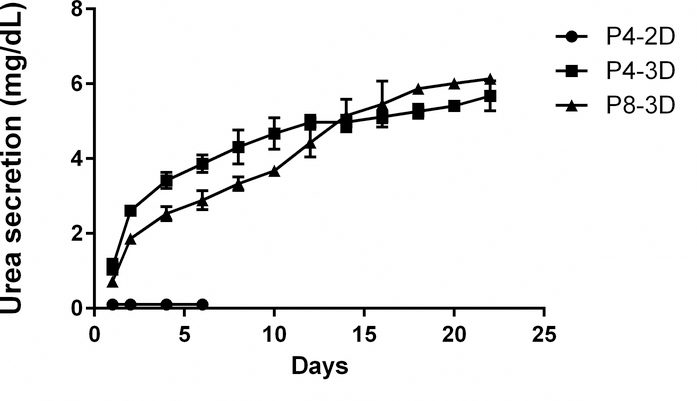
Urea secretion is one of the liver-specific functions. However, PHH from OTCD patients partially or completely failed to generate urea depending on the severity of OTC enzyme abnormality. It is well known that 2D cultures have more proliferation potential, compared to 3D cultures which have more differentiation potential.2 Culturing hepatic cell lines in 3D has been shown to change the gene expression, phenotype and cell surface receptor expression toward more liver-like properties.2 To characterize urea secretion in the OTCD-PHH, we measured the urea secretion of PHH in both 2D culture (F medium combined with CM) and 3D culture conditions. As a result, urea production of PHH in 2D culture was not detectable as expected. Then, we performed matrigel-based 3D culture for PHH at passage 4 and 8 for 22 days and measured urea secretion in the culture medium every other day.
Interestingly, under this culture condition, we found that PHH at passage 4 and 8 can secrete urea once they are seeded with the highest urea secretion detected on the second day (Figure 7). We observed that the urea secretion of PHH (about 6 mg/dL in 22 days) is much lower than HepG2 (35 mg/dL in 2 days under 2D culture condition, data not shown). We believe this is due to ornithine transcarbamylase deficiency-induced urea cycle disorder.
Figure 7.
Urea secretion of OTCD-PHH at passage 4 and 8 in 2D or 3D culture. 700,000 cells were seeded into 6-well plates which were dispersed into Matrigel. F medium combined with CM containing 10μM Y-27632 was added on the top of matrigel and medium was collected every other day and stored at −80°C for subsequence analysis of urea production. The concentration of urea in the culture medium was read from the standard curve. The urea secretion of cells was calculated by urea amount in culture medium subtracting urea amount in fresh medium. The results were obtained from two repeats. The medium we used in this experiment is phenol red-free.
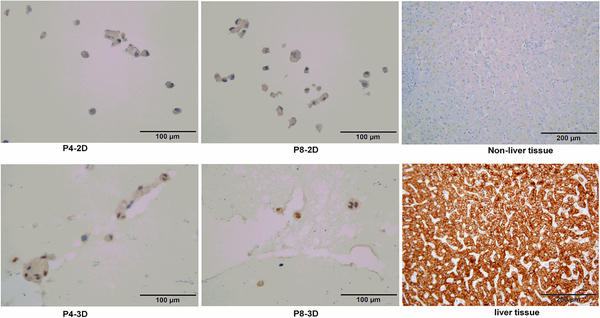
OTCD-PHH culture expresses hepatocyte marker
Since OTCD-PHH failed to be stained with hepatocyte marker under 2D culture condition, we used matrigel to culture OTCD-PHH to enhance the expression of hepatocyte marker. Figure 8 shows that under 3D culture condition OTCD-PHH can be strongly stained with hepatocyte marker, suggesting that 3D environment significantly induced the expression of hepatocyte marker in OTCD-PHH.
Figure 8.
Immunostaining of OTCD-PHH with hepatocyte marker antibody. Cells from 2D or 3D culture were fixed with 10% formalin overnight at 4 °C. Afterward, the cells were embedded, sliced and stained with monoclonal mouse anti-human hepatocyte antibody for 1h. Then, the slides were washed and incubated with biotin-conjugated goat anti-mouse secondary antibody for 30 minute. Magnification, 40×; bar, 100 μm.
Discussion and conclusions
In this study, we successfully established long-term cultured PHH from an OTCD patient using conditional programming, while maintaining proliferative capacity (Figure 2), genetic stability (Table 1) and hepatocyte-specific functions (Figure 3, 4, 5, 6 and 8). For the first time, we were able to exemplify the conditional reprogramming cell technology as a simple yet efficient cultivation technique that can be potentially applied to establish in vitro hepatocyte models to study liver diseases. Most strikingly, under 2D-culture conditions, the OTCD-PHH culture can be passaged for more than 120 days without obvious changes in morphology and fibroblast over-growth, whereas most commercial primary hepatocyte hardly survive several weeks.1 We found that the OTCD-PHH gradually lost mRNA expression and activity of CYP450 albumin secretion. Thus, the medium we used needs to be improved to maintain the liver functions for longer time culture.
There are several potential applications of the established OTCD-PHH culture in liver disease study. To our knowledge, this work provides the first in vitro hepatic cell model for studying OTCD. OTCD is X-linked genetic disorder characterized by complete or partial loss of the enzymatic activity of ornithine transcarbamylase, which is critically involved in urea cycle. Interestingly, we noticed that the cells secrete much less urea compared to HepG2 cells. This suggests that our cells may maintain urea biosynthesis defects seen in OTCD patients. Thus, these cells could be a valuable model to study the cell biology and genetics of OTCD and screen drugs to treat OTCD. In addition, these cells are highly suitable for toxicology study such as drug hepatotoxicity screening and metabolism analyses, because high CYP450 activities are maintained in early passages (up to passage 7). Furthermore, since most OTCD display mutation in the OTC gene only, these cells could be a powerful source for partial liver transplantation or to create bioartificial liver support systems which “bridge” patients with liver failure to transplantation or regeneration while waiting for the available donors.20
In summary, we successfully established a functional PHH culture derived from an infant with OTCD. The OTCD-PHH culture not only can be used to study liver diseases, particularly OTCD in infants, but also has diverse potential clinical and pharmaceutical applications.
Highlights.
First primary human hepatocyte derived from OTCD patient.
Successful establishment of long-term culture of primary human hepatocyte.
Liver specific functions are preserved during long-term culture of the cells.
Acknowledgment
The work presented in this paper is supported by National Institutes of Health (NIH) Grants U01CA185188 and R01GM123766 awarded to HWR. We would like to thank Dr. Lihui Wang for constructive discussions about this study. We thank the Histopathology and Tissue SahredResource (HTSR), the Tissue Culture and Biobanking Shared Resource (TCBSR), and the Genomics and Epigenomics Shared Resource(GESR) at Georgetown Lombardi Comprehensive Cancer Center for technical assistance. TheShared Resources are partially supported by NIH Grant P30CA051008.
Financial support
The work presented in this paper is supported by National Institutes of Health (NIH) Grants U01CA185188 and R01GM123766 awarded to HWR.
Abbreviations
- PHH
primary human hepatocyte
- CYP450
cytochromes P450
- CM
conditional medium
- STR
short tandem repeat
- OTC
ornithine transcarbamylase
- OTCD
ornithine transcarbamylase deficiency
- EMT
epithelial-mesenchymal transition
Footnotes
Conflicts of interest
All authors declared that they have no conflicts of interest.
Compliance with ethical standards
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2008. As the human liver tissue used in this study was considered a bio-waste, the requirement for informed consent was waived by the institutional review board.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shulman M & Nahmias Y in Epithelial cell culture protocols 287–302 (Springer, 2012). [Google Scholar]
- 2.Godoy P et al. Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch. Toxicol 87, 1315–1530 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ryan CM et al. Isolation and long-term culture of human hepatocytes. Surgery 113, 48–54 (1993). [PubMed] [Google Scholar]
- 4.Ploss A et al. Persistent hepatitis C virus infection in microscale primary human hepatocyte cultures. Proc. Natl. Acad. Sci. U. S. A 107, 3141–3145 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LeCluyse EL & Alexandre E Isolation and culture of primary hepatocytes from resected human liver tissue. Hepatocytes: Methods and Protocols, 57–82 (2010). [DOI] [PubMed] [Google Scholar]
- 6.LeCluyse EL Human hepatocyte culture systems for the in vitro evaluation of cytochrome P450 expression and regulation. European Journal of Pharmaceutical Sciences 13, 343–368 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Goral VN et al. Perfusion-based microfluidic device for three-dimensional dynamic primary human hepatocyte cell culture in the absence of biological or synthetic matrices or coagulants. Lab on a Chip 10, 3380–3386 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Meng Q Three-dimensional culture of hepatocytes for prediction of drug-induced hepatotoxicity. Expert opinion on drug metabolism & toxicology 6, 733–746 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Liu X et al. Conditional reprogramming and long-term expansion of normal and tumor cells from human biospecimens. Nat. Protoc 12, 439–451 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heng BC Effect of Rho-associated kinase (ROCK) inhibitor Y-27632 on the post-thaw viability of cryopreserved human bone marrow-derived mesenchymal stem cells. Tissue and Cell 41, 376–380 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Terunuma A, Limgala RP, Park CJ, Choudhary I & Vogel JC Efficient procurement of epithelial stem cells from human tissue specimens using a Rho-associated protein kinase inhibitor Y-27632. Tissue engineering Part A 16, 1363–1368 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am. J. Pathol 180, 599–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chapman S, Liu X, Meyers C, Schlegel R & McBride AA Human keratinocytes are efficiently immortalized by a Rho kinase inhibitor. J. Clin. Invest 120, 2619–2626 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su S et al. Long-term culture and characterization of patient-derived primary hepatocytes using conditional reprogramming. Exp. Biol. Med. (Maywood) 244, 857–864 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ & Schmittgen TD Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Kalluri R & Weinberg RA The basics of epithelial-mesenchymal transition. J. Clin. Invest 119, 1420–1428 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rendic S & Carlo FJD Human cytochrome P450 enzymes: a status report summarizing their reactions, substrates, inducers, and inhibitors. DrugMetab. Rev 29, 413–580 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Walgren JL, Mitchell MD & Thompson DC Role of metabolism in drug-induced idiosyncratic hepatotoxicity. Crit. Rev. Toxicol 35, 325–361 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Anzenbacher P & Anzenbacherová E Cytochromes P450 and metabolism of xenobiotics. Cellular and Molecular Life Sciences 58, 737–747 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park J & Lee D Bioartificial liver systems: current status and future perspective. Journal of bioscience and bioengineering 99, 311–319 (2005). [DOI] [PubMed] [Google Scholar]