Abstract
Two recent reports suggested that the less common, less virulent enterococcal species, Enterococcus gallinarum and E. casseliflavus, with low-level vancomycin resistance due to chromosomally encoded vanC1 and vanC2/3, may influence host immunity. We reported that peri-transplant gut colonization with E. gallinarum and E. casseliflavus is associated with lower mortality after allogeneic hematopoietic cell transplantation (HCT). Because most acute leukemia patients undergoing HCT have received intensive chemotherapy (usually requiring prolonged hospitalization) for their underlying disease before HCT, we hypothesized that some may have acquired vanC-positive enterococci during chemotherapy. Therefore, we evaluated the presence of the vanC gene cluster using vanC1 and vanC2/3 qPCR in thrice-weekly collected stool samples from 20 acute leukemia patients undergoing intensive chemotherapy. We found that an unexpectedly large proportion of patients have detectable vanC1 and vanC2/3 (15% and 35%, respectively) in at least one stool sample. Comparing qPCR results with 16S rRNA gene sequencing results suggested that E. gallinarum may reach high abundances, potentially persisting into HCT and influencing transplant outcomes.
Introduction
Hematopoietic cell transplantation (HCT) is a potentially curative approach for many patients with high-risk hematologic malignancies. After their initial anti-neoplastic treatment, often including intensive chemotherapy, such patients are referred for HCT. Gut colonization with antibiotic-resistant organisms is common during HCT [1–3]. The genus Enterococcus, frequently harboring antibiotic resistance genes (ARGs), is one of the most common gut colonizers in HCT patients [4,5]. The plasmid-encoded vanA gene is the most common enterococcal ARG conferring high-level vancomycin resistance to E. faecium and E. faecalis (VRE), with high transmissibility and propensity to cause major clinical infections [6,7]. In contrast, the vanC gene cluster is chromosomally encoded and confers low-level, intrinsic vancomycin resistance to the less commonly encountered E. casseliflavus (vanC2/3 gene) and E. gallinarum (vanC1 gene) [8,9].
We recently reported that colonization with E. casseliflavus or E. gallinarum is associated with lower transplant-related mortality and better survival [10]. The mechanisms underlying this association are unknown, but alterations in host immunity have been reported [11]. In addition, it is not known whether patients acquire E. casseliflavus and E. gallinarum during HCT or before HCT during the treatment of their underlying hematologic disorder. To determine when these species are acquired, we investigated the presence of vanC genes in the stool of acute leukemia patients undergoing intensive chemotherapy.
Methods
Patients and samples
We collected thrice-weekly stool samples between hospital admission and day 28 of chemotherapy (or discharge) from 20 acute leukemia patients receiving intensive chemotherapy. Day 0 was defined as the first day of chemotherapy. Our Institutional Review Board at the University of Minnesota approved the protocol (ID #2017NTLS052) and all patients provided written informed consent. Details of the protocol have been reported elsewhere [12]. Patients remained in private rooms for their entire hospitalization. Contact isolation was implemented if colonization or clinical infection with any vancomycin-resistant enterococci (including E. gallinarum or E. casseliflavus) was identified. We defined intensive chemotherapy as any regimen requiring a planned hospitalization of approximately 4 weeks. Patients did not enroll to the same study upon relapse. At our institution, we use levofloxacin for antibacterial, acyclovir for antiviral, and an azole for antifungal prophylaxis starting with chemotherapy until neutrophil recovery, and cefepime as the initial empiric antibiotic for neutropenic fever. Variations to this recommendation were allowed.
DNA extraction
Samples were stored at -80°C prior to DNA extraction. DNA was extracted from thawed fecal samples using the DNeasy® PowerSoil® kit (Qiagen, Hilden, Germany). Approximately 0.25 g fecal sample were added to the PowerBead tube and processed according to the manufacturer’s instructions using the automated QIAcube platform with the inhibitor removal technology protocol. A final volume of 100 μl DNA solution was obtained and the concentration of extracted DNA was quantified with a QubitTM 4 fluorometer and dsDNA HS Assay kit (Thermo Fisher Scientific, Waltham, MA, USA).
16S rRNA amplicon sequencing
The V4 hypervariable region of the 16S rRNA gene was sequenced on the Illumina MiSeq platform (Illumina Inc., San Diego, CA, USA). Raw sequence data are available under NCBI BioProject #SRP141394. Sequence data were processed as previously described [13]. Briefly, sequences were paired-end joined, trimmed for quality, and aligned against the SILVA database version 132 [14]. Sequences were subjected to a 2% pre-cluster step to remove likely errors [15], and chimeras were identified and removed using UCHIME ver. 4.2.40 [16]. Operational taxonomic units were classified at 97% sequence similarity using the furthest-neighbor algorithm, and taxonomic classifications were made against the version 16 release from the Ribosomal Database Project [17].
Quantitative PCR (qPCR)
qPCR assays targeting the vancomycin resistance genes vanC1 and vanC2/3 were used as described previously (Table 1) [18]. The primers were synthesized by Integrated DNA Technologies (IDT; Coralville, IA, USA) and diluted to 10 μM working stocks. Primer targets were cloned into a gBlocks plasmid standard (IDT), using previously published accession numbers [18]. Standard curves ranging from 106 to 101 gene copies were prepared from 10-fold serial dilutions of the plasmid standard. Each standard concentration was run in triplicate on each plate. Two negative (sterile water) controls were included on each plate. PCR reactions included 10 μl QuantiTect SYBR Green MasterMix (Qiagen), 1 μl of each primer (1.5 μl for vanC1), 6 μl nuclease-free water, and 2 μl DNA template. PCR was carried out using the LightCycler 480 II thermocycler (Roche, Basel, Switzerland) with the following two-step thermocycling conditions for vancomycin resistance genes: an initial denaturation at 95°C for 15 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. DNA from all samples was run in duplicate. Gene copies were extrapolated from the standard curve using the absolute quantification/2nd derivative max method with the LightCycler 480 software, for which the manufacturer recommends that error should be <0.2 for accurate quantitation. Data were reported as gene copies per ng DNA if both replicates showed quantifiable amplification. If only one replicate was quantifiable, results were reported as positive, but not quantifiable.
Table 1. Primers and performance characteristics for qPCR assays.
| Target gene | Primer | Sequence (5’-3’) | Size (bp) | Tm* (°C) | Efficiency (%) | Error |
|---|---|---|---|---|---|---|
|
vanC1 |
VanC1-F | TGCTTGTGATGCGATTTCTC | 204 | 83.57 ± 0.07 | 91.44 ± 1.41 | 0.05 ± 0.03 |
| VanC1-R | ATCGCTCCTTGATGGTGAC | |||||
| vanC2/3 | VanC23-F | GGGAAGATGGCAGTATCCAA | 102 | 80.34 ± 0.09 | 94.81 ± 3.09 | 0.04 ± 0.03 |
| VanC23-R | GCAGCAGCCATTTGTTCATA |
*Melting temperature (Tm), shown as mean ± standard error based on standard curves generated in this study.
Statistical analysis
Alpha diversity was measured by Shannon index, incorporating both richness and evenness of the abundances of bacterial community members [19]. We determined the correlation between Enterococcus relative abundance and vanC abundance by a Spearman’s correlation test.
Results
Detailed patient characteristics are shown in Table 2. We collected and analyzed 207 stool samples. There was a negative correlation between Shannon diversity and Enterococcus relative abundance (Spearman’s ρ = -0.50, P < 0.001; Fig 1A). Fig 1B shows the heatmap of the 15 most common genera among all samples. Enterococcus was the second most common genus, with an aggregate mean (range) relative abundance of 10 (0–99)%, following Bacteroides, with an aggregate mean (range) relative abundance of 25 (0–92)%. Fig 2 shows changes in Enterococcus relative abundance and vanC gene copies per ng DNA over time for each patient. vanC1 was detectable in 11 (5%) samples from 3 (15%) patients, with a median of 30,147 (range: 2–1,873,333) copies per ng DNA. vanC1 was quantifiable in all positive samples. vanC2/3 was detectable in 19 (9%) samples from 7 (35%) patients, with a median of 13 (range: 1–232) copies per ng DNA in quantifiable cases (13 samples). All three patients with vanC1 also had detectable vanC2/3.
Table 2. Patient, disease, and treatment characteristics.
| Patient | Age (y), Sex | Disease, Phase | Prior intensive chemotherapy | Interval from last intensive chemotherapy | Chemotherapy during study |
|---|---|---|---|---|---|
| 1 | 22, M | AML, Induction | - | - | 7+3 |
| 2 | 60, M | AML, Re-induction | Yes | 0.5 months | MEC |
| 3 | 68, F | AML, Re-induction | Yes | 7 months | Clo/Ara-C |
| 4 | 36, F | ALL, Induction | - | - | GRAAL [20] |
| 5 | 52, F | ALL, Induction | - | - | PETHEMA [21] |
| 6 | 35, M | AML, Re-induction | Yes | 7 months | Clo/Ara-C |
| 7 | 67, F | AML, Induction | - | - | 7+3 |
| 8 | 74, F | AML, Induction | - | - | 7+3 |
| 9 | 58, F | AML, Induction | - | - | 7+3 |
| 10 | 50, F | AML, Re-induction | Yes | 6 months | MEC |
| 11 | 65, M | AML, Re-induction | Yes | 4 months | MEC |
| 12 | 73, M | AML, Induction | - | - | 7+3 |
| 13 | 51, F | AML, Induction | - | - | 7+3 |
| 14 | 53, M | ALL, Induction | - | - | GRAAL |
| 15 | 23, M | ALL, Induction | - | - | PETHEMA |
| 16 | 52, F | AML, Induction | - | - | 7+3 |
| 17 | 52, F | AML, Re-induction | Yes | 10 months | MEC |
| 18 | 68, M | AML, Induction | - | - | 7+3 |
| 19 | 22, M | AML, Re-induction | Yes | 4 months | MEC |
| 20 | 61, F | AML, Induction | - | - | 7+3 |
7+3: Idarubicin + Cytarabine; ALL: Acute lymphoblastic leukemia; AML: Acute myeloid leukemia; Clo/Ara-C: Clofarabine + Cytarabine; F: Female; M: Male; MEC: Mitoxantrone + Etoposide + Cytarabine
Fig 1. Microbiota diversity and composition.
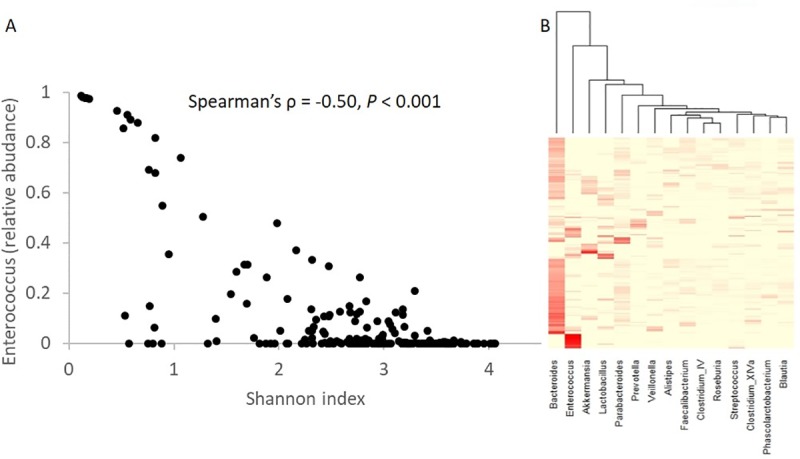
There is a negative correlation between Shannon diversity and Enterococcus relative abundance. Each point represents a sample. (B) Heatmap of microbiota relative abundances from 16S rRNA amplicon sequencing. Each column shows a genus and each row represents a sample. Only the 15 most abundant genera are shown.
Fig 2. Changes in Enterococcus relative abundance and vanC1 copies per ng DNA over time.
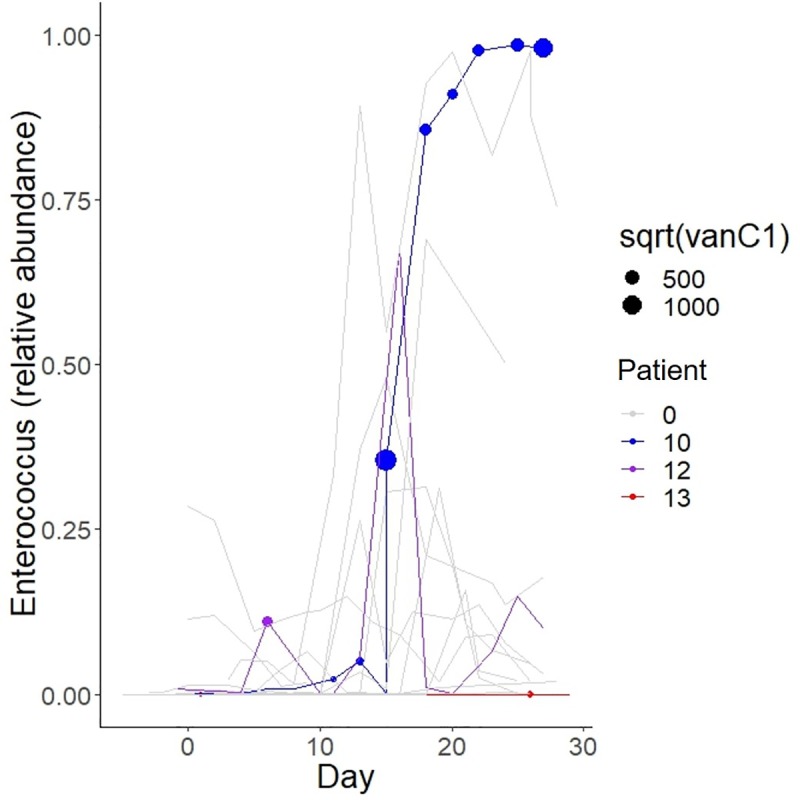
Each line represents samples from the same patient. Patients with detectable vanC1 (patients #10, 12, and 13) are shown in color while all other patients (labeled as #0) are grayed out for improved visibility. vanC1-positive samples are indicated with a circle, with a diameter proportional to the square root of vanC1 gene copies per ng DNA. Day 0 is the first day of chemotherapy.
We classified patients into 3 groups (Table 3). Group A (3 patients) had at least one vanC1-positive sample; group B (4 patients) had at least one vanC2/3-positive sample, but no vanC1-positive sample; group C (13 patients) had no vanC1- or vanC2/3-positive samples. No relationship was found between antibiotic exposure and the vanC group, though this observation was limited by the small sample size and high variability of antibiotic exposures. Thirteen patients received induction and seven received re-induction/salvage chemotherapy. Treatment phase was not associated with the vanC group (P = 0.80, Fisher’s exact test).
Table 3. vanC group, antibiotics, and clinical outcomes.
| Pt | Group1 | Pre-and post-vanC antibacterial antibiotics2 | NF | BSI | CDI | Relapse/ Progression |
Survival |
|---|---|---|---|---|---|---|---|
| 1 | B | Pre: Cefepime, Pip-Tazo Post: Cefepime, Pip-Tazo, Vanc |
D14 | - | D9 | - | Alive, 22m |
| 2 | B | Pre: Cefepime, FQN, Pip-Tazo, Vanc Post: Cefepime, Daptomycin |
D8 | D14 (VRE) | - | 13m | Alive, 22m |
| 3 | C | FQN, Pip-Tazo | D7 | - | - | 0.5m | Dead, 0.5m |
| 4 | C | Cephalexin, FQN | - | - | - | - | Dead, 22m |
| 5 | C | Cefepime, Ceftriaxone, Vanc | D26 | D26 (Strep. mitis) | - | - | Alive, 19m |
| 6 | B | Pre: FQN, Meropenem Post: Aztreonam, Daptomycin, FQN, Linezolid, Metronidazole, Vancomycin |
D13 | D14 (Strep. sanguinis) | - | 7m | Dead, 12m |
| 7 | C | FQN, Cefepime, Metronidazole | D8 | - | D7 | 9m | Dead, 20m |
| 8 | C | FQN, Cefepime, Vanc | D13 | D13 (MRSA) | - | 8m | Dead, 11m |
| 9 | B | Pre: FQN, Cefepime, Meropenem, Pip-Tazo Post: FQN, Meropenem, Metronidazole |
D5 | - | - | 8m | Dead, 9m |
| 10 | A | Pre: FQN, Cefepime Post: Azithromycin, Cefepime, FQN, Metronidazole, Vanc |
D10 | - | - | 13m | Dead, 20m |
| 11 | C | FQN, Cefepime, Ertapenem, Metronidazole, Pip-Tazo | D7 | - | - | 1m | Dead, 2m |
| 12 | A | Pre: Cefdinir Post: Azithromycin, Cefdinir, Cefepime, FQN, Metronidazole, Pip-Tazo, Vanc |
D10 | - | D10 | 5m | Dead, 13m |
| 13 | A | Pre: Cefdinir, Cefepime, FQN, Metronidazole, Vanc Post: Cefdinir, Cefepime, Doxycycline, Vanc |
D7 | - | - | 8m | Dead, 11m |
| 14 | C | FQN | - | - | - | - | Alive, 19m |
| 15 | C | FQN, Cefepime, Pip-Tazo, Vanc | D-1 | D-1 (Strep. mitis) | - | - | Alive, 19m |
| 16 | C | FQN, Cefepime, Meropenem, Vanc | D-3 | - | - | 1m | Dead, 4m |
| 17 | C | FQN, Cefepime, Pip-Tazo, Vanc | D9 | - | - | 5m | Dead, 12m |
| 18 | C | FQN, Cefepime | D15 | - | - | 8m | Dead, 15m |
| 19 | C | FQN, Pip-Tazo, Vanc | D9 | D9 (Strep. mitis) | - | 1m | Dead, 6m |
| 20 | C | Cefepime, Clindamycin, FQN, Metronidazole, Nitrofurantoin, Vancomycin | D5 | - | D12 | 5m | Dead, 5m |
1Groups: (i) group A, ≥1 vanC1-positive sample, (ii) group B, ≥1 vanC2/3-positive but no vanC1-positive sample, and (iii) group C, no vanC1- or vanC2/3-positive samples.
2For patients with at least one vanC-positive sample (groups A and B), pre and post antibiotics refer to those initiated before and after the first positive sample. For patients without any vanC-positive samples (group C), antibiotics during chemotherapy are shown. BSI: Bloodstream infection; CDI: Clostridium difficile infection; FQN: Fluoroquinolone; m: months; MRSA: Methicillin-resistant Staphylococcus aureus; NF: Neutropenic fever; Pip-Tazo: Piperacillin-tazobactam; Pt: Patient; Vanc: Vancomycin; VRE: Vancomycin-resistant E. faecium
We next evaluated whether vanC gene abundance correlates with Enterococcus relative abundance as determined by 16S rRNA gene amplicon sequencing. For vanC1, this correlation was strong (Spearman’s ρ = 0.78, P = 0.004) but there was no correlation between vanC2/3 and Enterococcus relative abundance (ρ = 0.31, P = 0.30). Similarly, vanC1 correlated negatively with Shannon diversity (ρ = -0.64, P = 0.04), but vanC2/3 did not (ρ = 0.23, P = 0.41). The primary species known to harbor vanC2/3 is E. casseliflavus; therefore, these results suggested that E. casseliflavus comprised only a minor proportion of enterococci in vanC2/3-positive samples, hence its relative abundance (expected to positively correlate with vanC2/3 gene abundance) did not substantially influence the relative abundance of all enterococci combined. Variations in Enterococcus relative abundance in these samples were likely largely driven by E. faecium and E. faecalis. In contrast, the strong correlation between vanC1 gene abundance, primarily present in E. gallinarum, and Enterococcus relative abundance suggested that E. gallinarum was an abundant species in vanC1-positive samples.
We next focused on the three patients whose stools had detectable vanC1. The first stool sample with detectable vanC1 was collected on day 11 (sample 6) in patient #10, day 6 (sample 4) in patient #12, and day 1 (sample 2) in patient #13. These findings suggested that patients #10 and #12 and possibly patient #13 did not have vanC1 in their stool at baseline but acquired it during hospitalization. Patient #10 continued to have detectable vanC1 in all stool samples following the first positive sample. Interestingly, vanC1 was detectable in stool samples collected from this patient during subsequent HCT. Two other patients in our cohort underwent HCT after completing chemotherapy. Patient #1 (with detectable vanC2/3 during chemotherapy) had detectable vanC2/3 in stool samples collected during HCT. Patient #14 did not have detectable vanC1 or vanC2/3 during chemotherapy or HCT. No patient developed a clinical infection with a vanC-positive Enterococcus sp. Within the limits of a small sample size, we found no obvious differences in clinical outcomes (Table 3).
Discussion
Gut colonization with E. casseliflavus and E. gallinarum is uncommon, occurring in only about 5% of individuals (hospitalized or non-hospitalized) [22,23]. Because of this rarity, expected non-transmissibility of chromosomally encoded vanC genes, and overall low pathogenicity, E. casseliflavus and E. gallinarum have received little attention in the medical literature. However, two recent studies provided evidence that E. casseliflavus and E. gallinarum may influence host immunity. In one study, E. gallinarum translocated from the gut into the organs and drove autoimmunity in genetically predisposed mice [11]. In the second study, we reported improved survival due to lower transplant-related mortality in allogeneic HCT recipients who had peri-transplant gut colonization with E. casseliflavus or E. gallinarum [10]. Because the source of these bacteria in the gut is unknown, we conducted the present study to determine whether patients with acute leukemia (the most common indication for allogeneic HCT) acquire E. casseliflavus and E. gallinarum during their ~4-week hospitalization for intensive chemotherapy.
We used vanC1 and vanC2/3 as surrogates for E. gallinarum and E. casseliflavus, respectively, because with rare exceptions [24,25], these genes are species-specific. Our results suggest a higher than expected incidence of colonization with E. gallinarum (15% of patients) and E. casseliflavus (35% of patients) in intensively treated patients with acute leukemia. vanC1 was detectable at much higher copy numbers than vanC2/3. Although we do not know the source of E. gallinarum and E. casseliflavus, vanC genes were not present in baseline stool samples collected from patients, suggesting that prolonged hospitalization, extensive healthcare contact, major dietary changes during intensive chemotherapy, and microbiota alterations contributed to the acquisition and expansion of E. gallinarum and E. casseliflavus to detectable levels. For example, disruptions of gut microbial communities due to antibiotics may have suppressed putative natural competitors of E. gallinarum and E. casseliflavus. The negative correlation we found between vanC1 copy number and Shannon diversity supports this hypothesis. Approximately one third of all vanC-positive rectal swabs in a previous study of intensively treated cancer patients tested positive after the first week of hospitalization, supporting the possibility of nosocomial acquisition [26].
Although still controversial [27,28], some previous studies have suggested that antibiotic-induced changes in the gut resistome are transient, with a rapid reversion of ARGs to baseline after discontinuation of antibiotics [29]. Since exposure to antibiotics is nearly universal during chemotherapy, patients receiving re-induction/salvage chemotherapy have prior exposure to antibiotics. This is in contrast to patients receiving induction therapy, who may not have a significant prior history of antibiotic exposure. Our observation of no association between treatment phase and vanC detectability argues against permanent changes in the gut resistome due to induction chemotherapy and its associated antibiotics. However, vanC genes were detectable during subsequent HCT in stool samples collected from both patients who underwent HCT and had detectable vanC during chemotherapy. One possibility is that antibiotic selective pressure is necessary to maintain vanC, which was lost after completion of chemotherapy (and discontinuation of antibiotics) but recurred during HCT due to repeated exposure to antibiotics. Another possibility is that vanC persisted between completion of chemotherapy and HCT. In the absence of samples between different phases of treatment, we cannot distinguish between these possibilities.
Because chemotherapy-induced gut barrier damage facilitates bacterial translocation, even a transient gut colonization with vanC-positive enterococci during chemotherapy could lead to systemic spread of these bacteria, their structural components, or metabolites. This could potentially influence host immunity and transplant outcomes in vanC-positive acute leukemia patients who proceed to allogeneic HCT. More research is needed to determine the incidence of persistent low-level bacteremia with vanC-positive bacteremia and whether it is mechanistically linked with immune and non-immune effects after HCT.
Acknowledgments
We thank Markas Welke, Andrea Hoeschen, Kevin Olson, and Carolyn Graiziger for coordinating sample collections. Sequence data were processed and analyzed using the resources of the Minnesota Supercomputing Institute.
Data Availability
Raw sequencing data are deposited under accession number SRP141394 at the NCBI SRA.
Funding Statement
This work was supported by grants from the University of Minnesota (Medical School Innovation award and Foundation grant for new faculty) to AR. In addition, funding from Achieving Cures Together and Hubbard Broadcasting Foundation supported this research. TK received partial support from the Masonic Cancer Center at the University of Minnesota. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bilinski J, Robak K, Peric Z, Marchel H, Karakulska-Prystupiuk E, Halaburda K, et al. Impact of Gut Colonization by Antibiotic-Resistant Bacteria on the Outcomes of Allogeneic Hematopoietic Stem Cell Transplantation: A Retrospective, Single-Center Study. Biol Blood Marrow Transplant. 2016;22: 1087–1093. 10.1016/j.bbmt.2016.02.009 [DOI] [PubMed] [Google Scholar]
- 2.Scheich S, Lindner S, Koenig R, Reinheimer C, Wichelhaus TA, Hogardt M, et al. Clinical impact of colonization with multidrug-resistant organisms on outcome after allogeneic stem cell transplantation in patients with acute myeloid leukemia. Cancer. 2018;124: 286–296. 10.1002/cncr.31045 [DOI] [PubMed] [Google Scholar]
- 3.Scheich S, Reinheimer C, Brandt C, Wichelhaus TA, Hogardt M, Kempf VAJ, et al. Clinical Impact of Colonization with Multidrug-Resistant Organisms on Outcome after Autologous Stem Cell Transplantation: A Retrospective Single-Center Study. Biol Blood Marrow Transplant. 2017;23: 1455–1462. 10.1016/j.bbmt.2017.05.016 [DOI] [PubMed] [Google Scholar]
- 4.Taur Y, Xavier JB, Lipuma L, Ubeda C, Goldberg J, Gobourne A, et al. Intestinal Domination and the Risk of Bacteremia in Patients Undergoing Allogeneic Hematopoietic Stem Cell Transplantation. Clin Infect Dis. 2012;55: 905–914. 10.1093/cid/cis580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, Samstein M, et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest. 2010;120: 4332–4341. 10.1172/JCI43918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faron ML, Ledeboer NA, Buchan BW. Resistance Mechanisms, Epidemiology, and Approaches to Screening for Vancomycin-Resistant Enterococcus in the Health Care Setting. Kraft CS, editor. J Clin Microbiol. 2016;54: 2436–2447. 10.1128/JCM.00211-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brodrick HJ, Raven KE, Harrison EM, Blane B, Reuter S, Török ME, et al. Whole-genome sequencing reveals transmission of vancomycin-resistant Enterococcus faecium in a healthcare network. Genome Med. 2016;8: 4 10.1186/s13073-015-0259-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Driscoll T, Crank CW. Vancomycin-resistant enterococcal infections: epidemiology, clinical manifestations, and optimal management. Infect Drug Resist. 2015;8: 217–30. 10.2147/IDR.S54125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burger T, Fry D, Fusco R, Luschini M, Mayo JB, Ng V, et al. Multihospital surveillance of nosocomial methicillin-resistant Staphylococcus aureus, vancomycin-resistant enterococcus, and Clostridium difficile: Analysis of a 4-year data-sharing project, 1999–2002. Am J Infect Control. 2006;34: 458–464. 10.1016/j.ajic.2005.08.010 [DOI] [PubMed] [Google Scholar]
- 10.Rashidi A, Ebadi M, Shields-Cutler RR, DeFor TE, Al-Ghalith GA, Ferrieri P, et al. Pretransplant Gut Colonization with Intrinsically Vancomycin-Resistant Enterococci (E. gallinarum and E. casseliflavus) and Outcomes of Allogeneic Hematopoietic Cell Transplantation. Biol Blood Marrow Transplant. 2018; 10.1016/j.bbmt.2018.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manfredo Vieira S, Hiltensperger M, Kumar V, Zegarra-Ruiz D, Dehner C, Khan N, et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science (80-). 2018;359: 1156–1161. 10.1126/science.aar7201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rashidi A, Kaiser T, Shields-Cutler R, Graiziger C, Holtan SG, Rehman TU, et al. Dysbiosis patterns during re-induction/salvage versus induction chemotherapy for acute leukemia. Sci Rep. 2019;9: 6083 10.1038/s41598-019-42652-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Staley C, Kaiser T, Vaugh BP, Graiziger CT, Hamilton MJ, Khoruts A, et al. Predicting recurrence of Clostridium difficile infection following encapsulated fecal microbiota transplantation. Microbiome. 2018;6: 166 10.1186/s40168-018-0549-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig WG, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35: 7188–7196. 10.1093/nar/gkm864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huse SM, Welch DM, Morrison HG, Sogin ML. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol. 2010;12: 1889–98. 10.1111/j.1462-2920.2010.02193.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27: 2194–200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37: D141–5. 10.1093/nar/gkn879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang H, Ohlsson AK, Ullberg M, Özenci V. Evaluation of species-specific PCR, Bruker MS, VITEK MS and the VITEK 2 system for the identification of clinical Enterococcus isolates. Eur J Clin Microbiol Infect Dis. 2012;31: 3073–3077. 10.1007/s10096-012-1667-x [DOI] [PubMed] [Google Scholar]
- 19.Shannon CE, Weaver W. The Mathematical Theory of Communication. Urbana, IL: The University of Illinois Press; 1949. [Google Scholar]
- 20.Chalandon Y, Thomas X, Hayette S, Cayuela J-M, Abbal C, Huguet F, et al. Randomized study of reduced-intensity chemotherapy combined with imatinib in adults with Ph-positive acute lymphoblastic leukemia. Blood. 2015;125: 3711–3719. 10.1182/blood-2015-02-627935 [DOI] [PubMed] [Google Scholar]
- 21.Ribera J-M, Oriol A, Morgades M, Montesinos P, Sarrà J, González-Campos J, et al. Treatment of high-risk Philadelphia chromosome-negative acute lymphoblastic leukemia in adolescents and adults according to early cytologic response and minimal residual disease after consolidation assessed by flow cytometry: final results of the PETHEMA ALL-AR-03 trial. J Clin Oncol. 2014;32: 1595–604. 10.1200/JCO.2013.52.2425 [DOI] [PubMed] [Google Scholar]
- 22.Tedim AP, Ruiz-Garbajosa P, Corander J, Rodríguez CM, Cantón R, Willems RJ, et al. Population biology of intestinal enterococcus isolates from hospitalized and nonhospitalized individuals in different age groups. Macfarlane GT, editor. Appl Environ Microbiol. 2015;81: 1820–31. 10.1128/AEM.03661-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toye B, Shymanski J, Bobrowska M, Woods W, Ramotar K. Clinical and epidemiological significance of enterococci intrinsically resistant to vancomycin (possessing the vanC genotype). J Clin Microbiol. 1997;35: 3166–70. Available: http://www.ncbi.nlm.nih.gov/pubmed/9399514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moura TM de, Cassenego APV, Campos FS, Ribeiro AML, Franco AC, d’Azevedo PA, et al. Detection of vanC1 gene transcription in vancomycin-susceptible Enterococcus faecalis. Mem Inst Oswaldo Cruz. 2013;108: 453–6. 10.1590/S0074-0276108042013009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwaiger K, Bauer J, Hörmansdorfer S, Mölle G, Preikschat P, Kämpf P, et al. Presence of the resistance genes vanC1 and pbp5 in phenotypically vancomycin and ampicillin susceptible Enterococcus faecalis. Microb Drug Resist. 2012;18: 434–9. 10.1089/mdr.2011.0227 [DOI] [PubMed] [Google Scholar]
- 26.Tschudin Sutter S, Frei R, Dangel M, Gratwohl A, Bonten M, Widmer AF. Not All Patients with Vancomycin‐Resistant Enterococci Need To Be Isolated. Clin Infect Dis. 2010;51: 678–683. 10.1086/655824 [DOI] [PubMed] [Google Scholar]
- 27.Andersson DI. The biological cost of mutational antibiotic resistance: any practical conclusions? Curr Opin Microbiol. 2006;9: 461–5. 10.1016/j.mib.2006.07.002 [DOI] [PubMed] [Google Scholar]
- 28.Andersson DI. Persistence of antibiotic resistant bacteria. Curr Opin Microbiol. 2003;6: 452–6. Available: http://www.ncbi.nlm.nih.gov/pubmed/14572536 [DOI] [PubMed] [Google Scholar]
- 29.MacPherson CW, Mathieu O, Tremblay J, Champagne J, Nantel A, Girard S-A, et al. Gut Bacterial Microbiota and its Resistome Rapidly Recover to Basal State Levels after Short-term Amoxicillin-Clavulanic Acid Treatment in Healthy Adults. Sci Rep. 2018;8: 11192 10.1038/s41598-018-29229-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw sequencing data are deposited under accession number SRP141394 at the NCBI SRA.


