Abstract
Microenvironment evaluation of intact tissue for analysis of cell infiltration and spatial organization are essential in understanding the complexity of disease processes. The principle techniques used in the past include immunohistochemistry (IHC) and immunofluorescence (IF) which enable visualization of cells as a snapshot in time using between 1 and 4 markers. Both techniques have shortcomings including difficulty staining poorly antigenic targets and limitations related to cross-species reactivity. IHC is reliable and reproducible, but the nature of the chemistry and reliance on the visible light spectrum allows for only a few markers to be used and makes co-localization challenging. Use of IF broadens potential markers but typically relies on frozen tissue due to the extensive tissue autofluorescence following formalin fixation. Flow cytometry, a technique that enables simultaneous labeling of multiple epitopes, abrogates many of the deficiencies of IF and IHC, however, the need to examine cells as a single cell suspension loses the spatial context of cells discarding important biologic relationships. Multiplex fluorescent immunohistochemistry (mfIHC) bridges these technologies allowing for multi-epitope cellular phenotyping in formalin fixed paraffin embedded (FFPE) tissue while preserving the overall microenvironment architecture and spatial relationship of cells within intact undisrupted tissue. High fluorescent intensity fluorophores that covalently bond to the tissue epitope enables multiple applications of primary antibodies without worry of species specific cross-reactivity by secondary antibodies. Although this technology has been proven to produce reliable and accurate images for the study of disease, the process of creating a useful mfIHC staining strategy can be time consuming and exacting due to extensive optimization and design. In order to make robust images that represent accurate cellular interactions in-situ and to mitigate the optimization period for manual analysis, presented here are methods for slide preparation, optimizing antibodies, multiplex design as well as errors commonly encountered during the staining process.
Keywords: Cancer Research, Issue 149, cancer, metastasis, immunofluorescence, multiplex fluorescent immunohistochemistry, microenvironment, cell-to-cell engagement, staining, fluorophore
Introduction
Visualization of an intact tumor microenvironment (TME) is essential in evaluating not only cellular infiltration in solid malignancies but cell to cell interactions as well. Multiplex fluorescent immunohistochemistry (mfIHC) has emerged as an effective tool for multi-antigen phenotyping of cells in the study of cancer and associated diseases1,2,3,4,5,6,7. This, in combination with novel software and programs designed to analyze large data sets, enables observation of complex interactions between cells1,2,4. The rate limiting factor in data acquisition is often the quality of the stained tissue prior to analysis.
Previous techniques used to phenotype cells in the TME include immunohistochemistry (IHC), immunofluorescence (IF) and flow cytometry all of which present significant limitations. IHC uses formalin fixed paraffin embedded (FFPE) tissue sections which are deparaffinized and rehydrated before being stained by most often one antibody. Use of a horseradish peroxidase (HRP) bound secondary antibody and a chemical reaction allows visualization of a single antigenic epitope8. While IHC is reliable and performed on FFPE tissue which is easy to work with, limitations to the visible light spectrum means only one or two markers can be reliable distinguished and colocalization of antigens quite difficult8. A way to expand available markers and therefore antigens that can be probed on a single section is to change to fluorescence which allows for use of a broader range of the visual spectrum. For IF, frozen or FFPE tissue is transferred onto slides and antibodies used that are conjugated to various fluorophores. While this increases the number of antigens that can be probed, there are several important limitations. First, because each antibody typically only has one fluorophore attached, the brightness is often not strong enough to overcome tissue autofluorescence. It is for this reason, most IF is performed on frozen tissue which is expensive to store and difficult to work with. A limited number of fluorescent tags are available for use due to spectral overlap and cross species reactivity particularly when non-conjugated antibodies are used. Flow cytometry, which consists of fresh tissue processing into a single cell suspension and labeling with fluorescent antibodies has been the gold standard for immunophenotyping for decades9,10. A benefit of flow cytometry is the ability to label multiple antibodies without concern for species cross reactivity as most are conjugated. Because the cells are “visualized” by a machine and not human eyes, there are far more fluorophores available but this comes with a cost. Compensation must be done manually which can significantly alter results producing false positive and negative populations. The most significant limitation of flow cytometry is that tissue architecture and subsequently all spatial information is lost by the necessity for single cells suspension.
Multiplex fluorescent immunohistochemistry (mfIHC) using an automated fluorescent microscope in combination with novel software combines the benefits of IHC, IF and flow cytometry by allowing multi-antigen tissue staining with signal amplification and retention of spatial relationships without the need for compensation. FFPE tissue is placed on charged slides which, after antigen retrieval, undergo a round of primary antibody application to the target antigen of interest followed by a secondary antibody with an HRP chemical tag, similar to IHC. After placement of the secondary antibody, an HRP specific reaction results in a fluorophore covalently binding to the epitope of interest11. Once the tissue is labeled, another round of heating the slides is completed removing the previously applied primary and secondary antibody complex leaving only the fluorescent tag bound to the tissue epitope11. This allows for multiple antibodies of any species to be reapplied without concern for cross-reactivity11,12. To minimize any need for manual compensation of multiple fluorescent dyes, a collection of fluorophores with little spectral overlap including a nuclear counter stain is used to complete the mfIHC. To account for the autofluorescence encountered with FFPE tissue, software subtracts the autofluorescence from the final image using an image from a blank slide which is possible because of the strength of antigen specific fluorescence following fluorophore signal amplification. Using novel programs designed for large data sets, cell locations can be identified and analyzed for spatial context1,2,4. The most significant limitation of this technique is optimization time. A detailed methodology with instructions for experimental design and staining and imaging strategy is found here. mfIHC will be useful for laboratories that do not currently have an optimized automated staining system that would like to better understand the spatial context of cell-to-cell interactions in intact FFPE tissue using the manual technique.
Protocol
All work has been approved by the University of Michigan’s internal review board.
- Optimizing primary antibodies and slide preparation
- Determine ideal concentration of antibodies for multiplex using conventional IHC.
- Test the antibodies for the multiplex by manual conventional immunohistochemistry (IHC)13.
- Use specific tissues that have an abundant cell type for each antibody tested such as using tonsil tissue for CD3 antibody testing.
- Use the reference concentration for IHC recommended by the company and then complete IHC with antibody concentrations at the recommended concentrations as well as concentrations above and below the recommended concentration.
- Prepare formalin fixed paraffin embedded tissue onto slides.
-
Using a microtome, cut blocks of FFPE tissue at a thickness between 4 and 6 μm and apply to charged slides.NOTE: Using distilled water ensures proper tissue mounting and adherence throughout the multiplex.
- Place the slides flat, tissue side up, and allow to dry at 37 °C overnight.
- Store slides in a slide box away from extreme temperature until ready to complete the multiplex.
-
- General staining method
- Preparation of wash and antigen retrieval solutions
- Prepare the wash solution of 0.1% TBST by mixing 9 L of deionized water, 1 L of tris buffered saline (TBS) and 10 mL of polysorbate 20.
- Prepare both antigen retrieval solutions (pH 6 and pH 9; Table of Materials) by diluting to 1x with deionized water.
- Deparaffinization and rehydration
- Bake slides, lying flat, tissue side up at 60 °C for 1 h in a hybridization oven1. Remove slides from the oven and allow to cool for at least 5–10 min then place in a vertical slide rack.
- Treat the slides in the following solutions sequentially: xylene in triplicate, followed by a single treatment of 100% ethanol, 95% ethanol, and lastly 70% ethanol for 10 min each using a slide staining set1.
- Wash the rack of slides for 2 min with deionized water by submersion in a plastic slide box followed by fixation by submersion in a plastic slide box filled with neutral buffered formalin for 30 min13. Wash the slides in deionized water for 2 min then proceed to antigen retrieval.
- Antigen retrieval
-
Place the rack of slides into a heat resistant box filled to the internal line (approximately 300 mL) with pH 6 or pH 9 antigen retrieval buffer.NOTE: Antigen retrieval buffer can be either pH 6 or pH 9 which may need to be optimized per epitope. However, start by using pH 9 for antibodies that bind to nuclear epitopes and pH 6 for all other antibodies.
-
Cover the box with plastic wrap and use a rubber band to secure. Place the box into the microwave at the edge of the rotating plate (microwave must be equipped with inverter technology for even heating). Heat the slides for 45 s at 100% power followed by 15 min at 20% power13.NOTE: Microwave treatment may need optimization depending on the microwave used.
- Let the slides cool for approximately 15–20 min.
-
- Prepare working solutions of antibodies and fluorophores while slides cool.
- Prepare the primary antibody diluent by dissolving 0.5 g of bovine serum albumin granules into 50 mL of TBST. Alternatively, use 50 mL of 1x phosphate buffered saline (PBS) to dissolve granules.
- Make each primary antibody working solution to the optimized concentration determined in section 1, in the primary antibody diluent. Estimate approximately 200 μL per slide.
-
Prepare the fluorophore working solution by diluting each fluorophore in the fluorophore diluent at a concentration of 1:100.NOTE: This may need to be optimized depending on the antibody. Estimate approximately 100 μL per slide.
- On the last day of staining, prepare the 4′,6-diamidino-2-phenylindole (DAPI) working solution by adding 3 drops of DAPI into 1 mL of TBST.
- Slide staining (washing and blocking)
- Remove the box from the microwave after cooling and take off the plastic wrap, wash the slides with deionized water for 2 min followed by a 2 min wash in a plastic slide box filled TBST13.
- After drying each slide around the tissue with a delicate task wipe careful not to let the tissue dry out, trace around the outside of the tissue using a hydrophobic barrier pen. Do not touch the tissue with the delicate task wipe or pen.
- Place each slide in the humidified chamber and add blocking solution (approximately 4 drops) to the tissue. Incubate for 10 min at room temperature (RT).
- Slide staining (primary antibody application)
- Take each slide and remove the blocking solution by tapping the side of the slide on a stack of paper towels and using a delicate task wipe to remove excess blocking agent from around the tissue.
- Lay the slide back into the humidified chamber and add approximately 200 μL of the working primary antibody. Incubate in the humidified chamber for 1 h at RT.
-
Wash with TBST for 2 min by submersion in triplicate13.NOTE: After each washing step, changing the TBST will help ensure a cleaner image.
- Slide staining (secondary antibody application)
-
Dab off the remaining TBST from each slide and apply approximately 3 to 4 drops of secondary antibody (mixture of rabbit and mouse secondary HRP conjugated antibodies). Incubate for 10 min in the humidified chamber at RT13.NOTE: If planning to use a primary antibody that requires a secondary antibody other than mouse or rabbit or choosing to use an alternative secondary antibody, apply the appropriate HRP conjugated secondary to the slides and incubate at RT for 45 min. Optimization of incubation time may be needed for the alternative secondary14.
- After incubation, wash with TBST for 2 min in triplicate13.
-
- Removal of antibodies
-
Microwave slides (45 s at 100% then 15 min at 20%) in the heat resistant box (see step 2.3.2) for removal of antibodies13,14.NOTE: This completes one round of the multiplex. This is the only point at which the protocol can be paused. To pause the protocol, leave the slides submerged in the antigen retrieval buffer overnight at room temperature.
-
- Continue additional rounds of staining. Repeat steps 2.5.1–2.9.1 for the rest of the antibody-fluorophore pairs. Once all antibodies and fluorophores have been used, proceed to section 2.11.
- DAPI application and mounting
- After the last staining round in the multiplex, remove the last antibody application with antigen retrieval solution pH 613. Wash with deionized water followed by TBST for 2 min each13.
- Apply approximately 150 μL of the working DAPI solution to slides and incubate in the humidified chamber for 10 min at RT1.
- Wash with TBST for approximately 30 s and mount coverslips with an antifade mountant. Apply clear fingernail polish at the four corners of the coverslip once the mounting media has dried to secure coverslip (optional).
- Details for library, monoplexes, and multiplex
- Choose slides for building a fluorophore library.
-
For a 7-color multiplex gather 7 immune cell rich tissue slides (i.e., tonsil, spleen, etc.) for the library.NOTE: Choose control slides to be used for a fluorophore library with each slide representing each unique fluorophore that will be used in the multiplex. Control slides should have an abundant epitope of choice and may be the same type of tissue for each fluorophore.
-
- Stain and image library slides for use with monoplexes and multiplexes.
- Follow steps 2.1–2.9.1 using the control slides and control tissue slides of choice. Place a different fluorophore on each slide at a concentration of 1:100; one slide should contain only DAPI.
- Proceed to section 2.11 except omit DAPI staining and instead use TBST and mount as described.
-
After letting slides dry overnight, image with all channels DAPI, CY3, CY5, CY7, Texas Red, semiconductor quantum dots, and fluorescein isothiocyanate (FITC) setting the exposure to 250 ms with the saturation protection feature1.NOTE: The exposure times can be set at 50–250 ms13.
- Upload the library images onto the software and use in the analysis of the monoplexes and multiplex.
- Choose slides for monoplexes.
-
Choose control slides that have an abundant epitope of choice based on optimized antibodies (section 1). For every round of staining to be done in the multiplex, select that number of control slides to create monoplexes.NOTE: The purpose of the monoplex is to determine the best position (order) for each antibody to be used in the multiplex.
-
- Stain monoplexes.
-
Follow sections 2.1–2.11 using the primary antibody at a different order (position) for each of the slides. Each slide should have only one primary antibody applied at an order (position) different than the other slides.NOTE: For each slide where the round calls for no primary antibody or fluorophore, instead use primary antibody diluent and fluorophore diluent13.
-
- Image slides and analyze the monoplex.
-
Using the microscope and setting the exposure time to 250 ms for all channels capture each image utilizing DAPI to focus.NOTE: The exposure times can be set at 50–250 ms14.
- Using the analysis software evaluate each monoplex slide by looking at the fluorescent intensity of the stained marker.
- Compare this intensity to the background. If it is at least 5–10 times higher than the background, the position of that slide is an optimal position to use in the final multiplex.
-
- Choose slides for the multiplex.
- Choose the slides of interest and add an extra slide to be used as a blank slide for subtraction of autofluorescence after staining and imaging.
- Stain the multiplex.
-
Based on the monoplex results, choose the appropriate order (positions) for each antibody based on step 3.5.3. Assign each fluorophore to an antibody.NOTE: This may need optimization; however, plan to use bright antibodies for less abundant markers1, co-localizing antibodies should be matched with fluorophores at far spectrums from each other13, and fluorophores with the spectrum 540 and 570 should not be co-localized due to spectral overlap issues1.
- Proceed with sections 2.1–2.11 ensuring the blank slide does not receive primary antibody, fluorophore, or DAPI, instead use antibody diluent, fluorophore diluent, and TBST respectively in its place.
-
- Image multiplex and analyze for integrity of staining
- Using the microscope and setting the exposure time to 250 ms for all channels, capture each image utilizing DAPI to focus.
-
Using the analysis software (Table of Materials), evaluate each multiplex by fluorescent false color.NOTE: A clear image should demonstrate each marker clearly. If a marker looks diffuse and grainy or is nonexistent, it is likely that the marker didn’t work and needs to be re-optimized.
- Using the analysis software, evaluate each multiplex by pathology view. Pathology view will confirm the specificity of each marker. Compare to IHC previously optimized (section 1).
Representative Results
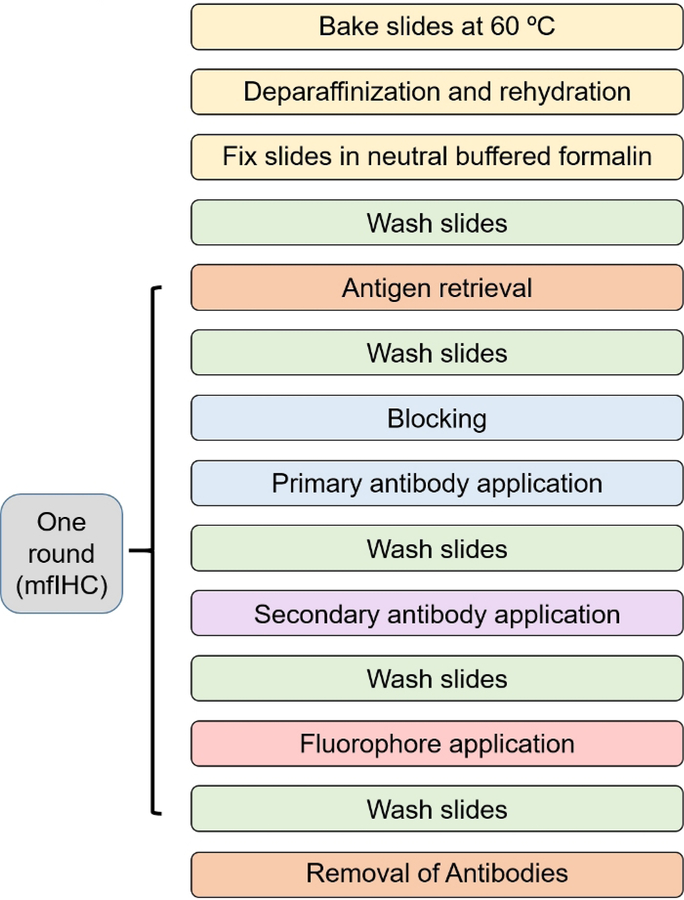
The overall process of obtaining a 7-color multiplex assay follows a repetitive pattern. Figure 1 describes the process in a diagrammatic form. Once slides are cut and dried or are received from the laboratory and baked in a hybridization oven at 60 °C for 1 h, then proceed to deparaffinization and rehydration, fix the slides in formalin again followed by antigen retrieval. Each round of multiplexing starts at antigen retrieval and finishes at antibody removal (Figure 1).
Figure 1: Staining workflow diagram.
Optimization of each step in the process is paramount to achieving a clear and useable image. Antibodies should be tested by IHC first using antigen retrieval buffers with pH 6 and pH 9 for each antibody to visualize which antigen retrieval buffer works best with that particular antibody. Generally, nuclear targets respond to antigen retrieval with buffer pH 9. For example, FOXP3 works best when antigen retrieval buffer of pH 9 is used as opposed to CD3 which works best with antigen retrieval buffer of pH 6. Images taken from the IHC process should be used to validate the specificity of the multiplex analysis.
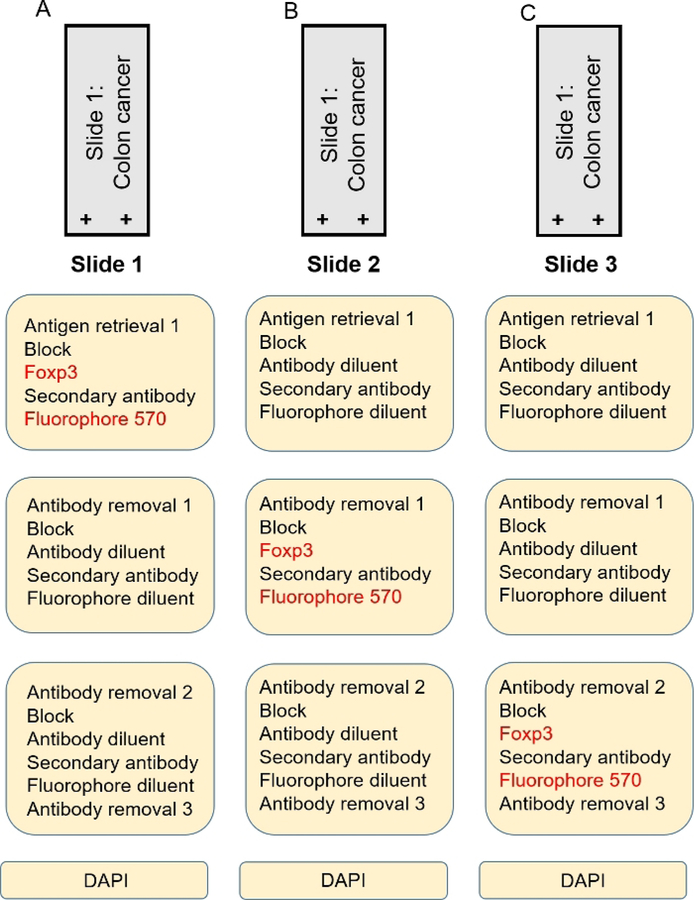
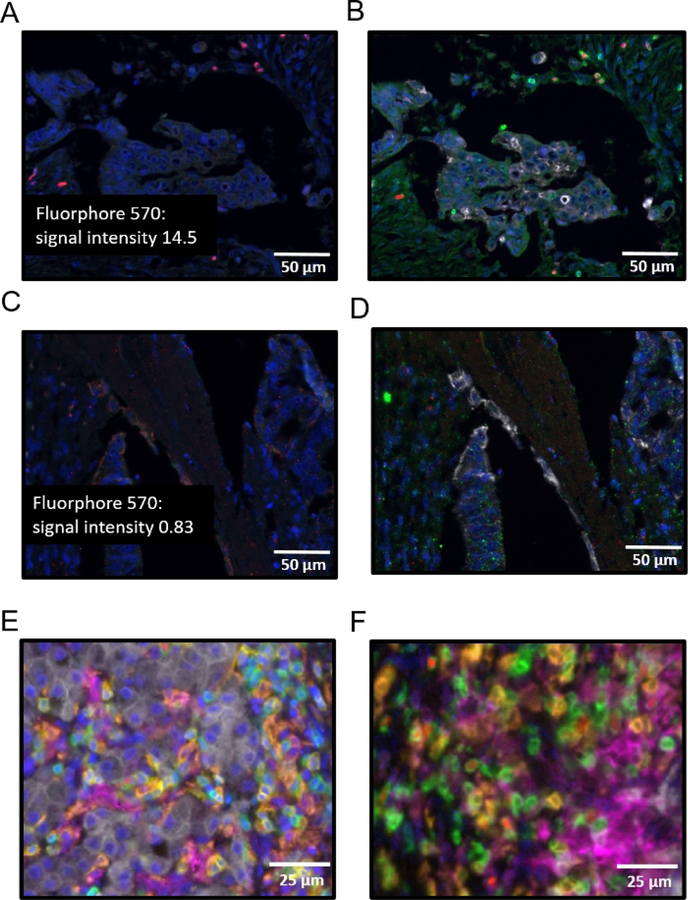
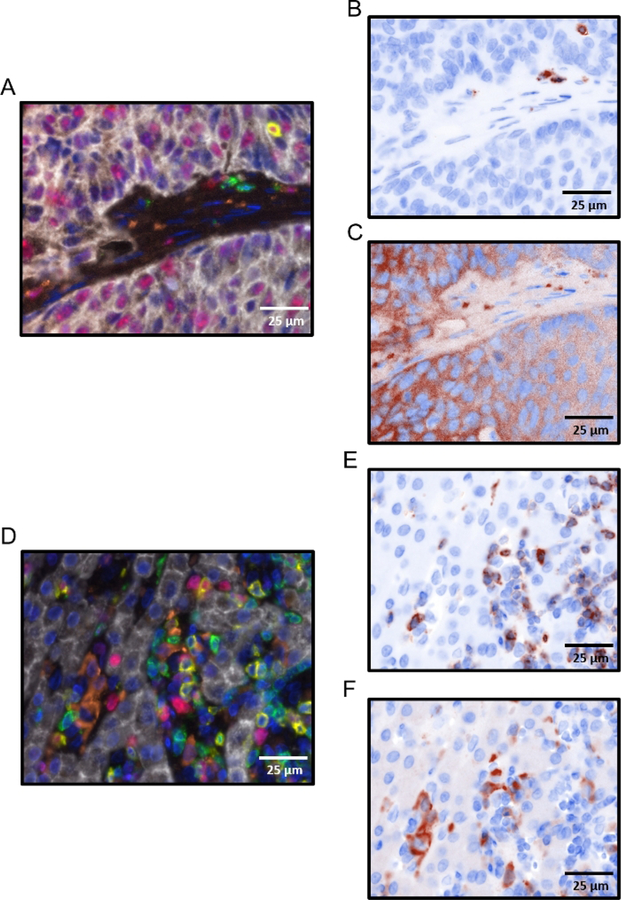
A monoplex is necessary to determine the order of each antibody in the multiplex. When using either a 4- or a 7-color kit, each antibody will have a preferred order in the array. For the number of antibodies that will be used, gather the appropriate control slides (i.e., for a 7-color multiplex where one color is DAPI, select 6 tissue specific representative control slides for the 6 other fluorophores). Figure 2A–C is a schematic representation of a monoplex assay using 3 slides with FFPE colon cancer tissue. Each slide has FOXP3 in a different position in the array using fluorophore 570 as its tag. DAPI is used as a counter stain to adequately visualize nuclei. In Figure 3, representative images of a monoplex and a multiplex with varying FOXP3 position are shown. In Figure 3A,B where FOXP3 is at the third position (corresponding to the order shown in Figure 2C), specific and robust staining of FOXP3 (red) is seen as demonstrated by bright nuclear staining Figure 3A and co-localization with CD3 Figure 3B. The fluorophore signal intensity confirms the specificity of FOXP3 without nonspecific staining elsewhere in the tissue Figure 3A. In Figure 3C, where FOXP3 is at the first position (corresponding to the order in Figure 2A), there is non-specific staining of FOXP3. Grainy diffuse areas of red cover most of the slide and fluorescent intensity at these areas are less than 1. Attempting to do a multiplex without completing a monoplex first to test the order of the staining results in a similar outcome and will waste reagents. Another critical step that cannot be overlooked in the multiplex process is DAPI staining. A working DAPI counterstain is the basis for cell identification and further spatial analysis using the analysis software. An important contrast can be seen in the composite image with working DAPI (blue) in Figure 3E and in the non-working DAPI in Figure 3F. The image in Figure 3F was not able to be used to identify cells in the analysis software. An attempt to rescue the multiplex and the tissue analysis, the coverslip maybe removed by soaking the slide in TBST followed by a microwaving step in pH 6 antigen retrieval buffer with an additional application of DAPI.
Figure 2: Monoplex development.
(A) Colon cancer slide with FOXP3 at position 1. (B) Colon cancer slide with FOXP3 at position 2. (C) Colon cancer slide with FOXP3 at position 3.
Figure 3: Successes and pitfalls of monoplex and multiplex.
(A) Monoplex assay of primary colon cancer with FOXP3 at position 3, fluorescent intensity label shown. (B) Multiplex assay of primary colon cancer with FOXP3 at position 3 and CD3 at position 2. (C) Monoplex assay of primary colon cancer with FOXP3 at position 1, fluorescent intensity label shown. (D) Multiplex assay of primary colon cancer with FOXP3 at position 1 and CD3 at position 2. (E) Multiplex assay of primary colon cancer with order of antibodies as follows: CD8 (yellow), CD3 (green), CD163 (orange), pancytokeratin (white), PD-L1 (magenta), FOXP3 (red), working DAPI (blue). (F) Multiplex assay of primary colon cancer with order of antibodies as follows: CD8 (yellow), CD3 (green), CD163 (orange), pancytokeratin (white), PD-L1 (magenta), FOXP3 (red), non-working DAPI (blue).
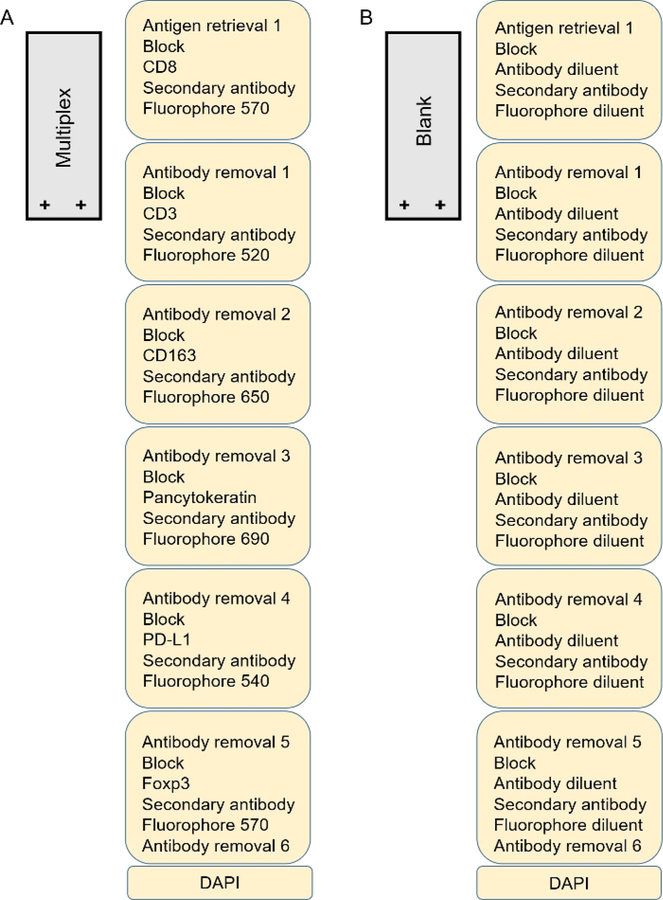
Once the monoplex assay has been completed and optimal antibody order confirmed, a multiplex strategy can be constructed Figure 4. For every target a different fluorophore must be chosen. The number associated with each fluorophore roughly represents the spectral fluorescent wavelength emitted after excitation, similar to flow cytometry. Care should be taken when matching proposed antibodies with fluorophores to limit spectral overlap and potential bleeding of markers. A good strategy is to choose wavelengths far from each other for co-localizing antibodies to reduce excess spectral overlap providing a crisp image and more reliable phenotyping13. For example, in the multiplex strategy displayed (Figure 4), CD8 is paired with fluorophore 570 while CD3 is paired with fluorophore 520. One should avoid use of fluorophore 540 and 570 in co-localized targets as these tend to be the most undiscriminating and can impair results. Fluorophore 620 tends to be very bright and should be used for antibodies that are not abundant in the tissue. A recent library used as a reference for each fluorophore also aids in reducing spectral overlap of fluorophores. A blank slide that undergoes all rounds of antigen retrieval is necessary to ensure autofluorescence extraction from the composite image. This blank slide will undergo the same treatment as the multiplex slides except in place of a working primary antibody and fluorophore, the antibody diluent and fluorophore diluent will be used respectively Figure 4B.
Figure 4: Multiplex development.
(A) Multiplex slide of colon cancer in a 7-color multiplex. (B) Blank slide in a 7-color multiplex using colon cancer tissue.
To ensure the multiplex design worked properly and the markers seen in the composite image are specific, use the “pathology image” in combination with the “brightfield” setting option using microscope software which creates a faux IHC image which can then be compared to the previous IHC assays discussed in section 1. Unfortunately, beautiful composite images may not necessarily reflect an accurate stain and this is an important step to quality control the process and prevent false positive results. Figure 5A demonstrates a composite image without initial concern. CD3 is visualized and shows specific staining (green) and is confirmed by the pathology brightfield image in Figure 5B. CD163 (orange) is seen in the composite image (Figure 5A); however, in evaluating the pathology brightfield view of CD163 (Figure 5C), the antibody is nonspecific. An example of an optimal multiplex image with specific staining is demonstrated in a composite image (Figure 5D). CD3 and CD163 specificity is confirmed using pathology brightfield view (Figure 5E and Figure 5F, respectively) and compares favorably to previous IHC assays performed using these antibodies (not shown).
Figure 5: Analyzing specificity of a 7-color multiplex.
(A) Multiplex composite image with antibodies as follows: CD8 (yellow), CD3 (green), CD163 (orange), pancytokeratin (white), Ki67 (red), DAPI (blue). (B) Pathology brightfield view of image in panel A on CD3 only view. (C) Pathology brightfield view of image in panel A on CD163 only view. (D) Multiplex composite image with antibodies as follows: CD8 (yellow), CD3 (green), CD163 (orange), pancytokeratin (white), PD-L1 (magenta), FOXP3 (red), DAPI (blue). (E) Pathology brightfield view of image in panel D on CD3 only view. (F) Pathology brightfield view of image in panel D on CD163 only view.
Discussion
Intact tissue specimens from solid tumor biopsy and surgical resection remain important diagnostic and predictive tools for disease analysis as well as patient prognosis. Multiplex fluorescent immunohistochemistry (mfIHC) is a novel technique that combines the benefits of immunohistochemistry (IHC), immunofluorescence (IF) and flow cytometry. Previous methods to probe cells in situ have allowed for evaluation of cell-to-cell arrangements in a tissue environment8, however, the low number of epitopes that can be probed limits complex analysis. Flow cytometry, although useful in analyzing many cell types in a specimen, loses spatial context by virtue of preparing samples as a single cell suspension9,10. mfIHC bridges these techniques by retaining the spatial context of the cellular microenvironment and increasing the number of epitopes that can be labeled while amplifying the signal per labeled epitope11. Previous research has used mfIHC for phenotyping the cellular infiltrate in cancer and non-cancer tissue specimens1,2,3,4,5,15. Post-image analysis has also been used to analyze spatial relationships of cells and structures within the tissue microenvironment1,2,4. For reliable analysis, a specific and accurate image must first be produced. The manual staining technique, as opposed to an automated staining system, enables smaller and cost-conscious laboratories access to this technology. Optimization time and staining time are among the largest barriers to achieving a reliable image for further spatial analysis.
Several general rules enhance the likelihood of successful generation of a reliable composite multiplex image and will save time and resources. Antibodies to be used in the multiplex should be chosen based on success with manual conventional IHC. Antigen retrieval buffers with pH 6 and pH 9 should be used for IHC in order to investigate the appropriate antigen retrieval for use in the multiplex. Monoplex development is necessary to identify the placement of antibodies within the multiplexing protocol. The rationale for this is complicated and varies between tissue specimens and epitopes. In some instances, the epitope can degrade with multiple microwaving steps and marker visualization is lost, as is often the case with CD3. FOXP3, however, becomes brighter with more microwaving steps and is barely visible if placed at the first or second round of multiplex13,14.
The staining process starts with FFPE tissue cut onto charged slides. The first problem that may be encountered is the tissue falling off of the slide after microwaving. When charged slides are not used, adherence of tissue to the glass surface is limited and the tissue will have either excessive folding or may slide off entirely. To further ensure the tissue adheres to the slides during the staining process, several techniques are performed. First while cutting the blocks onto slides, the purest form of water works best. For some laboratories, deionized water may be sufficient but distilled water has worked best. The slides should be dried flat immediately after cutting at 37 °C overnight. To further ensure adherence, the slides are then placed in a hybridization oven and baked lying flat at 60 °C for one hour just prior to use. Although formalin fixation has already occurred during the initial tissue preparation process, placing the slides in neutral buffered formalin for 30 min after the deparaffinization and rehydration process enhances the structural integrity of the mounted tissue1,13.
The manual staining process can take up to three, 8-hour days depending on the number of antibodies used. Avoiding pitfalls is essential in saving time and sparing reagents. The first common mistake often occurs in reagent preparation. Water differs among laboratories and can be the one single difference in results from one lab to the next. Using the same as well as a clean water source for all reactions and reagents ensures a consistent result. The antibodies, blocking solution, and fluorophores work best at room temperature. To avoid pitfalls in reagent and working solution preparation, use the same water for all reagents. Also use all of the working solutions and reagents at room temperature.
Troubleshooting of mfIHC is important and adherence to strict laboratory guidelines crucial to limit contamination of components and/or operator error. Production of suboptimal images will skew results and lead to false positive and negative cell populations which can negatively impact data. Because the ultimate analysis typically includes computer generated phenotypes based on machine learning, small errors in staining can be magnified to affect the entire data set. For example, although all the stains may work perfectly, an error or omission of DAPI will prevent future cell segmentation and counting rendering the experiment useless. Analysis software uses DAPI staining not only to identify cells but assigns x and y coordinates to each cell and if the multiplex has dim, poorly circumscribed, or diffuse DAPI staining, the image cannot be further used and the multiplex must be redone or attempted rescue and re-staining of DAPI. Use of software components such as the pathology and brightfield views will enhance confidence in sensitivity and specificity early in the optimization process and prevent mistakes early in the multiplex process.
The modifying aspects of the imaging process can also aid in image optimization. For example, taking an image using DAPI as the visual aid to focus helps to avoid blurry images. Adding the newly made fluorophore library to the software will ensure a clean image and less spectral overlap of each fluorophore during the analysis phase and using the blank slide to extract the autofluorescence intensifies the markers and enhances the image quality.
The use of tissue specimens has and will remain as an essential tool in investigating pathophysiology of disease. Emerging technology has increased our understanding of cell-to-cell interactions and mfIHC is a promising new player. Optimization and accuracy during the staining process in this technique is paramount for proper utilization and further analysis of cell-to-cell interactions.
Acknowledgments
The authors would like to thank Ed Stack, previously from Perkin Elmer, for his assistance with setup and optimization of original multiplex staining. The authors would also like to thank Kristen Schmidt from Akoya Biosciences for tips using the analysis software. Research reported in this publication was supported by the National Cancer Institutes of Health under Award Number P30CA046592, K08CA201581(tlf), K08CA234222 (js), R01CA15158807 (mpm), RSG1417301CSM (mpm), R01CA19807403 (mpm), U01CA22414501 (mpm, hc), CA170568 (hc). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Additional support was provided by Jeffery A. Colby Colon Cancer and Tom Liu Memorial Research Funds.
Footnotes
Video Link
The video component of this article can be found at https://www.jove.com/video/59915/
Disclosures
The authors have nothing to disclose.
References
- 1.Lazarus J et al. Spatial and phenotypic immune profiling of metastatic colon cancer. JCI Insight. 3 (22) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barua S et al. A Functional Spatial Analysis Platform for Discovery of Immunological Interactions Predictive of Low-Grade to High-Grade Transition of Pancreatic Intraductal Papillary Mucinous Neoplasms. Cancer Informatics. 17, 1176935118782880 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang L, Liu Z, Tan J, Dong H, Zhang X Multispectral imaging reveals hyper active TGF-beta signaling in colorectal cancer. Cancer Biology & Therapy. 19 (2), 105–112 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carstens JL et al. Spatial computation of intratumoral T cells correlates with survival of patients with pancreatic cancer. Nature Communications. 8, 15095 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steele KE, Brown C Multiplex Immunohistochemistry for Image Analysis of Tertiary Lymphoid Structures in Cancer. Methods In Molecular Biology. 1845, 87–98 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Gorris MAJ et al. Eight-Color Multiplex Immunohistochemistry for Simultaneous Detection of Multiple Immune Checkpoint Molecules within the Tumor Microenvironment. The Journal Of Immunology. 200 (1), 347–354 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Parra ER et al. Validation of multiplex immunofluorescence panels using multispectral microscopy for immune-profiling of formalin-fixed and paraffin-embedded human tumor tissues. Scientific Reports 7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katikireddy KR, O’Sullivan F Immunohistochemical and immunofluorescence procedures for protein analysis. Methods In Molecular Biology. 784, 155–167 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Adan A, Alizada G, Kiraz Y, Baran Y, Nalbant A Flow cytometry: basic principles and applications. Critical Reviews In Biotechnology. 37 (2), 163–176 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Zaritskaya L, Shurin MR, Sayers TJ, Malyguine AM New flow cytometric assays for monitoring cell-mediated cytotoxicity. Expert Review of Vaccines. 9 (6), 601–616 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stack EC, Wang C, Roman KA, Hoyt CC Multiplexed immunohistochemistry, imaging, and quantitation: a review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods. 70 (1), 46–58 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Zhang W et al. Fully automated 5-plex fluorescent immunohistochemistry with tyramide signal amplification and same species antibodies. Laboratory Investigation. 97 (7), 873–885 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Phenoptics Research Solutions. Perkin Elmer Manual. 32 (2016). [Google Scholar]
- 14.Opal Multiplex IHC Assay Development Guide. Perkin Elmer Manual. (2014).
- 15.Carey CD et al. Topological analysis reveals a PD-L1-associated microenvironmental niche for Reed-Sternberg cells in Hodgkin lymphoma. Blood. 130 (22), 2420–2430 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]