Abstract
Traditional approaches for chemical risk assessment cannot keep pace with the number of substances requiring assessment. Thus, in a global effort to expedite and modernize chemical risk assessment, New Approach Methodologies (NAMs) are being explored and developed. Included in this effort is the OECD Integrated Approaches for Testing and Assessment (IATA) program, which provides a forum for OECD member countries to develop and present case studies illustrating the application of NAM in various risk assessment contexts. Here, we present an IATA case study for the prediction of estrogenic potential of three target phenols: 4-tert-butylphenol, 2,4-di-tert-butylphenol and octabenzone. Key features of this IATA include the use of two computational approaches for analogue selection for read-across, data collected from traditional and NAM sources, and a workflow to generate predictions regarding the targets’ ability to bind the estrogen receptor (ER). Endocrine disruption can occur when a chemical substance mimics the activity of natural estrogen by binding to the ER and, if potency and exposure are sufficient, alters the function of the endocrine system to cause adverse effects. The data indicated that of the three target substances that were considered herein, 4-tert-butylphenol is a potential endocrine disruptor. Further, this IATA illustrates that the NAM approach explored is health protective when compared to in vivo endpoints traditionally used for human health risk assessment.
Keywords: New Approach Methodology (NAM); Integrated Approach for Testing and Assessment (IATA); Read-Across; 4-tert-butylphenol; 2,4-di-tert-butylphenol; octabenzone; Endocrine disruption; Estrogen; Applied dose equivalent (ADE); Bioactivity Exposure ratio (BER)
1. Introduction
It is well known that the traditional approaches for chemical testing are unable to keep pace with the number of substances that require assessment. Indeed, a global and overarching regulatory challenge is the everchanging chemical landscape with the continual introduction of new and more complex chemistries (Barton-Maclaren et al., 2017). For example, under the Canadian Environmental Protection Act (CEPA), Canada’s Chemicals Management Plan (CMP)2 has the aim of reducing the risk posed by 23,000 existing chemicals; 4,300 of which have been identified as priorities and are on target to be assessed by the year 2020. In the United States, new and existing chemicals are regulated under the authority of the Toxic Substances Control Act (TSCA). TSCA originally passed in 1976 requires a company to submit a premanufacture notice to EPA Office of Pollution Prevention and Toxics (OPPT) prior to either manufacturing or importing a new chemical, or to a Significant New Use Notice (SNUN) before initiating a new use of an existing chemical if it is subject to a Significant New Use Rule (SNUR). The 2016 Frank R Lautenberg Chemical Safety for the 21st Century Act (LCSA) amended TSCA to introduce mandatory deadlines and other statutory requirements (USA, 2016; US EPA, 2018). This changed the EPA OPPT’s authority to regulate both new and existing chemicals. The act also requires that OPPT promote the development and implementation of alternative test methods and strategies to reduce, refine or replace vertebrate animal testing under TSCA. In addition to evaluating new chemicals prior to their entry into US commerce, the LCSA requires that EPA OPPT evaluate risk to existing chemicals. The recently updated TSCA Inventory includes 86,228 chemicals of which 40,655 have been determined to be ‘active’ in US commerce in the past 10 years. The amended TSCA requires EPA OPPT to prioritize existing chemicals to determine which ones require risk evaluation or may be considered of low priority such that risk evaluation is not warranted at this time. This chemical landscape, together with significant data needs and aggressive assessment timelines, provides the opportunity to explore and integrate emerging technologies and novel approaches, or new approach methodologies (NAMs).
NAMs for chemical risk assessment that have been developed leverage methodologies that are high-throughput [e.g., ToxCast (Richard et al., 2016)], high-content [e.g., the DNA damage (TGx-DDI) toxicogenomic biomarker (Li et al., 2017)], and computational [e.g., the Endocrine Disruptor Screening Program, EDSP, estrogen receptor (ER) bioactivity model (Browne et al., 2015)]. However, the interpretation of these new data and endpoints, and their effective integration into regulatory chemical risk assessment continues to represent largely unfamiliar territory. The EDSP ER bioactivity model is an example of a NAM with an officially recognized path as an alternative to the in vivo uterotrophic assay for tier 1 screening for estrogenic substances (US-EPA, 2015). A barrier to the adoption of some NAMs in certain regulatory contexts is the lack of harmonised, internationally recognized guiding principles. To address this need, programmes such as the Accelerating the Pace of Chemical Risk Assessment (APCRA) (Kavlock et al., 2018), EU ToxRisk, and the Organisation for Economic Co-operation and Development (OECD) Integrated Approaches for Testing and Assessment (IATA) programme are using case studies to explore how to apply and interpret these new approaches and data for decision-making as well as document the impact based on decision context and jurisdiction (OECD, n.d.). The OECD’s IATA case studies programme is a mechanism by which different jurisdictions create case studies that highlight the use of NAMs and identify opportunities in revising existing or creating new guidance; thereby, forming an important stepping stone toward a more widespread acceptance of these methodologies.
Here we present the result of a case study created jointly by Health Canada and the US EPA that was developed as a contribution to the OECD IATA case studies programme (OECD, 2018a). Broadly, this case study aimed to support priority setting and hazard characterization for risk assessment of chemicals. More specifically, it was structured to address two objectives. The first, to provide support for the proposed hypothesis that predicting the estrogen receptor (ER) binding potential of substituted phenols is facilitated when the size, number and position relative to the hydroxyl group of the substituent(s) is considered; and, the second, to build confidence in the use of NAMs in human health risk assessment activities. In particular, we used predictive models and in vitro assays that measure/predict agonism and antagonism of the ER, which is considered herein to be the primary mode of action of estrogen and xenoestrogens.
Estrogen itself is a non-hindered phenol, which is a key structural feature that enables it to bind to the ER. This is because the hydroxyl group forms a hydrogen bond in the binding pocket of the ER. The three target chemicals that were selected for this study are also substituted phenols (i.e., they contain a benzene ring with a hydroxyl group; and, there are additional substituents on the benzene ring); when the substituents fall directly beside the hydroxyl group, the phenol is considered to be hindered (i.e., at the ‘R1’ and/or ‘R5’ positions; Table 1). Therefore, 4-tert-butylphenol is non-hindered, and 2,4-di-tert-butylphenol and octabenzone are partially-hindered. These three substituted phenols were selected for the case study because they are being addressed in the third phase of Canada’s CMP.
Table 1.
Chemical structures of a generic phenol scaffold, estrogen (the endogenous ligand of the ER), and the three target substances of this case study: 4-tert-butylphenol, 2,4-tert-butylphenol and octabenzone. R1 and R5 are the hindered positions on the phenol scaffold.
| Chemical Name | CAS RN | EPA DSSTox substance identifier3 | Chemical Structure |
|---|---|---|---|
| Phenol scaffold | -- | 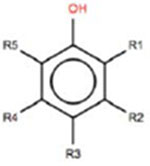 |
|
| Estrodiol (estrogen) (non-hindered) | 50-28-2 | DTXSID0020573 |  |
| Target substances | |||
| 4-Tert-Butylphenol (non-hindered) | 98-54-4 | DTXSID1020221 | 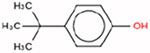 |
| 2,4-Di-Tert-Butylphenol (partially-hindered) | 96-76-4 | DTXSID2026602 | 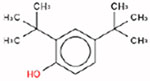 |
| Octabenzone (partially-hindered) | 1843-05-6 | DTXSID9027441 | 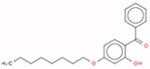 |
2. Methods
A schematic of the workflow developed and used for this case study is outlined in Figure 1 and described in greater detail below.
Figure 1:
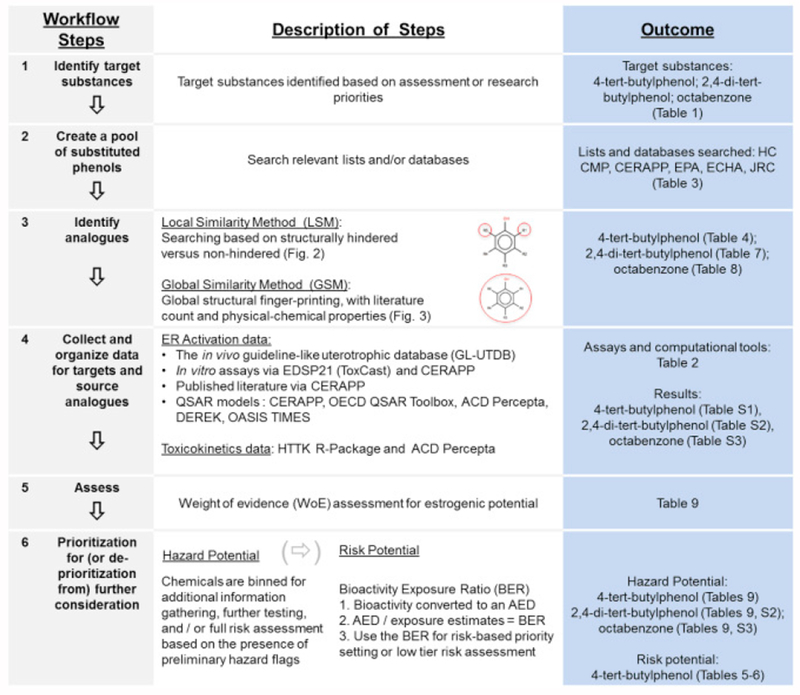
IATA Workflow for the prediction of estrogenicity of substituted phenols.
Data are summarized in a two-dimensional data matrix that was developed for each substance for hazard characterization (Tables S1–S3). In the horizontal direction of the matrix, read-across of the target phenol to the source analogues was performed for the purpose of data-gap filling, whereas in the vertical direction, data from different streams (traditional and NAM) were compared and contrasted, to evaluate concordance of orthogonal approaches for evaluating potential estrogenicity. The greater the degree of agreement in orthogonal approaches for determining bioactivity, the greater the confidence one has in using the collective results of such NAMs in hazard characterization of the target phenol.
2.1. Steps 1-2: Identify Targets and Create a Pool of Potential Analogues
Three target substituted phenols (4-tert-butylphenol, 2,4-di-tert-butylphenol, octabenzone) were selected during Step1 because they are being addressed in the third phase of Canada’s CMP. The purpose of Step2 was to create a pool of phenolic chemicals from which relevant source analogues could be identified. The pool was created by querying a number of international data inventories and chemical lists: the Canadian Chemicals Management Plan (CMP) Phase 3 list, the Collaborative Estrogen Receptor Activity Prediction Project (CERAPP), the Endocrine Disruptor Screening Program (EDSP) data availability list, the National Toxicology Program Interagency Center for Evaluation of Alternative Toxicological Methods (NICEATM) Uterotrophic database, the European Chemicals Agency (ECHA) Community Rolling Action Plan (CoRAP), the ECHA Public Activities Coordination Tool (PACT), ECHA Registration Dossiers, and the European Commission’s Endocrine Active Substances Information System (EASIS). These were selected based on the availability of associated, case-study-relevant data (most of these data are freely available online). The data sources were chosen that adequately covered the chemical space of interest and that were the most relevant for screening and assessment of endocrine disrupting chemicals. To be considered for inclusion into the aggregate pool, potential source analogues needed to have a CAS RN and/or DTXSID identifier and a discrete structure in the EPA CompTox Chemicals dashboard which is underpinned by the Distributed Structure-Searchable Toxicity (DSSTox) database (Williams et al., 2017; https://www.comptox.epa.gov).
2.2. Step 3: Identify Analogues
In Step 3, source analogues were identified from the chemical pool created during Step 2 using two different approaches, the Local Similarity Method (LSM) and Global Similarity Method (GSM), which are described in more detail elsewhere (OECD, 2018a) (Pradeep, et al. 2017). Since analogue identification and evaluation are critical aspects in a read-across approach, using two approaches provided an opportunity to compare and contrast the resulting source analogues identified for each target phenol and the relative impact this had on the subsequent assessment in the later steps of the workflow. The two approaches were considered complementary as they permitted a search for analogues based on postulated receptor interactions while at the same time investigating potentially unknown structural features that may influence activity. The local approach (LSM) was informed by the mechanistic knowledge that the hydroxyl group was essential for the ability of the substance to bind to the ER (Kiyama and Wada-Kiyama 2015). In contrast, the GSM identified analogues based on an initial global structural similarity basis with subsequent refinements. Analogues identified from both methods were carried forward to Step 4.
2.2.1. The Local Similarity Method (LSM)
The LSM has been described elsewhere (OECD, 2018a). Briefly, the LSM was developed around the hypothesis that the type of substituent(s) and their position(s) relative to the hydroxyl group influences the estrogenic potential and potency of a given phenol. Using a phenol scaffold, molecules were decomposed into R-positions about the aryl ring and a set of fragments at each position. For each R-position: (i) the unique set of fragments was determined; (ii) the fragments were fingerprinted; (iii) pair-wise similarity look-up tables (LUTs) were calculated; (iv) the full similarity matrices from the LUTs, including a similarity penalty for molecule pairs having different substitution patterns were filled; (v) the global similarity matrix was calculated; and, (vi) analogues were selected by filtering on global similarity. To perform these steps, a program was written in Python (https://www.python.org/) that made use of the Indigo cheminformatics library (http://lifescience.opensource.epam.com/indigo) for decomposition, fingerprinting, and fragment similarity calculation, and the Pandas library (http://pandas.pydata.org/) for handling matrices. This ‘local’ approach was informed by the mechanistic knowledge that the hydroxyl group is essential for the ability of the substance to bind to the ER (Kiyama and Wada-Kiyama, 2015). Thus, the model selects analogues by focusing only on the ortho substituents because these are the most important for facilitating or interfering with ER binding. The method is depicted schematically in Figure 2.
Figure 2:
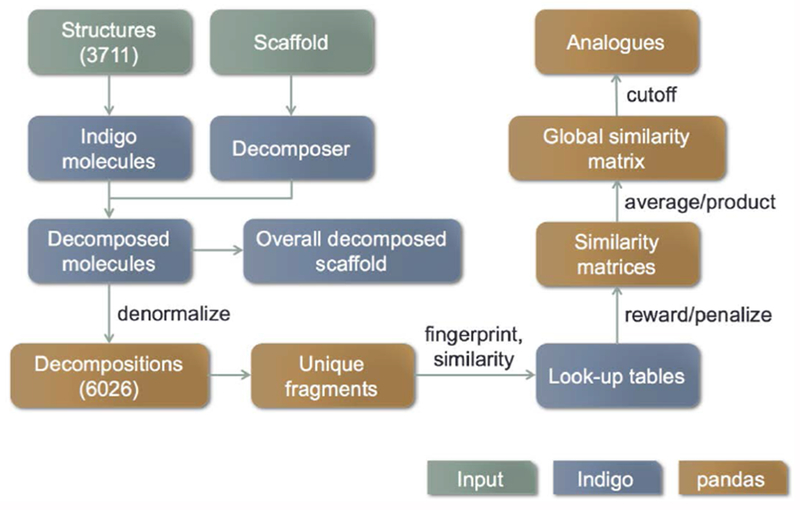
LSM analogue selection workflow
2.2.2. The Global Similarity Method (GSM)
Alternatively, the GSM considered the phenols on a global basis and did not consider the exact placement of substituents relative to the hydroxyl group. The GSM filtered analogues were informed by a data driven read-across approach that is described in more detail in Pradeep et al. (2017). In brief, this approach comprises a workflow that makes use of chemical fingerprints as a means to make initial read-across predictions of in vitro estrogenicity through to refinements that included calculating physicochemical properties considered to be relevant in driving ER activity as well as the availability of in vitro estrogenicity information for each analogue (applying a minimum threshold of at least four published studies for inclusion in the final list). The workflow is depicted in Figure 3.
Figure 3:

Summary of steps for the read-across analysis workflow adapted from Pradeep et al., 2017.
Briefly, PubChem fingerprints, a type of chemical structural fingerprint, were calculated for the target phenol and the entire source analogue pool using the KNIME analytics platform (version 2.11.3). The Jaccard distance or Tanimoto index (Tanimoto 1957) was then used to calculate pairwise similarity indices for all the chemicals as characterized by their PubChem fingerprints. For the three target chemicals, a similarity index of 0.7 was taken as a cut off for the purposes of identifying a pragmatic starting number of structurally similar source analogues. The threshold of 0.7 was selected based on the concordance analysis performed in Pradeep et al. (2017), where concordance is defined as the ratio of number of analogues with the same experimental outcome as the target and the total number of target-analogue pairs within each similarity threshold. In general, a marked increase in concordance (>80%) was observed at cut-offs above 0.7 along with a decrease in coverage (number of chemicals for which an analogue could be selected at the given similarity threshold) (discussed in more detail in Pradeep et al., 2017).
Literature reports on in vitro ER activity of each compound had been compiled as part of the overall CERAPP project (Mansouri et al., 2016). The reports arose from several different sources including the Tox21 data (Huang et al., 2014), the US FDA Estrogenic Activity Database (EADB; Shen et al., 2013), ChEMBL (Gaulton et al., 2012), and the METI database (METI, 2002). These are discussed in more detail in section 2.3. These data included an activity outcome (active/inactive) as well as 4 potency classes (very weak, weak, moderate, and strong) for active compounds. Each phenol in the CERAPP dataset was associated with a literature data count (how many independent reports of activity were found), which was used as an indicator of reproducibility. The expectation was that increasing numbers of consistent literature reports of ER activity, should give rise to more reliable estimates of true activity. Structural similarity based read-across predictions improved with the confidence in experimental data, but there was a threshold for optimal performance (as discussed in Pradeep et al., 2017). Given the impact that independent reproducibility had on analogue suitability in the case study, source analogues identified using the PubChem fingerprints were filtered to remove any that had fewer than 4 literature data sources.
Source analogues were also evaluated for their suitability using a selection of physicochemical properties that were expected to be relevant for ER binding as evidenced by other published QSAR studies (Lo Piparo and Worth, 2010). The evaluation was performed using a global filtering approach which relied upon several whole molecule properties characterising physicochemical, steric and electronic features (e.g., molecular volume, LogP, energy of the highest occupied molecular orbital (eHOMO), energy of the lowest unoccupied molecular orbital (eLUMO) etc.). Molecular properties were computed in MOE software (version 2015.10) using canonical SMILES strings. A Student’s t-test was then performed to test which properties were significantly different for the phenols that bind to the ER as compared to the non-binders. On the basis of the t-test p-values, three properties were identified as significant: LogP, molecular volume and number of hydrogen bond donors and acceptors. The distribution of each of the selected properties was used to guide the range of each property for the purposes of filtering source analogues.
The preliminary source analogues identified based on PubChem fingerprints were filtered if: (1) they did not have 4 or more literature data sources; (2) molecular volume was beyond ±100% of the molecular volume of the target chemical, if logP was beyond ±1 unit, and if the total number of hydrogen bond donors and acceptors were beyond ± 6 units. If an analogue did not meet the above criteria, it was discarded until N closest analogues were selected for each target substance within the 0.7 similarity threshold. If none of the analogues satisfied the acceptance thresholds, then the target was considered out of domain for analogue selection and any subsequent read-across predictions.
Each method has its own ‘Similarity Score’ metric that was used to rank potential analogues. Importantly, the numerical similarity values generated from the LSM and the GSM cannot be compared between methods. The LSM will always give a lower value than the GSM for the same target-analogue pair because: (1) the LSM excludes the phenol scaffold from the similarity calculation, while the GSM does not, resulting in values that will be lower than those from GSM, and (2) the multiplicative nature of LSM’s global similarity score results in lower values.
2.3. Step 4: Collect and Organize the Data
Data for the targets and analogues were collected and organized into data matrices (which are available in the Supplemental Information). Comprehensive, publicly available collections of curated data were used, which ensured efficient data collection of high quality data by leveraging the significant efforts on compiling data made by others. These data included in vivo and in vitro data as well as in silico predictions and were similar to tier 1 of the EDSP (US EPA, n.d.) and levels 1–3 of the OECD Conceptual Framework for Testing and Assessment of Endocrine Disrupters (OECD, 2018b). Data related to estrogenic activity were collected on the targets and source analogues from several sources that are listed in Table 2 and described in greater detail below. Data were extracted programmatically from many of the sources to facilitate efficient population of the OECD’s data matrix template (OECD, 2018c); details regarding the data sources used are summarized below.
Table 2.
Sources used to gather information on estrogenicity of the target and analogue chemicals.
| In vivo |
|
Assay: In vivo uterotrophic assay Endpoint(s): Increased uterine weight Data source: The GL-UTDB contains information from 400+ guideline-like studies extracted from 90+ publications, and encompasses data on 118 chemicals . This high quality database has been made freely available on the NTP website.1 (Kleinstreuer et al. 2016). |
| In vitro |
|
Assay: The results of 18 in vitro high-throughput screening (HTS) ToxCast assays were combined using a computational model to produce a single area under the curve (AUC) score for the prediction of estrogenicity of a substance (termed the EPA’s EDSP ToxCast ER model, or the ER defined approach (DA) (see OECD (2016) for general information on defined approaches) Endpoint(s): Mechanistic endpoints from the ER Pathway (e.g., ER binding, dimerization, chromatin binding, transcriptional activation and ER-dependent cell proliferation) Data source: US EPA ToxCast program and the US EPA EDSP 2 Computational model: (Browne, et al. 2015b, Judson, et al. 2015, Kleinstreuer, et al. 2017) |
|
Literature search: Literature collected under Collaborative Estrogen Receptor Activity Prediction Project (CERAPP) Endpoint: These data included an activity outcome (active / inactive) as well as a potency value for active compounds. Data source: (Mansouri, et al. 2016) |
| In silico |
|
Name: Collaborative Estrogen Receptor Activity Prediction Project (CERAPP) Model: Consensus ER QSAR model Source: (Mansouri, et al. 2016) |
|
Name: Advanced Chemistry Development (ACD) Percepta (v2015) – Estrogen Receptor Binding module Model: Statistical model for classifying ER binding affinities. Source: ACD/Percepta 2013 3 |
|
Name: Estrogen Receptor Binding Profiler – (in OECD QSAR Toolbox v3.4) Model: Scheme is based on structural and parametric rules extracted from literature sources and supported by experimental data. The ER-binding profiler classifies chemicals as non- binders or binders depending on molecular weight (MW) and structural characteristics of the chemicals. Source: OECD 2016 4 |
|
Name: US EPA rtER Expert System – (in OECD QSAR Toolbox v3.4) Model: Profiler consists of molecular definitions that mimic the structural criteria of chemical classes potential estrogen receptor-binders covered by US EPA Estrogen Receptor Expert System (ERES) The ERES profiler is an effects-based automated system used to predict estrogen receptor binding affinity. Source: OECD 2016 4 |
|
Name: Lhasa, Derek Nexus (v5.0.2) Model: Expert system used to profile category members for alerts related to estrogenicity. Source: Derek Nexus 2016 5 |
|
Name: OASIS TIMES (v2.27.19.13) Model: Estrogen Binding Affinity S9 activated (v4.04); QSAR with metabolic simulator for prediction of relative binding affinity for ER Source: TIMES 2016 6 |
|
Name: EPI Suite Model: The Estimation Programs Interface Suite (EPI Suite) is a predictive tool that estimates physical/chemical properties and environmental fate of chemical substances. Source: (US-EPA 2012) |
Abbreviations: AUC, area under the curve; CERAPP, Collective Estrogen Receptor Activity Prediction Project; EDSP, Endocrine Disruptor Screening Program; GL-UTDB , guideline-like uterotrophic assay database; HTS, high-throughput screening; NTP, U.S National Toxioclogy Program; NICEATM, NTP Interagency Center for the Evaluation of Alternative Toxiological Methods
Curated Database of Rodent Uterotrophic Bioactivity (https://ntp.niehs.nih.gov/pubhealth/evalatm/tox21-support/endocrine-disruptors/edhts.html)
EDSP21 Dashboard (https://actor.epa.gov/edsp21/)
ACD/ Percepta 2013. http://perceptahelp.acdlabs.com/help_v2014/index.php/Endocrine_System_Disruption
Derek Nexus 2016 https://www.lhasalimited.org/products/derek-nexus.htm
In vivo uterotrophic assay: EPA and OECD guidelines are available that describe the assay in more detail (EPA 2009, OECD 2007). A database of guideline-like uterotrophic studies (GL-UTDB) was previously created following an extensive literature search by the U.S National Toxicology Program (NTP) Interagency Center for the Evaluation of Alternative Toxicological Methods (NICEATM) (Kleinstreuer, et al. 2016). The GL-UTDB contains information from 400+ guideline-like studies extracted from 90+ publications and encompasses data on 118 chemicals. A study was determined to be “guideline-like” based on six criteria: group size, animal model, route of administration, necropsy timing, dosing interval, and number of dose groups. This database is freely available on the NTP website4.
In vitro high-throughput screening (HTS): The results of the 18 ToxCast and Tox21 assays as well as the ToxCast ER Model AUC scores for the target substances and potential analogues were extracted programmatically from the online US EPA Endocrine Disruptor Screening Program (EDSP) dashboard. Briefly, these assays, which measure mechanistic events related to the ER Pathway (i.e., ER binding, dimerization, chromatin binding, transcriptional activation and ER-dependent cell proliferation), are integrated into a computational model that can discriminate bioactivity from assay specific interference and responses related to cytotoxicity (Browne et al., 2015; Judson et al., 2015). The model provides an, ‘area under the curve’ (AUC) score relative to 17β-estradiol ranging from 0 (inactive substances) to 1 (bioactivity for 17β-estradiol). An AUC scores of 0 is considered to be inactive, an AUC score of >0 to <0.1 is considered to be inconclusive, and a score of ≥0.1 is considered to be active. The score is characterised by the AUC of the concentration-response profile, which is a consensus of the curves for the individual assays. The results of the individual ER related assays and the ER bioactivity model scores for tested chemicals are available online (https://actor.epa.gov/edsp21/).
In vitro studies collected under CERAPP: The Collaborative Estrogen Receptor Activity Prediction Project (CERAPP) was an international collaborative effort to predict estrogenic potential for tens of thousands of man-made chemicals found in the environment (Mansouri, et al. 2016). As part of the project, information on the estrogenic activity of chemicals was mined and curated to serve as a validation set for predictions of different models. Experimental data were collected from the following sources: HTS data from the Tox21 project, the U.S. FDA Estrogenic Activity Database (EADB), the METI (Ministry of Economy, Trade and Industry, Japan) database, and the ChEMBL database. Data are available for download on over 7000 substances on the U.S. EPA ToxCast™ Data website (CERAPP Experimental Data Evaluation Set5). All data entries were categorized into three assay classes: (a) binding, (b) reporter gene/transactivation, or (c) cell proliferation and potency levels for binding; agonist and antagonist activity were assigned based on these results. The assigned potency level and literature count from the CERAPP dataset were extracted and added to the corresponding target substance data matrices.
In silico data: Scores from the EPA’s EDSP ToxCast ER model (previously described in the in vitro HTS section) formed the basis of the training set for ER QSAR model development as part of the CERAPP project (Mansouri, et al. 2016). 48 individual ER QSAR models were derived from which a final consensus prediction was generated. These consensus predictions were then forward validated against the literature-derived ER activity data set described above. Overall, CERAPP predictions were made on approximately 32,000 unique chemical structures. The predictions for the target compounds and associated analogues were extracted from the EDSP21 Dashboard and added to the data matrices. Predictions were also generated with respect to ER binding potential from selected publicly available or commercial software tools, which are listed in Table 2. Finally, EPI Suite was used to form predictions of the following physical chemical properties: melting point, boiling point, vapour pressure and LogPow (the authors note that it is preferable to use measured physical-chemical data during a higher tier risk assessment when available, but that tools like EPI Suite are well suited for screening-level assessments and to fill data gaps when measured data are not available).
2.4. Step 5: Perform the Weight of Evidence Assessment
In Step 5, a weight of evidence assessment of the estrogenic activity of a given target chemical was then made using data for the target and its respective source analogues and expert judgement. Briefly, the results of the in vitro HTS assays (specifically, an AUC > 0.1) were used to bin substances as potentially estrogenic, or potentially not estrogenic. For more information on AUC scores, see Table 2 in Browne et al. (2015), which lists reference chemicals assigned agonist potency categories of strong, moderate, weak and very weak based on AC50 values and shows the correspondence with predicted AUC values (generally AUC values between 0.1 and 0.5 correspond to very weak and weak agonists whereas moderate and strong agonists have AUC values from 0.5 to 1). Binning based on estrogenic potential was then corroborated using the consensus of the calls from the QSAR models and CERAPP literature search. Lastly, this binning was compared to the results of the in vivo uterotrophic assay (when data were available). Data gaps were filled by reading across the data matrix to structurally similar analogues.
2.5. Step 6: Prioritization for (or de-prioritization from) Higher Tier Assessment
Finally, in Step 6, prioritization of the target chemicals occurs. Prioritization can be achieved in one of two ways: (1) by hazard potential, based on the weight of evidence assessment performed in Step 5; or, (2) by risk potential using a Bioactivity Exposure Ratio (BER) approach. The BER is a ratio of a hazard value to an exposure value (analogous to a Margin of Exposure (MOE) using traditional in vivo data in risk assessment). In the case of the BER, the hazard value is an administered equivalent dose (AED), which is obtained by transforming the in vitro bioactivity concentration (i.e., the ToxCast AC50) using high throughput toxicokinetic (HTTK) data and in vitro to in vivo extrapolation (IVIVE) per the methods described in Wetmore et al. (2012). It is important to note that while the BER is not intended to be predictive of specific endpoint toxicities, it is protective of human health when compared with point of departure values from traditional toxicity assays.
The BER approach builds from the exposure:activity profiling approach of Becker et al. (2015). The method used to calculate AED is described in detail in Wetmore et al. (2012). Briefly, plasma protein binding and hepatic metabolic clearance are measured using in vitro HTTK assays. These in vitro data are then used to parameterize a simple pharmacokinetic model to estimate expected steady state blood concentrations at a given dose (assuming 100% oral bioavailability). Monte Carlo simulation provides estimates of the variability of the steady-state blood concentration (Css) in a population of healthy individuals. For substances deemed to be active by the ToxCast AUC scoring method, the AUC score was then dropped, and the actual ToxCast assays dose response data for the substance was accessed and analyzed. Using reverse dosimetry, oral doses can then be estimated that would result in a steady-state blood concentration equivalent to the AC50 from the ToxCast assay. HTTK data has been generated for many of the ToxCast chemicals and the data along with the Wetmore et al. IVIVE modelling values have been made available in a publicly accessible httk R package (v1.8 from CRAN) to facilitate the conversion of in vitro bioactivity concentrations to AED (Pearce, et al., 2017). The AED required to achieve the upper 95th percentile Css values, equivalent to the AC50 value from the lowest active ER related assay, were estimated using the “get_wetmore_oral_equiv” function in the httk R package with the following options: the 95th quantile from the SimCYP Monte Carlo simulation (which.quantile = 0.95); selection of species (species = ‘Human’); input unit as μM (input.units = ‘uM’); output unit as mg/kg bw/day (output.units = ‘mg’); clearance assay concentration at 1 μM (clearance.assay.conc = 1). When calculating the AED for the respective median and max AC50 values from the active ER related assays the clearance assay concentration was set to 10 μM. Wetmore et al. conducted the primary human hepatocyte clearance assays using two concentrations of test chemical (1 or 10 μM) and the selection of the clearance assay concentration in the httk R package is based on the proximity to the respective AC50 value for which an AED is being derived. Finally, a BER is derived by taking the ratio of the lowest ER related AED value to the exposure estimate. In the case examples presented, the exposure estimate used was the upper limit of the 95% credible interval for predicted median exposures for the total US population from the US EPA ExpoCast program Systematic Empirical Evaluation of Models version 2 (SEEM2) model (Wambaugh et al., 2014).
3. Results
An initial pool of source analogues comprised approximately 3800 unique phenols (Step 2). There was considerable substance overlap of the selected inventories with the CERAPP chemical set (Table 3). This pool of substances was used to identify source analogues for each of the three target phenols. The results of Steps 3-6 are discussed for each target below
Table 3.
Inventories and chemical lists used to form the pool of phenolic substances.
| Inventory / Chemical List | Number of phenols (number of phenols not in CERAPP) |
|---|---|
| Canadian Chemicals Management Plan (CMP) Phase 3 List * | 128 (1) |
| Collective Estrogen Receptor Activity Prediction Project (CERAPP) | 3711 (0) |
| Endocrine Disruptor Screening Program (EDSP) Data Availability List | 672 (6) |
| National Toxicology Program (NTP) Interagency Center for Evaluation of Alternative Toxicological Methods (NICEATM) Uterotrophic database | 54 (0) |
| European Chemicals Agency (ECHA) Registration Dossiers | 441 (64) |
| ECHA Community rolling action plan (CoRAP) | 24 (0) |
| ECHA Public Activities Coordination Tool (PACT) | 25 (2) |
| European Commission Endocrine Active Substances Information System (EASIS) | 97 (17) |
3.1. 4-tert-butylphenol, CAS RN 98-54-4
3.1.1. Step 3 Identify Analogues (4-tert-butylphenol)
The LSM identified seven analogues; six with a variety of 3–5 carbon substituents at the para position (CAS 1805-61-4, 80-46-6, 1638-22-8, 99-71-8, 645-56-7 and 99-89-8), and one with a 10 carbon substituent at the R3 position (CAS 121158-58-5). The GSM identified nine analogues, four of which were also identified by the LSM (CAS 1805-61-4, 80-46-6, 1638-22-8, 99-71-8). Of the GSM analogues, seven had 3-6 carbon substituents at the R3 position (CAS 1805-61-4, 80-46-6, 1638-22-8, 99-71-8, 14938-35-3, 2409-55-4 and 92-69-3), one had two substituents at the R1 and R3 positions (CAS 2409-55-4) and one had methyl groups at both the R2 and R3 positions (CAS 95-65-8). The analogues are listed in Table 4.
Table 4:
Source analogues for 4-tert-butylphenol as identified by the LSM and GSM. CAS numbers and ToxCast ER agonist AUC model scores indicated when available.
Target: 4-tert-butylphenol CAS RN: 98-54-4 AUC = 0.161 
| |||||
|---|---|---|---|---|---|
| LSM and GSM Analogues | LSM Analogues | GSM Analogues | |||
| p-Isoamylphenol 1805-61-4 |  |
(1-Methylethyl)-phenol 25168-06-3 | 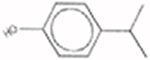 |
4-Pentylphenol 14933-35-3 | 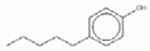 |
| 4-(2-Methylbutan-2-yl)phenol 80-46-6 AUC = 0.111 | 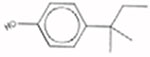 |
4-Propylphenol 645-56-7 AUC = 0.0275 | 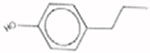 |
**2-tert-Butyl-4-methylphenol 2409-55-4 | 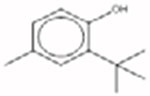 |
| 4-Butylphenol 1638-22-8 AUC = 0.282 | 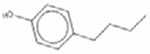 |
4-Isopropyl-phenol 99-89-8 AUC = 0.021 | 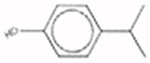 |
4-Cyclopentyl-phenol 2409-55-4 | 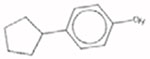 |
| 4-(Butan-2-yl)phenol 99-71-3 AUC = 0.163 |  |
Phenol, dodecyl-brarched 121158-58-5 | 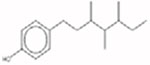 |
4-Phenylphenol 92-69-3 |  |
| 3,4-Dimethylphenol 95-65-8 | 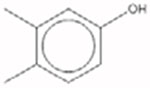 |
||||
= partially hindered
3.1.2. Step 4 Collect and Organize the Data (4-tert-butylphenol)
Data collected for 4-tert-butylphenol are summarized in Supplementary Table S1.
The ToxCast ER agonist AUC model score for 4-tert-butylphenol is 0.161, which is above the threshold to be considered active (active is defined as ≥0.1; Browne et al., 2015). Five analogues had also been tested in ToxCast. CAS RNs 80-46-6, 1638-22-8, 99-71-8 were identified by both the LSM and GSM, and were found to be active as ER agonists (AUC = 0.111, 0.282 and 0.163, respectively). These AUC scores correspond to positive control substances deemed “weak” or “very weak” in terms of their agonist potency and have lower AUC scores than the natural phytoestrogen genistein (deemed a “weak” agonist and predicted to have an AUC score of 0.54 by Browne et al., 2015). Two analogues, CAS RNs 645-56-7 and 99-89-8, found by the LSM were found not to be active (AUC = 0.0275 and 0.021, respectively). None of the analogues were found to be ER antagonists (AUC = 0–0.0002).
Predictions for both the target and source analogues were also made using the CERAPP QSAR consensus model, as well as selected in silico tools (OASIS TIMES, the OECD Toolbox, Derek Nexus and ACD Percepta). Briefly, the CERAPP consensus QSAR model predicted weak activity for ‘consensus binding’ and ‘consensus agonism’; and moderate to strong activity for ‘consensus antagonism’ (strong activity was associated with analogues with larger substituents of five or more carbons). There were no data for CAS 1805-61-4, and substances CAS 2409-55-4 and 95-65-8 had ‘suspicious inactive’ calls with this model. The ER binding model with OASIS TIMES gave predictions of ‘weak’ or ‘low’ activity for the target substance and its source analogues (with the exception of CAS 121158-58-5 and 2409-55-4, which had ‘not active’ calls). The OECD QSAR Toolbox (ER profiler and US EPA rtER expert system) also predicted weak binding across most of the substances, except CAS 121158-58-5 (strong binder) and 92-69-3 (moderate binder). ACD Percepta predicted weak or no binding to the ER-alpha for all substances. Finally, Derek Nexus generated ‘no alerts’ for nine of the substances, including the target, and an alert of ‘Mammal Plausible, Alkylphenol or precursor’ for five of the analogues (CAS 1805-61-4, 1638-22-8, 121158-58-5, 14938-35-3 and 1518-83-8). This alert indicates that these chemical structures fulfil the criteria of para substituted alkyl chain/saturated ring of four or more carbon atoms that are known to be associated with estrogenicity.
4-tert-butylphenol was found to be active in the in vivo uterotrophic assay (lowest effect level, LEL = 100 mg/kg-bw/day; route of administration: subcutaneous injection). Source analogue, 4-(Methylbutan-2-yl)phenol (CAS 80-46-6, identified by both methods), was also found to be active in the uterotrophic assay (LEL = 200 mg/kg-bw/day).
3.1.3. Step 5 Weight of Evidence Assessment (4-tert-butylphenol)
The data collected for 4-tert-butylphenol and its identified analogues are presented in Table S1; a high-level summary is presented in Table 9. As a non-hindered phenol, 4-tert-butylphenol was expected to be estrogenic. Overall, both the available NAM results and the available in vivo data indicate the potential for estrogenic activity. For ER agonism, integrating the results of the ToxCast data (as a single orthogonal assay using the AUC method) with the QSAR models, there is a high degree of concordance supporting weak estrogenic activity of the analogues and 4-tert-butylphenol. The CERAPP Consensus Antagonist model generally predicts activity ranging from inactive to active (strong); while the ToxCast antagonism model did not identify any of the analogues to be ER antagonists. Based on the data collected, the overall call for 4-tert-butylphenol was estrogenic.
Table 9.
Summary of data for the three target substances from the sources queried.
| 4-tert-butylphenol (98-54-4) | 2,4-di-tert-butylphenol (96-76-4) | Octabenzone (1843-05-6) | |
|---|---|---|---|
| In vivo | |||
| Uterotrophic assay | Active | Inactive | No data |
| In vitro | |||
| ToxCast | ER Agonism: Active ER Antagatonism: Inactive |
ER Agonism: Inactive ER Antagatonism: Inactive |
ER Agonism: Inactive ER Antagatonism: Inactive |
| CERAPP (literature sources) | ER binding: Active (vw) (15) ER Agonism: no data ER Antagonism: Inactive (6) |
ER binding: Active (vw) (6) ER Agonism: Inactive (4) ER Antagonism: no data |
ER binding: Inactive (12) ER Agonism: Inactive (6) ER Antagonism: Inactive (6) |
| QSAR | |||
| CERAPP | ER binding: Active (w) ER Agonism: Active (w) ER Antagonism: Active (s) |
ER binding: Inactive ER Agonism: Inactive ER Antagonism: Inactive |
ER binding: Active (vw) ER Agonism: Active (vw) ER Antagonism: Inactive |
| OASIS TIMES | Binding (P): Active (w) Binding (M): Active (low) |
Binding (P): Inactive Binding (M): Inactive |
Binding (P): Inactive Binding (M): Active (s) |
| OECD Toolbox | Binder (w) | Binder (s) | Binder (s) |
| Derek Nexus Expert System | No alert | No alert | No alert |
| ACD Percepta | Weak Binder ERα | Inactive | Weak Binder ERα |
| Overall call based on LSM analogues | Active | Inactive | Inactive |
| Overall call based on GSM analogues | Active | Active (with some discordant results) | Inactive (with some discordant results) |
Acronyms: ER=estrogen receptor; M=metabolite; P=parent; s=strong; vw=very weak; w=weak.
3.1.4. Step 6 Prioritization (4-tert-butylphenol)
Because Step 5 indicated that there was the potential for 4-tert-butylphenol to be estrogenic, the decision was made to use both hazard- and risk-based approaches to inform prioritization. The HTS ToxCast bioactivity data for 4-tert-butylphenol were used to estimate an administered equivalent dose (AED) through the application of reverse dosimetry. The AEDs were obtained by converting in vitro bioactivity levels (i.e., the ToxCast AC50s) using high-throughput toxicokinetic (HTTK) data coupled with in vitro to in vivo extrapolation (IVIVE). The range of AEDs for the ToxCast ER Pathway Assays was 11.22–288.77 mg/kg bw/day. A BER of 2340 was generated by dividing the lowest AED by an ExpoCast exposure estimate of 0.0048 mg/kg bw/day (Table 5). The available in vivo data for 4-tert-butylphenol were used to generate a margin of exposure (MOE) of over 14,000 for female reproductive effects observed in a two-generational study Table 6.
Table 5.
HTS ER related AC50 values, ADE values, ExpoCast exposure estimate, and BER for,4-tert-butylphenol (CAS RN 98-54-4).
| AC50 (µM) | ADE (mg/kg bw/day) | ExpoCast upper 95% CI on predicted exposure (mg/kg bw/day) for the total U.S. population | BER | |
|---|---|---|---|---|
| Lowest Activity for ER Pathway Assay † | 1.63ǂ | 11.22 | 0.0048 | 2340ǂ |
| Range of Activity for ER Pathway Assays | 1.63 – 58.05 Median 22.85 | 11.22 – 288.77 Median = 113.66 | 0.0048 | 237001 |
Assays measure mechanistic events related to the ER Pathway: ER binding, dimerization, chromatin binding, transcriptional activation and ER-dependent cell proliferation
Based on ER pathway assay with lowest AC50 (ATG_ERa_Trans_up), which refers to induction of mRNA transcription
Calculated using the median AED.
Acronyms: Active concentration at 50% (AC50); applied dose equivalent (ADE); bioactivity-exposure ratio (BER); credible interval (CI); Estrogen receptor (ER); High-throughput screening (HTS)
Table 6.
In vivo data available for 4-tert-butylphenol (CAS RN 98-54-4), ExpoCast exposure estimate and margin of exposure (MOE).
| Study Type | Study outcome | ExpoCast upper 95% CI on predicted exposure (mg/kg/day) for the total U.S. population (mg/kg bw/day) | MOE |
|---|---|---|---|
| Uterotrophic Assay |
Effect Level: LEL = 100 mg/kg-bw/day Result: 1.3 fold increase relative to control; subcutaneous injection (s.c.) over 3 days; Crj:CD(SD) Rat Source: NICEATM DB (Kleinstreuer et al. 2015) |
0.0048 | 20830ǂ |
| 2-Generation Reproductive Toxicity study (OECD 416) |
Effect Level: NOAEL/LOAEL: 70/200 mg/kg-bw/day Study design: S.D. rats; 0, 800, 2500 and 7500 ppm corresponding to 0, 70, 200 and 600 mg/kg bw/day in diet. Result: Increased vaginal epithelium atrophy, reduced relative weight of ovaries, and reduced relative weight of adrenal glands in females Source: (EU 2008) |
0.0048 | 14580 (NOAEL) 41670 (LOAEL) |
Note: This value is illustrative only. In practice, the uterotrophic assay would not typically be used to generate an MOE.
3.2. 2,4-di-tert-butylphenol, CAS RN 96-76-4
3.2.1. Step 3 Identify Analogues (2,4-di-tert-butylphenol)
Eighteen analogues were identified for 2,4-di-tert-butylphenol (Table 7), including one analogue that was identified by both methods (2,4-Diisopropylphenol). The target has tert-butyl substituents at both the R1 and R3 positions (refer to Table 1 for numbering of substituent positions). Similarly, the 10 analogues identified by the LSM had substituents at the R1 and R3 positions: five analogues have 1–5 carbon substituents (CAS 2934-05-6, 96-70-8, 120-95-6, 98-27-1, 1740-97-2 and 98-27-1), one analogue has two nine carbon substituents (CAS 137-99-5), two analogues have substituents with more complex structures including an additional aromatic ring at R3 (CAS 42479-88-9 and 79-96-9), and one analogue has a methyl ether at R3 (CAS 121-00-6). The GSM identified ten analogues. The substituents of the GSM analogues are distributed at a variety of positions: two analogues have substituents at R1 and R3 (like the target; CAS 79-97-0 and 2934-05-6), one analogue has a single substituent at R1 (partially-hindered; CAS 3884-95-5), two have substituents at R1 and R5 (fully-hindered; CAS 2078-54-8 and 5510-99-6), and five analogues have a single substituent at R3 (unhindered; CAS 140-66-9, 599-64-4, 80-05-7, 104-40-5 and 1806-26-4). The source analogues for 2,4-di-tert-butylphenol are listed in Table 7.
Table 7:
Source analogues for 2, 4-di-tert-butylphenol as identified by the LSM and GSM.
Target: 2,4-di-tert-butylphenol, 96-76-4†

| |||
|---|---|---|---|
| LSM Analogues | GSM Analogues | ||
| †2-tert-Butyl-4-ethylphenol 96-70-8 | 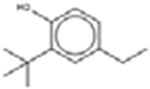 |
*† 4-[1,1,3,3-Tetramethylbutyl) phenol 140-66-9 |  |
| 2,4-Bis(2-methylbutan-2-yl)phenol 120-95-6 | 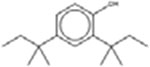 |
2-(1,1,3,3-tetramethylbutyl)-Phenol 3884-95-5 | 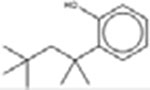 |
| 2,4-Dinonylphenol 137-99-5 | 3,3’-Dimethylbisphenol A 79-97-0 | 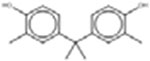 |
|
| 4-(T-Butyl)-2-cresol 98-27-1 | 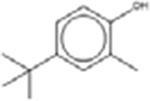 |
*† 4-Cumylphenol 599-64-4 | 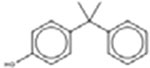 |
| 3,4’-bis(1,1-dimethylethyl)[1,1’-biphenyl]-4-ol 42479-88-9 | 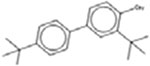 |
† Propofol 2078-54-8 | 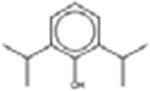 |
| 2-Methyl-4-isopropylphenol 1740-97-2 |  |
2,6-Di(butan-2-yl)phenol 5510-99-6 | 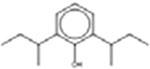 |
| † 2-tert-Butyl-4-methoxyphenol 121-00-6 | 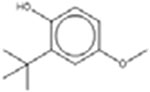 |
*† Bisphenol A 80-05-7 | 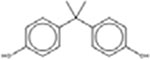 |
| Butylated hydroxyanisole 25013-16-5 |  |
*† 4-Nonylphenol 104-40-5 | |
| 4,4’-Propane-2,2-diylbis(2-tert-butylphenol) 79-96-9 | 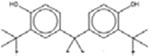 |
*† 4-Octylphenol 1806-26-4 | |
| †ǂ 2,4-Diisopropylphenol 2934-05-6 | 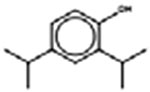 |
†, ǂ 2,4-Diisopropylphenol 2934-05-6 |  |
non-hindered
Tested in Toxcast
identified by both methods
3.2.2. Step 4 Collect and Organize the Data (2,4-di-tert-butylphenol)
Data collected for 2,4-di-tert-butylphenol are summarized in Supplementary Table S2.
The target, 2,4-di-tert-butylphenol (CAS 96-76-4), is inactive in the in vitro ToxCast battery for ER agonism (AUC=0) and ER antagonism (AUC=0). ToxCast data exists for ten of the analogues. One analogue (CAS 2934-05-6; identified by both methods) was found to be not active as an ER agonist or antagonist (AUC = 0); the three remaining LSM analogues were also found to be inactive for ER agonism and antagonism (AUC = 0.00528-0.0193; CAS 96-70-8, 120-95-6, 121-00-6). Therefore, the LSM analogues are good predictors of the activity of the target chemical. The six remaining GSM analogues had mixed results: four were active for ER agonism (AUC = 0.118–0.45; CAS 140-66-9, 599-64-4, 80-05-7 and 1806-26-4), two were not active (AUC = 0–0.088; CAS 2078-54-8 and 104-40-5), and all were not active for ER antagonism (AUC = 0). Importantly, the four GSM analogues that were found to be active are structurally different from the target because they are all unhindered phenols. Of the two that were found to be not active, one was fully hindered (CAS 2078-54-8) and one was unhindered (CAS 104-40-5).
Predictions from the CERAPP consensus QSAR models and other in silico tools were made for the target and analogues. The target chemical 2,4-di-tert-butylphenol was ‘inactive’ across these models. For the analogues, the CERAPP consensus QSAR models made predictions of ‘inactive’ or ‘activity’ (ranging from very weak to moderate) for ‘consensus binding’ and ‘consensus agonism’ across all 19 analogues. The calls for ‘consensus antagonism’ ranged from ‘inactive’ to ‘strong activity’; and, the two substances predicted to be strongly active (CAS 599-64-4 and 80-05-7; both identified by the GSM) also had the highest AUC scores for ER agonism (0.376 and 0.45), and the former was also active in the in vivo uterotrophic assay. For the LSM analogues, OASIS TIMES gave predictions of ‘not active’ or ‘weak activity’ for ER binding. For the GSM analogues, OASIS TIMES gave predictions of ‘not active’ or ‘weak/low/moderate activity’ for ER binding. Similarly, ACD Percepta gave predictions of ‘no binding’ or ‘weak binding’ for the LSM analogues; and, ‘weak’ or ‘strong’ binding for the GSM analogues. Finally, Derek Nexus generated ‘no alerts’ for the LSM analogues and the target. Alerts for the GSM substances included: ‘no alert’, ‘Mammal Plausible, Alkylphenol or precursor’ and ‘Mammal Plausible, bisphenol or precursor.’ These alerts indicate that these chemical structures fulfil the structural criteria of substances that are known to be associated with estrogenicity. Taken together, the in silico predictions for the LSM analogues were in better agreement with the predictions of the target substance than those for the GSM analogues.
2,4-di-tert-butylphenol has been tested and found to be inactive in the in vivo uterotrophic assay (maximum dose 1000 mg/kg-bw/day). Three of the analogues identified by GSM had in vivo data associated with them: 4-cumylphenol (CAS 599-64-4), 4-nonylphenol (CAS 104-40-5) and 4-octylphenol (CAS 1806-26-4). Each of these phenols is unhindered (single, large substituents are located at para position). Being unhindered, it was not anticipated that they would adequately predict the activity of the partially-hindered target substance, 2,4-di-tert-butylphenol (based upon what is known about the biochemistry of ER binding). Indeed, 4-cumylphenol was found to be active in the uterotrophic assay (LEL = 20 and 200 mg/kg-be/day; subcutaneous administration); 4-nonylphenol had mixed results based on nine iterations of the assay (Active: LEL = 75, 90, 100 and 200 mg/kg-bw/day; Not Active: Maximum dose = 2 and 200 mg/kg-bw/day); and, 4-octylphenol also had mixed results (Active: LEL = 100 mg/kg-bw/day; Not Active: LEL = 200 mg/kg-bw/day). These discordant outcomes are a known issue of the uterotrophic assay and have been described elsewhere (Kleinstreuer et al., 2016).
3.2.3. Step 5 Weight of Evidence Assessment (2,4-di-tert-butylphenol)
The data collected for 2,4-di-tert-butylphenol and its identified analogues are presented in Table S2; a high-level summary is presented in Table 9. The data from the LSM indicated that the overall call for estrogenicity should be inactive. Although the GSM analogues seemed to indicative the potential for estrogenic activity, there were discordant results in this evidence stream. 2,4-di-tert-butylphenol is not expected to be estrogenic due to partial hindrance of the hydroxyl group by a substituent. It was observed that analogues that shared this structural feature were more predictive of the known in vitro, in vivo and modeled estrogenic activity of the target. Expert judgment of the overall data is consistent with the hypothesis that 2,4-di-tert-butylphenol, like other hindered phenols, is expected not to be potentially estrogenic, whereas non-hindered phenols are.
3.2.4. Step 6 Prioritization (2,4-di-tert-butylphenol)
Based on lack of hazard potential regarding ER binding and estrogenicity, 2,4-di-tert-butylphenol would likely be deprioritized from further testing for estrogenic endocrine disruption. Therefore, no AED was derived for this substance, and no risk-based assessment (which would take both hazard and exposure potential into account) was done.
3.3. Octabenzone, CAS RN 1843-05-6
3.3.1. Step 3 Identify Analogues (Octabenzone)
Eleven analogues were identified for octabenzone. The LSM identified four analogues with large substituents at the R1 and R4 positions, which are the same positions as those on octabenzone (refer to Table 1 for numbering of substituent positions). Like octabenzone, the R1 substituent is always a ketone linkage to an aryl ring. Three analogues have an R4 substituent that is an ether linkage to an alkyl group (like octabenzone), and the fourth analogue has a nitrogen-containing compound at the R4 position. The GSM identified 7 analogues for the target. There were no shared structures between the two methods. The GSM analogues were also large and often complex; however, their locations on the phenolic ring were not limited to the R1 and R4 positions. Therefore, with respect to substituent position, the analogues produced by the LSM are structurally similar to the target, whereas the analogues produced by the GSM are more structurally diverse (Table 8).
Table 8:
Source Analogues for Octabenzone as identified by the LSM and GSM.
| Target: Octabenzone, 1843-05-6†
| |||
|---|---|---|---|
| LSM Analogues | GSM Analogues | ||
| † 2-Hydroxy-4-methoxybenzophenone 131-57-7 | 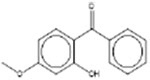 |
* Dodecylparaben 2664-60-0 | |
| 2,2’-Dihydroxy-4-methoxybenzophenone 131-53-3 | 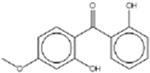 |
† Dodecyl gallate 1166-52-5 | |
| 2,2’-Dihydroxy-4,4’-dimethoxybenzophenone 131-54-4 | 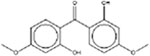 |
† Tiratricol 51-24-1 | |
| 2-[4-(Dibutylamino)-2-hydroxybenzoyl]benzoic acid 54574-82-2 | 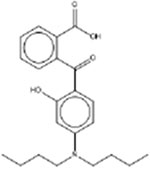 |
Siccanin 22733-60-4 |  |
| * Acolbifene 182167-02-8 | 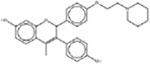 |
||
| a-Naphtholphthalein 595-01-0 | 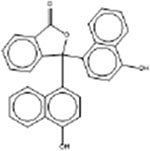 |
||
| * (Z)-4-Hydroxytamoxifen 68047-06-3 | 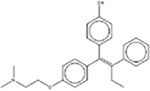 |
||
non-hindered
Tested in ToxCast
3.3.2. Step 4 Collect and Organize the Data (Octabenzone)
Data collected for octabenzone are summarized in Supplementary Table S3.
Three analogues had previously undergone in vitro HTS testing in ToxCast. 2-Hydroxy-4-methoxybenzophenone (LSM; CAS 131-57-7) and dodecyl gallate (GSM; 1166-52-5) are inactive for ER agonism. Whereas tiratricol (GSM; CAS 51-24-1) is active; its AUC score of 0.179 corresponds to positive control substances deemed “weak” or “very weak” (Browne et al., 2015). All three are inactive for ER antagonism.
Predictions from the CERAPP consensus QSAR models and in silico tools were made for the target and analogues. For CERAPP consensus binding, the target is “active (very weak)” for ER binding and agonism, and “in active” for antagonism. The LSM analogues typically had calls of inactivity (CAS 54574-82-2) or weak activity (CAS131-57-7, 131-53-3, 131-54-4); whereas, the GSM analogues had calls of weak activity (CAS 2664-60-0, 1166-52-5, 52-24-1, 22733-60-4, 182167-02-8) or a mixture of inactive to moderate activity (596-01-0, 68047-06-3). Results were similar for the OASIS TIMES ER binding QSAR; however, several of the analogues identified by the GSM were deemed to be ‘out of domain’ (CAS 2664-60-0, 51-24-2, 22733-60-4, 182167-02-8 and 596-01-0). Derek Nexus produced an alert for plausible estrogenicity for CAS 182167-02-8. Finally, ACD Percepta predicted that the target would be a weak ER binder; whereas all of the LSM analogues had a call of ‘no ER binding’ except CAS 54574-82-2, which was predicted to be a weak ER binder. The GSM analogues were predicted to be weak ER binders, except for CAS 182167-02-8 and 68047-06-3, which were predicted to be strong ER binders.
Octabenzone has not been tested in the in vivo uterotrophic assay and is negative in the ToxCast batteries for ER agonism and antagonism. Two analogues have in vivo data associated with them: 2-hydroxy-4-methoxybenzophenone (identified by LSM; CAS 131-57-7) and tiratricol (identified by GSM; CAS 54-24-1). The former is inactive in the uterotrophic assay (maximum dose >1000 mg/kg-bw/day, n = 2 assays); whereas the latter is active (LEL = 1000 mg/kg-bw/day, n = 1 assay). Of the two substances, 2-hydroxy-4-methoxybenzophenone is the nearest neighbor to the target; therefore, it would be considered the more appropriate choice for read-across.
3.3.3. Step 5 Weight of Evidence Assessment (Octabenzone)
The data collected for octabenzone and its identified analogues are presented in Table S3; a high-level summary is presented in Table 9. Octabenzone is a substance that does not have any in vivo data but has been tested in ToxCast. The results for octabenzone (AUC = 0) are consistent with the hypothesis that hindered phenols are unlikely to be estrogenic. One LSM analogue, CAS 131-57-7, has been tested in ToxCast and it exhibited an AUC score of 0.0645 but falls below the score (0.1) considered by EPA to be the lower limit for estrogenic bioactivity (US-EPA, 2014). Two GSM analogues have been tested in ToxCast, 1166-52-5 and 51-24-1. The former is not active (AUC = 0.0000527), while the latter is predicted to be active (AUC = 0.179). Taken together, source analogues with substituents in the same position as the target are more predictive of the target’s activity.
3.3.4. Step 6 Prioritization (Octabenzone)
Based on lack of hazard potential regarding ER binding and estrogenicity, octabenzone would likely be deprioritized from further testing for estrogenic endocrine disruption. Therefore, no AED was derived for this substance, and no risk-based assessment (which would take both hazard and exposure potential into account) was done.
3.4. Summary of Results
The data for the three target chemicals are summarized in Table 9. The data for the target chemicals and their source analogues are in the Supplemental Materials (Tables S1–S3).
4. Discussion
Endocrine disruption occurs when a chemical substance mimics the activity of a hormone, in this case by binding to the ER. If potency and exposure are sufficient, the substance can alter the function of the endocrine system to cause adverse effects. Because estrogen is a powerful hormone, both the quantity of estrogen and the timing of its release are under tight physiological control. Improper control of this hormone can lead to adverse effects such as reproductive, developmental and carcinogenic effects. However, the ability of an estrogen-active substance to produce an adverse effect depends upon the magnitude, duration and frequency of exposure, its pharmacokinetics and potency and the susceptibility of the organism (Borgert et al., 2012). The effects of estrogenic endocrine disruptors have been well studied and are reviewed elsewhere (Kiyama and Wada-Kiyama, 2015). In this case study, information from different NAMs was used to predict ER binding potential of the three target phenols: 4-tert-butylphenol, 2,4-di-tert-butylphenol, and octabenzone. These included in vitro HTS, predictive models based on the experimental data, and (Q)SAR approaches.
4.1. Mapping NAM data to the ER Mode of Action (MOA)
The ER is a ligand-activated nuclear receptor; when bound to estrogen it translocates to the nucleus and modifies gene expression. Chemical substances which have the potential to bind to the ER can trigger the same changes in gene expression. These changes can be transient and of little consequence, or, if the magnitude, duration and frequency of exposure is sufficient, they may have a variety of more profound effects on the biology of the cell, which can lead to effects in the organism as a whole (Kiyama and Wada-Kiyama, 2015). The data collected were aligned to the mode of action (MOA) described for estrogenic activity such that to the extent possible, each data stream characterised one or more key events within the MOA (Table 10). Ideally, this alignment would have been to an Adverse Outcome Pathway (AOP) published by the OECD; however, no suitable AOP could be identified on the AOP wiki (aopwiki.org) at the time of writing. A number of AOPs that are currently under development could be complementary to (or used instead of) this MOA once they have been completed. Patlewicz, et al (2015) discuss the challenges of using the AOP approach for predictive toxicology and present a scientific confidence framework case example of the ER AOP pathway. The framework includes the mapping of assays to key events, establishing the analytical performance of each assay, determining the ability of upstream key events to predict downstream key events, and justifying the use the AOP within a specific decision context.
Table 10.
Assays used to measure events in the estrogenic mode of action.
| Mode of Action | Description | Relevant Assay(s) and in silico models |
|---|---|---|
| 1. ER complex formation | Binding of estrogen (or estrogenic substance) to the ER | ToxCast cell-free protein binding assays, CERAPP, OASIS TIMES, ACD Percepta, OECD Toolbox |
| 2. DNA binding and trans-activation of gene expression | Binding of the ER-estrogen complex to the ERE causing changes in estrogen-dependent gene expression | ToxCast ERE reporter gene assays, OASIS TIMES, ACD Percepta |
| 3. Protein expression | Translation of mRNA to protein | Western blot; proteomics (not assessed herein) |
| 4. Altered levels of circulating hormones | Abnormal levels of circulating estrogen and related hormones | Enzyme-linked immunosorbant assay (ELISA; not assessed herein) |
| 5. Organ-level changes | E.g., a change in uterine weight | Uterotrophic assay (TG 440); Derek Nexus Expert System |
| 6. Organismal-level changes | E.g., reproductive or developmental effects | Two-generation reproductive toxicity assay (TG 416); Derek Nexus Expert System |
4.2. Data integration for Risk Estimation
Because there was an indication of hazard potential for 4-tert-butylpenol, risk-based BER and MOE values were generated (Table 5, Table 6). The lower limit of the ER-related AED for 4-tert-butylphenol (11.22 mg/kg-bw/day) was found to be more health protective than the No Observed Effect Level (NOAEL) from the in vivo two-generation reproductive toxicity study, which was 70 mg/kg-bw/day (LOAEL = 200 mg/kg-bw/day); it was also lower than the lowest effect level (LEL) from the uterotrophic assay, which was 100 mg/kg-bw/day. The median AED value (113 mg/kg-bw/day) fell in between the NOAEL and LOAEL for the two-generational study (70 and 200 mg/kg-bw/day) and was very similar to the uterotrophic LEL (Fig. 4). Overall, these data demonstrate consistency between in vitro and in vivo point of departure doses. However, the lower limit of the range of AEDs was considered to be health protective because it was determined to be a lower (more conservative) estimate of effect level. This indicates that using this approach may be a conservative estimate of potential risk to support screening level (lower tier) chemical risk assessment. Indeed, this finding is in keeping with the biology that is assumed to underlie AOPs, in which lower dose levels are expected to elicit response at the molecular and biochemical levels, whereas higher doses (or sustained exposures) are needed to produce the toxicological endpoint.
Figure 4.
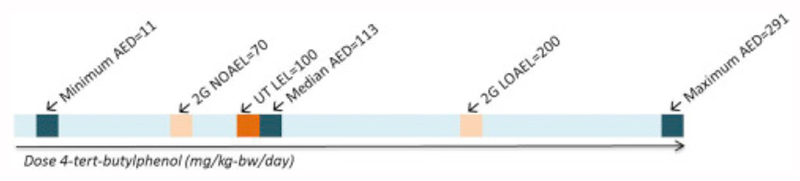
Points of departure for 4-tert-butylphenol. AEDs are derived from in vitro bioactivity results using IVIVE (see text for details). Blue = minimum, median and maximum AEDs; light orange = NOAEL and LOAEL for the two-generational study; dark orange = uterotrophic (UT) assay..
BER and MOE values were generated from the respective ADEs and in vivo effect levels in the analysis. For comparing exposure levels in humans to ADEs derived from HTS ToxCast data for estrogenic activity, the BER approach was used. This is similar to the approach used in the proof of concept for exposure:activity profiling of ToxCast positive estrogenic agents by Becker et al. (2015). The MOE approach for the uterotrophic assay is similar to the benchmark dose/internal exposure analysis of in vivo endocrine screening results of Becker et al. (2014). The magnitude of the BER and MOE is an indication of relative risk; the greater the BER or MOE, the lower the potential risk. Typically, MOEs exceeding 100-1000 indicate low concern for risk (US EPA, 2013). Interpretations of the level of adversity of the assay vary depending on the regulatory authority. The use of the BER is based on a data-driven framework for tiered toxicity testing for chemicals with selective modes of action (MOAs) published by Thomas et al. (2013). The BER approach has the potential to be used in streamlined screening level risk assessments. Further, it can be used as a high-throughput risk approximation/classification tool for priority setting, or as a supplemental line of evidence to support decision making in risk assessment. This is especially promising for jurisdictions with flexible regulatory frameworks that do not have explicit data submission requirements supporting the risk assessment program and when various sources of information are collected to inform a regulatory decision on substances or groups of substances using fit for purpose approaches (e.g., Existing Substances under CEPA in Canada). Further, it will facilitate timely, evidence-based decision making for data poor substances. The approach is being corroborated through further case studies of additional chemicals; for example, a complementary case study is currently underway under the ‘Advancing the Pace of Chemical Risk Assessment’ (APCRA) program; this case study is deriving BERs for a large number of substances (Thomas et al., 2019; Barton-Maclaren et al., 2019).
4.2. Comparison of the LSM and GSM approaches
The use of two approaches highlighted some of the challenges associated with analogue identification. The LSM was informed by the mechanistic knowledge that an unhindered hydroxyl group was essential for the ability of the substance to bind to the ER, and therefore searched for analogues with similarly placed substituents to estrogen. Whereas, the GSM used global chemical fingerprints in conjunction with whole molecular properties and number of literature sources. The LSM and GSM resulted in differing sets of analogues for each of the target chemicals, with some overlap. Both methods performed well in the case of 4-tert-butylphenol where the most common source analogues were identified by either approach. Alternatively, robust predictions for the partially-hindered phenols proved more challenging owing to the conflicting results from the source analogues identified by the two approaches. The LSM identified the most predictive analogues for the targets 2,4-di-tert-butylphenol and octabenzone because it was able to identify analogues with substituents in the same position on the aryl ring as those on the target substance.
The way in which source analogues were identified and evaluated played a large role in the read-across inferences that were derived. Custom searches that took into account any mechanistic knowledge about the endpoint and the structural implications gave rise to analogues that were ultimately more predictive. This provides a practical demonstration of how to couple knowledge of biological receptors and modes of action/AOPs with custom searches, and the impacts this can have when applied to endpoint specific read-across (also see: Patlewicz et al., 2018). Further, the case study also highlighted some of the difficulties in comparing and contrasting different analogue identification approaches given the similarity metrics were not on a common scale. Taken together, these observations demonstrate the value of incorporating expert knowledge during analogue searches. In particular, biochemical and mechanistic information should be considered during model building and analogue selection.
5. Conclusions
This IATA case study was developed as a contribution to the OECD IATA case studies programme in an effort to build experience and confidence in the interpretation and application of NAMs in the context of regulatory decision making as well as to identify opportunities for new or refined guidance. The case study was structured to address two objectives. The first, to provide support for the proposed hypothesis that predicting the estrogen receptor (ER) binding potential of substituted phenols is facilitated when the size, number and position relative to the hydroxyl group of the substituent(s) is considered; and, the second, to build confidence in the use of NAMs in human health risk assessment activities.
This case study represents an early step in the building of confidence in the use of NAMs for screening level chemical risk assessment. Data from in vivo and in vitro sources formed predictions that 4-tert-butylphenol was likely to be weakly active, whereas 2,4-di-tert-butylphenol and octabenzone were likely to be inactive for ER binding. An AED was generated for 4-tert-butylphenol by transforming its AC50 values using in vitro to in vivo extrapolation methods. The lower limit of the AED (11 mg/kg bw/day) was found to be lower than the available reported effect levels from a 2-generational reproductive toxicity study (NOAEL = 70 mg/kg-bw/day, LOAEL = 200 mg/kg-bw/day). The median AED value of 113 mg/kg bw/day was between these NOAEL and LOAEL values. Likewise, the BER was lower than the in vivo MOE values. The illustration of this risk-based approach for chemical screening suggests that this health protective approach could facilitate a substance’s prioritization or deprioritization for further action, including the need for comprehensive in vivo testing.
Supplementary Material
Supplementary Table S1: Data Matrix for CAS 98-54-4; attached as a separate Excel file.
Supplementary Table S2: Data Matrix for CAS 96-76-4; attached as a separate Excel file.
Supplementary Table S3: Data Matrix for CAS 1843-05-6; attached as a separate Excel file.
Highlights.
The use of New Approach Methodologies (NAMs) in chemical risk assessment is explored.
A workflow for the prediction of estrogen receptor activation by substituted phenols.
The workflow is applied to: 4-tert-butylphenol, 2,4-di-tert-butylphenol, octabenzone.
Two computational approaches for analogue selection were used and evaluated.
When compared to in vivo endpoints, the NAM approach was health protective.
Acknowledgments
Funding Sources
Prachi Pradeep is funded by Participation Program at the Office of Research and Development, U.S. Environmental Protection Agency, administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and EPA.
Abbreviations:
- ADE
Applied Dose Equivalent
- AOP
Adverse Outcome Pathway
- BER
Bioactivity Exposure Ratio
- CERAPP
Collaborative Estrogen Receptor Activity Prediction Project (U.S.)
- CEPA
Canadian Environmental Protection Act
- CMP
Chemicals Management Plan
- DSL
Domestic Substances List
- EDSP
Endocrine Disruptor Screening Program (U.S. EPA)
- EPA
Environmental Protection Agency (U.S.)
- ER
estrogen receptor
- ERE
estrogen response element
- HTTK
High Throughput Toxicokinetics
- HTS
High Throughput Screening
- IATA
Integrated Approaches for Testing and Assessment
- IPCS
International Programme on Chemical Safety
- IVIVE
In vitro to in vivo extrapolation
- MIE
Molecular Initiating Event
- MOA
mode of action
- NAM
New Approach Methodology
- NCCT
National Center for Computational Toxicology (U.S.)
- NICEATM
NTP Interagency Center for the Evaluation of Alternative Toxicological Methods (U.S.)
- NTP
National Toxicology Program (U.S.)
- OECD
Organisation for Economic Cooperation and Development
- OED
oral equivalent dose
- QSAR
Quantitative Structure-Activity Relationship
Footnotes
DTXSIDs are unique substance identifiers (described in more detail in https://jcheminf.springeropen.com/articles/10.1186/s13321-017-0247-6), where a substance can be any single chemical, mixture, polymer or chemical family that used in the EPA CompTox Chemicals Dashboard (see https://comptox.epa.gov/dashboard)
Publisher's Disclaimer: Disclaimer
The views expressed in this article are those of the authors and do not necessarily reflect the views or policies of the U.S. Environmental Protection Agency or Health Canada. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
References
- Barton-Maclaren TS, Westphal M, Sarwar E, Mattison D, Chiu WA, Dix D, Kavlock R, Krewski D, 2017. Challenges and opportunities in the risk assessment of existing substances in Canada: Lessons learned from the international community. Int. J. Risk Assess. Manage 20, 261–283. [Google Scholar]
- Becker RA, Friedman KP, Simon TW, Marty MS, Patlewicz G, Rowlands JC, 2015. An exposure:activity profiling method for interpreting high-throughput screening data for estrogenic activity--proof of concept. Regul. Toxicol. Pharmacol 71, 398–408. [DOI] [PubMed] [Google Scholar]
- Becker RA, Hays SM, Kirman CR, Aylward LL, Wise K, 2014. Interpreting estrogen screening assays in the context of potency and human exposure relative to natural exposures to phytoestrogens. Birth Defects Res. B. Dev. Reprod. Toxicol 101, 114–124. [DOI] [PubMed] [Google Scholar]
- Borgert CJ, Sargent EV, Casella G, Dietrich DR, McCarty LS, Golden RJ, 2012. The human relevant potency threshold: reducing uncertainty by human calibration of cumulative risk assessments. Regul Toxicol Pharmacol. 62, 313–328. [DOI] [PubMed] [Google Scholar]
- Browne P, Judson RS, Casey WM, Kleinstreuer NC, Thomas RS, 2015a. Screening Chemicals for Estrogen Receptor Bioactivity Using a Computational Model. Environ. Sci. Technol 49, 8804–8814. [DOI] [PubMed] [Google Scholar]
- Browne P, Judson RS, Casey WM, Kleinstreuer NC, Thomas RS, 2015b. Screening Chemicals for Estrogen Receptor Bioactivity Using a Computational Model. Environ. Sci. Technol 49, 8804–8814. [DOI] [PubMed] [Google Scholar]
- Burden N, Sewell F, Chapman K, 2015. Testing chemical safety: what is needed to ensure the widespread application of non-animal approaches? PLoS Biol. 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EPA, 2009. Endocrine disruptor screening program test guidelines OPPTS 890-1600: Uterotrophic Assay. 740-C-09-0010.
- EU, 2008. European Union Risk Assessment Report: P-TERT-BUTYLPHENOL, CAS No: 98-54-4 . EINECS No: 202-679-0.
- Judson RS, Magpantay FM, Chickarmane V, Haskell C, Tania N, Taylor J, Xia M, Huang R, Rotroff DM, Filer DL, Houck KA, Martin MT, Sipes N, Richard AM, Mansouri K, Setzer RW, Knudsen TB, Crofton KM, Thomas RS, 2015. Integrated Model of Chemical Perturbations of a Biological Pathway Using 18 In Vitro High-Throughput Screening Assays for the Estrogen Receptor. Toxicol. Sci 148, 137–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavlock RJ, Bahadori T, Barton-Maclaren T, Gwinn MR, Rasenberg M, Thomas RS, 2018. Accelerating the Pace of Chemical Risk Assessment. Chem. Res. Toxicol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyama R, Wada-Kiyama Y, 2015. Estrogenic endocrine disruptors: Molecular mechanisms of action. Environ. Int 83, 11–40. [DOI] [PubMed] [Google Scholar]
- Kleinstreuer NC, Ceger P, Watt ED, Martin M, Houck K, Browne P, Thomas RS, Casey WM, Dix DJ, Allen D, Sakamuru S, Xia M, Huang R, Judson R, 2017. Development and Validation of a Computational Model for Androgen Receptor Activity. Chem. Res. Toxicol 30, 946–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstreuer NC, Ceger PC, Allen DG, Strickland J, Chang X, Hamm JT, Casey WM, 2016. A Curated Database of Rodent Uterotrophic Bioactivity. Environ. Health Perspect 124, 556–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H−., Chen R, Hyduke DR, Williams A, Frötschl R, Ellinger-Ziegelbauer H, O’Lone R, Yauk CL, Aubrecht J, Fornace AJ, Cleaver JE, 2017. Development and validation of a high-throughput transcriptomic biomarker to address 21st century genetic toxicology needs. Proc. Natl. Acad. Sci. U. S. A 114, E10881–E10889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri K, Abdelaziz A, Rybacka A, Roncaglioni A, Tropsha A, Varnek A, Zakharov A, Worth A, Richard AM, Grulke CM, Trisciuzzi D, Fourches D, Horvath D, Benfenati E, Muratov E, Wedebye EB, Grisoni F, Mangiatordi GF, Incisivo GM, Hong H, Ng HW, Tetko IV, Balabin I, Kancherla J, Shen J, Burton J, Nicklaus M, Cassotti M, Nikolov NG, Nicolotti O, Andersson PL, Zang Q, Politi R, Beger RD, Todeschini R, Huang R, Farag S, Rosenberg SA, Slavov S, Hu X, Judson RS, 2016. CERAPP: Collaborative Estrogen Receptor Activity Prediction Project. Environ. Health Perspect 124, 1023–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OECD, 2007. Test no. 440: Uterotrophic Bioassay in Rodents. Organisation for Economic Co-operation and Development, Geneva, Switzerland. [Google Scholar]
- Patlewicz G, Simon TW, Rowlands JC, Budinsky RA, Becker RA, 2015. Proposing a scientific confidence framework to help support the application of adverse outcome pathways for regulatory purposes. Regul. Toxicol. Pharmacol 71, 463–477. [DOI] [PubMed] [Google Scholar]
- Pradeep P, Mansouri K, Patlewicz G, Judson R, 2017. A systematic evaluation of analogs and automated read-across prediction of estrogenicity: A case study using hindered phenols. Comput. Toxicol 4, 22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard AM, Judson RS, Houck KA, Grulke CM, Volarath P, Thillainadarajah I, Yang C, Rathman J, Martin MT, Wambaugh JF, Knudsen TB, Kancherla J, Mansouri K, Patlewicz G, Williams AJ, Little SB, Crofton KM, Thomas RS, 2016. ToxCast Chemical Landscape: Paving the Road to 21st Century Toxicology. Chem. Res. Toxicol 29, 1225–1251. [DOI] [PubMed] [Google Scholar]
- Russell W, Burch R, 1959. The Principles of Human Experimental Technique. Methuen, London. [Google Scholar]
- Tanimoto T, 1957. An Elementary Mathematical theory of Classification and Prediction. Internal IBM Technical Report. [Google Scholar]
- Thomas RS, Philbert MA, Auerbach SS, Wetmore BA, Devito MJ, Cote I, Rowlands JC, Whelan MP, Hays SM, Andersen ME, Meek ME, Reiter LW, Lambert JC, Clewell HJ 3rd, Stephens ML, Zhao QJ, Wesselkamper SC, Flowers L, Carney EW, Pastoor TP, Petersen DD, Yauk CL, Nong A, 2013. Incorporating new technologies into toxicity testing and risk assessment: moving from 21st century vision to a data-driven framework. Toxicol. Sci 136, 4–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USA, 2016. Frank R. Lautenberg Chemical Safety For the 21st Century Act.
- US-EPA, 2018a. Assessing and Managing Chemicals under TSCA: Alternative Test Methods and Strategies to Reduce Vertebrate Animal Testing. https://www.epa.gov/assessing-and-managing-chemicals-under-tsca/alternative-test-methods-and-strategies-reduce. 2018.
- US-EPA, 2018b. Reviewing New Chemicals under the Toxic Substances Control Act (TSCA): Statistics for the New Chemicals Review Program under TSCA.
- US-EPA, 2015. Use of High Throughput Assays and Computational Tools; Endocrine Disruptor Screening Program; Notice of Availability and Opportunity for Comment. https://www.federalregister.gov/documents/2015/06/19/2015-15182/use-of-high-throughput-assays-and-computational-tools-endocrine-disruptor-screening-program-notice. Federal Register: The Daily Journal of the US Government 2015-15182.
- US-EPA, 2014. Integrated Bioactivity and Exposure Ranking: A Computational Approach for the Prioritization and Screening of Chemicals in the Endocrine Disruptor Screening Program EPA-HQ-OPP-2014-0614-0003. [Google Scholar]
- US-EPA, 2012. Estimation Programs Interface Suite for Microsoft Windows.
- Wambaugh JF, Wang A, Dionisio KL, Frame A, Egeghy P, Judson R, Setzer RW, 2014. High throughput heuristics for prioritizing human exposure to environmental chemicals. Environ. Sci. Technol 48, 12760–12767. [DOI] [PubMed] [Google Scholar]
- Wetmore BA, Wambaugh JF, Ferguson SS, Sochaski MA, Rotroff DM, Freeman K, Clewell HJ 3rd, Dix DJ, Andersen ME, Houck KA, Allen B, Judson RS, Singh R, Kavlock RJ, Richard AM, Thomas RS, 2012. Integration of dosimetry, exposure, and high-throughput screening data in chemical toxicity assessment. Toxicol. Sci 125, 157–174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1: Data Matrix for CAS 98-54-4; attached as a separate Excel file.
Supplementary Table S2: Data Matrix for CAS 96-76-4; attached as a separate Excel file.
Supplementary Table S3: Data Matrix for CAS 1843-05-6; attached as a separate Excel file.


