Abstract
Glycolysis, the breakdown of glucose, is one of the most conserved and extensively studied biochemical pathways. Designing principles from chemistry and thermodynamics allow for energy production, biosynthesis and cellular communication. However, the kinetics or metabolic flux through the pathway also determines its function. Recently, there have been numerous developments that establish new allosteric interactions of glycolytic enzymes with small molecule metabolites and other mechanisms that may cooperate to allow for addition complex regulation of glycolysis. This review surveys these newfound sources of glycolysis regulation and discusses their possible roles in establishing kinetic design principles of glycolysis.
Keywords: Kinetics, glucose metabolism, network design principles, feedback regulation, feedforward regulation
Glycolysis: structure and function
Glycolysis, the breakdown of glucose, is a pathway that exemplifies the core principles of how metabolic biochemistry can achieve biological function[1,2] (Figure 1). It is the oldest studied biological pathway with much of its discovery dating back to the 1950s. Thus, it is remarkable and speaks to the complexity of biology that almost 70 years later, there is still much to learn about both its biochemical makeup and biological function. Recently, several studies have discovered new elements of biochemistry within the pathway that may function to create feedback and feedforward forms of interaction throughout the pathway. How these newfound interactions confer regulatory principles for glucose metabolism is largely unknown. This review will provide a brief overview of glycolysis and mention, non-exhaustively, some classic forms of glycolysis regulation. It will next discuss recent developments in defining new aspects of the structure of the pathway, through allosteric interactions and other means, and speculate on the possible roles for these additional layers of wiring. These mechanisms likely confer numerous additional properties to glycolysis regulation but our knowledge is still in its infancy. I ultimately hope to give the reader a sense of the new possibilities for glycolysis regulation that may be present in certain contexts.
Figure 1– New concepts of regulation in glycolysis.
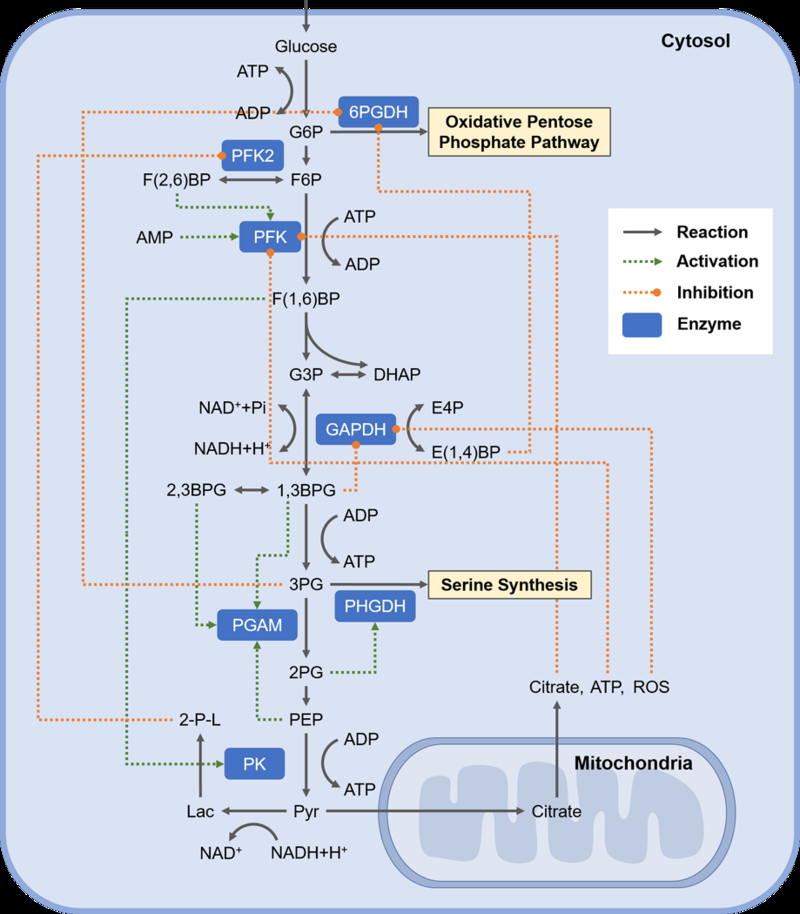
Overview of glycolysis and new mechanisms of feedforward and feedback regulation. The pathway of glycolysis begins with the uptake of glucose and ends with the production of lactate of CO2 in the mitochondria. Classic and newly identified feedback interactions in the pathway are highlighted. Abbreviations: G6P - Glucose-6-phosphate, F6P - Fructose-6-phosphate, F(1,6)BP - Fructose-1,6-bisphosphate, DHAP - Dihydroxyacetone phosphate, G3P - Glyceraldehyde-3-phopshate, 1,3BPG - 1,3-bisphosphoglycerate, 2,3BPG - 2,3-bisphosphoglycerate, 3PG - 3-phosphoglycerate, 2PG - 2-phosphoglycerate, PEP - phosphoenolpyruvate, PEP - phosphoenolpyruvate, Pyr - pyruvate, Lac - lactate, SAICAR - 2-[5-Amino-1-(5-phospho-D-ribosyl)imidazole-4-carboxamido] succinate), NAD+ - Nicotinamide adenine dinucleotide (oxidized), NADH - Nicotinamide adenine dinucleotide (reduced), ATP - Adenosine triphosphate, ADP - Adenosine diphosphate, AMP - Adenosine monophosphate, ROS - Reactive oxygen species, Acyl-coA - Acyl coenzyme A, 2-P-L 2-phospholactate, E4P - 4-phosphoerythronate, E(1,4)BP - 1,4-bisphosphoerythronate, HK - hexokinase, PFK - phosphofructokinase, PFK2 - phosphofructokinase 2, 6PGDH - 6-glucose-6-phosphate dehydrogenase, GAPDH - Glyceraldehyde phosphate dehydrogenase, PHGDH - Phosphoglycerate dehydrogenase, PGAM - Phosphoglycerate mutase, PK - pyruvate kinase.
Glycolysis involves the uptake and metabolism of glucose through a set of chemical reactions that together constitute one of the oldest and most broadly studied biochemical pathways. As a result, energy in the form of ATP is generated and electrons are passed from glucose to the cofactor NADH. The intermediates in the pathway can undergo additional chemistry to generate biomass in the form of nucleic acids, proteins, and lipids. The product of glycolysis, pyruvate, can be further oxidized in the mitochondria or reduced and thus fermented to produce lactic acid or lactate. The lactate produced rapidly exchanges with pyruvate to enter the mitochondria as well[3,4]. Furthermore, the process of metabolizing glucose also signals to other aspects of the cellular milieu such as the chromatin state by affecting histone acetylation or the activation of receptor tyrosine kinase pathways by changing the redox state that leads to oxidation of the catalytic cysteine and thus catalytic inactivation of protein phosphatases[5–7]. Altogether each of these fates of glucose confers numerous essential functions to cells many of which are evolutionarily conserved from the most primitive of organisms to the most complex of multicellular species.
The biochemical setup of glycolysis contains several design principles that allows for each step in the pathway to achieve a specific outcome. This topic has been reviewed and analyzed extensively[2,8,9]. These goals either achieve thermodynamic favorability, satisfy physiochemical constraints such as membrane permeability, or allow for the creation of intermediates that can initiate biosynthesis from enzymatically feasible chemistry. As a result, much and in some cases, all demand for biomass and energy can be met from the breakdown and chemical conversion of sugar alone.
While the structure (i.e. the chemical reactions and thermodynamic properties) of glycolysis enables numerous cellular functions, metabolism is never in thermodynamic equilibrium and the kinetic properties of metabolism determine the function of the pathway. For example, during normal tumor development, cells exhibit larger rates of glycolysis and this leads to overflow metabolism such as fermentation of lactate (i.e. the Warburg Effect). As a result, cells carrying out this Warburg Effect have dramatically different phenotypes because of this differential in the kinetics of glucose metabolism[5,10]. The mechanism for how the rate of glycolysis can influence biological outcomes is complex. For instance, glycolysis is coupled to the activity of nearly all other metabolic pathways. The flux through glycolysis is also coupled to the concentrations of glycolytic intermediates that are sensed by other cellular processes. Thus, cells have evolved intricate mechanisms to regulate the kinetics of glycolysis through both positive and negative feedback and feedforward mechanisms.
Regulation of glycolysis
Classic examples
Typically metazoan cells when supplied with an abundance of glucose maintain low rates of glycolysis due to the saturation of enzymes in glycolysis[11]. Thus mechanisms in addition to increasing nutrient availability are needed to increase the rate of glycolysis. This has been extensively studied during insulin signaling and in cancer cell biology where the Warburg Effect is induced by oncogenic signaling pathways and transcription factors which decrease the Km and increase the Vmax of glycolytic enzymes through posttranslational modifications and increased expression. For example master transcription factors such as MYC and HIF1 can increase the Vmax by several fold for multiple glycolytic enzymes at once[12].
Allosteric binding of small-molecules to glycolytic enzymes is another pervasive theme in positive glycolysis regulation. Examples include pyruvate kinase, the enzyme that catalyzes the final step in glycolysis, is binds to and is activated by fructose-1,6,-bisphosphate (F1,6BP) an early intermediate in the pathway[13]. This allows for the creation of a feedforward loop. When glycolysis is backed up and FBP accumulates, its binding to pyruvate kinase (PK) facilitates an increase in glycolytic rate and relief from accumulation of glycolytic intermediates. Also, nucleotides and amino acids derived from glycolytic intermediates including SAICAR and serine bind to and activate PK creating additional feedforward loops[14–16]. Another feedforward loop occurs through the generation of fructose-2,6,-bisphosphate (F2,6BP) from fructose-6-phosphate (F6P) by phosphofructokinase 2 (PFK2)[17]. F2,6BP allosterically activates phosphofructokinase 1 (which also is activated by AMP) to allow for additional layers of rate control at the phosphofructokinase (PFK) step.
In addition to these ways of increasing the rate of glycolysis, there are also numerous modes of negative regulation. Both citrate and ATP bind to and inhibit PFK and thus rising ATP of citrate levels that might occur from enhanced mitochondria TCA cycle activity provide negative feedback to PFK and glycolysis. Other negative feedback loops via other glycolytic intermediates exist as well.
New elements of glycolysis regulation
While many aspects of glycolysis regulation have been known for several decades, the surge of interest in metabolism has allowed for deeper investigations into glycolysis and as a result has uncovered many new elements of glycolysis regulation. Historically in mammals, much of the positive regulation has focused on the front end of glycolysis such as the trafficking of the glucose transporter from endomembranes to the cell surface during insulin stimulation[18] and the phosphorylation of glycolysis enzymes such as hexokinase by AKT[19]. More recently it is now appreciated that other steps in glycolysis are subjected to substantial positive regulation. Several of these mechanisms only recently discovered are highlighted below.
Glyceraldehyde phosphate dehydrogenase (GAPDH) catalyzes the fifth step in glycolysis that converts glyceraldehyde-3-phosphate (G3P) to 1,3-bisphosphoglycerate (1,3BPG). The reaction occurs through an oxidative phosphorylation mechanism using a catalytic cysteine that is coupled to the oxidation of G3P. It has long been considered an enzymatic step in glycolysis that lacks regulation. This conclusion has largely been based on reported estimates of the free energy of this reaction in red blood cells being near zero and thus the reaction is in equilibrium[9,20]. It is also generally thought that reactions farther away from equilibrium tend to be under more regulation, although recent analyses have questioned this assertion[21]. As can be seen in both the chemical and kinetic equations, the multiple cofactors and the transient nature of 1,3BPG due to its reactive acyl-phosphate groups, large variations in concentrations can alter the kinetics and also shift the free energy far from equilibrium. For example, the NAD+/NADH ratio can vary over 100-fold when cells undergo differing rates of glycolysis[22,23]. Inorganic phosphate and intracellular pH also are thought to vary over several orders of magnitude and regulate GAPDH activity[24]. As a result, regulated changes in these variables exert both positive and negative influences on the pace of glycolysis. Metabolic control analysis has shown that under certain conditions GAPDH exerts a surprising amount of control over glycolytic pathway flux[10,23–25]. For example, during the Warburg Effect, some recent calculations have placed GAPDH as having as much control of glycolysis as the enzymes placed in more canonical rate-determining steps such as PFK and pyruvate kinase[10,23,24].
A recent study using mathematical modeling, flux measurements, and single cell analysis reported in a subset of yeast cells that during adaptation to increases in glucose availability, an imbalance in glycolysis can be created that marked by differences in the flux through upper and lower glycolysis leading to an accumulation of FBP and depletion of ATP that leads to cytotoxicity[24]. Phosphate release from the vacuole and changes in pH affect the extent of this imbalance and can restore the overall rate of glycolysis. A form of control analysis showed that these results were largely connected to the activity of GAPDH that was mediated by nutrient availability.
Numerous other elements of negative regulation are now appreciated to also occur at the GAPDH step. 1,3BPG has recently been shown to covalently modify a lysine near the active site of GAPDH[26]. Thus, if this acylation reaction can occur with appreciable kinetics in cells, it establishes a negative feedback loop. Other products of metabolism downstream of glycolysis such as acyl-coenzyme A species derived from mitochondrial metabolism also react with GAPDH to covalently modify and inhibit its activity[27–29] although whether and how these metabolites generated in the mitochondria may react with GAPDH in the cytosol or nucleus is unclear.
Another aspect of negative regulation of GAPDH activity occurs through reactive oxygen species generation via hydrogen peroxide mediated oxidation and inactivation of the catalytic cysteine[30]. Thus oxidative stress and the generation of hydrogen peroxide that comes for example from uncoupling of the TCA cycle from or excessive activity in the electron transport chain in the mitochondria[31]. For example, an interesting and provocative concept for a cancer therapy is to overload cancer cells that can differentially uptake Vitamin C which at high concentrations acts as an oxidant and among other toxic effects to cells, inhibits GAPDH by inducing oxidative stress[30]. Given its potential effects on rate-control in glycolysis, oxidative inactivation or other covalent modifications to GAPDH even in small amounts could exert large reductions to the rate of glycolysis. Additionally, positive regulation of GAPDH has recently been discovered to occur via protein-protein interactions by glutathione S-transferase Pi 1 (GSTP1) as well[32].
Another enzyme that has historically been considered to have less regulatory capacity in glycolysis due to the thermodynamics of the reaction is phosphoglycerate mutase (PGAM)[20]. PGAM, the 7th step in glycolysis catalyzes the conversion of 3-phosphoglycerate (3PG) to 2-phosphoglycerate (2PG). 3PG binds to and inhibits, phosphogluconate dehydrogenase (6PGDH), an enzyme in the pentose phosphate pathway which branches from glycolysis[33]. Its product 2PG can bind to and activate phosphoglycerate dehydrogenase (PHGDH). Each of these interactions confer feedback and possibly feedforward regulation to the pathway. Inhibition of the PPP may increase the overall rate of glycolysis conferring either positive feedback or negative feedback (depending on the kinetics of the interaction) to glycolysis. Further, activation of serine synthesis which occurs through PHGDH possibly provides negative feedback to the pathway[33,34]. For its reaction mechanism, PGAM is first primed using a phosphorylated histidine on the enzyme to mediate the transfer of the phosphate. Thus the acceptance of a phosphate on the histidine which is required for its complete catalytic activity can result in positive regulation and possibly feedback if that phosphate is derived from an intermediate in glycolysis. Indeed, it has been shown that phosphoenolpyruvate (PEP) can prime this reaction which creates a positive feedback loop[35]. Additionally, a recent study has now demonstrated both 1,3BPG and another acylphosphate, 2,3-bisphosphoglycerate (2,3BPG), can prime PGAM[36]. Both thus provide feedforward regulation and therefore PGAM and related metabolites provide numerous aspects of positive and regulation to glycolysis.
Minor products in glycolysis as allosteric regulators
While enzymes are in principle thought to operate with unique specificity for a single substrate and product, their properties do allow for catalysis of alternative reactions from the same or alternative substrates yielding additional products. The relative extent of the generation of these products depends on the kinetic and thermodynamic properties associated with the reactions. In the case of glycolysis, the overall rate can be very high relative to other metabolic reactions in cells. Glycolysis enzymes are in high abundance in cells (up to 50% of all metabolic enzymes and 10% of the entire proteome in mammalian cells) and thus have high VMax values [37]. Thus, if side products are created at even very small extents, they can have biological functions. For example, lactate dehydrogenase (LDH), which predominantly reduces pyruvate to generate lactate, has recently been shown to produce S-2-hydroxyglutarate (2HG) as a minor product in quantities sufficient to affect biology[38]. Recently, a study also reported that two enzymes in glycolysis produce minor product that interact with other enzymes in glycolysis. LDH produces 2-phospholactate (2-P-L), and GAPDH 1,4-bisphosphoerythronate (E(1,4)BP) from 4-phosphoerythronate (4EP) [39]. 2-P-L inhibits PFK2 and 1,4PE was shown to bind to and inhibit 6PGDH in the Pentose Phosphate Pathway. These products were shown to be catalytically degraded by a newly identified enzyme phosphoglycolate phosphatase (PGP). Since these products are coupled to the rate of glycolysis, they provide interesting and unexplored possibilities for avenues of feedback regulation of glycolysis. Binding of 2-P-L to PFK2 creates a negative feedback loop whereas binding of 4PE creates a positive feedback. These possibilities are in addition to transcriptional regulation of PGP or epigenetic alterations by 2HG which have been extensively reported[40].
Outlook: from cataloguing interactions to mechanistic systems biology
This review has reported multiple newly characterized aspects feedback and feedforward interaction in glycolysis. Undoubtedly there are many discoveries left as the biochemistry thus far reported is by no means exhaustive. Nevertheless, with these many catalogued interactions, the challenge is to synthesize them collectively to understand what biological functions they might confer. For example, in the context of both cancer and development, high rates of glycolysis maintain numerous biological functions such as maintenance of chromatin structure and can have kinetic advantages over acquiring ATP from oxidative phosphorylation when competing for limited glucose[41–44]. Lowering the rate of glycolysis may also be advantageous when glucose is environmentally limited and thus negative regulation enhances the fitness of a multicellular community by providing more glucose to other cells[41–43]. Higher rates may also in some contexts have toxicities associated with increased ATP or ROS production or general mitochondrial overload[45]. Whether and how these mechanisms may be involved in any of these process remains to be determined.
In addition to increasing or decreasing the rate of glycolysis through feedback regulation, the kinetics of glucose metabolism is highly nonlinear. Nonlinear feedback results in more complex behavior. Positive feedback can result in bi- or multi- stability where cells can stably over long periods of time operate in multiple steady states and exhibit hysteresis or biochemical memory such that when one state (i.e. a set of fluxes and concentrations related to glycolysis) is reached the transitioning to another stable state is irreversibly slow. Modeling results of glycolysis have pointed to this bi- or multi-stability as having a regulatory function[46,47]. Thus, a high glycolytic rate that was established by for example the presence of insulin or a growth factor could be maintained long after removal of the stimuli that initiated the high rate of glycolysis. Such concepts of irreversibility due to bistability could be established through any of the feedback loops mentioned[48].
Negative feedback loops also can give rise to complex behavior. They can give rise to oscillations by establishing limit cycles[49]. Such circadian behavior could confer numerous biological properties to glycolysis. While the period of such oscillations would be commensurate with the kinetics of glycolysis thus rendering them unlikely to account for circadian rhythms which are slower, the accumulation of minor products that negatively feedback for example could occur over hours giving glycolysis a periodicity that would occur over similar timings as the mammalian circadian transcriptional networks[50]. These would allow for central carbon metabolism to be coupled to transcriptional processes in a multitude of ways. Finally, feedforward loops can also result in other complex kinetics including their ability to establish robustness or consistency of rates of glycolysis in the presence of changes in nutrient availability in the environment[51].
In summary, the consequences of these feedback loops remain largely unexplored and further research is needed to define their biological functions. Advances in technology such as flux analysis[52] and quantitative metabolomics[53] and single cell analysis[54,55] will enable further understanding of these iterations. Nevertheless, with the wealth of available data, the development of quantitative models that can understand the biological consequences of these phenomenon are likely imminent.
Research highlights.
-
–
Glycolysis is one of the most highly studied and evolutionary conserved studied pathways
-
–
Multiple newly defined small-molecule enzyme interactions create feedback and feedforward loops in glucose metabolism
-
–
These feedback and feedforward loops confer multiple additional layers of regulation to glucose metabolism
-
–
How these nonlinear interactions loops integrate changes to signals and nutrient availability to confer biological function is unknown
Acknowledgements
Support from the American Cancer Society (RSG-16–214-01-TBE) and National Institutes of Health (R01CA193256, R00CA168997, P30CA014236) are gratefully acknowledged. I would like to thank Ziwei Dai for preparation of the figure and the many colleagues who have inspired me to think about glucose metabolism over the years. I also apologize for any appropriate work that has been omitted in this article due to space limitations.
References
* Of special interest
** Of outstanding interest
- 1.Milo R, Last RL: Achieving diversity in the face of constraints: lessons from metabolism. Science 2012, 336:1663–1667. [DOI] [PubMed] [Google Scholar]
- 2.Bar-Even A, Flamholz A, Noor E, Milo R: Rethinking glycolysis: on the biochemical logic of metabolic pathways. Nat Chem Biol 2012, 8:509–517.** This important review elaborates on the design principles of the chemistry of glycolysis
- 3.Hui S, Ghergurovich JM, Morscher RJ, Jang C, Teng X, Lu W, Esparza LA, Reya T, Le Z, Yanxiang Guo J, et al. : Glucose feeds the TCA cycle via circulating lactate. Nature 2017, 551:115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faubert B, Li KY, Cai L, Hensley CT, Kim J, Zacharias LG, Yang C, Do QN, Doucette S, Burguete D, et al. : Lactate Metabolism in Human Lung Tumors. Cell 2017, 171:358–371 e359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liberti MV, Locasale JW: The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem Sci 2016, 41:211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Locasale JW: The consequences of enhanced cell-autonomous glucose metabolism. Trends Endocrinol Metab 2012, 23:545–551. [DOI] [PubMed] [Google Scholar]
- 7.Locasale JW, Cantley LC: Metabolic flux and the regulation of mammalian cell growth. Cell Metab 2011, 14:443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noor E, Eden E, Milo R, Alon U: Central carbon metabolism as a minimal biochemical walk between precursors for biomass and energy. Mol Cell 2010, 39:809–820. [DOI] [PubMed] [Google Scholar]
- 9.Rabinowitz JD, Vastag L: Teaching the design principles of metabolism. Nat Chem Biol 2012, 8:497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liberti MV, Dai Z, Wardell SE, Baccile JA, Liu X, Gao X, Baldi R, Mehrmohamadi M, Johnson MO, Madhukar NS, et al. : A Predictive Model for Selective Targeting of the Warburg Effect through GAPDH Inhibition with a Natural Product. Cell Metab 2017, 26:648–659 e648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vander Heiden MG, Plas DR, Rathmell JC, Fox CJ, Harris MH, Thompson CB: Growth factors can influence cell growth and survival through effects on glucose metabolism. Mol Cell Biol 2001, 21:5899–5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim JW, Dang CV: Cancer’s molecular sweet tooth and the Warburg effect. Cancer Res 2006, 66:8927–8930. [DOI] [PubMed] [Google Scholar]
- 13.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC: Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature 2008, 452:181–186. [DOI] [PubMed] [Google Scholar]
- 14.Chaneton B, Hillmann P, Zheng L, Martin ACL, Maddocks ODK, Chokkathukalam A, Coyle JE, Jankevics A, Holding FP, Vousden KH, et al. : Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature 2012, 491:458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan M, Chakravarthy S, Tokuda JM, Pollack L, Bowman GD, Lee YS: Succinyl-5-aminoimidazole-4-carboxamide-1-ribose 5’-Phosphate (SAICAR) Activates Pyruvate Kinase Isoform M2 (PKM2) in Its Dimeric Form. Biochemistry 2016, 55:4731–4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keller KE, Tan IS, Lee YS: SAICAR stimulates pyruvate kinase isoform M2 and promotes cancer cell survival in glucose-limited conditions. Science 2012, 338:1069–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adadi R, Volkmer B, Milo R, Heinemann M, Shlomi T: Prediction of microbial growth rate versus biomass yield by a metabolic network with kinetic parameters. PLoS Comput Biol 2012, 8:e1002575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.James DE, Strube M, Mueckler M: Molecular cloning and characterization of an insulin-regulatable glucose transporter. Nature 1989, 338:83–87. [DOI] [PubMed] [Google Scholar]
- 19.Majewski N, Nogueira V, Robey RB, Hay N: Akt inhibits apoptosis downstream of BID cleavage via a glucose-dependent mechanism involving mitochondrial hexokinases. Mol Cell Biol 2004, 24:730–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lehninger AL, Nelson DL, Cox MM: Lehninger principles of biochemistry edn 6th New York: W.H. Freeman; 2013. [Google Scholar]
- 21.Reznik E, Christodoulou D, Goldford JE, Briars E, Sauer U, Segre D, Noor E: Genome-Scale Architecture of Small Molecule Regulatory Networks and the Fundamental Trade-Off between Regulation and Enzymatic Activity. Cell Rep 2017, 20:2666–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hung YP, Albeck JG, Tantama M, Yellen G: Imaging cytosolic NADH-NAD(+) redox state with a genetically encoded fluorescent biosensor. Cell Metab 2011, 14:545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shestov AA, Liu X, Ser Z, Cluntun AA, Hung YP, Huang L, Kim D, Le A, Yellen G, Albeck JG, et al. : Quantitative determinants of aerobic glycolysis identify flux through the enzyme GAPDH as a limiting step. Elife 2014, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Heerden JH, Wortel MT, Bruggeman FJ, Heijnen JJ, Bollen YJ, Planque R, Hulshof J, O’Toole TG, Wahl SA, Teusink B: Lost in transition: start-up of glycolysis yields subpopulations of nongrowing cells. Science 2014, 343:1245114.** This study develops a quantitative model of glycolysis to study the Warburg Effect
- 25.Bakker BM, Westerhoff HV, Opperdoes FR, Michels PA: Metabolic control analysis of glycolysis in trypanosomes as an approach to improve selectivity and effectiveness of drugs. Mol Biochem Parasitol 2000, 106:1–10.** This study identifies imbalances in glycolysis that can be regulated by GAPDH activity
- 26.Moellering RE, Cravatt BF: Functional lysine modification by an intrinsically reactive primary glycolytic metabolite. Science 2013, 341:549–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishida Y, Rardin MJ, Carrico C, He W, Sahu AK, Gut P, Najjar R, Fitch M, Hellerstein M, Gibson BW, et al. : SIRT5 Regulates both Cytosolic and Mitochondrial Protein Malonylation with Glycolysis as a Major Target. Mol Cell 2015, 59:321–332.* These studies (24-26) together report acylation modifications that can result from carbon overload from the mitochondria may have regulatory roles in glycolysis
- 28.Wagner GR, Bhatt DP, O’Connell TM, Thompson JW, Dubois LG, Backos DS, Yang H, Mitchell GA, Ilkayeva OR, Stevens RD, et al. : A Class of Reactive Acyl-CoA Species Reveals the Non-enzymatic Origins of Protein Acylation. Cell Metab 2017, 25:823–837 e828.* These studies (24-26) together report acylation modifications that can result from carbon overload from the mitochondria may have regulatory roles in glycolysis
- 29.Wagner GR, Hirschey MD: Nonenzymatic protein acylation as a carbon stress regulated by sirtuin deacylases. Mol Cell 2014, 54:5–16.* These studies (24-26) together report acylation modifications that can result from carbon overload from the mitochondria may have regulatory roles in glycolysis
- 30.Yun J, Mullarky E, Lu C, Bosch KN, Kavalier A, Rivera K, Roper J, Chio II, Giannopoulou EG, Rago C, et al. : Vitamin C selectively kills KRAS and BRAF mutant colorectal cancer cells by targeting GAPDH. Science 2015, 350:1391–1396.* These studies (24-26) together report acylation modifications that can result from carbon overload from the mitochondria may have regulatory roles in glycolysis
- 31.Sabharwal SS, Schumacker PT: Mitochondrial ROS in cancer: initiators, amplifiers or an Achilles’ heel? Nat Rev Cancer 2014, 14:709–721.* This provocative study reports the use of Vitamin C as an oxidant that may target glycolysis
- 32.Louie SM, Grossman EA, Crawford LA, Ding L, Camarda R, Huffman TR, Miyamoto DK, Goga A, Weerapana E, Nomura DK: GSTP1 Is a Driver of Triple-Negative Breast Cancer Cell Metabolism and Pathogenicity. Cell Chem Biol 2016, 23:567–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hitosugi T, Zhou L, Elf S, Fan J, Kang HB, Seo JH, Shan C, Dai Q, Zhang L, Xie J, et al. : Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth. Cancer Cell 2012, 22:585–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, et al. : Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet 2011, 43:869–874.** This important study identifies two additonal feedback loops in glycolysis
- 35.Vander Heiden MG, Locasale JW, Swanson KD, Sharfi H, Heffron GJ, Amador-Noguez D, Christofk HR, Wagner G, Rabinowitz JD, Asara JM, et al. : Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science 2010, 329:1492–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oslund RC, Su X, Haugbro M, Kee JM, Esposito M, David Y, Wang B, Ge E, Perlman DH, Kang Y, et al. : Bisphosphoglycerate mutase controls serine pathway flux via 3-phosphoglycerate. Nat Chem Biol 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madhukar NS, Warmoes MO, Locasale JW: Organization of enzyme concentration across the metabolic network in cancer cells. PLoS One 2015, 10:e0117131.* This study reports the discovery of an additonal feedforward loop in glycolysis
- 38.Intlekofer AM, Wang B, Liu H, Shah H, Carmona-Fontaine C, Rustenburg AS, Salah S, Gunner MR, Chodera JD, Cross JR, et al. : L-2-Hydroxyglutarate production arises from noncanonical enzyme function at acidic pH. Nat Chem Biol 2017, 13:494–500.** This interesting study identifies two additional novel feedback loops in glycolysis
- 39.Collard F, Baldin F, Gerin I, Bolsee J, Noel G, Graff J, Veiga-da-Cunha M, Stroobant V, Vertommen D, Houddane A, et al. : A conserved phosphatase destroys toxic glycolytic side products in mammals and yeast. Nat Chem Biol 2016, 12:601–607. [DOI] [PubMed] [Google Scholar]
- 40.Losman JA, Kaelin WG Jr.: What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev 2013, 27:836–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang CH, Qiu J, O’Sullivan D, Buck MD, Noguchi T, Curtis JD, Chen Q, Gindin M, Gubin MM, van der Windt GJ, et al. : Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell 2015, 162:1229–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ho PC, Bihuniak JD, Macintyre AN, Staron M, Liu X, Amezquita R, Tsui YC, Cui G, Micevic G, Perales JC, et al. : Phosphoenolpyruvate Is a Metabolic Checkpoint of Anti-tumor T Cell Responses. Cell 2015, 162:1217–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pfeiffer T, Schuster S, Bonhoeffer S: Cooperation and competition in the evolution of ATP-producing pathways. Science 2001, 292:504–507. [DOI] [PubMed] [Google Scholar]
- 44.Cluntun AA, Huang H, Dai L, Liu X, Zhao Y, Locasale JW: The rate of glycolysis quantitatively mediates specific histone acetylation sites. Cancer Metab 2015, 3:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muoio DM: Metabolic inflexibility: when mitochondrial indecision leads to metabolic gridlock. Cell 2014, 159:1253–1262.* This review highlights negative consequences of increased glycolysis
- 46.Mulukutla BC, Yongky A, Daoutidis P, Hu WS: Bistability in glycolysis pathway as a physiological switch in energy metabolism. PLoS One 2014, 9:e98756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mulukutla BC, Yongky A, Grimm S, Daoutidis P, Hu WS: Multiplicity of steady states in glycolysis and shift of metabolic state in cultured mammalian cells. PLoS One 2015, 10:e0121561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferrell JE Jr.: Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr Opin Cell Biol 2002, 14:140–148. [DOI] [PubMed] [Google Scholar]
- 49.Strogatz SH: Nonlinear dynamics and chaos : with applications to physics, biology, chemistry, and engineering edn Second edition Boulder, CO: Westview Press, a member of the Perseus Books Group; 2015. [Google Scholar]
- 50.Asher G, Sassone-Corsi P: Time for food: the intimate interplay between nutrition,metabolism, and the circadian clock. Cell 2015, 161:84–92. [DOI] [PubMed] [Google Scholar]
- 51.Zhang C, Tsoi R, Wu F, You L: Processing Oscillatory Signals by Incoherent Feedforward Loops. PLoS Comput Biol 2016, 12:e1005101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dai Z, Locasale JW: Understanding metabolism with flux analysis: From theory to application. Metab Eng 2017, 43:94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu X, Locasale JW: Metabolomics: A Primer. Trends Biochem Sci 2017, 42:274–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Single-cell biology. Nature 2017, 547:19. [DOI] [PubMed] [Google Scholar]
- 55.Onjiko RM, Moody SA, Nemes P: Single-cell mass spectrometry reveals small molecules that affect cell fates in the 16-cell embryo. Proc Natl Acad Sci U S A 2015, 112:6545–6550.* This study demonstrates feasibility of single cell metabolomics


