TP53 mutations occur in nearly 10% of adults with de novo acute myeloid leukaemia (AML); however, these mutations are encountered at higher frequency in therapy-related AML (t-AML) (~35%) and AML with complex cytogenetics (50–60%) (Ok, et al 2015, Rucker, et al 2012). The presence of the TP53 mutation (TP53m) conveys an extremely poor prognosis (Grossmann, et al 2012, Rucker, et al 2012). Patients with TP53m AML are generally older in age (Rucker, et al 2012), and their physical fitness, especially in those with t-AML, is poor (Granfeldt Ostgard, et al 2015). The clinical response of TP53m AML to standard induction regimens has been disappointing, and if achieved, is usually short-lived (Grossmann, et al 2012, Rucker, et al 2012).
Venetoclax (VEN) has shown encouraging activity when combined with hypomethylating agents (HMAs) in both newly diagnosed and relapsed/refractory (r/r) AML (Aldoss, et al 2018, DiNardo, et al 2019). The combination is well-tolerated even in frail patients and is associated with low treatment-related mortality (TRM) (DiNardo, et al 2019). Apoptosis mediated by venetoclax appears to be TP53-independent (Anderson, et al 2016), and VEN/HMA activity was observed across various high-risk leukaemia genetics (Aldoss, et al 2018, DiNardo, et al 2019).
We conducted a retrospective analysis of AML patients treated with the combination of VEN/HMA at our institution between June 2016 and April 2019. We identified TP53m utilizing an in-house next generation sequencing panel. The study was approved by the City of Hope Institutional Review Board. Response was defined as either complete remission (CR) or CR with incomplete count recovery (CRi). In univariate analysis, descriptive statistics were used to summarize the covariates for both response and non-response groups. The correlations between response to VEN/HMA and these covariates were assessed by the Pearson Chi-square test for categorical variables. A logistic multivariable regression model was applied to the three covariates with P values not exceeding 0.1; odds ratios and 95% confidence intervals (CI) are listed in Table SI. The median leukaemia-free survival (LFS) and overall survival (OS) were summarized for the response group only. All analyses were performed with R 3.5.1 (http://www.R-project.org) or SAS 9.4 (SAS Institute Inc, Cary, NC).
We identified 32 adults with TP53m AML who were treated with VEN/HMA. One patient with r/r AML was unevaluable for response because of death from sepsis 10 days after initiating therapy. The median age was 68 years (22–85). Sixteen (52%) patients had r/r AML while 15 (48%) patients were newly diagnosed. The majority of patients had complex cytogenetics (n= 24, 77%). The median variant allele frequency (VAF) for TP53m was 56% (range, 3–95%), and 9 (29%) patients had more than one TP53 mutation. Nineteen (61%) patients had additional somatic mutations. Decitabine was the HMA used in the majority of cases (90%) and was more frequently given as a 5-day schedule rather than a 10-day schedule (52% vs. 39%).
Among the 31 evaluable patients, 16 (52%) experienced a response, including 7 CR and 9 CRi. Response was achieved after a median of 2 (range, 1–3) cycles. Minimal residual disease (MRD), defined as >0.01% by multiparameter flow cytometry in a reference laboratory, was evaluated in 10 of the responders, and 7 (70%) achieved MRD negativity. Prior HMA monotherapy was the only factor observed to be associated with a lower response compared to that in patients who were HMA-naive (14% vs. 63%, P= 0.025). There was a trend toward a higher CR/CRi rate in patients with more than one TP53 mutation (78% vs. 41%, P= 0.062) and in those who were treated with VEN/HMA in the frontline setting (67% vs. 38%, P= 0.1). Response was comparable across patient age, sex, AML type, receipt of prior allogeneic haematopoietic cell transplantation (alloHCT), cytogenetics, presence of additional mutations and type and duration of HMA therapy. Table I depicts patient characteristics and response rates. In multivariate analysis of response, prior exposure to HMA was associated with a non-significant trend toward lower response (odds ratio= 0.12; 95%CI: −1.69–1.27; P=0.092) (Table SII).
Table I.
Patient characteristics and response to VEN/HMA
| Evaluable patients | CR/CRi | P-value | |
|---|---|---|---|
| Number | 31 | 16 (50) | |
| Age, years; median (range) | 68 (22–85) | 0.376 | |
| ≤ 65 years | 14 (45) | 6 (43) | |
| >65 years | 17 (55) | 10 (59) | |
| Sex | 0.2 | ||
| Male | 14 (45) | 9 (64) | |
| Female | 17 (55) | 7 (41) | |
| AML setting | 0.104 | ||
| Newly diagnosed | 15 (48) | 10 (67) | |
| R/R | 16 (52) | 6 (38) | |
| AML type | 0.213 | ||
| De novo | 13 (42) | 5 (38) | |
| Secondary/therapy-related | 18 (58) | 11(61) | |
| Prior HMA treatment | 0.025 | ||
| Yes | 7 (23) | 1(14) | |
| No | 24 (77) | 15 (63) | |
| Prior allogeneic HCT | 0.571 | ||
| Yes | 5 (16) | 2 (40) | |
| No | 26 (84) | 14 (54) | |
| Cytogenetics | 0.562 | ||
| Complex | 24 (77) | 13 (54) | |
| with MK | 16 (52) | 8 (50) | |
| without MK | 8 (26) | 5 (63) | |
| Normal karyotype | 2 (6) | 1 (50) | |
| Others | 5 (16) | 2 (40) | |
| Additional mutations | 0.552 | ||
| No | 12 (39) | 7 (58) | |
| Yes | 19 (61) | 9 (47) | |
| TP53 allele frequency, %; median (range) | 56 (3–95) | 56 (9–92) | |
| TP53 mutations (n) | 0.062 | ||
| 1 | 22 (71) | 9 (41) | |
| ≥2 | 9 (29) | 7 (78) | |
| HMA type/schedule | 1 | ||
| Decitabine | 28 (90) | 14 (50) | |
| 5 days | 16 (52) | 8 (50) | |
| 10 days | 12 (39) | 6 (50) | |
| Azacitadine | 3 (10) | 2 (67) |
Abbreviations: AML, acute myeloid leukaemia; CR, complete remission; CRi, complete remission with incomplete count recovery; HCT, haematopoietic cell transplantation; HMA, hypomethylating agent; MK, monosomal karyotype; N, number; VEN, venetoclax.
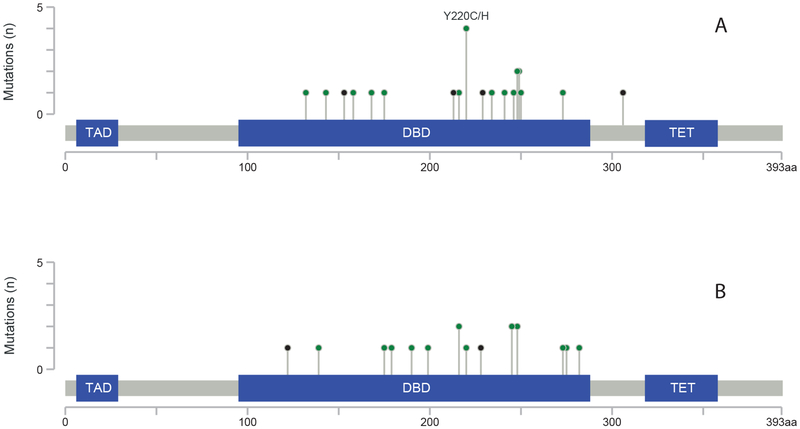
Response to VEN/HMA in TP53m AML according to associated additional mutations is shown in Table SI. The location and pattern of TP53 mutations for responders and non-responders is depicted in Figure 1. The median TP53 VAF was comparable for responders and non-responders (56% vs. 52%). The median LFS and OS for responders was 234 days (95% CI: 101–329) and 329 days (95% CI: 284-not reached), respectively. Five responders (31%) underwent alloHCT in CR.
Figure 1. Location of TP53 mutations in a) responders and b) non-responders.
DBD: DNA-binding domain; TAD: topologically associating domain; TET: tetramerization domain.m
Consistent with previous reports (DiNardo, et al 2019), we illustrate that the response to this regimen in the frontline setting is encouraging for this high-risk group, especially considering the fact that the majority of patients with TP53m AML carried complex cytogenetics. Furthermore, we also show a promising CR/CRi rate (38%) in patients with r/r TP53m AML, which includes some patients relapsing after prior alloHCT. Deep responses were observed, and a proportion of responders underwent alloHCT.
Prior exposure to HMA predicted lower response to VEN/HMA in this cohort, in contrast to another report from our institution that did not restrict analysis to TP53m AML (Aldoss, et al 2018). Most patients in our cohort received decitabine as their HMA, and this preference may have been influenced by results of a study utilising 10-day courses of decitabine in TP53m myeloid neoplasms and yielding a 100% response rate (Welch, et al 2016). In our cohort, the response rate was comparable between patients treated with either a 5- or 10-day course of decitabine. The VAF of TP53m was comparable in responders and non-responders, in contrast to a myelodysplastic syndrome study which indicated an impact of TP53 VAF on survival outcomes among patients treated with HMA (Sallman, et al 2016). The majority of TP53 mutations were present in the DNA-binding domain in both groups, consistent with COSMIC data and in line with the fact that TP53 acts as a tumor suppressor primarily through its function as a DNA-binding transcription factor regulating the expression of genes that control cell cycle arrest and apoptosis (Tate, et al 2019).
Although our preliminary results with VEN/HMA appear favourable compared to the dismal outcomes reported in TP53m AML patients when treated with conventional combination chemotherapy (Grossmann, et al 2012, Kadia, et al 2016, Rucker, et al 2012), the median LFS was relatively short, and relapses occurred frequently. The combination of VEN/HMA may serve as a well-tolerated backbone to which other novel agents could be added in order to further improve response rates and remission duration in TP53m AML and thereby safely bridge more patients to transplantation.
Supplementary Material
Acknowledgments
The research reported in this publication included work performed in the Pathology and Biostatistics Cores supported by the National Cancer Institute of the National Institutes of Health under award number P30CA033572. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Disclosure and competing interests
IA serves on an advisory board with Agios and the speakers’ bureau with Jazz Pharmaceuticals, and is a consultant Autolus Therapeutics. RN serves on advisory boards with Merck and Celgene and has a research collaboration with Jazz Pharmaceuticals. AS is on the speakers’ bureau for Amgen, Celgene and Stemline. VP has served on the advisory boards for Abbvie and Jazz Pharmaceuticals and is member of speakers’ bureau for Jazz Pharmaceuticals, Amgen, Novartis and Abbvie. GM is on the speakers’ bureau for Abbvie. The remaining authors have no relevant conflicts of interest to declare.
References
- Aldoss I, Yang D, Aribi A, Ali H, Sandhu K, Al Malki MM, Mei M, Salhotra A, Khaled S, Nakamura R, Snyder D, O’Donnell M, Stein AS, Forman SJ, Marcucci G & Pullarkat V (2018) Efficacy of the combination of venetoclax and hypomethylating agents in relapsed/refractory acute myeloid leukemia. Haematologica, 103, e404–e407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MA, Deng J, Seymour JF, Tam C, Kim SY, Fein J, Yu L, Brown JR, Westerman D, Si EG, Majewski IJ, Segal D, Heitner Enschede SL, Huang DC, Davids MS, Letai A & Roberts AW (2016) The BCL2 selective inhibitor venetoclax induces rapid onset apoptosis of CLL cells in patients via a TP53-independent mechanism. Blood, 127, 3215–3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNardo CD, Pratz K, Pullarkat V, Jonas BA, Arellano M, Becker PS, Frankfurt O, Konopleva M, Wei AH, Kantarjian HM, Xu T, Hong WJ, Chyla B, Potluri J, Pollyea DA & Letai A (2019) Venetoclax combined with decitabine or azacitidine in treatment-naive, elderly patients with acute myeloid leukemia. Blood, 133, 7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granfeldt Ostgard LS, Medeiros BC, Sengelov H, Norgaard M, Andersen MK, Dufva IH, Friis LS, Kjeldsen E, Marcher CW, Preiss B, Severinsen M & Norgaard JM (2015) Epidemiology and Clinical Significance of Secondary and Therapy-Related Acute Myeloid Leukemia: A National Population-Based Cohort Study. J Clin Oncol, 33, 3641–3649. [DOI] [PubMed] [Google Scholar]
- Grossmann V, Schnittger S, Kohlmann A, Eder C, Roller A, Dicker F, Schmid C, Wendtner CM, Staib P, Serve H, Kreuzer KA, Kern W, Haferlach T & Haferlach C (2012) A novel hierarchical prognostic model of AML solely based on molecular mutations. Blood, 120, 2963–2972. [DOI] [PubMed] [Google Scholar]
- Ok CY, Patel KP, Garcia-Manero G, Routbort MJ, Peng J, Tang G, Goswami M, Young KH, Singh R, Medeiros LJ, Kantarjian HM, Luthra R & Wang SA (2015) TP53 mutation characteristics in therapy-related myelodysplastic syndromes and acute myeloid leukemia is similar to de novo diseases. J Hematol Oncol, 8, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rucker FG, Schlenk RF, Bullinger L, Kayser S, Teleanu V, Kett H, Habdank M, Kugler CM, Holzmann K, Gaidzik VI, Paschka P, Held G, von Lilienfeld-Toal M, Lubbert M, Frohling S, Zenz T, Krauter J, Schlegelberger B, Ganser A, Lichter P, Dohner K & Dohner H (2012) TP53 alterations in acute myeloid leukemia with complex karyotype correlate with specific copy number alterations, monosomal karyotype, and dismal outcome. Blood, 119, 2114–2121. [DOI] [PubMed] [Google Scholar]
- Sallman DA, Komrokji R, Vaupel C, Cluzeau T, Geyer SM, McGraw KL, Al Ali NH, Lancet J, McGinniss MJ, Nahas S, Smith AE, Kulasekararaj A, Mufti G, List A, Hall J & Padron E (2016) Impact of TP53 mutation variant allele frequency on phenotype and outcomes in myelodysplastic syndromes. Leukemia, 30, 666–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, Boutselakis H, Cole CG, Creatore C, Dawson E, Fish P, Harsha B, Hathaway C, Jupe SC, Kok CY, Noble K, Ponting L, Ramshaw CC, Rye CE, Speedy HE, Stefancsik R, Thompson SL, Wang S, Ward S, Campbell PJ & Forbes SA (2019) COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res, 47, D941–D947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JS, Petti AA, Miller CA, Fronick CC, O’Laughlin M, Fulton RS, Wilson RK, Baty JD, Duncavage EJ, Tandon B, Lee YS, Wartman LD, Uy GL, Ghobadi A, Tomasson MH, Pusic I, Romee R, Fehniger TA, Stockerl-Goldstein KE, Vij R, Oh ST, Abboud CN, Cashen AF, Schroeder MA, Jacoby MA, Heath SE, Luber K, Janke MR, Hantel A, Khan N, Sukhanova MJ, Knoebel RW, Stock W, Graubert TA, Walter MJ, Westervelt P, Link DC, DiPersio JF & Ley TJ (2016) TP53 and Decitabine in Acute Myeloid Leukemia and Myelodysplastic Syndromes. N Engl J Med, 375, 2023–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.