Abstract
Spermidine/spermine N1-acetyltransferase 1 (SAT1), the rate limiting enzyme in polyamine catabolism, has broad regulatory roles due to near ubiquitous polyamine binding. We describe a novel function of SAT1 as a gene-specific transcriptional regulator through local polyamine acetylation. SAT1 expression is elevated in aggressive brain tumors and promotes resistance to radiotherapy. Expression profiling in glioma cells identified SAT1 target genes that distinguish high and low grade tumors, in support of the prognostic utility of SAT1 expression. We further discovered mechanisms of SAT1-driven tumor aggressiveness through promotion of expression of both DNA damage response pathways as well as cell cycle regulatory genes. Mechanistically, SAT1 associates specifically with the promoter of the MELK gene, which functionally controls other SAT1 targets, and leads biologically to maintenance of neurosphere stemness in conjunction with FOXM1 and EZH2. CRISPR knockin mutants demonstrate the essentiality of the polyamine acetyl transferase activity of SAT1 for its function as a transcriptional regulator. Together, the data demonstrate that gene-specific polyamine removal is a major transcriptional regulatory mechanism active in high grade gliomas that drives poor outcomes.
Keywords: SAT1, SSAT, polyamines, MELK, brain tumors, glioma, transcription
Introduction
Polyamines putrescine, spermidine, and spermine are small, positively-charged molecules present in millimolar amounts in cells that bind acidic macromolecules, and broadly regulate functions such as replication, translation, chromatin condensation. Polyamines are both essential for life, and facilitators of cell death, and are thus tightly regulated through synthesis, uptake, and degradation/secretion to maintain homeostatic levels15. SAT1 is the global polyamine rheostat, serving as the rate-limiting catabolic enzyme that acetylates spermine and spermidine, and primes them for either back-oxidation (spermine to spermidine, and spermidine to putrescine) or cellular efflux. SAT1 itself is highly regulated by polyamine concentrations, underscoring significant cellular investment in controlling polyamines14.
Polyamines and SAT1 are present in all cellular compartments8, but specific roles in different locations are not well-desribed. SAT1 has a role at the cell membrane, controlling motility by regulating a polyamine-sensitive potassium channel, and is recruited by interaction with α9β1 integrin6. SAT1 has also been found to interact with the membrane-bound diamine transporter SLC3A219, and cytoplasmic HIF1α1 and eIF5A13; the latter of which is a target of SAT1 acetylation controlling translation. While SAT1 is found in the nucleus, and polyamine catabolism is critical for unwinding DNA8, 22, no localized nuclear functions have been thus far ascribed.
SAT1 has been implicated in cancer, affecting cell migration20, proliferation and response to ionizing radiation (IR)2. Our group found SAT1 through an shRNA screen to cause resistance to radiation in glioblastoma (GBM), and showed that SAT1 is critical for tumor growth. We found that SAT1 controls expression of BRCA1 through a transcriptional mechanism. The observation helped explain why SAT1-elevated tumors respond less well to radiotherapy/temozolomide than SAT1-reduced tumors. However, the finding also led to the question of how SAT1 controls gene expression, and what other genes are controlled by SAT1. In the present study, we performed expression profiling on SAT1 competent and deficient brain tumor cells and identified the breadth of SAT1-dependent genes. We confirmed regulation in multiple neurosphere lines, and evaluated expression of the genes in TCGA data of 677 brain tumors. Our studies delineate the mechanism of SAT1-mediated gene expression, uncovering an unknown function of SAT1 in general, but also exposing SAT1 as a veritable therapeutic target in GBM that controls tumor cell stemness and aggressiveness.
Results
SAT1 regulates gene programs controlling cell cycle and DNA dynamics
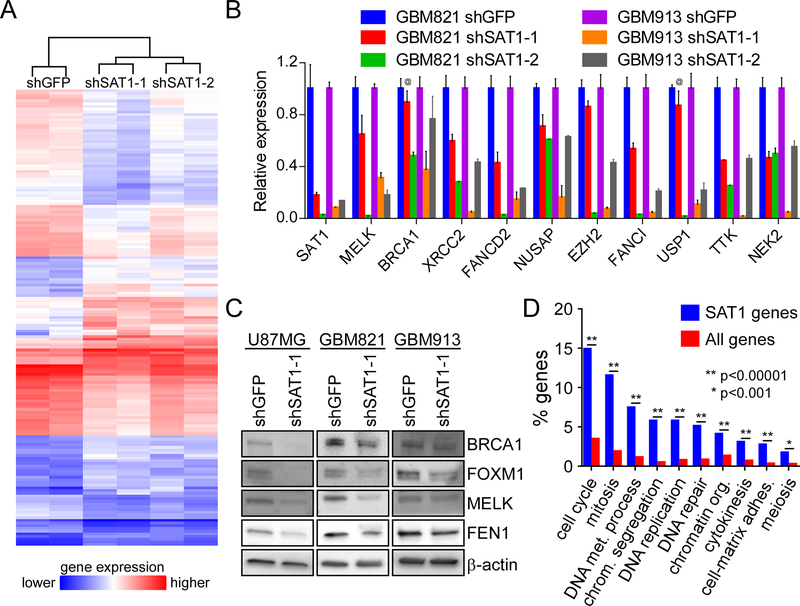
In order to assess the extent of genes regulated by SAT1 in brain tumors, we performed gene expression profiling of U87MG cells with two shRNAs to SAT1 using Affymetrix microarrays covering 67,528 gene transcripts. U87MG cells were stably transduced with lentiviruses expressing shRNAs to either the green fluorescent protein (GFP) or SAT1, and RNA was harvest within 7 days of infection. RNA was reverse transcribed into cDNA, labelled with biotin and hybridized onto the arrays for analyses. Hierarchical clustering of duplicate experiments correctly segregated samples according to sample identity (i.e. shGFP clustered together, shSAT1–1 clustered together, and shSAT1–2 clustered together; with the latter two separately branching from shGFP) (Figure 1A). At a fold-difference of ≥2.5, and an ANOVA p value of ≤0.07, 169 genes were differentially expressed between control and the shRNA cell lines (Supplementary Table 1), including previously identified BRCA1. At a threshold of 2.0, 332 genes were identified.
Figure 1: SAT1 regulates cell cycle and DNA regulatory genes in GBM cells:
A) Hierarchical clustering of differentially expressed genes from microarray data from U87MG cells expression shGFP or one of two shSAT1 shRNAs. Gene names are included in supplementary table 1. B) Validation of SAT1 target genes in two neurosphere lines by qRT-PCR. p<0.05 for all genes in the knockdown experiments except for those indicated by “@.” C) Validation of SAT1 target genes by Western blot. D) Gene ontology classification of SAT1 target genes (blue) into functional groups compared to all genes in the genome (red). Statistically significant differences are indicated.
To validate gene expression changes in U87MG cells, we tested SAT1 knock down on a cohort of genes in two neurosphere lines (GBM821 and GBM913) that retain characteristics of tumor stem cells in vivo7. All of the genes demonstrated significant reductions in expression upon knock down of SAT1 (Figure 1B). We also validated that reduction of transcript resulted in decreased protein for several of the genes by Western blot (Figure 1C). Finally, to gain insight into the functions of the genes affected by SAT1, we performed gene ontology-based functional classification and found significant over-representation of genes involved in both cell cycle/mitosis/cytokinesis and DNA metabolism/replication/repair over all other categories (Figure 1D). Since we found previously that SAT1 regulates DNA repair and tumor growth, the data support a broad effect of SAT1 on genes driving aggressive tumor biology.
SAT1 target genes are enriched in high grade gliomas
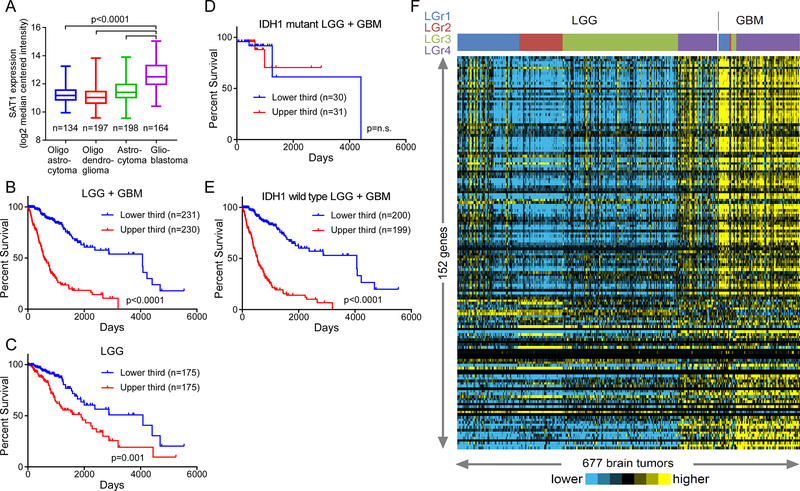
To glean clinical relevance for SAT1 target genes we queried TCGA and assessed expression in 677 gliomas. SAT1 is significantly overexpressed in high-grade glioblastoma compared to all subtypes of low-grade gliomas (Figure 2A). Furthermore, SAT1 expression correlates with poor outcome in the LGG + GBM (Low-Grade Glioma and Glioblastoma) cohort with a high level of significance (p<0.001, log rank test) (Figure 2B), largely because when ranked by SAT1 expression levels, 80% of the glioblastomas segregate to the upper-third of the tumors, and only 4.9% fall to the lower-third. Considering only the low-grade gliomas, high SAT1 tumors again perform significantly worse than those with lower SAT1 levels (p=0.001, log rank test) (Figure 2C). Notably, SAT1 expression was only prognostic in IDH1 wild type tumors, which tend to have poorer outcomes than IDH1 mutant (Figure 2D–E). Thus, SAT1 expression predicts poor outcome in human gliomas.
Figure 2: SAT1 target genes are elevated in aggressive brain tumors:
A) The expression of SAT1 in low and high grade gliomas from the TCGA LGG+GBM database. Pairwise statistical comparisons of low grade glioma to GBM are indicated (student’s t-tests). B) Kaplan-Maier survival plot for low and high grade gliomas dichotomized by SAT1 expression. C) Kaplan-Maier survival plot for only low grade gliomas dichotomized by SAT1 expression. D) Kaplan-Maier survival plot for low and high grade IDH1 mutant gliomas dichotomized by SAT1 expression. E) Kaplan-Maier survival plot for low and high grade IDH1 wild type gliomas dichotomized by SAT1 expression. F) Expression of SAT1 target genes in 677 brain tumors of both low and high grades, organized by LGr classifications.
We next assessed the expression of the 152 SAT1 target genes (2.5-fold genes) that mapped to the TCGA dataset in each of the tumors in the LGG+GBM cohort that were clustered according a recent integrative analysis producing four grade-independent subtypes (LGr1–4)3. We noted strikingly that expression of the SAT1 genes was independent of subtype and distinctly higher in the GBM subset compared to the LGG subsets. Further among LGG, some of the LGr4 samples appeared more similar to the GBM samples, which are themselves comprised mostly of LGr4 (Figure 2D). Together the data demonstrate that SAT1 and its target genes are expressed in more aggressive gliomas (i.e. GBM) and portend poor prognosis in the LGG groups.
SAT1 regulates MELK and EZH2 by direct interaction with chromatin
Intriguingly, MELK, FOXM1, EZH2, NEK2, and PLK1 are SAT1 target genes. Recent studies have demonstrated a role for the maternal embryonic leucine zipper kinase (MELK) in brain tumors. MELK was shown to phosphorylate the FOXM1 transcription factor, leading to expression of EZH2, the catalytic subunit of the polycomb repressive complex PRC211. All three proteins are necessary for stem cell maintenance, tumor growth, and resistance to IR. FOXM1 regulates proliferation by controlling polo-like kinase 1 (PLK1) during mitosis10, and NIMA-related kinase 2 (NEK2) is a regulatory binding partner of EZH2 in glioma stem cells21. Together, the MELK network promotes tumor aggressiveness.
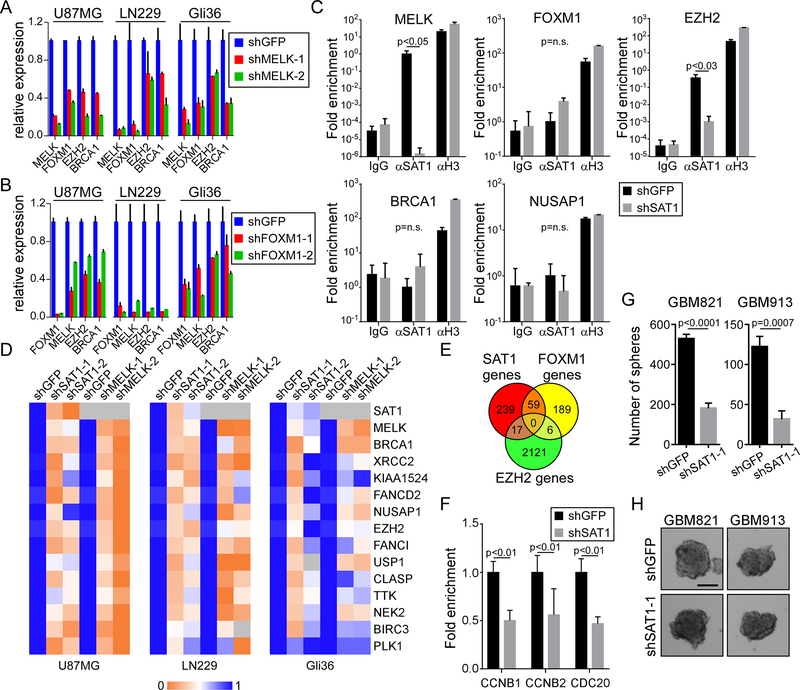
We explored the interaction of MELK, FOXM1 and EZH2 at the gene expression level in three established cell lines (U87MG, LN229, and Gli36), and observed that knockdown of MELK caused reductions in expression of FOXM1, EZH2, and BRCA1 (Figure 3A). We next targeted FOXM1 by shRNA and found similarly that MELK, EZH2 and BRCA1 were again reduced as a consequence (Figure 3B). These data support published findings of a functional dependence/interaction between MELK and FOXM110, and also that MELK and FOXM1 control each other at the transcriptional level11. We next investigated how SAT1 regulates expression of MELK and FOXM1, and performed chromatin immunoprecipitation (ChIP) to ask if SAT1 interacts with the proximal promoter regions of several of its targets. Using shGFP U87MG and shSAT1 U87MG, we found an association of SAT1 with both the MELK and EZH2 promoters, but not FOXM1, BRCA1, or NUSAP1 (Figure 3C). This observation led to the hypothesis that MELK and EZH2 are major nodes of SAT1 gene regulation, and the data in Figure 3A would suggest MELK precedes EZH2. We then tested the effect of MELK knockdown in comparison to SAT1 knockdown over a range of identified target genes across the three cell lines. We found in all genes tested, MELK knockdown with two shRNAs phenocopied SAT1 knockdown (Figure 3D). Interestingly, ectopic expression of MELK induced SAT1 target genes, but in an SAT1-dependent manner (Supplementary figure 1); suggesting MELK is necessary but not sufficient to drive SAT1 genes. To gain a broader view of the roles of MELK/FOXM1 and EZH2 in SAT1 driven gene expression programs, we analyzed recently published ChIPseq data for both FOXM14 and H3K27me317 (the EZH2-directed histone modification) in cancer cells, and compared their target genes to SAT1 target genes. Roughly one fifth (59 of 315, or 18.7%) of the SAT1 genes (≥2 change) were identified to have FOXM1 binding (Figure 3E, and Supplementary Table 2); and one twentieth (17 of 315, or 5.4%) of the published EZH2 targets were found on the SAT1 gene list. To verify the effect of SAT1 knockdown on FOXM1 occupancy, we performed FOXM1 ChIP on three of its identified targets (CCNB1, CCNB2, and CDC20) and found reduced association of FOXM1 (Figure 3F). Finally, because MELK, FOXM1 and EZH2 have been associated with stemness of glioblastoma cells, we assessed the ability of SAT1 knockdown primary glioblastoma lines to form neurospheres in culture. In both the GBM821 and GBM913 lines, we found that SAT1 depletion led to significant reductions in the ability to form neurospheres (Figure 3G–H). Therefore, the data demonstrate that MELK and EZH2 are key drivers of SAT1-mediated gene regulation and biology, promoting brain tumor stemness.
Figure 3: MELK and EZH2 are SAT1 direct target nodes:
A) Assessment of FOXM1, EZH2, and BRCA1 in control (shGFP) and MELK knockdown U87MG, LN229 and Gli36 cells by qRT-PCR. p<0.05 for all genes in the knockdown experiments. B) Assessment of MELK, EZH2, and BRCA1 in shGFP and shFOXM1 U87MG, LN229 and Gli36 cells by qRT-PCR. p<0.05 for all genes in the knockdown experiments. C) Input normalized SAT1 ChIP assay on U87MG shGFP and shSAT1 cells on the MELK, FOXM1, EZH2, BRCA1, and NUSAP1 promoters. Histone H3 ChIP is a positive control. D) Heat map of expression of SAT1 targets measured by RTPCR in shSAT1 U87MG, LN229 and Gli36 cells compared to shMELK cells. E) Venn diagram comparing SAT1 target genes with published FOXM1 and EZH2 targets. F) FOXM1 ChIP in shGFP and shSAT1 U87MG cells. G) Neurosphere formation assay in GBM821 and GBM913 lines with shGFP or shSAT1–1. H) Photomirographs of neurospheres. Scale bar 100 μm
SAT1 regulates transcription of specific targets through polyamine catabolism
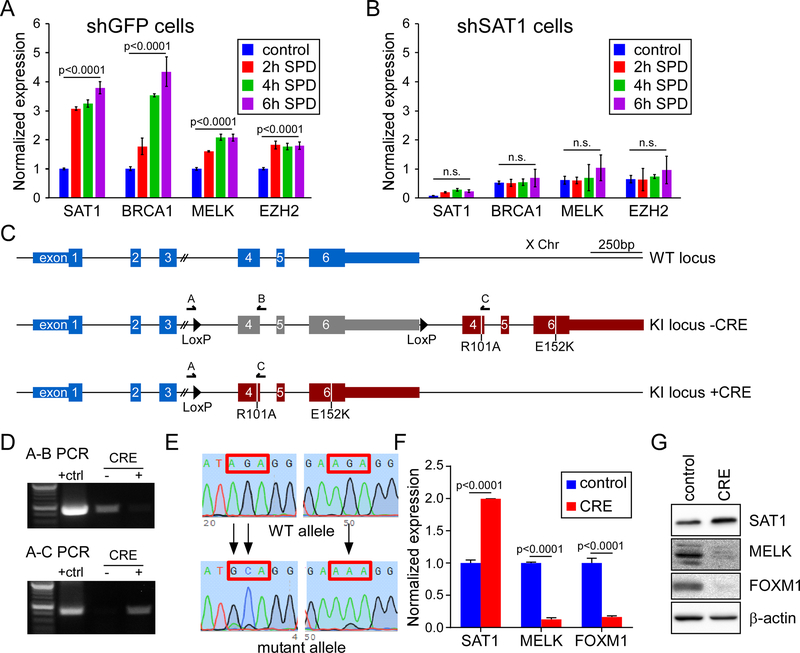
SAT1 is a polyamine catabolic enzyme. To assess the role of polyamines in SAT1-directed gene expression, we treated U87MG cells with 100 μM spermidine (SPD) for 2, 4, or 6 hours, and harvested RNA for analysis. Notably, SAT1 itself is highly responsive to excess polyamines14, and we observed rapid induction of SAT1 mRNA (Figure 4A). Similarly, we saw an induction of SAT1 targets BRCA1, MELK and EZH2. To determine if SAT1 was required for induction of the genes, or whether polyamines were responsible independently to induce transcription, we tested SAT1 knockdown cells and found a loss of responsiveness for all genes. The data argue that polyamine stimulation of SAT1 is required, but do not delineate whether catabolism of polyamines is involved. To determine if SAT1 polyamine acetylation activity is required for target gene activation, we created a conditional CRISPR/Cas9 knockin cell line in which wild type SAT1 remains expressed by its endogenous promoter until CRE recombinase induces excision of wild type exons 4–6, resulting in inclusion of mutant exons 4–6 with point mutations in both the acetyl Co-A and polyamine binding sites (R101A and E152K, respectively) (Figure 4C). The mutations have been shown to abrogate polyamine catabolism of SAT15. To validate the cell line, adenoviral mediated expression of CRE recombinase was used, and genomic PCR was performed using primer sets A-B and A-C, as indicated in Figure 4C. The A-B primer pair only amplifies the target prior to recombination, while A-C amplifies after recombination. As seen in Figure 4D, virtually complete recombination was evidenced by the lack of signal in the A-B PCR post-recombination. We also validated that the resulting locus contained the designed mutations by Sanger sequencing the PCR products (Figure 4E). We then assessed the transcriptional activity of SAT1 mutants. As see in Figure 4F–G, CRE recombination resulted in a moderate increase in SAT1 expression compared to control cells, but a dramatic loss of expression of both MELK and FOXM1 at both the mRNA and protein levels. The increase of SAT1 expression conforms with the production of non-functional SAT1, to which the cells respond by attempting to elevate expression. The loss of MELK and FOXM1 expression demonstrates that polyamine catabolism is required for transcriptional activation.
Figure 4: Polyamine catabolism is necessary for SAT1 transcriptional activity.
A, B) Measurement of SAT1 target genes after exposure to spermidine (SPD) by qRTPCR in shGFP cells (A) or shSAT1 cells (B). C) Schematic of the SAT1 locus on the X Chromosome (top) and the modified locus after CRISPR/Cas9 induced knockin pre (middle) and post (lower) CRE recombination. D) PCR validation of recombination using primers indicated in the schematic. E) Sanger sequencing of the wild type and two mutations sites in the SAT1 after CRE recombination. F, G) Measurement of SAT1, MELK, and FOXM1 by qRTPCR (F) and Western blot (G) in control or CRE adenovirus infected cells.
Discussion
We have uncovered a novel function for SAT1 as a gene-specific transcriptional modulator. Depletion of SAT1 from cells revealed altered gene expression programs that include cell cycle regulation and DNA repair, which portend poor prognosis in brain tumors. Physical association of SAT1 with the MELK and EZH2 promoters, and the requirement of SAT1 enzymatic function to regulate genes support a model in which localization of SAT1 to target genes, and acetylation and removal of polyamines from chromatin promote transcriptional activation. The studies highlight an expanding role for polyamine metabolism in cancer, and support pathway modulators as potential therapeutics.
How SAT1 is recruited to specific sites on DNA, and whether SAT1 transcriptional programs in different tissues are unique are open questions; analysis of SAT1 binding partners in different contexts will likely lend insight. The only reported mechanism of functional localization of SAT1 is for cell motility, wherein SAT1 has been shown to interact with α9β1 integrin. As SAT1 does not have an identifiable DNA binding domain, protein-protein interaction is the likely mechanism to interact with chromatin. Additionally recent studies have highlighted differential affinities of DNA sequences for polyamines, such that AT-rich regions bind more strongly than GC-rich regions; and polyamine binding stabilizes DNA duplexes22. The significance of this is that SAT1-mediated acetylation and removal of polyamines could be sequence-dependent, offering a potential mode of specificity for transcriptional activity, and a differential role of SAT1 on different genes. Interestingly, while we found MELK inhibition to phenocopy SAT1, and that ectopic expression of MELK elevated SAT1 target genes, MELK was insufficient to restore function in absence of SAT1. This suggests SAT1 function is still required beyond the control of MELK. Understanding exactly where SAT1 is needed in chromatin could illuminate a novel mechanism of epigenetic-like regulation of chromatin compaction and relaxation.
For brain tumors, the data presented argue that SAT1 function controls two major pathways, both of which are implicated in tumor aggressiveness and therapeutic resistance. Roughly 25% of SAT1 genes are direct targets of FOXM1 or EZH2. Some of the targets are transcription factors themselves, including DEPDC19 and TSHZ216; thus it is conceivable that FOXM1/EZH2 account for virtually all of the SAT1 targets. With the negative association of SAT1 expression and patient outcome, focusing therapeutic strategies on MELK, FOXM1, and EZH2, for which inhibitors currently exist, may be therapeutically advantageous. Whether inhibition of SAT1 rather than these three targets would be more efficacious will require further study, but expanding the list of viable therapeutic targets is an advance in a disease in need of novel approaches.
Materials and Methods
Cells and Neurospheres
U87MG and LN229 (ATCC), and Gli36 (gift of Dr. Susanne Brady-Kalnay) cells were maintained in DMEM 10%FCS. GBM821 and GBM9137 neurospheres were grown in neuro stem cell media7. shRNAs were acquired from Sigma (supplementary material), For neurosphere quantification, the media contained 1.5% methyl-cellulose; spheres were scored after 12 days with Metamorph software. pCSII-IB-MELK was a gift of Dr. Takeuchi18.
Microarray analyses
RNA was processed and hybridized at the CWRU Integrated Genomics Shared Resource. Analyses were performed using Transcriptome Analysis Console (Affymetrix), the PANTHER Classification System (www.pantherdb.org), and the Xena Functional Genomics Explorer (xenabrowser.net).
Statistics
Statistical tests were performed with Graphpad Prism 7.0 as indicated for each figure. All assays were repeated with both technical and biological replicates (n≥3).
ChIP/RT-PCR/Western Blotting
ChIP was described previously12. For antibodies and RT-PCR primers, see supplementary material.
CRISPR
Knockin vectors were constructed in pBSKSII+ with a BSR cassette flanking mutated exons 4–6. The gRNA sequence targeting the PAM sequence GGGAATACGTCAGGTTTACACGG within intron 3 of SAT1 was used to design an sgRNA by in vitro transcription, and was transfected with CAS9 and linearized vector into U87MG cells. Individual blasticidin-resistant clones were screened by PCR. Recombination was induced with Ad5CMVCre-eGFP (University of Iowa).
Supplementary Material
Acknowledgments
This work was supported by P30CA125123 (CJC); and R01CA187053 (SMW).
Footnotes
Conflict of interest statement: The authors declare no conflicts of interest.
References
- 1.Baek JH, Liu YV, McDonald KR, Wesley JB, Zhang H, Semenza GL. Spermidine/spermine N(1)-acetyltransferase-1 binds to hypoxia-inducible factor-1alpha (HIF-1alpha) and RACK1 and promotes ubiquitination and degradation of HIF-1alpha. The Journal of biological chemistry 2007; 282: 33358–33366. [DOI] [PubMed] [Google Scholar]
- 2.Brett-Morris A, Wright BM, Seo Y, Pasupuleti V, Zhang J, Lu J et al. The polyamine catabolic enzyme SAT1 modulates tumorigenesis and radiation response in GBM. Cancer research 2014; 74: 6925–6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell 2016; 164: 550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen X, Muller GA, Quaas M, Fischer M, Han N, Stutchbury B et al. The forkhead transcription factor FOXM1 controls cell cycle-dependent gene expression through an atypical chromatin binding mechanism. Molecular and cellular biology 2013; 33: 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coleman CS, Huang H, Pegg AE. Structure and critical residues at the active site of spermidine/spermine-N1-acetyltransferase. The Biochemical journal 1996; 316 ( Pt 3): 697–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.deHart GW, Jin T, McCloskey DE, Pegg AE, Sheppard D. The alpha9beta1 integrin enhances cell migration by polyamine-mediated modulation of an inward-rectifier potassium channel. Proc Natl Acad Sci U S A 2008; 105: 7188–7193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer research 2004; 64: 7011–7021. [DOI] [PubMed] [Google Scholar]
- 8.Holst CM, Nevsten P, Johansson F, Carlemalm E, Oredsson SM. Subcellular distribution of spermidine/spermine N1-acetyltransferase. Cell biology international 2008; 32: 39–47. [DOI] [PubMed] [Google Scholar]
- 9.Huang L, Chen K, Cai ZP, Chen FC, Shen HY, Zhao WH et al. DEPDC1 promotes cell proliferation and tumor growth via activation of E2F signaling in prostate cancer. Biochemical and biophysical research communications 2017; 490: 707–712. [DOI] [PubMed] [Google Scholar]
- 10.Joshi K, Banasavadi-Siddegowda Y, Mo X, Kim SH, Mao P, Kig C et al. MELK-dependent FOXM1 phosphorylation is essential for proliferation of glioma stem cells. Stem cells 2013; 31: 1051–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim SH, Joshi K, Ezhilarasan R, Myers TR, Siu J, Gu C et al. EZH2 protects glioma stem cells from radiation-induced cell death in a MELK/FOXM1-dependent manner. Stem cell reports 2015; 4: 226–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krieg AJ, Hammond EM, Giaccia AJ. Functional analysis of p53 binding under differential stresses. Molecular and cellular biology 2006; 26: 7030–7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee SB, Park JH, Folk JE, Deck JA, Pegg AE, Sokabe M et al. Inactivation of eukaryotic initiation factor 5A (eIF5A) by specific acetylation of its hypusine residue by spermidine/spermine acetyltransferase 1 (SSAT1). The Biochemical journal 2011; 433: 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pegg AE. Spermidine/spermine-N(1)-acetyltransferase: a key metabolic regulator. American journal of physiology Endocrinology and metabolism 2008; 294: E995–1010. [DOI] [PubMed] [Google Scholar]
- 15.Pegg AE, Casero RA Jr. Current status of the polyamine research field. Methods in molecular biology 2011; 720: 3–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santos JS, Fonseca NA, Vieira CP, Vieira J, Casares F. Phylogeny of the teashirt-related zinc finger (tshz) gene family and analysis of the developmental expression of tshz2 and tshz3b in the zebrafish. Dev Dyn 2010; 239: 1010–1018. [DOI] [PubMed] [Google Scholar]
- 17.Sharma V, Malgulwar PB, Purkait S, Patil V, Pathak P, Agrawal R et al. Genome-wide ChIP-seq analysis of EZH2-mediated H3K27me3 target gene profile highlights differences between low- and high-grade astrocytic tumors. Carcinogenesis 2017; 38: 152–161. [DOI] [PubMed] [Google Scholar]
- 18.Takeuchi H, Saito H, Noda T, Miyamoto T, Yoshinaga T, Terahara K et al. Phosphorylation of the HIV-1 capsid by MELK triggers uncoating to promote viral cDNA synthesis. PLoS pathogens 2017; 13: e1006441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uemura T, Yerushalmi HF, Tsaprailis G, Stringer DE, Pastorian KE, Hawel L 3rd et al. Identification and characterization of a diamine exporter in colon epithelial cells. The Journal of biological chemistry 2008; 283: 26428–26435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Veeravalli KK, Ponnala S, Chetty C, Tsung AJ, Gujrati M, Rao JS. Integrin alpha9beta1-mediated cell migration in glioblastoma via SSAT and Kir4.2 potassium channel pathway. Cellular signalling 2012; 24: 272–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, Cheng P, Pavlyukov MS, Yu H, Zhang Z, Kim SH et al. Targeting NEK2 attenuates glioblastoma growth and radioresistance by destabilizing histone methyltransferase EZH2. The Journal of clinical investigation 2017; 127: 3075–3089. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Yoo J, Kim H, Aksimentiev A, Ha T. Direct evidence for sequence-dependent attraction between double-stranded DNA controlled by methylation. Nat Commun 2016; 7: 11045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.