Abstract
Methane-producing archaea are among a select group of microorganisms that utilize tetrahydromethanopterin (H4MPT) as a one-carbon carrier instead of tetrahydrofolate. In H4MPT biosynthesis, β-ribofuranosylaminobenzene 5′-phosphate (RFAP) synthase catalyzes the production of RFAP, CO2, and pyrophosphate from p-aminobenzoic acid (pABA) and phosphoribosyl-pyrophosphate (PRPP). In this work, to gain insight into amino acid residues required for substrate binding, RFAP synthase from Methanothermobacter thermautotrophicus was produced in Escherichia coli, and site-directed mutagenesis was used to alter arginine 26 (R26) and aspartic acid 19 (D19), located in a conserved sequence of amino acids resembling the pABA binding site of dihydropteroate synthase. Replacement of R26 with lysine increased the KM for pABA by an order of magnitude relative to wild-type enzyme without substantially altering the KM for PRPP. Although replacement of D19 with alanine produced inactive enzyme, asparagine substitution allowed retention of some activity, and the KM for pABA increased about threefold relative to wild-type enzyme. A molecular model developed by threading RFAP synthase onto the crystal structure of homoserine kinase places R26 in the proposed active site. In the static model, D19 is located close to the active site, yet appears too far away to influence ligand binding directly. This may be indicative of the protein conformational change predicted previously in the Bi-Ter kinetic mechanism and/or formation of the active site at the interface of two subunits. Due to the vital role of RFAP synthase in H4MPT biosynthesis, insights into the mode of substrate binding and mechanism could be beneficial for developing RFAP synthase inhibitors designed to reduce the production of methane as a greenhouse gas.
Keywords: methanogenesis, methanopterin, RFAP synthase, site-directed mutagenesis, substrate binding
1. Introduction
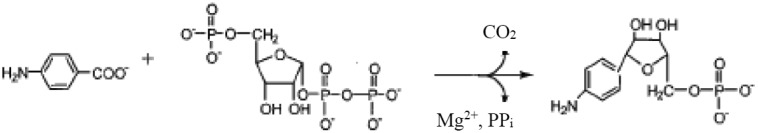
β-Ribofuranosylaminobenzene 5′-phosphate (RFAP) synthase catalyzes the first committed step in the biosynthesis of tetrahydromethanopterin (H4MPT), a tetrahydrofolate-like cofactor originally discovered in methane-producing microorganisms (methanogens) [1]–[6]. The coenzyme H4MPT is essential not only for the microbial production of methane but also for the survival of all known methanogenic organisms [7]–[10]. RFAP synthase catalyzes the transfer of a phosphoribosyl group from phosphoribosylpyrophosphate (PRPP) to the aminobenzene moiety of p-aminobenzoic acid (pABA), forming RFAP, CO2, and inorganic pyrophosphate (Figure 1). The decarboxylation of pABA catalyzed by RFAP synthase gives H4MPT the main chemical feature that distinguishes it from tetrahydrofolate, which retains the carboxyl group attached to the benzene ring [11].
Figure 1. Reaction catalyzed by RFAP synthase.
RFAP synthase is distinctive among phosphoribosyltransferases in that the phosphoribosyl moiety of the product is bound to a carbon atom rather than a nitrogen atom [1],[2]. Detailed kinetic analysis of RFAP synthase from the hyperthermophilic methanogen Methanocaldococcus jannaschii predicts an ordered Bi-Ter mechanism in which PRPP binds first, followed by pABA [12]. During catalysis, the order of product release is CO2, RFAP, and then PPi. In the mechanism proposed by Dumitru and Ragsdale [12] the formation of this unusual bond involves the production of a negatively charged species at the C-1 position of pABA. This carbon, rather than the amino nitrogen, appears to act as the nucleophile to attack an oxocarbenium phosphoribosyl intermediate formed after the loss of PPi from PRPP. The proposed mechanism also predicts that PRPP binding may induce a protein conformational change that is critical for catalysis.
Kinetics studies using N-substituted analogs of pABA have revealed that selected substrate analogs can inhibit RFAP synthase activity in vitro with Ki values as low as 7.5 µM [13]. This work has provided the basis for designing agrichemicals that may decrease the levels of methane as a greenhouse gas emitted from microorganisms in the digestive tracts of domestic livestock such as cattle [13]. Interestingly, an abundance of methane-producing microorganisms in the digestive tracts if humans and other animals has also been correlated with obesity [14],[15]. Based on these studies, increased knowledge of the pABA-binding site of RFAP synthase and inhibition of RFAP synthase activity by N-substituted pABA analogs may deepen our understanding of microbial factors that contribute to obesity in humans.
At present, only limited information is available regarding the structure of RFAP synthase and the roles that specific amino acids play in substrate binding and catalysis. Computational molecular modeling of RFAP synthase from M. jannaschii has predicted that the substrate binding site lies at the interface between identical subunits of the RFAP synthase homodimer [12]. In a separate study, alanine-screening mutagenesis of RFAP synthase from the moderate thermophile Methanothermobacter thermautotrophicus (MTH0830) has identified several charged amino acids that may play roles in substrate binding or catalysis [16]. In particular, the substitution of alanine for aspartic acid at position 19 (D19A) or arginine at position 26 (R26A) resulted in the production of inactive protein. However, due to the lack of detectable enzyme activity in the altered proteins, the specific functions of residues D19 and R26 were not further investigated.
The purified MTH0830 enzyme has shown optimal activity at pH 7, nearly two pH units higher than the optimal pH values for RFAP synthases from Archaeoglobus fulgidus and M. jannaschii [3],[12],[17]. This difference in pH optimum is likely to alter the KM for the charged substrate pABA. In this work, to provide insight into specific amino acids required for RFAP synthase activity, site-directed mutagenesis was used to produce altered enzymes of the M. thermautotrophicus RFAP synthase with more conservative substitutions (D19N and R26K), and the wild-type and altered proteins were analyzed by enzymatic assay, isothermal titration calorimetry, and computational modeling.
2. Materials and methods
2.1. Chemicals
Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs (Beverly, MA). Pfu DNA polymerase was from Stratagene (Agilent Technologies, Santa Clara, CA). Luria-Bertani broth mix (Becton Dickinson, Franklin Lakes, NJ) was purchased from Fisher Scientific (Suwanee, GA). Deoxyribonuclease I, citrate, MgCl2, 4-aminobenzoic acid (pABA), 5-phospho-D-ribose 1-diphosphate sodium salt (PRPP) and N-(1-naphthyl) ethylenediamine were purchased from Sigma-Aldrich Chemical Company, Inc. (St. Louis, MO). Bradford protein assay reagent and polyacrylamide gels were from Bio-Rad (Hercules, CA). Gases were obtained from Strate Welding (Gainesville, FL) and Oxygen Services (Orange, CA). All other chemicals were of reagent grade and purchased from Fisher Scientific.
2.2. Recombinant expression of the MTH0830 RFAP synthase genes in E. coli
MTH0830 has been previously identified as the gene encoding the H4MPT biosynthetic enzyme RFAP synthase in the methanogen M. thermautotrophicus [3]. For the heterologous production of the enzyme, cells of the E. coli expression strain KB1 [BL21(DE3) cells with the chaperone-encoding plasmid pG-Tf2 and the MTH0830 expression plasmid pED2] [17] were grown, induced with 1 mM isopropylthiogalactoside, and harvested as described previously [17]. For expression of RFAP synthase variants, plasmids with the mutated genes were transformed as described for the wild-type into BL21(DE3) cells containing plasmid pG-Tf2 [17].
2.3. Site-directed mutagenesis of recombinant RFAP synthase
Plasmid pED2 containing the RFAP synthase gene (MTH0830) (17) was purified using the Spin MiniPrep plasmid isolation kit from Qiagen (Valencia, CA) and was used as the template for site-directed mutagenesis. Residues R26 and D19 of the M. thermautotrophicus RFAP synthase were altered in a non-conservative and conservative manner using the QuikChange XL kit (Stratagene, Agilent Technologies, Santa Clara, CA). The thermocycling conditions were denaturation at 95 °C for 30 s, primer annealing at 55 °C for 60 s, and extension for 12 min at 68 °C through 16 cycles. The sequences of the forward primers were 5′-CTCAACGGTGAGAGGGGCAAGCTTGACGGTGGAGTTGG for R26K and 5′-CCACCTGACCCTCATAAACCTCAACGGTGAGAGGG for D19N. Dideoxy sequencing [18] was used to verify the mutations (Interdisciplinary Center for Biotechnology Research, University of Florida, Gainesville, FL). The plasmids with the mutated genes were transferred into XL-10 Gold cells, purified, and used to transform competent BL21(DE3) cells containing the chaperone plasmid pG-Tf2. The cells were grown, and the MTH0830 variant genes were expressed in the same manner as the wild type.
2.4. Purification of recombinant wild-type RFAP synthase
All purification steps were performed at ambient temperature in the presence of 2 mM dithiothreitol (DTT) or 15 mM beta-mercaptoethanol. Cells were lysed using a French press at 20,000 psi, and the cell lysate was centrifuged at 31,000 x g for 1 hour to separate the cell-free extract (CFE, supernatant) from the insoluble fraction (pellet).
For wild-type protein, CFE (50 mL) was heat-treated at 65°C for 15 min and centrifuged at 13,000 x g for 15 min to remove precipitated proteins. Heat-treated CFE (25 mL) was loaded onto a 40-mL ceramic hydroxyapatite column (2.5 × 11 cm) equilibrated with 50 mM TES [N-tris (hydroxymethyl) methyl-2-aminoethanesulfonic acid], 2 mM DTT, pH 6.8. The column was washed with 80 mL of the same buffer, and proteins were eluted using a 400 mL linear gradient from 0 to 125 mM NaH2PO4 in equilibration buffer at pH 6.8 collecting 20 mL fractions. Active RFAP synthase was found in fractions eluting between 60 and 80 mM NaH2PO4.
The most active hydroxyapatite fractions were loaded onto a Mono Q HR 5/5 anion exchange column (Amersham-Pharmacia Biotech-Piscataway, NJ) equilibrated with 50 mM TES, 5 mM MgCl2, 2 mM DTT pH 6.8. The column was washed with 300 mM NaCl in the equilibration buffer, followed by a 20 mL linear gradient from 300 mM to 600 mM NaCl. Active RFAP synthase was found in fractions eluting between 375 and 390 mM NaCl. The active Mono Q fractions were combined, concentrated, and loaded onto a Superdex 75 gel filtration column equilibrated with 50 mM TES, 150 mM KCl, and 2 mM DTT. The active pure RFAP synthase enzyme eluted after 19 min corresponding to a molecular mass of 68.7 kDa.
2.5. Determination of optimal enzyme assay conditions
RFAP synthase activity was determined by producing the azo-dye derivative of the product RFAP as described previously [1],[17]. To determine the temperature optimum, the reaction velocity of hydroxyapatite-purified enzyme was measured using 35 mM pABA, 8.8 mM PRPP and 30 mM MgCl2, 150 mM NaCl, 50 mM MES (pH 6.8), at temperatures ranging from 37 °C to 80 °C. To determine the effect of enzyme concentration on the activity of RFAP synthase, the activity of enzyme (approximately 95% pure) was determined using 35 mM pABA and 8.8 mM PRPP with various amounts of enzyme ranging from 0.012 to 0.200 nmol (1 to 25 µg). From this point on, the amount of enzyme used in activity assays was within the linear range of 8 to 12 µg. For determination of the KM for pABA, the PRPP concentration was kept constant at 8.8 mM with 17.6 mM MgCl2, while the pABA concentration was varied from 0.18 to 35 mM. All assays were carried out under optimal enzymatic conditions in 70 mM PIPES (pH 6.8), 2 mM DTT at 70 °C for 1 h.
2.6. Partial purification of wild-type RFAP synthase and variants for kinetic comparison
The RFAP synthase variants showed less heat stability than the wild-type enzyme. Therefore, for comparison, the variant and wild-type enzymes were partially purified from CFE (approximately 500 mg protein) by hydroxyapatite chromatography without heat treatment. CFE was loaded onto a 10 ml ceramic hydroxyapatite column equilibrated with 50 mM TES (pH 6.8), 2 mM DTT buffer and washed with 20 ml of the same buffer. A 100 ml linear gradient was applied to the column from 0 mM to 400 mM NaH2PO4 in equilibration buffer at pH 6.8, and 5 ml fractions were collected. Active wild-type RFAP synthase eluted between 150 mM and 200 mM NaH2PO4, while active variants eluted between 80 mM and 100 mM NaH2PO4.
The most active hydroxyapatite fractions were loaded onto a Mono Q HR 5/5 anion exchange column equilibrated with 50 mM TES, 5 mM MgCl2, 2 mM DTT pH 6.8, and the column was washed with 130 mM NaCl in the equilibration buffer, followed by a 30 ml linear gradient from 130 to 330 mM NaCl. Active RFAP synthase variants eluted between 120 and 130 mM NaCl. For comparison with the variants altered by site-directed mutagenesis, the recombinant wild type enzyme was partially purified using this modified protocol.
2.7. Kinetic analyses of recombinant RFAP synthase variants
To determine the KM for pABA for the RFAP synthase variants using the enzyme assay, the specific activities of the Mono Q purified proteins were determined in the presence of 8.8 mM PRPP and 17.6 mM MgCl2, with a range of pABA concentrations from 181 µM to 67 mM for the D19N variant, and 3.8 mM to 130 mM for the R26K variant. Due to the inability of the variants to function at 70 °C, the enzymatic assays comparing variant and wild-type kinetics were run in 70 mM PIPES (pH 6.8) with 2 mM DTT at 50 °C for three hours using a single time-point assay. This is the time period over which the enzyme activities were linear at the lower temperature of 50 °C. For these kinetic studies, the wild-type enzyme was purified in the same manner as the variants and was also examined at 50 °C for three hours.
2.8. Gel electrophoresis and protein concentration measurements
Protein purity was determined using reducing sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) [19] with Coomassie Brilliant Blue R-250 as the stain (Bio-Rad). Protein concentrations were determined using the Bradford protein assay (Bio-Rad, Hercules, CA) [20] with bovine serum albumin as the standard.
2.9. Isothermal titration calorimetry (ITC) of wild type and variant RFAP synthases
The ITC conditions previously established for M. jannaschii RFAP synthase [12] were modified for the RFAP synthase from M. thermautotrophicus. Partially purified wild-type (heat-treated at 65 °C) and variant (heat-treated at 50 °C) RFAP synthases (2–5 mL) were dialyzed against two changes of buffer (250 mL each of 50 mM Tris, 100 mM NaCl, pH 7.6). The dialysis buffer was used to create a 200 mM PRPP stock solution or a 48 to 200 mM pABA solution. All ITC measurements were carried out using a NANO-ITC isothermal titration calorimeter (TA Instruments, New Castle, DE).
For determination of the KD for PRPP, a 20 µM solution of dialyzed RFAP synthase (250 µL) was equilibrated in the sample cell at 35 °C, and 50 µL of 200 mM PRPP was loaded into the injection syringe. Following an 1800 s delay, 10 injections of 5 µL of PRPP each were added every 240 seconds with stirring at 300 rpm. Because a previous study demonstrated ordered binding with PRPP binding first [12], pABA binding in this work was measured in the presence of a saturating concentration of PRPP. For pABA binding, the sample cell was equilibrated at 35 °C with 200 µL of 20 µM dialyzed RFAP synthase and 50 µL of 200 mM PRPP. For the wild-type enzyme, 48 mM pABA was added in ten 5 µL aliquots to a total of 50 µL. For the R26K variant, the stock solution of pABA was 200 mM. The binding constants for pABA and PRPP were estimated by plotting the area under each peak of the resultant heat profile against the concentration of pABA or PRPP and by fitting to a theoretical curve using the NanoAnalyze software provided with the instrument (TA instruments, New Castle, NE).
2.10. Molecular modeling of RFAP synthase
All molecular modeling and docking were performed using a stochastic global energy optimization procedure in Internal Coordinate Mechanics (ICM) using the MolSoft ICM-Pro package version 3.7 (Molsoft, San Diego, CA) [21],[22]. An alignment was generated between the M. thermautotrophicus RFAP synthase and the sequence of homoserine kinase using an adaptation of the Needleman and Wunsch algorithm [23],[24]. Homoserine kinase, the closest available homologous crystal structure in the Protein Databank (PDB 1FWK), was previously predicted to be a good model template for RFAP synthase from M. jannaschii [12].
The initial RFAP synthase model was produced by threading the sequence of M. thermautotrophicus RFAP synthase onto the template crystal structure of the homoserine kinase (PDB code: 1FWK subunit A). The model was refined by globally optimizing the side-chains and annealing the backbone. The iterative refinement procedure contained three main steps: (i) random sampling of the dihedral angles according to the biased probability Monte Carlo method [22], (ii) a local minimization step, and (iii) application of the Metropolis criterion [25] to accept or reject a conformation. A non-homologous loop region in the vicinity of the predicted ligand binding region (residues 23 to 30 of RFAP synthase) was modeled by searching a database of loop fragments to identify loop templates with matching ends and homologous sequence and refining the subsequent model. The final model was selected based on an ICM calculated energy profile [22],[26].
The MTH0830 RFAP ligand binding pocket was calculated using the ICMPocketFinder algorithm [27], and the pocket was analyzed using ICM-Pro. The PRPP ligand was docked to the pocket using the ICM-Docking method [21],[22].
3. Results
3.1. Purification of M. thermautotrophicus RFAP synthase produced in E. coli
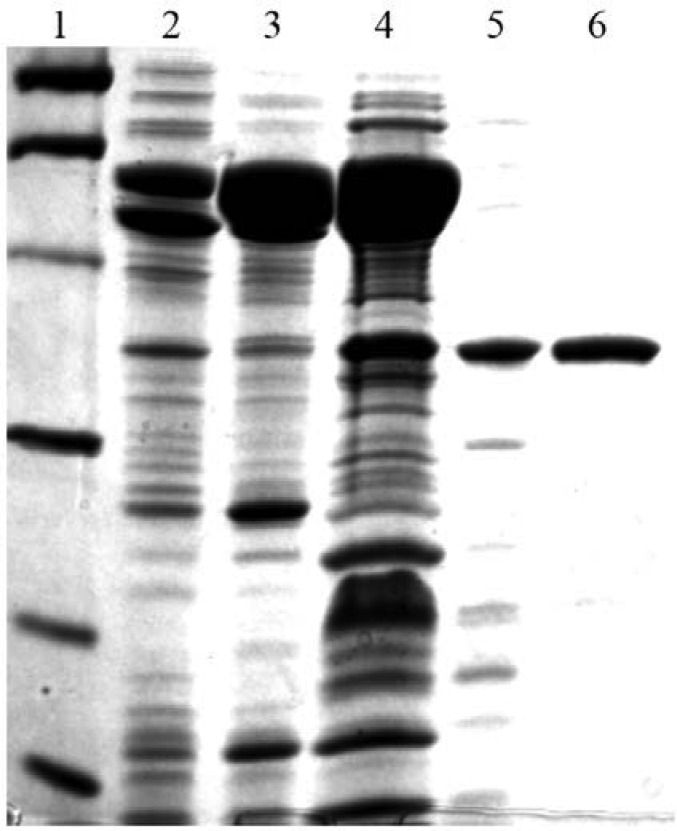
In this work, a recombinant form of M. thermautotrophicus RFAP synthase was purified for analysis by enzyme assay, site-directed mutagenesis, and isothermal titration calorimetry. The enzyme was purified 2800 fold to homogeneity with a specific activity of 360 nmol/min/mg of enzyme (Table 1 and Figure 2). The 20 fold increase in specific activity following the Mono Q step (Table 1) may be due to the removal of the abundant chaperone proteins (Figure 2), which could have detrimentally affected the specific activity by binding to RFAP synthase in the early purification steps. Based on gel filtration using Superdex 75 medium and SDS-PAGE analysis, the native molecular mass of the purified enzyme was estimated to be 69 kDa, consistent with a homodimer of 38 kDa subunits.
Table 1. Purification of wild-type M. thermautotrophicus RFAP synthase produced in E. coli.
| Purification step | Total activity1 (nmol per min) | Specific activity Yield (nmol per min per mg) | Fold (%) | purification |
| Cell-free extract | 690 | 0.13 | 10 | 1 |
| Heat-treated | 390 | 0.79 | 57 | 6 |
| Hydroxyapatite | 260 | 14 | 38 | 110 |
| Mono Q | 110 | 290 | 16 | 2200 |
| Superdex 75 | 30 | 360 | 4.2 | 2800 |
RFAP synthase assays were performed at 70 °C at pH 6.8 for one hour as described in the Material and Methods
Figure 2. SDS-PAGE gel of RFAP synthase purification. Protein samples were boiled in the presence of 7.5% 2-mercaptoethanol in SDS-PAGE sample buffer and loaded onto a 12% polyacrylamide gel. The gel was stained with Coomassie Brilliant Blue R-250 (Bio-Rad). Lane 1, Molecular mass markers. Lane 2, CFE (8 µg). Lane 3, heated CFE (10.2 µg). Lane 4, hydroxyapatite fraction (17 µg). Lane 5, Mono Q fraction (2 µg). Lane 6, Superdex 75 fraction (2 µg). The bands in lane 1 indicate the positions of the following molecular mass markers: phosphorylase b (97.4 kDa), bovine serum albumin (66.2 kDa), ovalbumin (45 kDa), carbonic anhydrase (31 kDa), soybean trypsin inhibitor (21.5 kDa), and lysozyme (14.4 kDa).
3.2. Determination of optimal enzymatic conditions
To determine the optimal enzymatic conditions for recombinant methanogen RFAP synthase, the activity of hydroxyapatite-purified RFAP synthase was tested at different temperatures and pH values. The highest enzyme activity was found at 70 °C and pH 7.0, and the optimal MgCl2 concentration was 1.5 to 3 fold higher than the PRPP concentration used (data not shown). The activity of hydroxyapatite-purified RFAP synthase remained constant up to an ionic strength of 200 mM, and thus from this point, the activity of wild-type M. thermautotrophicus RFAP synthase was measured using these optimal conditions. For enzyme concentration, a non-linear concave relationship was observed between the amount of enzyme used and the reaction velocity until about 0.1 nmol (6.5 µg) of dimeric enzyme, at which point the relationship became linear until at least 0.2 nmol of enzyme (data not shown).
3.3. Kinetic analyses of recombinant RFAP synthases
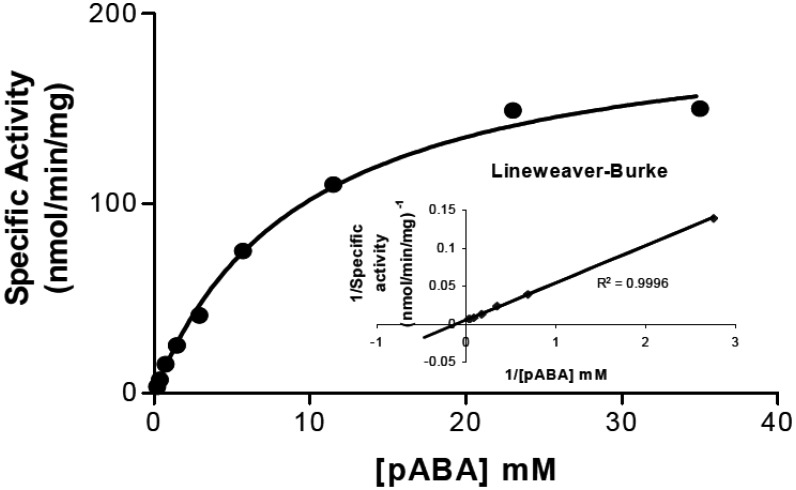
During the course of the purification and biochemical analysis of the recombinant enzyme, it was observed that purification of the enzyme beyond the Mono Q anion exchange step (about 80% pure) resulted in loss of enzyme stability during storage. Thus, the KM for the substrate pABA was measured using enzyme purified through the Mono Q step. When the enzyme activity was measured at 8.8 mM PRPP and 17.6 mM MgCl2, the KM for pABA was estimated to be approximately 4.5 mM (Figure 3).
Figure 3. Determination of pABA KM for recombinant RFAP synthase. RFAP synthase activity was measured at a constant PRPP concentration of 8.8 mM while varying the pABA concentration from 181 µM to 36 mM. The inset shows the Lineweaver-Burke plot derived from the Michaelis-Menton plot. The estimated KM for pABA was 4.5 mM and the Vmax at 70 °C was approximately 190 nmol/min/mg protein.
3.4. Bioinformatic analysis of RFAP synthase
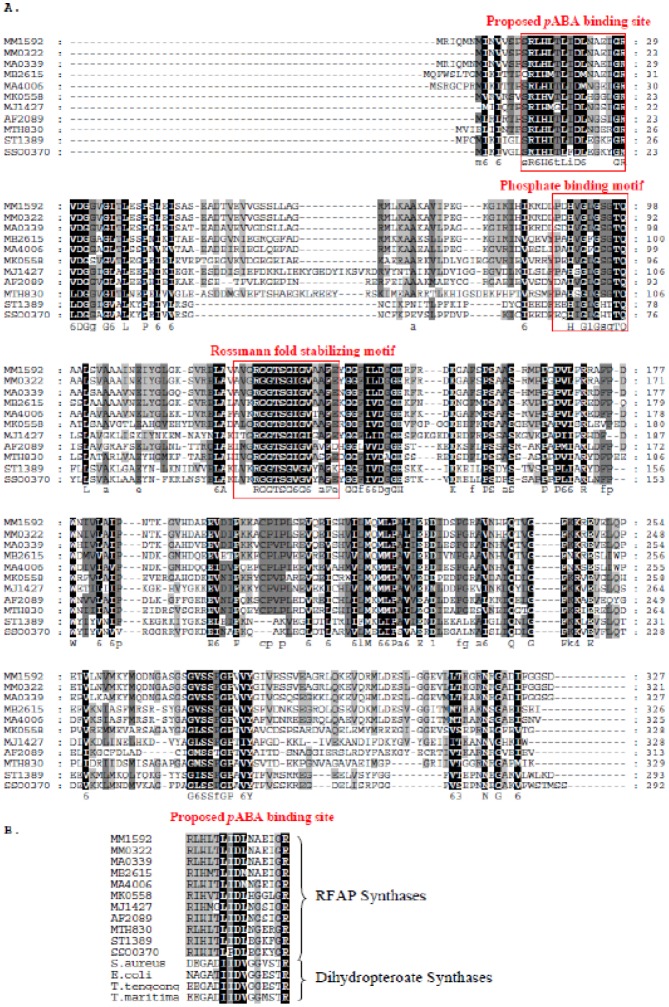
RFAP synthase homologs have previously been divided into three groups based on phylogenetic analysis [3]. Group 1 homologs, including RFAP synthases from methanogenic archaea and Sulfolobus, share conserved regions that are less prominent in the remaining two groups. Groups 2 and 3 include other non-methanogenic archaea and the bacterial sequence from Methylobacterium extorquens. Multiple sequence alignment of group 1 RFAP synthases (Figure 4A) has shown two conserved regions with significant similarity to glycine-rich amino acid motifs that bind phosphoribosyl-containing substrates [28],[29]. The first region has high homology to the phosphate binding loop of mevalonate kinase, which is usually composed of a loop with the consensus sequence PX3GLGSSAA connecting a β-strand to an α-helix [30]. In mevalonate kinase, this binding loop is also known to interact with the pyrophosphate tail of ATP [28],[30]. The presence of a similar region in the RFAP synthase sequences suggests a role for this conserved region in binding the pyrophosphate of PRPP.
Figure 4. Bioinformatic analysis of group I RFAP synthase genes. A. Multiple amino acid sequence alignment of Group I RFAP synthases using the alignment program GeneDoc [43]. Possible phosphate binding motifs are indicated by brackets. The protein abbreviations are as follows: Methanosarcina mazei, MM1592 and MM0322; Methanosarcina acetivorans, MA4006 and MA0339; Methanosarcina barkeri, MB2615; Methanopyrus kandleri, MK0558; Methanocaldococcus jannaschii, MJ1427; Archaeoglobus fulgidus, AF2089; Methanothermobacter thermautotrophicus, MTH0830; Sulfolobus tokodaii, ST1329; Sulfolobus sulfataricus, SSO0370. B. Multiple amino acid sequence alignment showing homology between the conserved region of RFAP synthase and a predicted pABA-binding region of dihydropteroate synthase (DHPS) from Staphylococcus aureus, Escherichia coli; Thermoanaerobacter tengcongensis, and Thermotoga maritima.
The second highly conserved region has significant homology to the consensus sequence [V/I]XGX1-2GX2GX3[G/A], known to stabilize β-strand and α-helix interactions for Rossmann fold proteins that bind FAD and NAD(P) (Figure 4A) [29]. In these proteins the β-strand/α-helix loop not only stabilizes the Rossmann fold but also interacts with the nucleotide near the phosphoribosyl moiety. The presence of this sequence in RFAP synthase homologs provides evidence that this region likely binds to the phosphoribosyl portion of PRPP and possibly works together with the first putative phosphate binding loop to form an overall PRPP binding region within the active site of the enzyme. It is interesting to note however, that this particular conserved region is not present in the groups 2 and 3 RFAP synthase homologs [31].
A third conserved region (amino acids 11 to 26 in M. thermautotrophicus) has a number of charged residues, including D19 and R26 (Figure 4A and 4B). Bioinformatic analysis showed that these amino acids share up to 40% identity with a region of the enzyme dihydropteroate synthase that forms part of a channel that binds pABA [32]. These similarities provided support for the proposal that the region containing D19 and R26 in RFAP synthase may play a role in binding pABA.
3.5. Site-directed mutagenesis
In a previous study, to begin to identify amino acid residues critical for substrate binding and catalysis, RFAP synthase from M. thermautotrophicus (MTH0830) was studied using alanine screening mutagenesis of highly conserved charged amino acids, including arginine 26 (R26) and aspartic acid 19 (D19) [16]. Two variants (R26A and D19A) produced in E. coli were partially purified, revealing that R26A and D19A were soluble, but lacked detectable RFAP synthase activity. The non-conservative replacement of R26 and D19 with alanine removed the charged functional groups, which may explain the lack of activity in spite of the production of soluble protein. In the current work, more conservative substitutions of R26 and D19 were constructed. R26 was replaced with a lysine (R26K) to maintain a positive charge on the side chain, and D19 was replaced with an asparagine (D19N) to maintain side chain polarity.
Initial characterization of the R26K and D19N variants showed that they were soluble and had low, but detectable, enzyme activity at 70 °C, which is the temperature optimum for the wild-type enzyme. The temperature optima for the R26K and D19N variants, however, were found to be significantly lower (50 °C and 60 °C, respectively) than that of the wild-type. Thus, for consistency, the activities of the variant and wild-type enzymes were studied at 50 °C. At this temperature, the wild-type enzyme purified by hydroxyapatite and Mono Q chromatography (without the 65 °C heat-step), exhibited an apparent KM for pABA of approximately 3.9 mM at 50 °C, similar to the wild-type value obtained at 70 °C (4.5 mM). However, the apparent Vmax for the wild-type enzyme at 50 °C (1.45 nmol/min/mg protein) was lower, most likely due to the lower temperature used and partial purification (Table 2).
Table 2. Kinetic values determined for wild-type and altered RFAP synthases assayed at 50 °C.
| Conservative substitution | Temperatureoptimum (°C) | KM for pABA1(mM) | Vmax1(nmol/min/mg) | |
| Wild-type | 70 | 3.9 | 1.5 | |
| R26K | 50 | 38 | 5.0 | |
| D19N | 60 | 12 | 0.5 | |
Assays for KM determination were conducted at 50 °C for 3 h.
When the R26K and D19N variants were partially purified in the same way and assayed at 50 °C, R26K showed an apparent KM for pABA of 38 mM, which is nearly ten-fold higher than the KM for the wild-type under similar conditions (Table 2). This result would be consistent with a role for R26 in the binding of pABA. Under the conditions studied, the Vmax for the R26K variant was approximately 5.0 nmol/min/mg, about 3-fold higher than that of the wild-type.
The estimated KM for the D19N variant was 12 mM, which is three-fold higher than the wild-type, while the Vmax for the D19N variant was approximately 0.45 nmol/min/mg (Table 2). The higher KM might also be consistent with a role for D19 in the binding of pABA, although D19 would appear to play a more subtle role than R26 in substrate binding. The differences between the wild-type and variant Vmax values could suggest that altering the amino acids may slightly affect the catalytic properties as well.
3.6. Isothermal titration calorimetry (ITC)
Given the modest three-fold effect of the D19N alteration on the KM for pABA, we sought an additional means of assessing the effect of the asparagine substitution on pABA binding. It was also of interest to determine the effect of the amino acid substitutions on the binding constant for PRPP. Previously, Dumitru and Ragsdale [12] used ITC to determine the KD values for PRPP binding to the hyperthermophilic RFAP synthase from the M. jannaschii. An advantage of this technique is that ITC can measure the heat released upon binding of individual substrates, even if the enzyme is catalytically incompetent.
Using ITC, the estimated KD values of PRPP and pABA were 2.1 mM and 0.96 mM, respectively, for the wild-type MTH0830 RFAP synthase. Based on triplicate ITC determinations, the estimated KD for PRPP binding to the D19N variant was 2.0 mM, similar to the wild type value of 2.1 mM (Table 3) and to the value obtained previously for RFAP synthase from M. jannaschii [12]. This result indicates that replacing the carboxylic acid side chain of D19 with an amide has no detectable effect on PRPP binding. In contrast, the apparent KD for pABA was (3.7 mM), about 4 times higher than the KD of the wild type enzyme (0.96 mM). This four-fold increase in the KD for pABA estimated by ITC is similar to the three-fold increase in KM estimated by enzymatic assay (Table 2) and provides evidence for only a subtle role of D19 in pABA binding.
Table 3. Isothermal titration calorimetry of wild type and site-directed variants of MTH0830 RFAP synthase.
| RFAP Synthase Variant | KD for PRPP (mM) | Apparent KD for pABA (mM) |
| Wild-type | 2.1 ± 0.11 | 0.96 ± 0.65 |
| D19N | 2.0 ± 0.30 | 3.7 ± 0.45 |
| R26K | 2.4 ± 0.25 | 24 ± 2.2 |
For R26K, the estimated KD for PRPP using ITC was 2.4 mM, which was comparable to the wild type KD of 2.1 mM (Table 3). However, the apparent KD for pABA (24 mM) was approximately 25 times higher than that of the wild-type under the ITC conditions. These data are consistent with the results using enzymatic assays (Table 2), which showed a dramatic increase in KM when R26 was substituted with lysine. Combined with the previous report that the R26A variant shows no detectable enzyme activity, these results indicate that the positive charge of the R26 side chain may play an important function in the binding of pABA.
3.7. Molecular modeling of M. thermautotrophicus RFAP synthase (MTH0830)
The crystal structure for homoserine kinase was used previously as the template to generate an overall model for RFAP synthase from M. jannaschii [12]. To gain additional insight into the potential roles of R26 and D19 in pABA binding in MTH0830, a molecular model of RFAP synthase from M. thermautotrophicus was generated by threading the MTH0830 sequence onto the crystal structure of homoserine kinase from M. jannaschii complexed with ADP (PDB 1FWK) (Figure 5).
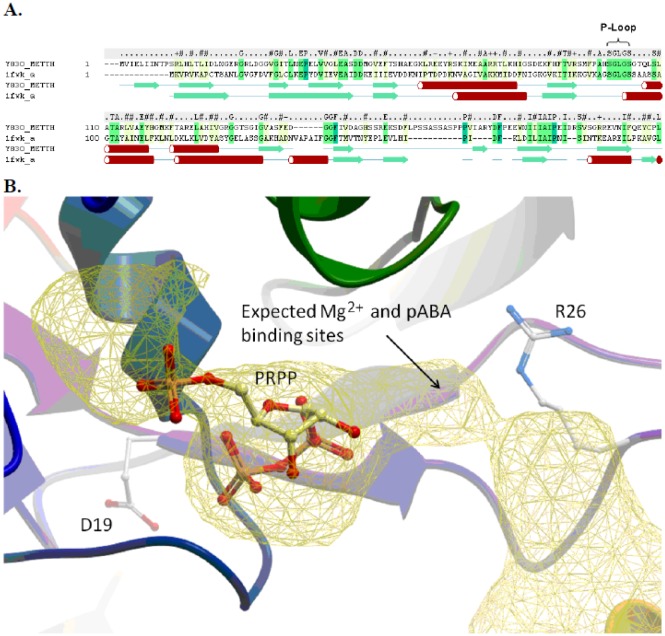
Figure 5. Computational molecular model of MTH0830 RFAP synthase. A: Secondary structure and sequence alignment of the N-terminus of MTH0830 (Y830_METTH) (from residue 1 to 215) and homoserine kinase template structure (pdb code: 1fwk) [44]. The secondary structure elements are colored green (beta Sheet) and red (alpha helix). The secondary structure of MTH0830 was calculated using the DSSP method [45], and the secondary structure of 1fwk is shown as defined in the Protein Data Bank file. B: The predicted PRPP and pABA binding pocket. The MTH0830 model is displayed in ribbon colored from the N to C terminus (blue – red), residues D19 and R26 are displayed as a stick representation. The docked PRPP ligand is displayed in stick representation with carbon atoms colored yellow. The binding pocket is displayed as a yellow mesh, and the predicted sites for pABA and Mg2+ are annotated.
Comparison of the amino acid sequences of MTH0830 and homoserine kinase showed 36% similarity and 21% identity. The suitability of this template for modeling was further supported by (i) the predicted secondary structure of MTH0830 RFAP synthase which showed good alignment with homoserine kinase in the N-terminal region and (ii) perfect conservation in the SGLGS sequence corresponding to the P-Loop motif of nucleotide binding proteins (Figure 5A).
The previous model of RFAP synthase from M. jannaschii [12] predicted that the active site of the homodimer is located at the interface between the two subunits. Figure 5B illustrates the predicted active site for one subunit of the M. thermautotrophicus RFAP synthase.
Secondary structure analysis predicted that D19 occurs at the C-terminal end of a beta sheet, while the region surrounding R26 (residues 23 to 30) forms a non-homologous loop, which was modeled by searching a database of loop fragments and identifying loop templates with matching ends and homologous sequence followed by refinement.
Figure 5B shows the most energetically favorable pose for PRPP in the substrate binding pocket. PRPP was predicted to interact noncovalently with the side chains of lysine 69 and histidine 98 as well as the backbone of histidine 98 and residues 101 to 104. The latter two amino acids are part of a conserved phosphate-binding loop motif previously identified among RFAP synthases (Figure 4).
In the molecular model (Figure 5B), R26 occupies a key position in the predicted pABA-binding pocket on the side distal from PRPP. R26 has no interaction with PRPP, but is in a position to interact with pABA possibly by forming a salt bridge. The formation of a salt bridge with arginine or lysine could explain why the alanine variant (R26A) is inactive and unable to bind pABA [16]. In contrast, R26K retains partial pABA-binding ability and catalytic competence.
With PRPP docked to the RFAP synthase monomer, D19 appears to occupy a position in the active site where it is unlikely to make direct contact with pABA. As no specific interaction with pABA could be predicted, it is possible that D19 may participate in the overall structure or conformational shape of the pABA binding pocket without affecting PRPP binding affinity. This positioning could also be indicative of the protein conformational change predicted previously for the Bi-Ter kinetic mechanism and/or the formation of the active site by the interaction of two subunits [12].
4. Discussion
The formation of RFAP from pABA and PRPP is the committed step in the H4MPT biosynthetic pathway. Moreover, the resulting decarboxylation of pABA by RFAP synthase produces the chemical feature that distinguishes H4MPT from tetrahydrofolate [1],[11],[33]. Due to the low abundance of RFAP synthase present in methanogenic archaea [3],[17], it was necessary to develop a recombinant expression system for the RFAP synthase gene from M. thermautotrophicus (MTH0830) to further analyze the biochemical properties of the enzyme. Previous reports have suggested that inhibitors of RFAP synthase may help to mitigate the production of methane by methanogenic microorganisms in the digestive tracts of both livestock and humans, showing potential for future application to the development of future agrichemical and pharmaceutical chemicals [13],[14]. The homolog from the moderate thermophile M. thermautotrophicus was selected for the current studies because the growth temperature optimum for this organism is closer than the optima for RFAP synthases from hyperthermophiles to the growth temperature of animals [3],[12]. In addition, the complete genome for this methanogen is readily available [34], and alanine-screening mutagenesis has been previously carried out on the M. thermautotrophicus enzyme.
M. thermautotrophicus RFAP synthase was produced in E. coli and purified to homogeneity (Figure 2) with a final specific activity of 360 nmol/min/mg protein (Table 1). This value is slightly higher than the rates obtained for partially purified recombinant RFAP synthases from the hyperthermophilic archaea M. jannaschii (248 nmol/min/mg protein) [12] and Archaeoglobus fulgidus (290 nmol/min/mg protein) [3]. The purified recombinant M. thermautotrophicus RFAP synthase was a dimer of identical subunits when separated on a Superdex 75 gel filtration column, consistent with the quaternary structure reported for other active RFAP synthases [3],[12].
When the recombinant M. thermautotrophicus RFAP synthase was analyzed biochemically, some interesting differences emerged. In contrast to the hyperthermophilic enzymes, the enzyme from M. thermautotrophicus showed high activity at the physiological pH of 7, while the hyperthermophilic enzymes showed pH optima around pH 5 [3],[12]. For MTH0830, enzymatic assays estimated that the KM for pABA was about 4 mM (Figure 3 and Table 2), and this value is substantially higher than the apparent KM for pABA determined for RFAP synthases from M. thermophila and M. jannaschii (58 and 150 µM, respectively) [2],[12]. To rule out the possibility that the high KM value was an artifact of heterologous protein production in E. coli, native RFAP synthase was partially purified from the original organism M. thermautotrophicus (a gift from Dr. Ralph Wolfe, University of Illinois). The apparent KM for the native enzyme from M. thermautotrophicus cells was 3.9 mM (data not shown), similar to the recombinant enzyme, providing additional support that the KM for pABA of the M. thermautotrophicus homolog is substantially different from previously studied methanogen RFAP synthases. The differences in pH optima may account in part for the higher KM for pABA observed in the M. thermautotrophicus enzyme. The KM differences could reflect differing ionization environments within the active sites due to amino acid variation between the enzymes. Alternatively, since pABA has two acidic pKa values of 4.65 and 4.80 [35], the higher KM for the M. thermautotrophicus enzyme at its pH optimum of 7, may indicate that the protonation state of either the amino or carboxylic acid group of pABA plays an important role in substrate binding affinity. Our hypothesis is that the acidic (protonated) form of pABA, rather than the deprotonated form, is the actual substrate for the reaction. Thus, when we used a pH of 6.8 instead of 5.3 as for the previously published enzymes (2, 3, 12), a large proportion of the pABA would have been deprotonated. At near neutral pH, the concentration of protonated pABA would be lower by 1–2 orders of magnitude, resulting in a higher apparent KM. This is consistent with the 26 fold higher KM we observed for the M. thermautotrophicus enzyme under near neutral conditions when compared to previous homologs with a pH optimum at 5.3 (2, 3, 12). The effect of pH would be a significant consideration for inhibiting methanogenesis under physiological conditions in cattle rumen, where the pH is typically neutral.
Bioinformatic analysis of conserved regions among the RFAP synthase sequences revealed two phosphate-binding motifs, which are likely to interact with the phosphates of the substrate PRPP [30] (Figure 4A). A second region (amino acids 16 and 26) showed similarity to a predicted pABA-binding site in dihydropteroate synthase (Figure 4B). The structure of dihydropteroate synthase is made up of a TIM-barrel fold composed of eight helices surrounding eight central β-strands with 8 loops connecting the α-helices and β-strands playing critical roles in catalysis [32]. In dihydropteroate synthase from Mycobacterium tuberculosis, it is thought that loop 2 aids in the overall formation of a pABA binding site within a solvent inaccessible channel [32]. The similarity of the loop containing R26 in RFAP synthase to the loop 2 region of dihydropteroate synthase, including the presence of conserved arginine and aspartic acid residues (Figure 4), could indicate a similar role in pABA binding for this region in RFAP synthase.
To test this hypothesis, site-directed mutagenesis of two conserved charged amino acids (R26 and D19) in the putative pABA-binding region was undertaken. The non-conservative replacement of these residues to alanine was previously shown to produce soluble enzymes that are devoid of detectable enzyme activity [16], supporting a role for R26 and D19 in the activity of RFAP synthase. However, the inactivity of the variants did not allow for the determination of the specific role of these amino acids. In the current study, the more conservative substitutions of R26 with lysine (R26K) and D19 with asparagine (D19N) produced enzymes that retained partial activity. Based on isothermal calorimetry, the binding constant (KD) for PRPP for the wild-type, R26K, and D19N enzymes were similar to the value of 2 mM reported for the M. jannaschii enzyme [12]. In contrast, using either enzymatic assay or ITC, the KM for the second substrate pABA increased at least 10 fold for R26K and 3–4 fold for D19N (Tables 2 and 3). These kinetic results are consistent with the prediction that R26 and D19 affect enzyme interactions with pABA but not PRPP.
It is of interest to note that the apparent KD for wild-type pABA binding obtained by ITC experiments (0.96 mM, Table 3) was lower than the KM determined using the enzymatic assay at pH 7 (3.9 mM, Table 2), but higher than the value obtained previously for RFAP synthase from M. thermophila (58–150 µM at pH 4.8) [2],[3]. We suspect two reasons for these differences. First, consistent with previous studies [12], the ITC studies were performed in the absence of the cofactor Mg2+, and the absence of this ion may have affected pABA affinity. Second, the final pH of the ITC assays in this study was 6, intermediate between the pH values of 4.8 and 7.0 used in the enzymatic assays. Although the wild-type KM and KD values for pABA differ using the ITC versus enzymatic assays, both data show that substitution of R26 with lysine substantially lowered the affinity of the enzyme for pABA without affecting the affinity for PRPP.
A model of MTH0830 RFAP synthase was generated by threading the sequence of MTH0830 RFAP synthase onto the template crystal structure of the homoserine kinase. Docking studies predicted that the PRPP binds first within the predicted active site where a key nucleotide binding motif is conserved (Figure 5). This is consistent with the Bi-Ter mechanism proposed for RFAP synthase from M. jannaschii, which predicts a requirement for PRPP binding prior to pABA (12). Computational modeling of RFAP synthase predicted that the amino acid D19 is located close to the active site, but not directly interacting with it and somewhat distant from the predicted pABA binding site. In this position, D19 could have an affect on the overall conformation of the binding pocket without interacting directly with pABA. This can explain why a conservative change from aspartic acid to asparagine had only a subtle effect on the apparent KM or KD of pABA. On the other hand, the model places the guanidinium of arginine 26 close to the predicted position for pABA (Figure 5) in an orientation that could allow a noncovalent interaction with pABA.
In dihydropteroate synthase [32],[36], the conserved arginine corresponding to R26 is predicted to contribute its guanidinium group for hydrogen bonding as well as its main chain for hydrophobic interactions during the binding of pABA. In the computational model for RFAP synthase, R26 is in a position that allows hydrogen bonding with pABA (Figure 5), which is supported by the site-directed mutagenesis results. While substituting R26 with alanine eliminated enzyme activity, conservative replacement with lysine (R26K) resulted in active RFAP synthase with substantially lower affinity for pABA (Tables 2 and 3). The increase in the KM for pABA for the R26K variant, along with the previously proposed role for the corresponding arginine in dihydropteroate synthase is consistent with a role for the amino group or positive charge of the guanidinium of R26 in pABA binding.
In RFAP synthase, the replacement of D19 with asparagine resulted in the production of a soluble, active D19N variant with only a threefold increase in the KM for pABA (Table 2). The small increase in the KM for pABA suggests less of a direct role in substrate binding, but does not rule out a possible a role in catalysis or overall structure of the active site. The presence of a conserved carboxylate residue surrounded by hydrophobic residues has previously been proposed to be involved in the stabilization of a carboxonium ion intermediate formed during one of the proposed mechanisms of RFAP synthase, as well as in other phosphoribosyl transferase (PRTase) enzymes [2],[37].
Bioinformatic analysis of the RFAP sequences showed two phosphate-binding motifs. Distinct differences in the active site structures of PRTases have resulted in two evolutionarily distinct types of PRTases. The active site of type I PRTases are made up of an α/β core structure with a characteristic Rossmann fold and a solvent exposed active site that is flanked by hood and loop structures which close over the active site during catalysis [38]–[40]. The active structure of type II PRTases, solely represented by quinolinate phosphoribosyltransferase, is made up of a seven stranded α/β barrel open sandwich structure, which serves as a cap for the active site of the other subunit [38]. The data presented here along with the identification of a characteristic Rossmann fold and an apparent phosphate binding loop in RFAP synthase suggests a model for similar to that of type I PRTases [38]–[40]. The absence of this characteristic Rossmann fold in the two remaining groups of RFAP synthase homologs suggests not only structural differences, but also differences in substrate binding modes between the groups of RFAP synthases. This proposed model for the active site structure and catalytic mechanism of RFAP synthase, like that of other type I PRTases, would involve the formation of a slightly hydrophobic active site that would be inaccessible to bulk solvent water upon the sequential or random binding of the substrates [38]–[40]. The ability of the active site of RFAP synthase to form a solvent inaccessible active site would be crucial for the enzyme to catalyze the phosphoribosyl transfer event, as bulk solvent water can serve as a nucleophile hydrolyzing the activated substrate PRPP [32],[41],[42]. This model would also accommodate a previously proposed two-step SN1-like mechanism for RFAP synthase, which involves the formation of a carboxonium ion intermediate of PRPP that may be stabilized by carboxylate residues [12].
The work presented here contributes to the fundamental understanding of ligand-binding interactions in an important enzyme of biological methanogenesis. Increased knowledge of this enzyme could have multiple practical impacts for elucidating the structure and function of this enzyme and developing more specific RFAP synthase inhibitors to mitigate methane production related to agricultural production of greenhouse gases [13] as well as human health [14].
Acknowledgments
This research was supported by National Science Foundation grant numbers MCB-0420766, MCB-1020200, and CHE-1508801. The authors are grateful for the research contributions of Rosemarie Garcia and Alex Nguyen.
Abbreviations
- CFE
Cell-free extract
- DTT
dithiothreitol
- H4MPT
tetrahydromethanopterin MES, 2-(N-morpholino) ethanesulfonic acid
- ITC
isothermal titration calorimetry
- pABA
p-aminobenzoic acid
- PIPES
piperazine-N,N′-bis (2-ethanesulfonic acid)
- PRPP
5-phospho-D-ribose 1-diphosphate
- PRTase
phosphoribosyl transferase
- RFAP
ribofuranosylaminobenzene 5′-phosphate
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- TES
N-[Tris(hydroxymethyl)methyl]-2-aminoethanesulfonic acid
Footnotes
Conflict of interest: All authors declare no conflicts of interest in this paper.
References
- 1.White RH. Biosynthesis of methanopterin. Biochemistry. 1996;35:3447–3456. doi: 10.1021/bi952308m. [DOI] [PubMed] [Google Scholar]
- 2.Rasche ME, White RH. Mechanism for the enzymatic formation of 4-(beta-D-ribofuranosyl) aminobenzene 5′-phosphate during the biosynthesis of methanopterin. Biochemistry. 1998;37:11343–11351. doi: 10.1021/bi973086q. [DOI] [PubMed] [Google Scholar]
- 3.Scott JW, Rasche ME. Purification, overproduction, and partial characterization of beta-RFAP synthase, a key enzyme in the methanopterin biosynthesis pathway. J Bacteriol. 2002;184:4442–4448. doi: 10.1128/JB.184.16.4442-4448.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leigh JA. Levels of water-soluble vitamins in methanogenic and non-methanogenic bacteria. Appl Environ Microbiol. 1983;45:800–803. doi: 10.1128/aem.45.3.800-803.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vanbeelen P, Labro JFA, Keltjens JT, et al. Derivatives of methanopterin, a coenzyme involved in methanogenesis. Eur J Bioche. 1984;139:359–365. doi: 10.1111/j.1432-1033.1984.tb08014.x. [DOI] [PubMed] [Google Scholar]
- 6.DiMarco AA, Bobik TA, Wolfe RS. Unusual coenzymes of methanogenesis. Annu Rev Biochem. 1990;59:355–394. doi: 10.1146/annurev.bi.59.070190.002035. [DOI] [PubMed] [Google Scholar]
- 7.Thauer RK, Hedderich R, Fischer R. Reactions and enzymes involved in methanogenesis from CO2 and H2. In: Ferry J.G., editor. Methanogenesis, 5Eds. New York: Chapman Hall; 1993. pp. 209–252. [Google Scholar]
- 8.White D. The Physiology and Biochemistry of Prokaryotes. New York: Oxford University Press; 2000. C1 metabolism; pp. 348–350. [Google Scholar]
- 9.Keltjens JT, Vogels GD. Methanopterin and methanogenic bacteria. Biofactors. 1988;1:95–103. [PubMed] [Google Scholar]
- 10.Deppenmeier U, Muller V, Gottschalk G. Pathways of energy conservation in methanogenic archaea. Arch Microbiol. 1996;165:149–163. [Google Scholar]
- 11.Maden BEH. Tetrahydrofolate and tetrahydromethanopterin compared: functionally distinct carriers in C1 metabolism. Biochem J. 2000;350:609–629. [PMC free article] [PubMed] [Google Scholar]
- 12.Dumitru RV, Ragsdale SW. Mechanism of 4 (β-D-ribofuranosyl) aminobenzene 5′-phosphate synthase, a key enzyme in the methanopterin biosynthetic pathway. J Biol Chem. 2004;279:39389–39395. doi: 10.1074/jbc.M406442200. [DOI] [PubMed] [Google Scholar]
- 13.Dumitru R, Palencia H, Schroeder SD, et al. Targeting methanopterin biosynthesis to inhibit methanogenesis. Appl Environ Microbiol. 2003;69:7236–7241. doi: 10.1128/AEM.69.12.7236-7241.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;44:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 15.Samuel BS, Hansen EE, Manchester JK, et al. Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proc Natl Acad Sci USA. 2007;104:10643–10648. doi: 10.1073/pnas.0704189104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rasche ME. Outcomes of a research-driven laboratory and literature course designed to enhance undergraduate contributions to original research. Biochem Molec Biol Education. 2004;32:101–107. doi: 10.1002/bmb.2004.494032020313. [DOI] [PubMed] [Google Scholar]
- 17.Bechard ME, Chhatwal S, Garcia RE, et al. Application of a colorimetric assay to identify putative ribofuranosylaminobenzene 5′-phosphate synthase genes expressed with activity in Escherichia coli. Biol Proced Online. 2003;5:69–77. doi: 10.1251/bpo48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook J, Russell DW. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 19.Garfin D. One-dimensional gel electrophoresis. Meth Enzymol. 1990;182:425–441. doi: 10.1016/0076-6879(90)82035-z. [DOI] [PubMed] [Google Scholar]
- 20.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 21.Abagyan R, Orry A, Raush E, et al. ICM User Guide 3.7. La Jolla, CA: Molsoft LLC; 2010. [Google Scholar]
- 22.Abagyan R, Totrov M. Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J Mol Biol. 1994;235:983–1002. doi: 10.1006/jmbi.1994.1052. [DOI] [PubMed] [Google Scholar]
- 23.Abagyan RA, Batalov S. Do aligned sequences share the same fold? J Mol Biol. 1997;273:355–368. doi: 10.1006/jmbi.1997.1287. [DOI] [PubMed] [Google Scholar]
- 24.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 25.Metropolis N, Rosenbluth AW, Rosenbluth MN, et al. Equation of state calculations by fast computing machines. J Chem Phys. 1953;21:1087–1092. [Google Scholar]
- 26.Maiorov V, Abagyan R. Energy strain in three-dimensional protein structures. Fold. 1998;3:259–269. doi: 10.1016/S1359-0278(98)00037-6. [DOI] [PubMed] [Google Scholar]
- 27.An J, Totrov M, Abagyan R. Comprehensive identification of ‘druggable’ protein ligand binding sites. Genome Inform. 2004;15:31–41. [PubMed] [Google Scholar]
- 28.Fu Z, Wang M, Potter D, et al. Structure of a binary complex between a mammalian mevalonate kinase and ATP. J Biol Chem. 2002;277:18134–18142. doi: 10.1074/jbc.M200912200. [DOI] [PubMed] [Google Scholar]
- 29.Kleiger G, Eisenberg D. GXXG and GXXA motifs stabilize FAD and NAD(P)-binding Rossmann Folds through Ca-H····O hydrogen bonds and Van der Waals interactions. J Mol Biol. 2002;323:69–76. doi: 10.1016/s0022-2836(02)00885-9. [DOI] [PubMed] [Google Scholar]
- 30.Yang D, Shipman LW, Roessner CA, et al. Structure of the Methanococcus jannaschii mevalonate kinase, a member of the GHMP kinase superfamily. J Biol Chem. 2002;277:9462–9467. doi: 10.1074/jbc.M110787200. [DOI] [PubMed] [Google Scholar]
- 31.Bechard ME. Thesis. University of Florida; 2004. Purification, characterization, and site-directed mutagenesis of a methanogen ribofuranosylaminobenzene 5′-phosphate synthase. [Google Scholar]
- 32.Baca AM, Sirawaraporn R, Turley S, et al. Crystal structure of Mycobacterium tuberculosis 6-hydroxymethyl-7,8-dihydropteroate synthase in complex with pterin monophosphate: new insight into the enzymatic mechanism and sulfa-drug action. J Mol Biol. 2000;302:1193–1212. doi: 10.1006/jmbi.2000.4094. [DOI] [PubMed] [Google Scholar]
- 33.White RH. Biosynthesis of the methanogenic cofactors. Vitam Horm. 2001;61:299–337. doi: 10.1016/s0083-6729(01)61010-0. [DOI] [PubMed] [Google Scholar]
- 34.Smith DR, Doucette-Stamm LA, Deloughery C, et al. Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Budavari S. The Merck Index. 12th ed. New Jersey: Merck & Co; 1996. p. 438. [Google Scholar]
- 36.Achari A, Somers DO, Champness JN, et al. Crystal structure of the anti-bacterial sulfonamide drug target dihydropteroate synthase. Nat Struc Biol. 1997;4:490–497. doi: 10.1038/nsb0697-490. [DOI] [PubMed] [Google Scholar]
- 37.Scapin G, Ozturk DH, Grubmeyer C, et al. The crystal structure of the orotate phosphribosyltransferase complexed with orotate and alpha-D-5-phosphoribosyl-1-pyrophosphate. Biochemistry. 1995;34:10744–10754. doi: 10.1021/bi00034a006. [DOI] [PubMed] [Google Scholar]
- 38.Cao H, Pietrak BL, Grubmeyer C. Quinolinate phosphoribosyltransferases: kinetic mechanism for a type II PRTases. Biochemistry. 2002;41:3520–3528. doi: 10.1021/bi012148g. [DOI] [PubMed] [Google Scholar]
- 39.Eads JC, Scapin G, Xu YM, et al. The crystal-structure of human hypoxanthine guanine phosphoribosyltransferase with bound GMP. Cell. 1994;78:325–334. doi: 10.1016/0092-8674(94)90301-8. [DOI] [PubMed] [Google Scholar]
- 40.Smith JL. Forming and inhibiting PRT active sites. Nat Struc Biol. 1999;6:502–504. doi: 10.1038/9266. [DOI] [PubMed] [Google Scholar]
- 41.Heroux AELW, Ross LJ, Davis RL, et al. Crystal structure of Toxoplasma gondii hypoxanthine-guanine phosphoribosyltransferase with XMP, pyrophosphate, and two Mg2+ ions bound: insights into the catalytic mechanism. Biochemistry. 1999;38:14495–14506. doi: 10.1021/bi990508i. [DOI] [PubMed] [Google Scholar]
- 42.Shi W, Li CM, Tyler PC, et al. The 2.0 A structure of human hypoxanthineguanine phosphoribosyltransferase in complex with a transition-state analog inhibitor. Nat Struc Biol. 1999;6:588–593. doi: 10.1038/9376. [DOI] [PubMed] [Google Scholar]
- 43.Nicholas KB, Nicholas HB, Deerfield DW., II GeneDoc: analysis and visualization of genetic variation. Embnew News. 1997;4:14. [Google Scholar]
- 44.Zhou T, Daugherty M, Grishin NV, et al. Structure and mechanism of homoserine kinase: prototype for the GHMP kinase superfamily. Structure. 2000;8:1247–1257. doi: 10.1016/s0969-2126(00)00533-5. [DOI] [PubMed] [Google Scholar]
- 45.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]