Abstract

To thrive as a global civilization, food production must meet the demands of our ever-growing population. There are more than a billion people on the planet suffering from malnutrition through poor quality or lack of food. Nutrient content of food can be determined by a variety of methods, which have issues such as slow analysis or sample destruction. Near-infrared (NIR) spectroscopy is a long-standing alternative to these methods. In this work, we demonstrated that Raman spectroscopy (RS), another spectroscopic method, can also be used to assess the nutrient content of maize (Zea mays), one of the most widely cultivated grains in the world. Using a handheld Raman spectrometer, we predicted the content of carbohydrates, fibers, carotenoids, and proteins in six different varieties of maize. This analysis requires only a single maize kernel and is fast (1s), portable, noninvasive, and nondestructive. Moreover, we showed that RS in combination with chemometric methods can be used for highly accurate (approximately 90%) spectroscopic typing of maize, which is important for plant breeders and farmers. Finally, we demonstrate that Raman-based approach is as accurate as NIR analysis. These findings suggest that portable Raman systems can be used on combines and grain elevators for autonomous control of grain quality.
Introduction
Food quality has a direct impact on the economic growth of nations and global food security. Therefore, food quality is strictly regulated in many developed countries in the world.1 There are many parameters, such as nutrient content, texture, flavor, and visual appearance, which are used to evaluate the quality of food. However, two of them, the presence of pathogens2,3 and nutrient content, have major impacts on human well-being.
The nutrient content of food is determined by the amount of carbohydrates, proteins, fibers, and oils, as well as by vitamins and minerals present in it. Several colorimetric and chromatographic methods can be used for quantitative assessment of these nutrients. These methods are destructive, time-consuming, and labor-intensive. Also, these assays require sophisticated laboratory equipment, which is often not available in developing countries.4 Development of near-infrared (NIR) analyzers enabled completely noninvasive and nondestructive assessment of nutrient content in agricultural crops.5−9 Some nutritional qualities assayable by NIR include moisture, starch, protein, and many others.10 This method is also capable of determining the nutrient content of single grains of crops such as maize or wheat.11−13
A potential alternative or complementary method to NIR is Raman spectroscopy (RS). The Raman effect is based on the inelastic scattering of photons by sample molecules that are being excited to higher vibrational or rotational states.14 Thus, RS provides information about molecular vibrations and consequently the structure and composition of the analyzed sample. Our group recently demonstrated that RS could be used for confirmatory, noninvasive, and nondestructive detection and identification of fungal diseases in wheat, maize, and sorghum seeds.15,16 We have also shown that RS could detect insects inside intact cowpeas with high statistical accuracy.17 Also, using RS, we were able to diagnose nutrient deficiency and asymptomatic Huanglongbing disease on orange and grapefruit trees.18
In this study, we investigate whether RS could be used for noninvasive, nondestructive, confirmatory assessment of the nutrient content of maize, one of the most widely cultivated cereals in the world.19 The commercial impact of maize exceeds 50 billion U.S. dollars.20 Maize has a wide variety of applications, including as livestock feed, raw material in industry, biofuel, and a human food. We show that Raman-based approach is fast and self-sufficient for the rapid assessment of nutrients in food. We also show that in addition to nutrient content assessment, RS can be used for the identification or typing of maize.
Results and Discussion
We collected more than 600 Raman spectra from six different varieties of maize (Zea mays). Maize varieties that have been chosen for our experiment had a distinctly different phenotypic appearance, Figure 1. Specifically, we have chosen three light color (small yellow (SY), large yellow (LY), and small white (SW)) and three dark color (purple (PP), blue (BL), and red (RD)) maize varieties.
Figure 1.

Photographs of small white (SW), small yellow (SY), extra-large (XL), blue (BL), purple (PP), and red (RD) maize kernels.
Raman-Based Assessment of Nutrient Content
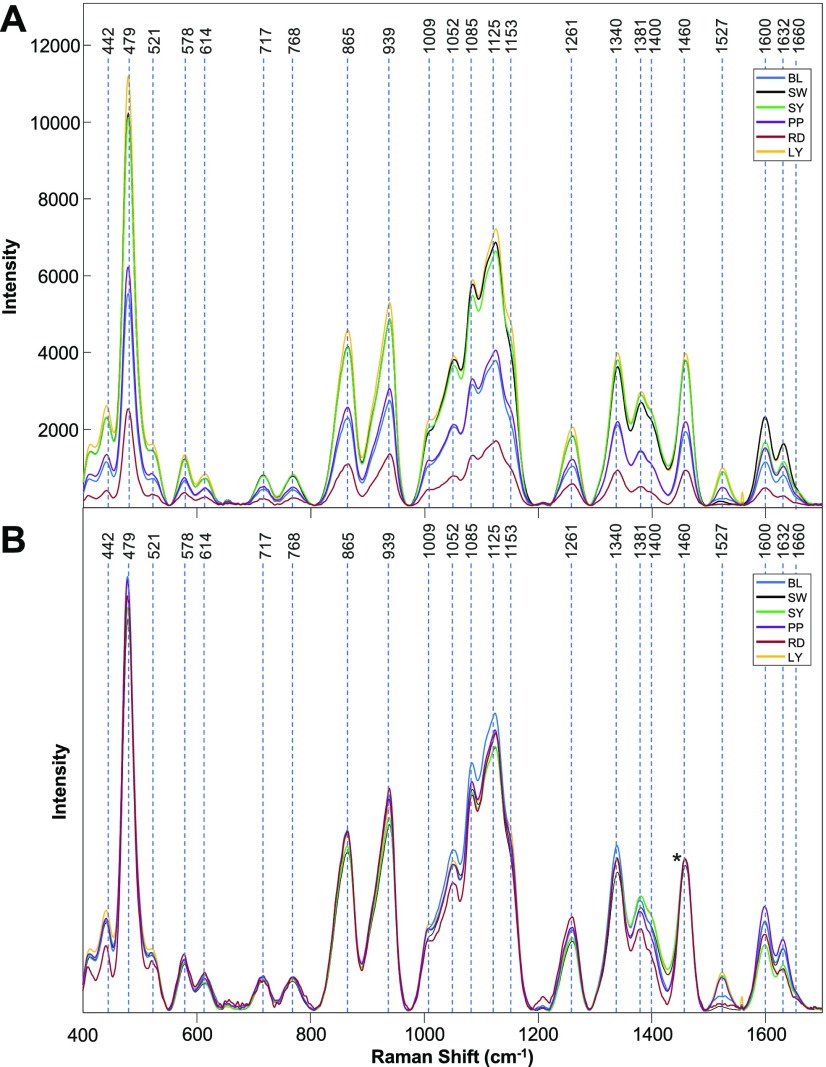
We have found that all six varieties of maize had similar spectral profiles with different intensities of vibrational bands, which can be assigned to carbohydrates, carotenoids, fibers (lignin), and proteins, Figure 2A and Table 1. Interestingly, the spectra collected from colored maize varieties (BL, PP, RD) exhibited a much lower intensity of these vibrational bands compared to the spectra acquired from yellow or pale color (SY, LY, SW) kernels. One can imagine that dark maize kernels absorb more and consequently scatter less light compared to the yellow or pale color kernels. Since RS is based on inelastic light scattering, dark color maize varieties should produce less intense spectra (under the same experimental conditions) compared to the light color maize varieties. Therefore, observed variations in spectral intensities likely originate from different light absorption and scattering properties of these maize kernels.
Figure 2.
Baseline-corrected (A) and normalized (B) Raman spectra of BL, SW, SY, PP, RD, and LY maize kernels. The 1458 cm–1 peak, which was used for spectral normalization, is indicated by an asterisk (*).
Table 1. Vibrational Bands and Their Assignments for Maize Kernels.
| band | vibrational mode | assignment |
|---|---|---|
| 1660 | C=O stretching (amide I) | proteins14 |
| 1632 | C=C–C (ring) | lignin24 |
| 1600 | ν(CC)ring + σ(CH) | lignin23 |
| 1527 | –C=C– (in-plane) | carotene25 |
| 1460 | δ(CH) + δ(CH2) + δ(C–O–H) CH, CH2, and COH deformations. | carbohydrates26 |
| 1400 | δ(C–C–H) | carbohydrates26 |
| 1381 | δ(C–O–H) - coupling of the CCH and COH deformation modes | carbohydrates26 |
| 1340 | ν(C–O); δ(C–O–H) | carbohydrates26 |
| 1261 | δ(C–C–H) + δ(O–C–H) + δ(C–O–H) | carbohydrates26,27 |
| 1153 | C–C stretching; ν(COC), ν(CC) in glycosidic linkage, asymmetric ring breathing | carotenoids.28 carbohydrates29 |
| 1125 | ν(C–O) + ν(C–C) + δ(C–O–H) | carbohydrates26 |
| 1085 | ν(C–O) + ν(C–C) + δ(C–O–H) | carbohydrates26 |
| 1052 | ν(C–O) + ν(C–C) + δ(C–O–H) | carbohydrates26 |
| 1009 | phenylalanine ring stretching mode | proteins14 |
| 939 | δ(C–O–C) + δ(C–O–H) + ν(C–O) α-1,4 glycosidic linkages | carbohydrates26 |
| 865 | δ(C–C–H) + δ(C–O–C) glycosidic bond; anomeric region | carbohydrates26 |
| 768 | δ(C–C–O) | carbohydrates26 |
| 717 | δ(C–C–O) related to glycosidic ring skeletal deformations | carbohydrates26 |
| 576 | δ(C–C–O) + τ(C–O) | carbohydrates26 |
| 614 | δ(C–C–C) | carbohydrates26 |
| 521 | S–S gauche-gauche-trans | protein30 |
| 479 | CCO and CCC deformations; related to glycosidic ring skeletal deformations δ(C–C–C) + τ(C–O) Scissoring of C–C–C and out-of-plane bending of C–O | carbohydrates26 |
| 442 | skeletal modes of pyranose ring | carbohydrates26 |
This observation indicates that raw spectra collected from colored maize kernels cannot be used for direct assessment of their nutrient content. For instance, the intensity of the 479 cm–1 band, which can be used to estimate the carbohydrate content in the maize, appears lower in the spectra collected from PP maize compared to that in LY or SY maize. However, this low intensity of this band originates from the poor scattering properties of PP maize kernels relative to those of LY or SY maize. This problem can be solved by spectral normalization. However, normalizing spectra without an internal standard is a challenging task. Normalization on one particular band that can be assigned to the specific class of molecules, such as carbohydrates, may not be appropriate. Such normalization would bias spectral interpretation in regard to the nutrient content of that class of molecules. Because the 1458 cm–1 band, assigned to CH2 vibrations, cannot be associated with any specific class of biomolecule, we have chosen to normalize our spectra to 1458 cm–1.
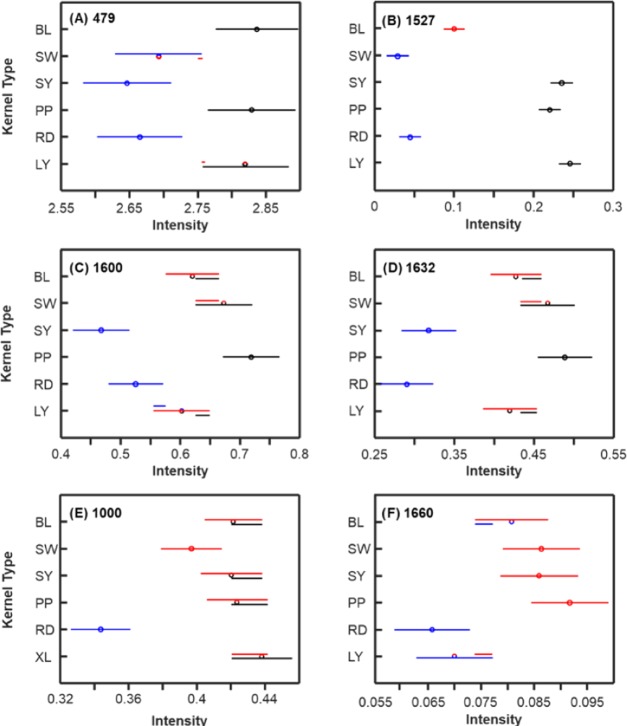
This unbiased spectral normalization can be used to access the nutrient content of maize varieties. The analysis of variance (ANOVA) of the 479 cm–1 band, which can be assigned to carbohydrates such as monomeric sugars and starch, revealed two statistically significant groups of maize (Figure 3). We found that the SY and RD carbohydrate contents were significantly different from those of BL and PP. At the same time, SW and LY were not significantly different from the first and second groups of maize, respectively.
Figure 3.
Means (circles) and confidence intervals for the intensities of the maize kernel spectra, normalized to 1458 cm–1, at the indicated Raman shift. Colors indicate significantly different groups. Multiple colors indicate a member of a group that has overlap between two separate groups. Each of (A)–(F) corresponds to a different selected Raman shift discussed in the text.
Carotenoids exhibit bands around 1530 cm–1, which can be assigned to in-plane −C=C– vibrations. ANOVA revealed three statistically significant groups of maize based on the intensities of these carotenoid bands. SW and RD had the lowest carotenoid content and could be assigned to the first group. The carotenoid content of this group is statistically different from that of BL. Finally, SY, PP, and LY belong to the third group with the highest carotenoid content. From Figure 3B, it is apparent that the darkness of a kernel is not correlated with the intensity of the carotenoid band. Our results support a previous hypothesis that the color of maize kernels is determined by anthocyanins.21,22
Fibers are polyphenolic molecular assemblies that have two distinct vibrational bands in Raman spectra.23,24 The band at 1600 cm–1 can be assigned to C–C ring stretching and symmetric C–H vibration.23 The 1632 cm–1 band originates from C=C aromatic ring vibration.24 ANOVA revealed at least two groups of maize based on the fiber content. Specifically, SY and RD can be assigned to the first group with the lowest fiber content. PP showed the highest fiber content that is statistically significant from all other groups. Based on the intensity of 1600 cm–1 band, the fiber content in BL, SW, and LY is higher compared to that in SY and RD, but lower than in PP. However, it is not statistically different from SY, RD, and PP. At the same time, the analysis of the intensity of 1632 cm–1 band showed that the fiber content of LY is significantly different from both first (SY and RD) and second (PP) groups, whereas BL and SW were not significantly different from PP.
Proteins exhibit a carbonyl vibration of the peptide bond at 1640–1670 cm–1, known as the amide I band, as well as a band at 1000 cm–1, which can be assigned to phenylalanine.14 Both of these bands were observed in the Raman spectra collected from maize kernels. At the same time, the band around 1650 cm–1 could be attributed to C=C bond of unsaturated fatty acids, whereas 1000 cm–1 could originate from carotenoid vibrations.25,31 Our results reveal a very similar pattern for both 1000 and 1660 cm–1 bands indicating that these bands represent the same chemical structure. Since the ANOVA pattern of both 1000 and 1660 cm–1 bands is distinctly different from the ANOVA pattern that was observed from 1527 cm–1 band, which can be unambiguously assigned to carotenoids, we can conclude that both 1000 and 1660 cm–1 bands in the Raman spectra of maize can be used to reveal the protein content. We found that RD maize has the lowest protein content, whereas BL, SW, SY, and PP have statistically higher protein content.
These results clearly demonstrate that RS can be used for fast (1s spectral acquisition), noninvasive, and nondestructive assessment of carbohydrates, proteins, fibers, and carotenoids in the intact grain. When coupled to ANOVA, RS provides a highly accurate content of these four classes of nutrients. Also, handheld spectrometer enables the performance of this analysis directly in the field with only one kernel required for quantitative elucidation of the grain nutrient content.
The question to ask is how this information can be related to nutrient content analysis made by already established techniques. To answer this question, we performed the analysis of the same grain samples using NIR and determined protein and carbohydrate contents using the Dumas combustion method32 and megazyme total starch content assay (subsequently megazyme assay),33 respectively (Tables 2 and 3).
Table 2. Results of NIR Analysis of Nutrient Content of Six Different Maize Varieties.
| starch (%) | protein (%) | moisture (%) | oil (%) | ash (%) | |
|---|---|---|---|---|---|
| BL | 63.1 | 10.0 | 13.4 | 4.6 | 0.8 |
| SW | 62.7 | 10.7 | 13.2 | 3.4 | 0.8 |
| SY | 63.1 | 11.9 | 13.0 | 4.0 | 0.9 |
| PP | 62.2 | 11.4 | 13.5 | 3.5 | 0.8 |
| RD | 60.2 | 12.0 | 14.1 | 3.7 | 0.9 |
| LY | 62.4 | 10.6 | 13.1 | 3.8 | 0.9 |
Table 3. Results of Megazyme Assay for Total Starch and Dumas Combustion Analysis for Six Different Maize Varieties.
| starch (%) | protein (%) | |
|---|---|---|
| BL | 58.0 | 10.4 |
| SW | 54.6 | 11.7 |
| SY | 59.3 | 11.6 |
| PP | 56.6 | 11.3 |
| RD | 54.8 | 11.9 |
| LY | 58.4 | 10.4 |
According to NIR analysis, the amount of starch was found to vary in the maize varieties from 60.2 to 63.1%. Considering the fact that the accuracy of such analysis is ±5%,34 NIR analysis suggests that there is no significant difference in starch content among the maize varieties. According to the results of megazyme assay, starch content varies from 54.6 to 59.3% (±3%).33 These results can also be used to reference Raman readings to the starch content. Thus, 2.55–2.9 scale in the Raman can be assigned to 57% of starch by the dry weight. Although RS reveals two statistically different groups of maize based on their starch content (Figure 3), the lack of accuracy expressed by already established methods does not allow us to prove that RS is more accurate in the prediction of starch content than NIR and megazyme assay.
Our NIR results indicated that the amount of protein in the analyzed samples varied from 10.0 to 12.0%. It is expected that reported values have a precision ranging from 3 to 6%.32 Dumas combustion analysis revealed similar values of protein content in the analyzed maize kernels (10.4–11.9%). However, the accuracy of this method ranges between 2 and 4%.32 These results suggest that 0.05–0.1 scale in Raman corresponds to 11.2% of protein. Thus, the same as in the case of starch, the lack of accuracy expressed by both NIR and Dumas assays does not allow us to prove that RS is more accurate in the prediction of protein content.
Raman-Based Typing of Maize Varieties
We conducted partial least-squares discriminant analysis (PLS-DA)35 to demonstrate that RS can be used for typing of these maize varieties. The model was built using all 613 spectra with the mean offset removed at each wavenumber and cross-validated with the same data partitioned by the venetian blinds method.36 This model contained 19 latent variables (LVs) and was used to generate a misclassification table (Table 4). The first three LVs explain 94.6, 1.7, and 0.7% of the variation, respectively. The misclassification table reports the true positive rate (accuracy) of the model; green cells in the table indicate members of a class that were correctly assigned to their class during cross-validation. Overall, we can see that the model performs with an 89% classification accuracy at minimum.
Table 4. Misclassification Table of Cross-Validation for the PLS-DA Model.
| members | correct (%) | BL | SW | SY | PP | RD | LY | |
|---|---|---|---|---|---|---|---|---|
| BL | 113 | 98 | 111 | 0 | 0 | 0 | 2 | 0 |
| SW | 100 | 98 | 0 | 98 | 2 | 0 | 0 | 0 |
| SY | 98 | 94 | 0 | 0 | 92 | 1 | 2 | 3 |
| PP | 100 | 89 | 5 | 2 | 0 | 89 | 1 | 3 |
| RD | 100 | 99 | 0 | 0 | 1 | 0 | 99 | 0 |
| LY | 102 | 94 | 0 | 0 | 5 | 1 | 0 | 96 |
Conclusions
Our findings indicate that RS in combination with advanced statistical analysis can be used to predict the nutrient content of carbohydrates, carotenoids, fibers, and protein in intact maize kernels. We have also shown that RS is capable of highly accurate typing of maize grain. Thus, this study shows that RS is a highly efficient multifunctional method for the analysis of grain.
Materials and Methods
Maize
Six different varieties of maize (Zea mays) were purchased from Amish Country Popcorn (Berne, Indiana) and used as received.
Raman Spectroscopy
Raman spectra were collected with a handheld Resolve Agilent spectrometer equipped with an 831 nm laser and a spectral resolution of 15 cm–1. The following experimental parameters were used for all collected spectra: 1s acquisition time, 495 mW power, surface scanning mode, and baseline spectral subtraction by device software. Spectra shown in the manuscript are baseline-corrected by the instrument software without smoothing. One spectrum was collected per one maize kernel.
Spectral Processing
Spectral processing (described below) and averaging were conducted using PLS_Toolbox 8.6.2 (Eigenvector Research, Inc., Manson, WA).
Statistical Analysis
Raman spectra were imported into Matlab (Mathworks) and assigned a class based on their visual phenotype (color). For ANOVA, spectra were normalized to the 1458 cm–1 band. For PLS-DA, the mean offset was removed at each wavenumber before analysis.
NIR Analysis
PerkinElmer DA 7250 NIR analyzer was used to determine the amount of protein, starch, oils, moisture, and ash in six varieties of maize. For each maize variety, ∼30 g of material was submitted to the analyzer.
Chemical Content Analysis
Phenolic content was measured according to the Folin–Ciocalteu method.37 Total protein concentration was determined by combustion using LECO instrument and a nitrogen-to-protein conversion factor of 5.7.37 Total starch content was found using the megazyme assay kit, based on the RTS-NaOH procedure from the AACC 76.13 and AOAC 999.11 methods (K-TSTA; Megazyme, Bray, Ireland).
Acknowledgments
The authors are grateful to Joseph M. Awika and Audrey Girard from the Department of Soil and Crop Sciences of Texas A&M University for their help with NIR analysis of maize varieties. This study was supported by funds from Texas A&M AgriLife Research, Texas A&M University Governor’s University Research Initiative (GURI) grant program of 12-2016/M1700437.
Author Contributions
§ M.K. and C.F. contributed equally to this work.
The authors declare no competing financial interest.
References
- Strengthening Sector Policies for Better Food Security and Nutrition Results. In Food Systems for Healthy Diets; Food and Agriculture Organization of the United Nations; FAO: Rome, Italy, 2018. [Google Scholar]
- Williams J. H.; Phillips T. D.; Jolly P. E.; Stiles J. K.; Jolly C. M.; Aggarwal D. Human Aflatoxicosis in developing countries: a review of toxicology, exposure, potential health consequences, and interventions. Am. J. Clin. Nutr. 2004, 80, 1106–1122. 10.1093/ajcn/80.5.1106. [DOI] [PubMed] [Google Scholar]
- Norred W. P.; Voss K. A. Toxicity and Role of Fumonisins in Animal Diseases and Human Esophageal Cancer. J. Food Prot. 1994, 57, 522–527. 10.4315/0362-028X-57.6.522. [DOI] [PubMed] [Google Scholar]
- Damodaran S.; Parkin K. L.. Fennema’s Food Chemistry, 5th ed.; CRC Press, 2017 [Google Scholar]
- Osborne B. G.; Fearn T.; Hindle P. H.; Hindle P. T.. Practical NIR Spectroscopy with Applications in Food and Beverage Analysis; Longman Scientific & Technical, 1993 [Google Scholar]
- Kim Y.; Singh M.; Kays S. E. Near-infrared spectroscopic analysis of macronutrients and energy in homogenized meals. Food Chem. 2007, 105, 1248–1255. 10.1016/j.foodchem.2007.03.011. [DOI] [Google Scholar]
- Stubbs T. L.; Kennedy A. C.; Fortuna A.-M. Using NIRS To Predict Fiber and Nutrient Content of Dryland Cereal Cultivars. J. Agric. Food Chem. 2010, 58, 398–403. 10.1021/jf9025844. [DOI] [PubMed] [Google Scholar]
- Berardo N.; Brenna O. V.; Amato A.; Valoti P.; Pisacane V.; Motto M. Carotenoids concentration among maize genotypes measured by near infrared reflectance spectroscopy (NIRS). Innovative Food Sci. Emerging Technol. 2004, 5, 393–398. 10.1016/j.ifset.2004.03.001. [DOI] [Google Scholar]
- Baranska M.; Schütze W.; Schulz H. Determination of Lycopene and β-Carotene Content in Tomato Fruits and Related Products: Comparison of FT-Raman, ATR-IR, and NIR Spectroscopy. Anal. Chem. 2006, 78, 8456–8461. 10.1021/ac061220j. [DOI] [PubMed] [Google Scholar]
- Cen H.; He Y. Theory and application of near infrared reflectance spectroscopy in determination of food quality. Trends Food Sci. Technol. 2007, 18, 72–83. 10.1016/j.tifs.2006.09.003. [DOI] [Google Scholar]
- Caporaso N.; Whitworth M.; Fisk I. Application of calibrations to hyperspectral images of food grains: example of wheat falling number. J. Spectral Imaging 2017, 10.1255/jsi.2017.a4. [DOI] [Google Scholar]
- Caporaso N.; Whitworth M. B.; Fisk I. D. Protein content prediction in single wheat kernels using hyperspectral imaging. Food Chem. 2018, 240, 32–42. 10.1016/j.foodchem.2017.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendin K.; Manley M.; Baeten V.; Fernández Pierna J. A.; Williams P. J. Near Infrared Hyperspectral Imaging for White Maize Classification According to Grading Regulations. Food Anal. Methods 2019, 12, 1612–1624. 10.1007/s12161-019-01464-0. [DOI] [Google Scholar]
- Kurouski D.; Van Duyne R. P.; Lednev I. K. Exploring the structure and formation mechanism of amyloid fibrils by Raman spectroscopy: a review. Analyst 2015, 140, 4967–4980. 10.1039/C5AN00342C. [DOI] [PubMed] [Google Scholar]
- Farber C.; Kurouski D. Detection and Identification of Plant Pathogens on Maize Kernels with a Hand-Held Raman Spectrometer. Anal. Chem. 2018, 90, 3009–3012. 10.1021/acs.analchem.8b00222. [DOI] [PubMed] [Google Scholar]
- Egging V.; Nguyen J.; Kurouski D. Detection and Identification of Fungal Infections in Intact Wheat and Sorghum Grain Using a Hand-Held Raman Spectrometer. Anal. Chem. 2018, 90, 8616–8621. 10.1021/acs.analchem.8b01863. [DOI] [PubMed] [Google Scholar]
- Sanchez L.; Farber C.; Lei J.; Zhu-Salzman K.; Kurouski D. Noninvasive and Nondestructive Detection of Cowpea Bruchid within Cowpea Seeds with a Hand-Held Raman Spectrometer. Anal. Chem. 2019, 91, 1733–1737. 10.1021/acs.analchem.8b05555. [DOI] [PubMed] [Google Scholar]
- Sanchez L.; Pant S.; Xing Z.; Mandadi K.; Kurouski D. Rapid and noninvasive diagnostics of Huanglongbing and nutrient deficits on citrus trees with a handheld Raman spectrometer. Anal. Bioanal. Chem. 2019, 411, 3125–3133. 10.1007/s00216-019-01776-4. [DOI] [PubMed] [Google Scholar]
- National Corn Growers Association. World of Corn 2017, 2017.
- International Grain Council. Grain Market Report, 2017.
- Cevallos-Casals B. A.; Cisneros-Zevallos L. Stoichiometric and kinetic studies of phenolic antioxidants from Andean purple corn and red-fleshed sweetpotato. J. Agric. Food Chem. 2003, 51, 3313–3319. 10.1021/jf034109c. [DOI] [PubMed] [Google Scholar]
- Khoo H. E.; Azlan A.; Tang S. T.; Lim S. M. Anthocyanidins and anthocyanins: colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food Nutr. Res. 2017, 61, 1361779 10.1080/16546628.2017.1361779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang L.; Wang K.; Li X.; Zou B. High Pressure Structural Investigation of Benzoic Acid: Raman Spectroscopy and X-ray Diffraction. J. Phys. Chem. C 2016, 120, 14758–14766. 10.1021/acs.jpcc.6b05001. [DOI] [Google Scholar]
- Pompeu D. R.; Larondelle Y.; Rogez H.; Abbas O.; Pierna J. A. F.; Baeten V. Characterization and discrimination of phenolic compounds using Fourier transformation Raman spectroscopy and chemometric tools. Biotechnol. Agron. Soc. Environ. 2017, 22, 13–28. 10.25518/1780-4507.16270. [DOI] [Google Scholar]
- Adar F. Carotenoids - Their Resonance Raman Spectra and How They Can Be Helpful in Characterizing a Number of Biological Systems. Spectroscopy 2017, 32, 12–20. [Google Scholar]
- Almeida M. R.; Alves R. S.; Nascimbem L. B.; Stephani R.; Poppi R. J.; de Oliveira L. F. Determination of amylose content in starch using Raman spectroscopy and multivariate calibration analysis. Anal. Bioanal. Chem. 2010, 397, 2693–2701. 10.1007/s00216-010-3566-2. [DOI] [PubMed] [Google Scholar]
- Cael J. J.; Koenig J. L.; Blackwell J. Infrared and raman spectroscopy of carbohydrates. Part 4: Normal coordinate analysis of V-amylose. Biopolymers 1975, 14, 1885–1903. 10.1002/bip.1975.360140909. [DOI] [Google Scholar]
- Jehlička J.; Edwards H. G. M.; Osterrothová K.; Novotná J.; Nedbalová L.; Kopecký J.; Němec I.; Oren A. Potential and limits of Raman spectroscopy for carotenoid detection in microorganisms: implications for astrobiology. Philos. Trans. R. Soc., A 2014, 372, 20140199 10.1098/rsta.2014.0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiercigroch E.; Szafraniec E.; Czamara K.; Pacia M. Z.; Majzner K.; Kochan K.; Kaczor A.; Baranska M.; Malek K. Raman and infrared spectroscopy of carbohydrates: A review. Spectrochim. Acta, Part A 2017, 185, 317–335. 10.1016/j.saa.2017.05.045. [DOI] [PubMed] [Google Scholar]
- Rygula A.; Majzner K.; Marzec K. M.; Kaczor A.; Pilarczyk M.; Baranska M. Raman spectroscopy of proteins: a review. J. Raman Spectrosc. 2013, 44, 1061–1076. 10.1002/jrs.4335. [DOI] [Google Scholar]
- Jamieson L. E.; Li A.; Faulds K.; Graham D. Ratiometric analysis using Raman spectroscopy as a powerful predictor of structural properties of fatty acids. R. Soc. Open Sci. 2018, 5, 181483 10.1098/rsos.181483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaljev Ž.; Jakšić S.; Prica N. B.; Ćupić ŽN.; Baloš M. Ž. Comparison of the Kjeldahl method, Dumas method and NIR method for total nitrogen determination in meat and meat products. J. Agroaliment. Processes Technol. 2015, 21, 365–370. [Google Scholar]
- Zhu T.; Jackson D. S.; Wehling R. L.; Geera B. Comparison of Amylose Determination Methods and the Development of a Dual Wavelength Iodine Binding Technique. Cereal Chem. 2008, 85, 51–58. 10.1094/CCHEM-85-1-0051. [DOI] [Google Scholar]
- Pojić M.; Mastilović J.; Majcen N. In Infrared Spectroscopy Life and Biomedical Sciences;Theophile T., Ed.; InTech: Rijeka, Croatia, 2012; pp 167–184. [Google Scholar]
- Eriksson L.; Byrne T.; Johansson E.; Trygg J.; Vikstrom C.. Multi- and Megavariate Data Analysis Basic Principles and Applications, 3rd revised ed.; Umetrics: Malmö, Sweden, 2013; Vol. 1. [Google Scholar]
- Ballabio D.; Consonni V. Classification tools in chemistry. Part 1: linear models. PLS-DA. Anal. Methods 2013, 5, 3790–3798. 10.1039/c3ay40582f. [DOI] [Google Scholar]
- Dunn K. L.; Yang L.; Girard A.; Bean S.; Awika J. M. Interaction of Sorghum Tannins with Wheat Proteins and Effect on in Vitro Starch and Protein Digestibility in a Baked Product Matrix. J. Agric. Food Chem. 2015, 63, 1234–1241. 10.1021/jf504112z. [DOI] [PubMed] [Google Scholar]