Abstract
Cutaneous squamous cell carcinoma (cSCC) is a common malignancy initiated by keratinocytes of the epidermis, which are able to invade the dermis and its periphery. Although most patients with cSCC present with curable localized tumors, recurrence, metastasis and mortality occasionally occur. In the present study, nicotinamide N-methyltransferase (NNMT) was identified as an upregulated protein in the SCC12 cell line, which has high invasive potential compared with the SCC13 cell line. The effects of NNMT knockdown on proliferation, migration and invasion were investigated using SCC cells. shRNA-mediated downregulation of NNMT expression levels inhibited the proliferation and density-dependent growth of SCC12 cells. In addition, the results of a cell motility assay showed that the migration and invasion of SCC cells were markedly decreased in NNMT-knockdown cells. The assessment of epithelial-mesenchymal transition (EMT)-associated gene expression using PCR array analysis revealed that high NNMT expression levels were accompanied by high expression levels of EMT-associated genes, and that NNMT knockdown effectively suppressed the expression of matrix metalloproteinase 9, osteopontin, versican core protein and zinc finger protein SNAI2 in SCC12 cells. These results revealed that the upregulation of NNMT induced cellular invasion via EMT-related gene expression in SCC cells.
Keywords: nicotinamide N-methyltransferase, squamous cell carcinoma, epithelial-mesenchymal transition, invasion, metastasis
Introduction
Cutaneous squamous cell carcinoma (cSCC) is one of the primary types of skin cancer characterized by the malignant proliferation of keratinizing cells of the epidermis (1). Worldwide, cSCC is the second most common non-melanoma skin carcinoma (NMSC), following basal cell carcinoma (BCC) (2). Despite early stage diagnosis in most cases, cSCC tumors account for the majority of NMSC-associated mortalities (3), and ~5% of cSCC tumors metastasize to regional lymph nodes or distant sites (4). Although there has been considerable progress in understanding the various etiologies of cSCC, the molecular mechanisms of invasiveness have not been clarified.
Nicotinamide N-methyltransferase (NNMT) is an S-adenosyl-L-methionine-dependent cytoplasmic enzyme that catalyzes the N-methylation of nicotinamide, pyridines and their structural analogs (5,6). NNMT is predominantly expressed in the liver, where its activity leads to the metabolism of drugs and xenobiotic compounds (5,6). Elevated levels of NNMT expression have been identified in various tumors, suggesting that NNMT serves an important role in the progression of these cancer types (7–16). Moreover, increased NNMT expression levels have been detected in bodily fluids including the serum (14,17), saliva (18) and urine (19). These findings suggest that NNMT may act as a potential non-invasive cancer biomarker. Previous studies have also indicated that NNMT dysregulation may affect the proliferative, migratory, invasive and differentiation profiles of various cancer types (16,20,21). Notably, the upregulation of NNMT in cancer has been linked to poor prognosis and treatment resistance (10,15,22,23). However, the expression profile and biological functions of NNMT in cSCC are yet to be elucidated.
Enhanced metastasis and rapid tumor growth are associated with poor prognosis in cSCC (24,25). Epithelial-mesenchymal transition (EMT) is a transcriptional process that is activated in embryonic development and wound healing, and contributes to cancer initiation and progression, particularly during metastasis (26,27). During EMT, epithelial cells lose their polarity and intercellular adhesion properties, the cytoskeleton is reorganized, and mesenchymal characteristics are gained, including migratory activity. Invasion and metastasis are among the principal characteristics of cancer (28), and cancerous cells routinely acquire an invasive phenotype following EMT. Various signaling pathways have been found to regulate EMT, enhancing cell motility and invasiveness (29). Considering that it is necessary for skin healing and can also contribute to tumor invasion and metastasis, an understanding of the underlying mechanisms of EMT is crucial.
The present study aimed to identify differentially expressed proteins in an invasive, compared with those in a non-invasive cSCC cell line, and to elucidate the mechanism of invasiveness associated with specific genes, particularly NNMT. NNMT was revealed to be upregulated in SCC12 cells, and NNMT-targeted shRNA suppressed cell growth and invasion via the downregulation of EMT-related genes.
Materials and methods
Cell culture
The human skin squamous carcinoma cell lines SCC12 and SCC13 were kindly donated by Dr James Rheinwald (Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA). The cells were cultured in growth medium containing a 3:1 ratio of Dulbecco's modified Eagle's medium (DMEM) and Ham's F-12 Nutrient Mixture (Gibco; Thermo Fisher Scientific, Inc.), supplemented with 10% fetal bovine serum (FBS), 5 µg/ml insulin, 0.5 µg/ml hydrocortisone and 10 ng/ml epidermal growth factor. The cells were maintained at 37°C (5% CO2) in a humidified incubator.
Protein extraction and two-dimensional gel electrophoresis (2-DE)
Cells were lysed on ice in lysis buffer (8 M urea, 4% CHAPS, 40 mM Tris-base and 100 mM dithiothreitol) supplemented with a protease inhibitor cocktail (Sigma-Aldrich; Merck KGaA). The lysate was centrifuged at 13,000 × g for 10 min at 4°C to remove insoluble cell debris, followed by trichloroacetic acid precipitation. The precipitated proteins were resuspended in lysis buffer containing 2% (w/v) ampholyte and quantified using a Bradford protein assay kit (Bio-Rad Laboratories, Inc.). Equal amounts of protein (30 µg) were subjected to first-dimension isoelectric focusing (pH 4.0–7.0) and second-dimension gradient sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 7.5–17.5%).
Gel visualization and analysis
After 2-DE, the gels were fixed in methanol:acetic acid:water (50:12:38) for 1.5 h, pretreated with 0.02% Na2S2O3 for 1 min and washed twice with deionized water. The gels were soaked in a solution containing 0.2% silver nitrate and 0.075% (v/v) formaldehyde, and incubated in a developing solution [2% sodium bicarbonate, 0.0004% sodium sulfoxide and 0.06% (v/v) formaldehyde]. The silver-stained gels were scanned with a high-resolution scanner (GS-800 Calibrated Imaging Densitometer; Bio-Rad Laboratories, Inc.) and all images were analyzed using PDQuest™ software (version 8.0; Bio-Rad Laboratories, Inc.).
Protein identification using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS)
Protein spots of interest were excised from the 2-DE gels and digested with trypsin (Promega Corp.). Subsequently, the peptides were extracted using ammonium hydrogen carbonate buffer with 5% formic acid. The extracted peptides were vacuum-dried and reconstituted in a solution containing water, acetonitrile and trifluoroacetic acid (93:5:2 by volume). The protein digest spectrum was acquired in reflector mode and positive ion mode using the Voyager-DE STR MALDI-TOF mass spectrometer (Applied Biosystems; Thermo Fisher Scientific, Inc.). The following settings were used: acceleration voltage 20 kV, guide wire voltage 0.01%, grid voltage 76% and delay time 150 nsec. Protein identification was performed using the search program MS-FIT (http://prospector.ucsf.edu/prospector/mshome.htm).
Short hairpin RNA (shRNA)-mediated NNMT gene silencing
The shRNA target sequence for NNMT (5′-CATGATTGGTGAGCAGAAG-3′) was cloned into the pENTR™/U6 plasmid (Invitrogen; Thermo Fisher Scientific, Inc.). The cloned pENTR-NNMT plasmid was used to generate an adenovirus backbone by homologous recombination with pAd/BLOCK-iT™-DEST. The target construct, subjected to antibiotic-resistance screening and PCR confirmation, was termed pAd-shNNMT, and the adenoviral plasmid containing the GFP sequence was termed pAd-GFP. After endonuclease digestion with PacI, linearized pAd-shNNMT and pAd-GFP were transfected into 293A cells using Lipofectamine® 3000. Adenovirus was collected from the culture supernatants and showed a cytopathic effect of >80%. Adenovirus amplification was performed by infecting 293A cells with the harvested adenoviral stocks. To confirm NNMT gene-silencing, SCC12 cells were seeded into 6-well plates (1×105 cells per well). At 70–80% confluence (24 h), the cells were infected with Ad-shNNMT or Ad-GFP adenovirus at a multiplicity of infection (MOI) of 20, and harvested 72 h post-infection for protein extraction and immunoblotting.
Cell viability
The effect of NNMT knockdown on the viability of SCC12 cells was determined using the RealTime-Glo™ MT Cell Viability Assay (Promega Corp.) in an opaque-walled assay plate, according to the manufacturer's instructions but with minor modifications. Briefly, SCC12 cells were seeded into 6-well plates (1×105 cells per well) and incubated for 24 h at 37°C. The cells were then infected with Ad-GFP or Ad-shNNMT adenovirus at an MOI of 20. At 24 h post-infection, the growth medium was replaced with the aforementioned normal culture medium. After an additional 48 h incubation, the infected cells were detached with trypsin/EDTA and replated in 96-well plates (5×102 cells per well), and viability was assessed using the RealTime-Glo™ MT Cell Viability Assay (Promega Corp.). To continuously monitor viability, the pro-substrate and luciferase were added at the time of cell replating. The luminescence intensity at the desired time points was measured using the Glomax Multi Detection System (Promega Corp.), and each sample was analyzed at least in triplicate.
Colony formation assay
Ad-GFP and Ad-shNNMT adenovirus-infected SCC12 cells were seeded into 6-well plates in triplicate (5×102 cells per well). The culture plates were incubated at 37°C for 7 days in the aforementioned growth medium, which was replaced every 3 days. The resulting colonies were stained with crystal violet (1%, w/v; Sigma-Aldrich; Merck KGaA) for 2 h at room temperature and counted under a microscope (Eclipse 80Ti, Nikon Corporation).
Gap closure assay
Migration assays were performed using wound assay chambers (Ibidi GmbH). Ad-GFP and Ad-shNNMT adenovirus-infected SCC12 cells were trypsinized and resuspended in growth medium containing 10% FBS. The cells were seeded (4×104 cells per well) in the insert and allowed to adhere overnight. On the following day, the inserts were removed and light microscopy images were acquired (three for each sample). The cells were then cultured in serum-free cuture medium for the indicated time, and images were subsequently acquired using an inverted microscope equipped with an image capture system (Nikon Corporation). The images were analyzed using automated image analysis software (Nikon NIS Elements version 5.1 software; NIS-Elements Advanced Research).
Invasion assay
Cells infected with Ad-GFP or Ad-shNNMT adenovirus (1×105 cells per well) were seeded into the upper chamber of Matrigel-coated (BD Biosciences) Transwell inserts with 8-µm pore polycarbonate filters (Costar; Corning, Inc.). The lower chamber was filled with DMEM/F12 containing 10% FBS, and the inserts were incubated at 37°C for 24 h. Invaded cells were fixed in 4% paraformaldehyde and stained with DAPI. The cells were counted in five random fields and images were obtained using a fluorescence microscope (magnification, ×100).
Western blot analysis
Cell extracts were prepared using RIPA lysis buffer (Santa Cruz Biotechnology, Inc.) supplemented with protease and phosphatase inhibitor cocktails (Thermo Fisher Scientific, Inc.). The cells were sonicated for 2 min and centrifuged at 12,000 × g for 10 min at 4°C to remove insoluble cell debris, and the protein concentrations were determined using a bichinconinic acid protein assay kit (Pierce; Thermo Fisher Scientific, Inc.). A total of 30 µg protein per well were separated by 10% SDS-PAGE, and transferred to a nitrocellulose membrane (EMD Millipore). After blocking and washing following a standard protocol, the target proteins were detected by incubation with primary antibodies against NNMT (dilution 1:1,000; cat. no. AP13775C; Abgent, Inc.), E-cadherin (dilution 1:1,000; cat. no. sc-21791; Santa Cruz Biotechnology, Inc.), vimentin (dilution 1:1,000; cat. no. sc-6601; Santa Cruz Biotechnology), zinc finger E-box-binding homeobox 1 (ZEB1; dilution 1:1,000; cat. no. NBP1-05987; Novus biologicals, Ltd.), Snail (dilution 1:1,000; cat. no. NBP1-80022; Novus Biologicals, Ltd.), Slug (dilution 1:1,000; cat. no. ab128485; Abcam), MMP9 (dilution 1:1,000; cat. no. ab104686; Abcam), SPP1 (dilution 1:500; cat. no. sc-73631; Santa Cruz Biotechnology, Inc.) and β-actin (dilution 1:5,000; cat. no. A5441; Sigma-Aldrich; Merck KGaA). Protein bands were visualized using an enhanced chemiluminescence detection reagent (Pierce; Thermo Fisher Scientific, Inc.) and images were captured using the ChemiDoc Touch Imaging System (Bio-Rad Laboratories, Inc.).
Reverse-transcription-quantitative (RT-q)PCR array
The RNA expression levels of 84 EMT-related genes were simultaneously analyzed using the RT2 Profiler PCR Array (cat. no. PAHS-090ZA; Qiagen, Inc.). Total RNA was purified from SCC cells using the RNeasy kit (Qiagen, Inc.) according to the manufacturer's protocol, and residual genomic DNA was removed using the RNase-Free DNase Set (Qiagen, Inc.). Total RNA (5 µg) was reversed transcribed into cDNA in a 10-µl reaction volume using the RT2 First Strand Kit (Qiagen, Inc.). The cDNA was diluted with RT2 SYBR® Green Mastermix (Qiagen, Inc.) and distilled water to a total volume of 4,200 µl. Then, 10 µl cDNA solution was used for each primer set in the pathway-specific RT2 Profiler PCR Array, according to the manufacturer's protocol. Amplification reactions were performed using the Applied Biosystems ViiA7 Real-Time PCR System with automated baseline and threshold cycle detection. The threshold cycle number for each group of 84 genes was normalized to four built-in housekeeping gene controls using the RT2 Profiler PCR Array Data Analysis web-based software (SABiosciences Corp.).
Immunohistochemistry
Nine human skin cancer samples were collected at the time of surgical resection at the Gyeongsang National University Hospital (Jinju, Republic of Korea) during the period 2009–2015. There were 5 males and 4 females, with a median age of 70.5 (range, 47–89 years). A 5-µm section was cut from the formalin-fixed, paraffin-embedded tissue blocks of 9 patients with skin cancer. Each section was mounted on charged slides, deparaffinized and rehydrated in a graded alcohol series. To block endogenous peroxidase activity, each slide was incubated with 0.3% hydrogen peroxide for 30 min at room temperature. Antigen retrieval was conducted by boiling in 10 mM sodium citrate for 30 min, and immunostaining was performed using a rabbit polyclonal NNMT antibody (dilution 1:200) with a labeled streptavidin-biotin kit (Dako; Agilent Technologies, Inc.) according to the manufacturer's instructions. The slides were lightly counterstained with Mayer's hematoxylin and mounted.
RT-qPCR
Total RNA was extracted from SCC cells using Trizol reagent (Thermo Fisher Scientific, Inc.), and first-strand cDNA was generated using random hexamer primers provided in a first-strand cDNA synthesis kit (Bio-Rad Laboratories, Inc.), according to the manufacturer's instructions. The primers and probes (GAPDH; cat. no. Hs02758991-g1; NNMT; cat. no. Hs00196287) were obtained commercially (TaqMan® Gene Expression Assay; Thermo Fisher Scientific, Inc.) and the cDNA was amplified using the TaqMan® Gene Expression Master Mix (Thermo Fisher Scientific, Inc.). Amplification was performed in 96-well plates using the ViiA™ 7 Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling conditions were as follows: 50°C for 2 min and 95°C for 10 min, followed by 40 cycles at 94°C for 15 sec and 60°C for 1 min. GAPDH was used as the internal control, and three repetitive experiments were conducted. The comparative Ct (2−ΔΔCq) method (30) was used to calculate the relative levels of EMT-related genes.
Statistical analysis
Each experiment was conducted at least three times independently, and the data are presented as the mean ± standard deviation. The difference between two groups was assessed using the two-tailed Student's t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
Migration and invasion capacities of SCC12 cells
Cell migration and invasion are integral to the metastatic process. To characterize the phenotypic differences between two cutaneous SCC cell lines, the cell migration and invasion capacities of SCC12 and SCC13 cells were determined using wound healing and Matrigel invasion assays, respectively. SCC12 cells exhibited a progressive increase in the rate of cell migration (Fig. 1A), and the migration ability was considerably greater than that of SCC13 cells. In addition, the invasive ability of SCC12 cells was increased compared with that of SCC13 cells (Fig. 1B). During cultivation, the phenotype of cancerous cells is altered. However, the present study revealed higher motility and invasive potential in SCC12 cells compared with SCC13 cells. Therefore, these cell lines represent an ideal model for characterizing genomic and proteomic features that result in a metastatic phenotype.
Figure 1.
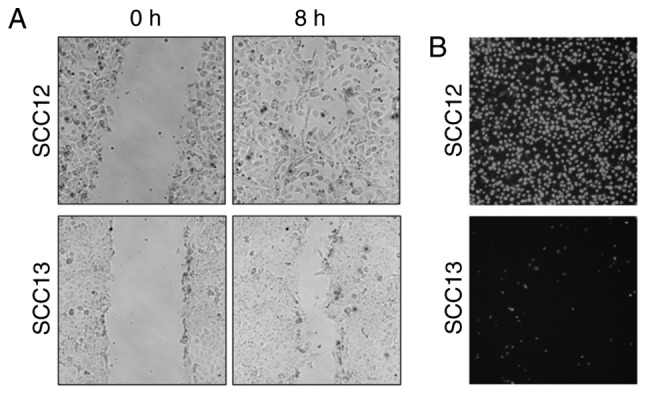
Migration and invasion assay of human skin squamous carcinoma cell lines SCC12 and SCC13. (A) Comparison of the migrational ability of SCC12 and SCC13 cells. A wound was created in a confluent SCC cell monolayer using a sterile micropipette tip, and the cells were incubated for 8 h. Migratory cells in the denuded zone were photographed. Magnification, ×100. (B) Invasion through Matrigel-coated Transwell inserts after 16 h. Nuclei were stained using DAPI, and images of the invaded cells were captured. Compared with SCC13 cells, SCC12 cells exhibited increased migration and invasion capacities, with metastatic potential. Magnification, ×200. SCC, squamous cell carcinoma.
Identification of molecular markers for metastasis-related genes in SCC cells using proteomic analysis
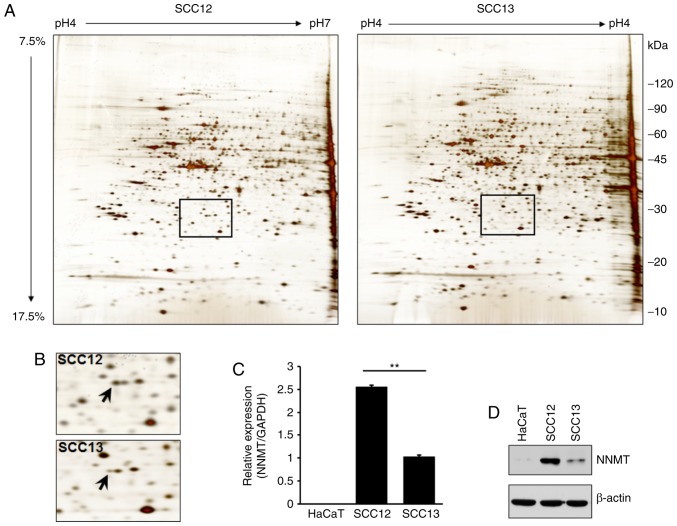
2-DE and LC-MS were used to analyze the differential expression levels of total proteins in two cSCC cell lines. A number of proteins were differentially expressed in SCC12 cells compared with SCC13 cells (Fig. 2A and Table I). Specifically, a protein spot identified as NNMT by mass spectrometry was upregulated in SCC12 cells (Fig. 2B). To verify the results of 2-DE, RT-qPCR and western blot analyses were also performed using the total RNA or cell lysates isolated from SCC12 and SCC13 cells, and the keratinocyte HaCaT cell line. Similarly, the NNMT mRNA expression level in SCC12 cells was 2.5-fold higher than that in the SCC13 cells (Fig. 2C). The corresponding level of NNMT protein expression was also higher in SCC12 than in SCC13 cells (Fig. 2D).
Figure 2.
Proteomics analysis of SCC cell lines. (A) Representative 2-DE maps of SCC12 and SCC13 cell proteins. Total proteins (50 µg) extracted from SCC12 and SCC13 cells were analyzed using a two-dimensional gel (focusing at pH 4.0–7.0, followed by 7.5–17.5% SDS-PAGE) and stained with silver nitrate. (B) Magnified images of the area shown in the rectangular box in (A) NNMT protein (indicated by an arrow) was upregulated in SCC12, compared with SCC13 cells. (C) Expression levels of NNMT in cutaneous SCC cells and HaCaT cells (an epidermal keratinocyte-derived non-tumorigenic cell line) were analyzed by RT-qPCR. GAPDH was used as the control. **P<0.01. (D) The NNMT protein expression level in the HaCaT, SCC12 and SCC13 cell lines was determined by western blot analysis using anti-NNMT antibodies. β-actin was used as the control for protein loading. SCC, squamous cell carcinoma; 2-DE, two-dimensional gel electrophoresis; NNMT, nicotinamide N-methyltransferase.
Table I.
Differentially expressed proteins in SCC12 cellsa.
| Spot# | Protein name | Accession no. | MW (kDa)/pI | No. of matched peptides | Seq. coverage (%) | P-valueb |
|---|---|---|---|---|---|---|
| 2010 | Cytokeratin 19 | P08727 | 44.1/5.04 | 14/23 | 32 | 0.0002 |
| 4008 | Cytokeratin 19 | P08727 | 44.1/5.04 | 10/17 | 22 | 0.0036 |
| 4825 | Tropomyosin 1α chain | P09493 | 32.7/4.69 | 5/13 | 15 | 0.0173 |
| 5008 | HSP27 | O14530 | 26.5/5.61 | 6/15 | 23 | 0.0004 |
| 6112 | Cytokeratin 8 | P05787 | 53.5/5.52 | 6/16 | 19 | 0.0125 |
| 3704 | Cytokeratin 8 | P05787 | 53.5/5.52 | 6/12 | 17 | 0.0081 |
| 5405 | Cytokeratin 8 | P05787 | 53.5/5.52 | 10/18 | 26 | 0.0133 |
| 2010 | Adenosylhomosysteinase | P23526 | 47.6/5.92 | 8/17 | 18 | 0.0007 |
| 4216 | Nicotinamide N-methyltransferase (NNMT) | P40261 | 29.6/5.56 | 9/22 | 42 | 0.0002 |
| 2010 | Glutathione S-transferase omega 1 | P78417 | 27.6/6.24 | 5/17 | 21 | 0.0015 |
A relative change (2-fold or more) in a protein highly expressed in SCC12 cells compared with one in SCC13 cells.
P-values were determined from the density of each pair spot obtained from PDQuest analyses of three independent experiments. MW, molecular weight; pI, protein isoelectric point.
Immunohistochemical examination
The expression levels of NNMT in healthy, SCC, basal cell carcinoma (BCC) and malignant melanoma (MM) tissues were examined using immunohistochemistry. NNMT protein was strongly expressed in SCC and MM tissues, and mildly expressed in BCC tissues; however, it was not expressed in healthy tissues (Fig. 3).
Figure 3.
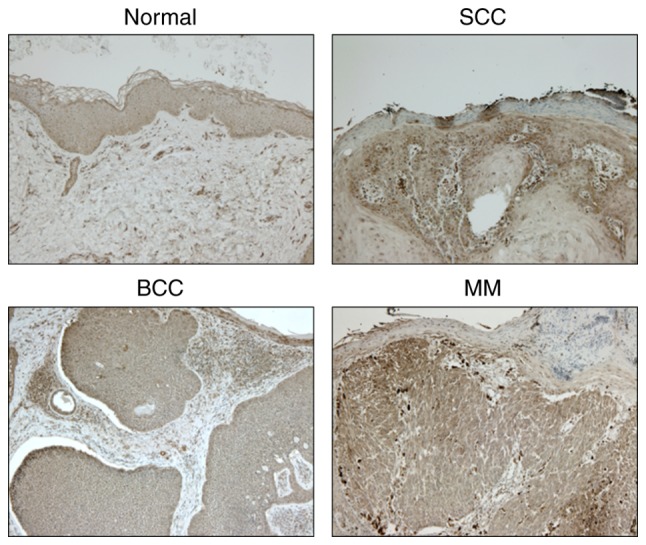
Immunohistochemical staining of NNMT in healthy, SCC, BCC and MM tissues. Magnification, ×200. NNMT, nicotinamide N-methyltransferase; SCC, squamous cell carcinoma; BCC, basal cell carcinoma; MM, malignant melanoma.
Evaluation of EMT-related gene expression in invasive and non-invasive SCC cells
To determine whether NNMT was able to regulate human SCC metastasis, the expression levels of EMT mediators were assessed in the HaCaT, SCC12 and SCC13 cell lines using western blotting. SCC12 cells showed a high expression level of the mesenchymal marker vimentin, and a low expression level of the epithelial marker E-cadherin, consistent with a migratory phenotype (Fig. 4A). To identify genes associated with the invasive phenotype of SCC12 cells, changes in the expression of EMT-related genes were determined using the RT2 Profiler PCR Array for 84 epithelial and mesenchymal-associated genes. As shown in Fig. 4B, 27 EMT-related genes were upregulated and suppressed in SCC12 cells with certain genes exhibiting more than 2-fold changes in expression levels compared with SCC13 cells. Several genes, including EMT-related vimentin, cytokeratin 19 (KRT19) (upregulated), cytokeratin 7 (KRT7) (upregulated), bone morphogenetic protein 7 (BMP7) (downregulated) and matrix metalloproteinase 2 (MMP2) (downregulated) were differentially expressed (Fig. 4B).
Figure 4.
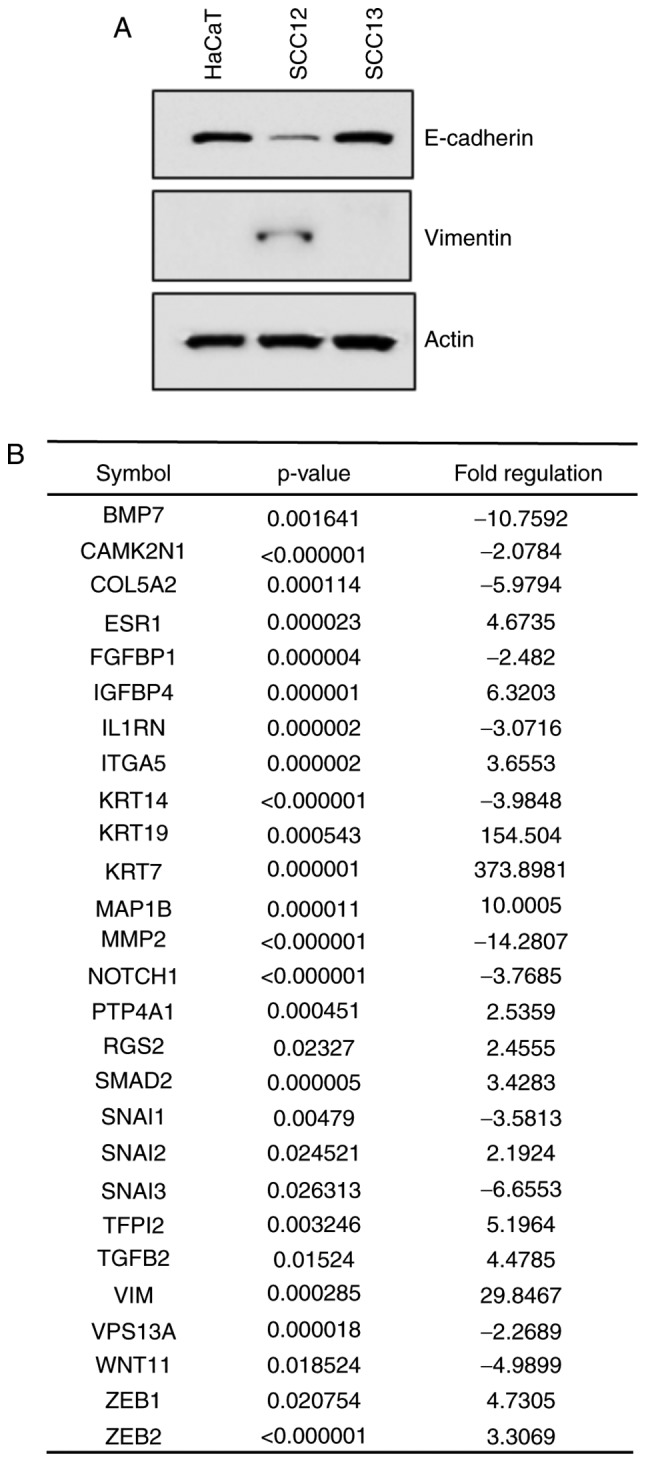
Western blot and RT-qPCR analyses. (A) Western blot analysis of the expression levels of E-cadherin and vimentin in HaCaT, SCC12 and SCC13 cells. β-actin was used as a loading control. (B) Evaluation of differentially expressed EMT-related genes in invasive and non-invasive SCC cells using the RT2 Profiler PCR Array. Among 84 epithelial and mesenchymal genes, a ≥2-fold difference in mRNA expression level in SCC12 cells compared with one in SCC13 cells was used as the cut-off to determine the significance of regulatory effects on genes involved in tumor metastasis. EMT, epithelial-mesenchymal transition; SCC, squamous cell carcinoma.
Effect of NNMT knockdown on cell proliferation and density-dependent growth
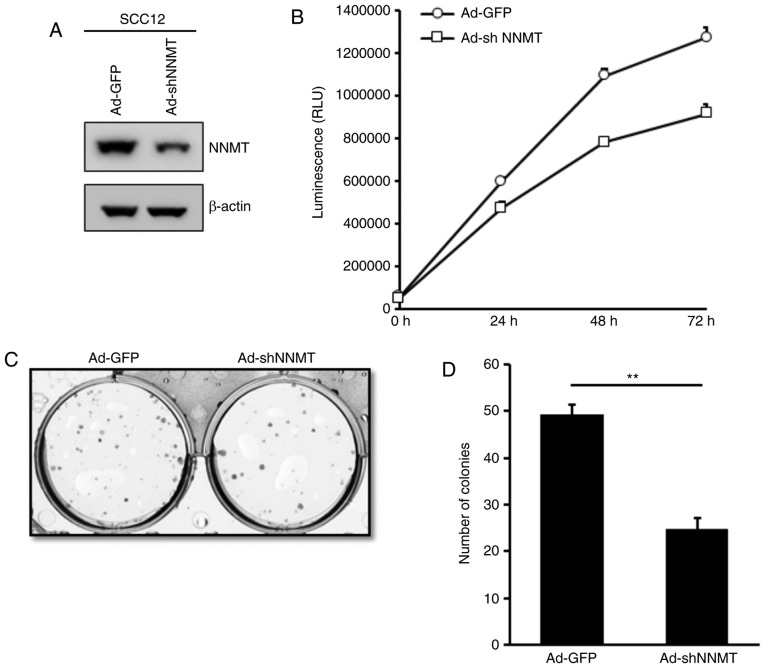
To investigate the effects of NNMT overexpression on SCC cells, NNMT knockdown was performed in SCC12 cells using shRNA-expressing adenovirus. The protein expression level of NNMT was determined and was markedly decreased in the Ad-shNNMT-infected, compared with the Ad-GFP-infected SCC12 cells (Fig. 5A). To assess the effects of NNMT-silencing on the proliferation of SCC12 cells, a bioluminescence assay was performed using the RealTime-Glo™ MT Cell Viability Assay. The proliferation of Ad-shNNMT-infected SCC12 cells was considerably impaired compared with that of the corresponding Ad-GFP-infected control cells (Fig. 5B). These results indicate that NNMT may promote the proliferation of SCC cells.
Figure 5.
Inhibition of the proliferation and density-dependent growth of SCC12 cells using NNMT knockdown. (A) Cells were infected with Ad-GFP or Ad-shNNMT adenovirus at an MOI of 20. At 24 h post-infection, the culture medium was replaced with the complete growth medium. After an additional 48 h of incubation, the expression of NNMT was determined by western blot analysis. β-actin served as the loading control. (B) Cells infected with Ad-GFP or Ad-shNNMT adenovirus for 72 h were harvested and replated in 96-well plates. Cell viability was examined using the RealTime-Glo™ MT Cell Viability Assay for the indicated time intervals. (C) Anchorage-dependent growth of Ad-GFP-infected or Ad-shNNMT-infected SCC12 cells was analyzed by colony formation assay. At 10 days post-seeding, colonies were fixed with 70% ethanol, stained with crystal violet and quantified. (D) Data are presented as the mean ± standard deviation of three independent experiments. **P<0.01. SCC, squamous cell carcinoma; NNMT, nicotinamide N-methyltransferase; MOI, multiplicity of infection; RLU, relative luminometer units.
Furthermore, the effect of NNMT knockdown on the anchorage-dependent growth of SCC12 cells was investigated using a colony formation assay. A similar inhibitory effect was observed; the mean colony number was 49.3 in the control group and 24 in the Ad-shNNMT group (Fig. 5C and D), indicating that NNMT knockdown reduced the colony forming capacity of SCC12 cells, and suggesting an inhibitory effect on cell proliferation.
Effect of NNMT knockdown on cell migration and invasion
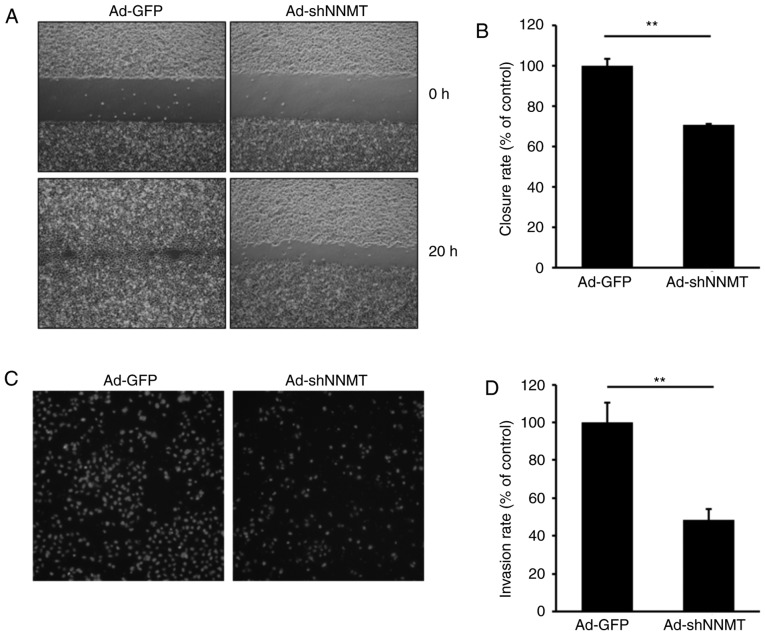
An increase in cell motility is known to affect the metastatic potential of cancer cells. Therefore, the potential effect of NNMT knockdown on cell motility was investigated in SCC12 cells, a cell line with high metastatic potential. A wound healing assay was subsequently performed on SCC12 cells infected with Ad-GFP or Ad-shNNMT adenovirus. Gap closure was predominantly due to cell migration rather than proliferation, at least at the early time points. The wound was ~90% closed by the Ad-GFP-infected cells, though only ~50% closed by Ad-shNNMT-infected cells (Fig. 6A and B), suggesting that NNMT knockdown inhibited the motility of SCC12 cells and resulted in reduced gap closure.
Figure 6.
Suppression of the migration and invasion of SCC12 cells by NNMT knockdown. (A) Confluent monolayers of Ad-GFP- and Ad-shNNMT-infected cells were scratched using a sterile pipette tip. Following a 20-h incubation period, the cells were photographed using a phase-contrast microscope (magnification, ×100) equipped with a CCD camera. (B) Wound closure rate was quantified as the percentage of gap closure compared with the 0 h time point. Columns, an average of three independent measurements; bars, standard deviation. **P<0.01. (C) Cell invasion was assessed over 16 h using Matrigel assays. The relative cell invasion rate of the Ad-shNNMT group was normalized to that of the Ad-GFP control group. (D) Results are presented as the mean ± standard deviation. **P<0.01. SCC, squamous cell carcinoma; NNMT, nicotinamide N-methyltransferase; CCD, charge-coupled device.
The potential effects of NNMT knockdown on SCC12 cell invasiveness were also investigated. As shown in Fig. 6C and D, the depletion of NNMT using Ad-shNNMT markedly reduced the invasive ability of SCC12 cells. The number of invaded knockdown cells was ~47% that of the control cells, suggesting that NNMT may promote SCC cell invasion.
Effect of NNMT knockdown on EMT-related gene expression
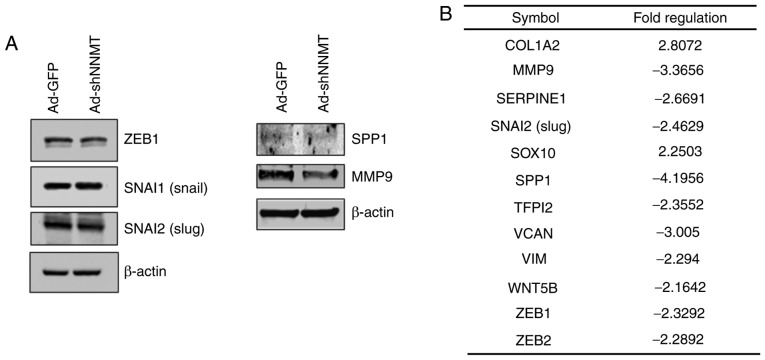
The role of NNMT in EMT regulation was further investigated by detecting the protein expression levels of key EMT effectors, including ZEB1, Slug and Snail. Notably, NNMT knockdown did not have a considerable effect on ZEB1 and Snail, but the expression level of Slug, MMP9 and osteopontin (SPP1) in NNMT-knockdown cells was notably altered (Fig. 7A). To identify genes associated with the invasiveness of SCC cells, and that are regulated by NNMT, the expression of EMT-related genes was assessed using the RT2 Profiler PCR Array. NNMT knockdown was found to downregulate 10 of the 84 EMT-related genes assessed (Fig. 7B), negatively affecting the expression levels of MMP9, SPP1 and versican core protein (VCAN), which are involved in the regulation of extracellular matrix (ECM) structure and function. In addition, the mRNA expression level of Slug was inhibited by NNMT knockdown. Collectively, these results indicate the metastatic potential of SCC12 cells.
Figure 7.
NNMT-mediated regulation of EMT effectors, including ZEB1, Slug and Snail. (A) SCC12 cells were infected with Ad-shNNMT or Ad-GFP (control) adenovirus at an MOI of 20. At 72 h post-infection, the cells were harvested and subjected to western blot analysis. β-actin served as the loading control. (B) Altered expression of several genes associated with EMT activation following NNMT knockdown. Relative changes >2-fold were observed for proteins expressed in the Ad-shNNMT-infected SCC12 cells, compared with those expressed in Ad-GFP-infected SCC12 cells. EMT, epithelial-mesenchymal transition; NNMT, nicotinamide N-methyltransferase; ZEB1, zinc finger E-box-binding homeobox 1; SCC, squamous cell carcinoma; MOI, multiplicity of infection.
Discussion
Despite considerable progress in reducing its incidence and mortality rates, and improving the survival of affected patients, cancer remains a major threat to public health worldwide. Metastasis to several organs, including the peritoneum and lymph nodes, is the cause of >90% of all cancer-associated mortalities (30). An EMT-induced increase in the invasive ability of cancer cells is an important process in carcinogenesis (26,29). Metastasis, the process by which cancer cells leave the primary tumor, disseminate to anatomically distant sites and form secondary tumors, is a complex multistep process that results from an intricate molecular cascade (29). Numerous studies have provided insights into the molecular components that facilitate this process, and understanding the molecular mechanisms that regulate metastasis will allow the development of novel therapeutic strategies. In the present study, differentially expressed genes involved in tumor metastasis in cSCC cells were identified using quantitative proteomics approaches. After separating the proteins by 2-DE, LC-MS analysis was performed to identify differentially expressed proteins between SCC12 and SCC13 cells. A total of 39 proteins whose expression was increased or decreased in SCC12 compared with SCC13 cells (>2-fold increase or decrease; P<0.05) were identified. Among these proteins, cytokeratin 19, tropomyosin 1 α chain, heat shock protein β-1, cytokeratin 8, adenosylhomocysteinase, NNMT and glutathione S-transferase omega 1 were significantly upregulated (P<0.05; Fig. 2 and Table I). In particular, a protein spot identified as NNMT by mass spectrometry, was upregulated at the mRNA and protein levels in the invasive SCC12 cells, compared with non-invasive SCC13 cells (Fig. 2).
NNMT catalyzes the N-methylation of nicotinamide and other pyridines, regulating the metabolism of drugs and xenobiotics (5,6). Various studies have revealed that NNMT serves important roles in a wide range of biological processes (7–16), suggesting that it may serve as a diagnostic and prognostic marker, and a therapeutic target for various tumors. Additionally, NNMT upregulation is involved in tumor cell migration, and it may be used as a potential biomarker for predicting tumor metastasis (16,20). However, its role in cSCC has not previously been described. In the present study, NNMT was found to be upregulated in an invasive, compared with a non-invasive cSCC cell line. To further evaluate the role of NNMT in invasive SCC12 cells, the effect of NNMT depletion on various biological functions was assessed. This demonstrated that NNMT knockdown suppressed the proliferation, colony formation, migration and invasion capacities of SCC12 cells. Therefore, NNMT may possess a tumor-promoting function in cSCC cells.
EMT is activated at the transcriptional level during embryonic development and wound healing, and contributes to the initiation and progression of cancer, especially during tumor metastasis (26,27). During EMT, polarized epithelial cells lose their polarity and intercellular adhesion properties, resulting in cytoskeleton reorganization and increased mesenchymal properties. Invasion and metastasis are two of the defining features of cancer (28). Cancer cells characteristically adopt an invasive phenotype through the process of EMT, and several signaling pathways reportedly regulate EMT, which enhances cell motility and invasiveness (29). An understanding of the underlying mechanisms of EMT are crucial, considering its vital involvement in skin healing and its contribution to tumor invasion and metastasis. In the present study, changes in the expression of EMT-related genes were profiled using the RT2 Profiler PCR Array. Several EMT-associated genes were found to be differentially expressed in SCC12 cells, including the upregulation of vimentin, KRT19 and KRT7, and the downegulation of BMP7 and MMP2 (Fig. 4B). SCC12 and SCC13 cells differ in their expression levels of MMP2 and MMP9. Low activity of MMP2 were revealed in SCC12 cells analyzed by zymography (data not shown); therefore, the differences in the expression and activity of MMP2 and MMP9 in the two cell lines suggests that they may be involved in cancer progression, and that different MMPs may be active in different cell types. A recent study reported that NNMT promoted EMT in gastric cancer cells (31); the present study revealed that NNMT silencing increased the mRNA expression levels of collagen α-2(I) chain (COL1A2) and the transcription factor SOX10, as well as suppressing those of MMP9, plasminogen activator inhibitor 1 (SERPINE1), SNAI2, SPP1, tissue factor pathway inhibitor 2 (TFP12), versican (VCAN), vimentin (VIM), WNT5B, ZEB1 and ZEB2 (Fig. 7). NNMT knockdown negatively impacted the expression of genes that regulate ECM structure and function, which included MMP9, SPP1 and VCAN (formerly CSPG2). These genes are reportedly involved in the regulation of SCC progression (32). The present study indicated that NNMT could facilitate the EMT of cSCC cells, suggesting that NNMT may be a promising prognostic indicator of invasive potential.
Although abnormal NNMT expression has been identified in several types of cancer, the biological roles of NNMT expression and activity in pathological conditions, including cancer-cell migration and invasion, remain poorly understood. Tang et al (20) demonstrated the crucial role of NNMT in the promotion of cellular invasion in clear cell renal cell carcinoma (ccRCC) cell lines; Akt inhibitor IV markedly attenuated the NNMT-induced invasion of ccRCC cells, indicating that activation of the PI3K/Akt signaling pathway is required for NNMT-dependent invasion. This finding suggests a potential mechanism in which NNMT acts upstream of the PI3K/Akt pathway. Nevertheless, how EMT-related gene expression is regulated in an NNMT-dependent manner remains unclear, in addition to how NNMT-induced EMT is directly associated with tumor cell metastasis.
In conclusion, the present study indicated that NNMT was upregulated in invasive SCC12 cells, and that it may serve as a potential biomarker of invasive tumor cells. NNMT knockdown inhibited tumor cell proliferation and invasion, and NNMT facilitated the EMT of cSCC cells by regulating EMT-related genes. Therefore, NNMT may present a novel prognostic biomarker and therapeutic target for patients with cSCC.
Acknowledgements
Not applicable.
Funding
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2018R1D1A1B07050577 and NRF- 2017R1A2B2005612).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
EPH and TJY conceived and designed the present study. YSH and HYC performed the experiments and collected the data. SYJ and YSP analyzed and interpreted the data. YSH and EPH drafted the manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Gyeongsang National University Hospital. Samples were taken from Gyeongsang National University Hospital with official written ethical consent from the patients.
Patient consent for publication
All patients provided their written informed consent for Publication and agreed to the publication of their associated data and any accompanying images as appropriate.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lomas A, Leonardi-Bee J, Bath-Hextall F. A systematic review of worldwide incidence of nonmelanoma skin cancer. Br J Dermatol. 2012;166:1069–1080. doi: 10.1111/j.1365-2133.2012.10830.x. [DOI] [PubMed] [Google Scholar]
- 2.Karia PS, Han J, Schmults CD. Cutaneous squamous cell carcinoma: Estimated incidence of disease, nodal metastasis, and deaths from disease in the United States 2012. J Am Acad Dermatol. 2013;68:957–956. doi: 10.1016/j.jaad.2012.11.037. [DOI] [PubMed] [Google Scholar]
- 3.Alam M, Ratner D. Cutaneous squamous-cell carcinoma. N Engl J Med. 2001;344:975–983. doi: 10.1056/NEJM200103293441306. [DOI] [PubMed] [Google Scholar]
- 4.O'Hara J, Ferlito A, Takes RP, Rinaldo A, Strojan P, Shaha AR, Rodrigo JP, Paleri V. Cutaneous squamous cell carcinoma of the head and neck metastasizing to the parotid glandea review of current recommendations. Head Neck. 2011;33:1789–1795. doi: 10.1002/hed.21583. [DOI] [PubMed] [Google Scholar]
- 5.Rini J, Szumlanski C, Guerciolini R, Weinshilboum RM. Human liver nicotinamide N-methyltransferase: Ion-pairing radiochemical assay, biochemical properties and individual variation. Clin Chim Acta. 1990;186:359–374. doi: 10.1016/0009-8981(90)90322-J. [DOI] [PubMed] [Google Scholar]
- 6.Aksoy S, Szumlanski CL, Weinshilboum RM. Human liver nicotinamide N-methyltransferase. cDNA cloning, expression, and biochemical characterization. J Biol Chem. 1994;269:14835–14840. [PubMed] [Google Scholar]
- 7.Xu J, Moatamed F, Caldwell JS, Walker JR, Kraiem Z, Taki K, Brent GA, Hershman JM. Enhanced expression of nicotinamide N-methyltransferase in human papillary thyroid carcinoma cells. J Clin Endocrinol Metab. 2003;88:4990–4996. doi: 10.1210/jc.2002-021843. [DOI] [PubMed] [Google Scholar]
- 8.Lim BH, Cho BI, Kim YN, Kim JW, Park ST, Lee CW. Overexpression of nicotinamide N-methyltransferase in gastric cancer tissues and its potential post-translational modification. Exp Mol Med. 2006;38:455–465. doi: 10.1038/emm.2006.54. [DOI] [PubMed] [Google Scholar]
- 9.Kim DS, Choi YP, Kang S, Gao MQ, Kim B, Park HR, Choi YD, Lim JB, Na HJ, Kim HK, et al. Panel of candidate biomarkers for renal cell carcinoma. J Proteome Res. 2010;9:3710–3719. doi: 10.1021/pr100236r. [DOI] [PubMed] [Google Scholar]
- 10.Yao M, Tabuchi H, Nagashima Y, Baba M, Nakaigawa N, Ishiguro H, Hamada K, Inayama Y, Kishida T, Hattori K, et al. Gene expression analysis of renal carcinoma: Adipose differentiation-related protein as a potential diagnostic and prognostic biomarker for clear-cell renal carcinoma. J Pathol. 2005;205:377–387. doi: 10.1002/path.1693. [DOI] [PubMed] [Google Scholar]
- 11.Sartini D, Muzzonigro G, Milanese G, Pierella F, Rossi V, Emanuelli M. Identification of nicotinamide N-methyltransferase as a novel tumor marker for renal clear cell carcinoma. J Urol. 2006;176:2248–2254. doi: 10.1016/j.juro.2006.07.046. [DOI] [PubMed] [Google Scholar]
- 12.Iacobuzio-Donahue CA, Maitra A, Shen-Ong GL, van Heek T, Ashfaq R, Meyer R, Walter K, Berg K, Hollingsworth MA, Cameron JL, et al. Discovery of novel tumor markers of pancreatic cancer using global gene expression technology. Am J Pathol. 2002;160:1239–1249. doi: 10.1016/S0002-9440(10)62551-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsuchiya A, Sakamoto M, Yasuda J, Chuma M, Ohta T, Ohki M, Yasugi T, Taketani Y, Hirohashi S. Expression profiling in ovarian clear cell carcinoma: Identification of hepatocyte nuclear factor-1 beta as a molecular marker and a possible molecular target for therapy of ovarian clear cell carcinoma. Am J Pathol. 2003;163:2503–2512. doi: 10.1016/S0002-9440(10)63605-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roessler M, Rollinger W, Palme S, Hagmann ML, Berndt P, Engel AM, Schneidinger B, Pfeffer M, Andres H, Karl J, et al. Identification of nicotinamide N-methyltransferase as a novel serum tumor marker for colorectal cancer. Clin Cancer Res. 2005;11:6550–6557. doi: 10.1158/1078-0432.CCR-05-0983. [DOI] [PubMed] [Google Scholar]
- 15.Kim J, Hong SJ, Lim EK, Yu YS, Kim SW, Roh JH, Do IG, Joh JW, Kim DS. Expression of nicotinamide N-methyltransferase in hepatocellular carcinoma is associated with poor prognosis. J Exp Clin Cancer Res. 2009;28:20. doi: 10.1186/1756-9966-28-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Y, Siadaty MS, Berens ME, Hampton GM, Theodorescu D. Overlapping gene expression profiles of cell migration and tumor invasion in human bladder cancer identify metallothionein 1E and nicotinamide N-methyltransferase as novel regulators of cell migration. Oncogene. 2008;27:6679–6689. doi: 10.1038/onc.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomida M, Mikami I, Takeuchi S, Nishimura H, Akiyama H. Serum levels of nicotinamide N-methyltransferase in patients with lung cancer. J Cancer Res Clin Oncol. 2009;135:1223–1229. doi: 10.1007/s00432-009-0563-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sartini D, Pozzi V, Renzi E, Morganti S, Rocchetti R, Rubini C, Santarelli A, Lo Muzio L, Emanuelli M. Analysis of tissue and salivary nicotinamide N-methyltransferase in oral squamous cell carcinoma: Basis for the development of a noninvasive diagnostic test for early-stage disease. Biol Chem. 2012;393:505–511. doi: 10.1515/hsz-2012-0112. [DOI] [PubMed] [Google Scholar]
- 19.Sartini D, Muzzonigro G, Milanese G, Pozzi V, Vici A, Morganti S, Rossi V, Mazzucchelli R, Montironi R, Emanuelli M. Upregulation of tissue and urinary nicotinamide N-methyltransferase in bladder cancer: Potential for the development of a urine-based diagnostic test. Cell Biochem Biophys. 2013;65:473–483. doi: 10.1007/s12013-012-9451-1. [DOI] [PubMed] [Google Scholar]
- 20.Tang SW, Yang TC, Lin WC, Chang WH, Wang CC, Lai MK, Lin JY. Nicotinamide N-methyltransferase induces cellular invasion through activating matrix metalloproteinase-2 expression in clear cell renal cell carcinoma cells. Carcinogenesis. 2011;32:138–145. doi: 10.1093/carcin/bgq225. [DOI] [PubMed] [Google Scholar]
- 21.Emanuelli M, Santarelli A, Sartini D, Ciavarella D, Rossi V, Pozzi V, Rubini C, Lo Muzio L. Nicotinamide N-Methyltransferase upregulation correlates with tumour differentiation in oral squamous cell carcinoma. Histol Histopathol. 2010;25:15–20. doi: 10.14670/HH-25.15. [DOI] [PubMed] [Google Scholar]
- 22.Chen C, Wang X, Huang X, Yong H, Shen J, Tang Q, Zhu J, Ni J, Feng Z. Nicotinamide N-methyltransferase: A potential biomarker for worse prognosis in gastric carcinoma. Am J Cancer Res. 2016;6:649–663. [PMC free article] [PubMed] [Google Scholar]
- 23.Xie X, Liu H, Wang Y, Zhou Y, Yu H, Li G, Ruan Z, Li F, Wang X, Zhang J. Nicotinamide N-methyltransferase enhances resistance to 5-fluorouracil in colorectal cancer cells through inhibition of the ASK1-p38 MAPK pathway. Oncotarget. 2016;7:45837–45848. doi: 10.18632/oncotarget.9962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hesse K, Satzger I, Schacht V, Köther B, Hillen U, Klode J, Schaper K, Gutzmer R. Characterisation of prognosis and invasion of cutaneous squamous cell carcinoma by podoplanin and E-cadherin expression. Dermatology. 2016;232:558–565. doi: 10.1159/000450920. [DOI] [PubMed] [Google Scholar]
- 25.Cañueto J, Martín-Vallejo J, Cardeñoso-Álvarez E, Fernández-López E, Pérez-Losada J, Román-Curto C. Rapid growth rate is associated with poor prognosis in cutaneous squamous cell carcinoma. Clin Exp Dermatol. 2018;43:876–882. doi: 10.1111/ced.13570. [DOI] [PubMed] [Google Scholar]
- 26.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial- mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 27.Valastyan S, Weinberg RA. Tumor metastasis: Molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 29.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Liang L, Zeng M, Pan H, Liu H, He Y. Nicotinamide N-methyltransferase promotes epithelial-mesenchymal transition in gastric cancer cells by activating transforming growth factor-β1 expression. Oncol Lett. 2018;15:4592–4598. doi: 10.3892/ol.2018.7885. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Kashyap MK, Marimuthu A, Kishore CJ, Peri S, Keerthikumar S, Prasad TS, Mahmood R, Rao S, Ranganathan P, Sanjeeviah RC, et al. Genomewide mRNA profiling of esophageal squamous cell carcinoma for identification of cancer biomarkers. Cancer Biol Ther. 2009;8:36–46. doi: 10.4161/cbt.8.1.7090. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.