Abstract
3D spheroids are built by heterogeneous cell types in different proliferative and metabolic states and are enriched in cancer stem cells. The main aim of the study was to investigate the usefulness of a novel metastatic renal cell carcinoma (RCC) 3D spheroid culture for in vitro cancer stem cell physiology research and drug toxicity screening. RCC cell lines, Caki-1 (skin metastasis derived) and ACHN (pleural effusion derived), were efficiently cultured in growth-factor/serum deprived, defined, StemXvivo and Nutristem medium on laminin-coated or poly-D-lysine-coated plates. In optimal 3D culture conditions, ACHN cells (StemXVivo/poly-D-lysine) formed small spheroids with remaining adherent cells of an epithelial phenotype, while Caki-1 cells (StemXVivo/laminin) formed large dark spheroids with significantly reduced cell viability in the center. In the 3D structures, expression levels of genes encoding stem transcription factors (OCT4, SOX2, NES) and RCC stem cell markers (CD105, CD133) were deregulated in comparison to these expression levels in traditional 2D culture. Sunitinib, epirubicin and doxycycline were more toxic to cells cultured in monolayers than for cells in 3D spheroids. High numbers of cells arrested in the G0/G1 phase of the cell cycle were found in spheroids under sunitinib treatment. We showed that metastatic RCC 3D spheroids supported with ECM are a useful model to determine the cancer cell growth characteristics that are not found in adherent 2D cultures. Due to the more complex architecture, spheroids may mimic in vivo micrometastases and may be more appropriate to investigate novel drug candidate responses, including the direct effects of tyrosine kinase inhibitor activity against RCC cells.
Keywords: renal cell cancer, cancer stem cells, 3D, spheroid, sunitinib
Introduction
Cancer stem cells (CSCs) are characterized by the potential to self-renew, high tumorigenicity in nude mice and the ability to efficiently reconstitute all tumor subpopulations and original tumor histology (1–3). CSCs are suggested to be responsible for cancer development, as well as recurrence, disease progression, cancer aggressiveness, metastatic spread and resistance to chemotherapy, targeted therapies and radiotherapy. CSCs are also expected to be more resistant to all types of therapies than differentiated tumor cells (4,5). In renal cell cancer (RCC), CSCs are characterized by expression of cell surface markers including CD105 (6,7), CD44 (8), or C-X-C chemokine receptor type 4 (CXCR-4) (9), as well as high expression of aldehyde dehydrogenase (ALDH1) (8,10). We recently showed that CD105+ subpopulations of cells may be found in multiple RCC cell lines, including metastatic tumor-derived ACHN and Caki-1 cell lines cultured in RPMI-1640/GlutaMax medium (11). High CD105 protein expression was also found in ACHN and Caki-1 cells cultured in DMEM-HG (12) and these CD105+ subpopulations are likely to represent RCC-CSCs. At the same time, sphere and colony formation assays may be used for the functional identification of CSCs (13,14). Although the number of CSCs is underestimated by standard experimental methods in RCC (2), the number of CSCs is increased in 3D culture including spheres (15). In fact in sphere-promoting culture conditions [serum-free, fibroblast growth factor (FGF), epidermal growth factor (EGF) and B27-enriched DMEM medium], ACHN cells form large 3D aggregates that fuse over time (16). The first aim of the present study was to develop a metastatic RCC cell line-based 3D culture model enriched in CSCs which was defined as CD105-expression promoting conditions. Optimal media supporting the CSC phenotype in metastatic RCC were to be selected. Extracellular matrix (ECM) further promoting cell growth was to be incorporated in this model. The optimized culture system (specifically selected cell line with media and ECM type) will be therefore referred to as the biomimic culture.
The second aim of the study was to investigate the usefulness of metastatic RCC 3D spheroids as a platform for effective in vitro anticancer drug screening. Our study was triggered by the fact that recently we demonstrated that RCC-CSCs are also potential therapeutic targets and are in fact targeted by tyrosine kinase inhibitors (TKIs) (e.g. sunitinib) (17–19). At the same time the first attempts to develop anticancer drugs targeting CSCs were carried out in an acute lymphoblastic leukaemia model (20), and subsequently in glioblastoma multiforme (21) by other research groups. The first group of drugs tested on CSCs, that are not classical cytostatics, the nonsteroidal anti-inflammatory drugs (NSAIDs), were tested in a colon cancer model. In preliminary research it was shown that NSAIDs effectively eliminate cancer cells from colon crypts, especially cells that have aberrant WNT signaling and represent the stem population (22). Moreover, N-[3,4-dimethoxycinnamoyl]-anthranilic acid (tranilast, INN, brand name Rizaben®; Kissei Pharmaceuticals, Japan) was tested in a breast cancer model. It was proven that this H1-receptor antagonist decreases the number of mammospheres that are formed by stem cells, decreases the number of colonies (in a colony forming assay), and decreases the expression of surface markers and has a direct anti-proliferative effect on CSCs (23). On the one hand, 3D/spheroid/sphere tests may provide in vitro tests with a higher predictive value of in vivo activity. At the same time, drug screens on 3D cultures, enriched in CSCs, can lead to development of novel effective treatments targeted to the elimination of these cells. Currently only a few trials are being run with specific CSC-toxic compounds (ClinicalTrials.gov) in solid tumors. Resveratrol is being tested in colon cancer (targeting Wnt signaling); or GDC-0449 and BMS-833923 in other tumors (targeting Hedgehog). Altogether there are approximately 10 trials that are being conducted to target CSCs, and only one drug is being tested in RCC. In the RCC targeted trial, CSC Notch signaling is targeted by RO4929097 (University Health Network, Toronto, Canada; no. NCT01141569) (24). Furthermore, TKI and mTOR kinase inhibitor pre-clinical and clinical trials in RCC did not include CSC analysis (25–27), as an appropriate model was not available. Therefore, the present study was designed with an aim to develop a new anti-RCC-CSC drug testing model and to investigate selected drug activities to prove its utility in the evaluation of different types of compounds (TKI, cytostatic, small-molecule). We believe that anti-CSC targeted therapies in RCC, and other solid tumors, represent a new direction for basic science exploration and potential subsequent clinical investigation in order to provide effective, advanced cancer care (28). We also believe that sunitinib is a benchmark TKI compound that may be used for drug testing model validation, as its activity has clearly been described in clinical trials, as well as in molecular reports. Sunitinib (SU11248) is a multi-targeted inhibitor of tyrosine kinases including vascular endothelial growth factor receptor (VEGFR)1, VEGFR2 and VEGFR3, platelet-derived growth factor receptor β (PDGFR-β), stem cell growth factor receptor (SCFR-c-KIT), fms-like tyrosine kinase 3 (Flt3) and 73 kinases in addition to its main targets (29,30). Sunitinib inhibits cancer growth primarily through an anti-angiogenic mechanism by inhibiting endothelial cell proliferation and also halting their motility and inhibiting cancer stem cell endothelial differentiation (31,32). Sunitinib also inhibits the growth of cancer cells that is stimulated by VEGF, SCF, or PDGF and induces cancer cell apoptosis (33,34). Most recently, the elucidated mechanisms of sunitinib action include targeting of CSCs, as we have shown in our previous research (32,35). Recent reports have suggested the direct effect of sunitinib on CSCs in vivo and in vitro, which further supports our model selection. In xenograft RCC models, sunitinib was shown to generate resistance to its own therapeutic mechanism due to induction of hypoxia in the tumor. On the other hand, in prostate cancer xenograft studies, sunitinib reduced tumor hypoxia and angiogenesis, with a radiosensitizing effect on stem-like cells (36). At the same time, in renal cancer xenografts, the number of CD133/CXCR4 co-expressing CSCs was found to be higher under hypoxia, in peri-necrotic tumor areas, and their tumorigenic potential was increased, while their sensitivity to sunitinib was decreased (37). Moreover, nuclear factor (NF)-κB/interleukin-6 (IL-6) activation was found to be responsible for p21-activated kinase 1 (PAK1)-mediated stem-like phenotype following sunitinib treatment in RCC (38). It is also worth emphasizing that sunitinib suppresses the proliferation, migration, apoptosis resistance, tumor angiogenesis and growth of triple-negative breast cancers but increases the percentage of breast cancer stem cells in tumors (39), which is in agreement with the fact that sunitinib was recently shown in preclinical models to increase invasive and metastatic properties of breast cancer cells (40). Surprisingly, recently, several groups have reported that antibiotics, including those routinely used in cell culture, can interfere with the sphere-forming abilities of cancer cells, and therefore their impact on CSCs may be expected (41,42). Selective toxic effects on CSCs have been reported for azithromycin, chloramphenicol and doxycycline (41). In light of these CSC-related data, we aimed to verify the effect of sunitinib, as well as the aforementioned antibiotics, on RCC cells in our novel 3D-extracellular matrix (ECM) supported cell culture model. As a control we decided to use the cytotoxic drug epirubicin [Epi; DNA-intercalating topoisomerase II inhibitor (43)] and to compare its effects on 3D culture with sunitinib [Su; tyrosine kinase inhibitor (TKI) anticancer activity in RCC (44)] to verify the anticancer cell specificity of selected in vitro treatments in the developed novel cell culture system.
In summary, the present study was designed to evaluate the effect of various 3D culture conditions on RCC cell pathophysiology, including promotion of a CSC phenotype defined by putative marker CD105 expression. Our objectives were to characterize gene expression and RCC cell proliferation changes in 3D culture-promoting conditions as compared to traditional monolayer 2D growth. We also aimed to describe the susceptibility of RCC cells grown in 3D to TKIs and chemotherapeutics in order to evaluate the applicability of our biomimic 3D RCC culture model for anticancer drug testing. We believe that to facilitate TKI testing in vivo in the future, the biomimic culture model is required for more relevant drug in vitro pre-screening. This culture model is needed in order to decrease the number of molecules ineffectively tested in in vivo animal experiments. The 3D cell culture model resembles micrometastases by complex spatial cell-cell interactions, intratumoral hypoxia in spheroids, and enrichment with cells presenting stem cell phenotype as we have shown for papillary RCC previously (45). Therefore, we believe that complex cell cultures, in defined media with ECM support, facilitate drug activity analysis and provide more relevant data, that are more likely to be replicated later in in vivo models. In general, the first goal of the present study was to provide such a biomimic model for the metastatic RCC field. The second goal of this study was to analyze the biological characteristics of RCC cells in this models including their drug sensitivity.
Materials and methods
Cell culture
Established human RCC cell lines were purchased from ATCC: Caki-1 (human kidney, epithelial, derived from skin metastasis; ATCC® HTB-46™) and ACHN (human kidney, epithelial, derived from pleural effusion metastasis; ATCC® CRL-1611™) (46). For initial passages, cells were cultured in penicillin/streptomycin (P/S; cat. no. L0022; Biowest) containing medium (RPMI-1640; L0498; Biowest) supplemented with 10% FBS (cat. no. A3160802; Gibco/Thermo Fisher Scientific, Inc.) to prepare early stocks; cells used for the experiments were not older than the 10th passage since purchase. Prior to the present study, the cells were passaged at least twice in antibiotic-free medium and then cultured in RPMI-1640 medium supplemented with 10% FBS (2D medium) until 80% confluence, and then detached with Accutase (cat. no. L0950; Biowest) to obtain single-cell suspensions, washed 2 times with serum-free and antibiotic-free DMEM (product code, L0501; Biowest) to remove remaining FBS and used in experiments. All cell lines cultured in the laboratory were screened for Mycoplasma contamination on a regular basis with Mycoplasma Detection Kit (cat. no. PP-401L; Jena Bioscience) and only negative passages were used for the presented experiments.
Extracellular matrix (ECM) coating testing
Laminin Corning™ BioCoat™ (cat. no. 08-774-84; Thermo Fisher Scientific, Inc.), Poly-D-lysine Corning™ BioCoat™ (cat. no. 08-775-30; Thermo Fisher Scientific, Inc.) and Poly-D-lysine + Laminin Corning™ BioCoat™ (cat. no. 08-774-169; Thermo Fisher Scientific, Inc.) in 24-well format were used for extracellular matrix dependence testing.
Stem cell medium testing
RPMI-1640 from Biowest (cat. no. L0501) was used as a reference control medium. NutriStem™ XF/FF Culture Medium for Human iPS and ES Cells (cat. no. 01-0005; Stemgent), StemXVivo Serum-Free Tumorsphere Media + StemXVivo EMT Inducing Media Supplement (cat. no. CCM012 and CCM017; R&D Systems) were used for 3D spheroid formation induction.
3D cell culture
For 3D structure formation, Caki-1 cells were cultured in laminin-coated (Lam; cat. no. 354412; Corning) and ACHN cells in poly-D-lysine-coated (PDL; cat. no. 354619; Corning) 24-well plates in StemXvivo or Nutristem (cat. no. 01-0005; Stemgent) media. A total of 10,000 cells per well in 1 ml of medium were seeded and cultured for 3 days to allow sphere formation. Then, drug treatment was applied as it was previously shown that a different response to each drug can be observed depending on whether the treatment was on a monolayer culture or directly to spheres (47); tested drugs were added in 100 µl of serum-free DMEM and after 3 additional days of culture, the cells were isolated for downstream assays. Alternatively, drugs were added 6 h after seeding and the cells were cultured for a total of 6 days. The 3D culture models employed are a subject of Polish patent application no. P.420002. Parallel 2D culture control was performed in standard tissue culture-treated plates (TC; cat. no. 10062-896; VWR) and RPMI supplemented with 10% FBS with culture time and drug treatment as above.
Microscopic observations
Cells were visualized using an inverted microscope Olympus CKX41 with camera UC30 and Olympus Entry Cell Sense 1.8.1. software (Olympus, Tokyo, Japan). The numbers of 3D structures formed were counted on the whole surface of the well.
Drug treatment
Stock concentrations of drugs were prepared in sterile PBS (pH=7.2; cat. no. 404.200; Bioshop) or DMSO (cat. no. EMR385100; EuroClone) as in Table I. For IC50 determination, a serial dilution of the drugs was prepared in serum-free DMEM in the range of 0.1–500 µg/ml or 0.01–50 µM final (in cell culture) concentrations, with DMEM not exceeding 10% of the total culture volume. As diluents, both PBS and DMSO did not alter the cell viability in the used concentrations in preliminary studies, and they were used in maximum concentrations only and served as controls. Penicillin/streptomycin solution was added to a final concentration used in standard culture (streptomycin, 100 µg/ml; penicillin, 100 U/ml).
Table I.
Preparation of the tested agents.
| Drug | Stock concentration | Diluent |
|---|---|---|
| Epirubicin (5909990796373, Accord) | 3 mM | PBS |
| Sunitinib (PZ0012, Sigma-Aldrich/Merck KGaA) | 10 mM | DMSO |
| Azithromycin (PZ0007, Sigma-Aldrich/Merck KGaA) | 20 mg/ml | DMSO |
| Chloramphenicol (C3175, Sigma-Aldrich/Merck KGaA) | 50 mg/ml | PBS |
| Doxycycline (D9891, Sigma-Aldrich/Merck KGaA) | 20 mg/ml | PBS |
PBS, phosphate-buffered saline; DMSO, dimethyl sulfoxide.
IC50 determination
Cells were seeded as above, in a 96-well format; 2,500 cells per well either in 100 µl of RPMI supplemented with 10% FBS (2D culture medium) or StemXvivo medium (cat. no. CCM004, R&D Systems; 3D-promoting medium as established in preliminary studies). After three days of culture, the drug dilutions described above were added, in triplicate in a volume of 10 µl to the wells. After an additional 3 days of culture, Alamar Blue reagent (cat. no. DAL1100; Molecular Probes/Thermo Fisher Scientific, Inc.) was added to the final concentration of 10% and incubated for 3 h (for adherent cultures; RPMI) or 24 h (for 3D cultures; StemXvivo medium). Absorbance was measured in a plate spectrophotometer (MultscanGO; Thermo Fisher Scientific, Inc.) at 570 and 600 nm, and the percentage of Alamar Blue reduction was calculated according to the manufacturer's instructions. Cell viability is represented as a change in Alamar Blue reduction in comparison to the untreated cells (% of control).
Cell isolation
Cells were cultured as described above and aggregates were collected from wells, washed with PBS and incubated for 10 min with Accutase solution to obtain single-cell suspensions. Then, the cells were washed and re-suspended in different buffers (depending on the assay) and used in further analyses.
Cell viability
Cells were suspended in 0.2 ml of serum-free DMEM and viability was measured with the Muse® Count & Viability Assay Kit (cat. no. MCH100102; Merck KGaA) according to the manufacturer's instructions. Briefly, 50 µl of cell suspension was mixed with 450 µl of viability stain and incubated for 5 min. Then, 2,000 cells were acquired on the Muse Cell Analyzer (cat. no. 0500-3115; Merck KGaA) and the percentage of live and dead cells was calculated.
Cell cycle analysis
Cells were pelleted to remove PBS, suspended by vortexing and pipetting and then 1 ml of 70% ice-cold ethanol (113964200; Chempur) was slowly added. Cells were incubated at −20°C for at least 3 h. Before analysis, alcohol was removed completely by two PBS washes and then 100 µl of Muse® Cell Cycle Reagent (The Muse® Cell Cycle Kit, cat. no. MCH100106; Merck KGaA) was added for 30 min. For acquisition, additionally 150 µl of PBS was added. At least 2,500 cells were acquired on the Muse Cell Analyzer (cat. no. 0500-3115; Merck KGaA) and analyzed for the percentage of cells in G0/G1, S and G2 phases, based on DNA content.
RNA isolation
Total RNA was isolated with Cells-to-Ct kit (cat. no. AM1728; Ambion); a total of 5,000 cells were suspended to remove excess PBS and 50 µl of DNAse containing Lysis buffer was pipetted to the sample. After a 5-min incubation, the reaction was stopped by gently mixing in 5 µl of stop solution. The resulting RNA isolates were stored at −80°C and then used for reverse transcription using the above mentioned kit. cDNA was obtained by adding 2X RT Buffer and 20X RT Enzyme mix to 10 µl of RNA isolates; reaction was adjusted to 25 µl with water and run for 60 min at 37°C, followed by 5 min inactivation at 95°C in a LifeECO thermal cycler (cat. no. BYQ6078; Bioer). Resulting cDNA was stored at −20°C and used in gene expression analysis.
Gene expression analysis
Real-time PCR was performed using the Applied Biosystems™ TaqMan™ Gene Expression Master Mix (cat. no. 4369010; Thermo Fisher Scientific, Inc.) with multiplex TaqMan primer/probe sets (Thermo Fisher Scientific, Inc.; listed in Table II) in 20 µl reactions in triplicate with 2 µl of 2X diluted cDNA reaction as a template. Two gene TaqMan sets were used in each reaction; compatibility of the kits was confirmed before experiments and only kits that gave the same signal when run in single or double reactions were used in the study. Also, -RT (minus reverse transcription) controls were performed to confirm no product on genomic DNA. Reactions were run in 8-well strips on LightCycler® 96 machine (cat. no. 05815916001; Roche). Data were calculated with using the 2(−ΔC(q)) method (48), with normalization to geometrical mean expression of PPIA (peptidylprolyl isomerase A) and GUSB (beta-glucuronidase) as housekeeping gene controls (49).
Table II.
List of paired TaqMan assays.
| Gene (FAM stain) | Assay ID | Gene (VIC stain) | Assay ID |
|---|---|---|---|
| HIF1 | Hs00153153_m1 | HIF2 | Hs01026149_m1 |
| VEGF | Hs00900055_m1 | PAX2 | Hs01565576_m1 |
| VHL | Hs00184451_m1 | CDH2 | Hs00983056_m1 |
| CDH1 | Hs01023894_m1 | C-MET | Hs01565576_m1 |
| CD133 | Hs01009257_m1 | NANOG | Hs04399610_g1 |
| NESTIN | Hs04187831_g1 | CD105 | Hs00923996_m1 |
| SOX2 | Hs01053049_s1 | OCT4 | Hs04260367_gH |
| PPIA | Hs01565699_g1 | GUSB | Hs00939627_m1 |
OCT4 or POU5F1, POU class 5 homeobox 1; CD105 or ENG, endoglin; CD133 or PROM1, prominin 1; CDH1, cadherin 1; HIF1, hypoxia-inducible factor 1-alpha; HIF2, hypoxia-inducible factor 2-alpha; MET, MET proto-oncogene, receptor tyrosine kinase; CDH2, cadherin 2; NESTIN, nestin; PAX2, paired box 2; SOX2, SRY-box 2; VEGF, vascular endothelial growth factor; PPIA, peptidylprolyl isomerase A; GUSB, glucuronidase β.
Statistical analysis
All experiments were performed in at least three biological repetitions, and one representative study is shown. Results are presented as means ± SD from duplicates or triplicates (3 wells). Differences between groups were determined using Mann-Whitney U test and considered significant if P<0.05 and marked with the symbol *on the respective graphs. For gene expression analysis non-parametric Kruskal-Wallis test was used and P-values are presented in Table III and on graphs or in the text.
Table III.
P-values of Mann-Whitney test for the comparisons between each 3D culture condition with 2D control for the gene expression analysisa.
| 2D vs. NutriStem | 2D vs. StemXvivo | |||
|---|---|---|---|---|
| Gene name | ACHN cells | Caki-1 cells | ACHN cells | Caki-1 cells |
| OCT4 | 0.081 | 0.081 | 0.081 | 0.081 |
| CD105 | 0.081 | 0.190 | 0.081 | 0.081 |
| CD133 | 0.081 | 0.081 | 0.081 | 0.081 |
| CDH1 | 0.081 | 0.081 | 0.081 | 0.081 |
| HIF1 | 0.081 | 0.077 | 0.081 | 0.077 |
| HIF2 | 0.081 | 0.139 | 0.081 | 0.139 |
| MET | 0.081 | 0.139 | 0.081 | 0.081 |
| CDH2 | 0.081 | 0.081 | 0.081 | 0.081 |
| NESTIN | 0.081 | 0.081 | 0.081 | 0.663 |
| PAX2 | 0.081 | ND | 0.081 | ND |
| SOX2 | 0.063 | 0.081 | 0.081 | 0.081 |
| VEGF | 0.081 | 0.081 | 0.081 | 0.081 |
Real-time PCR results as 2(−ΔC(q)) values. OCT4 or POU5F1, POU class 5 homeobox 1; CD105 or ENG, endoglin; CD133 or PROM1, prominin 1; CDH1, cadherin 1; HIF1, hypoxia-inducible factor 1-alpha; HIF2, hypoxia-inducible factor 2-alpha; MET, MET proto-oncogene, receptor tyrosine kinase; CDH2, cadherin 2; NESTIN, nestin; PAX2, paired box 2; SOX2, SRY-box 2; VEGF, vascular endothelial growth factor. ND, not detected.
Results
The mesenchymal phenotype is induced in 3D RCC spheroids
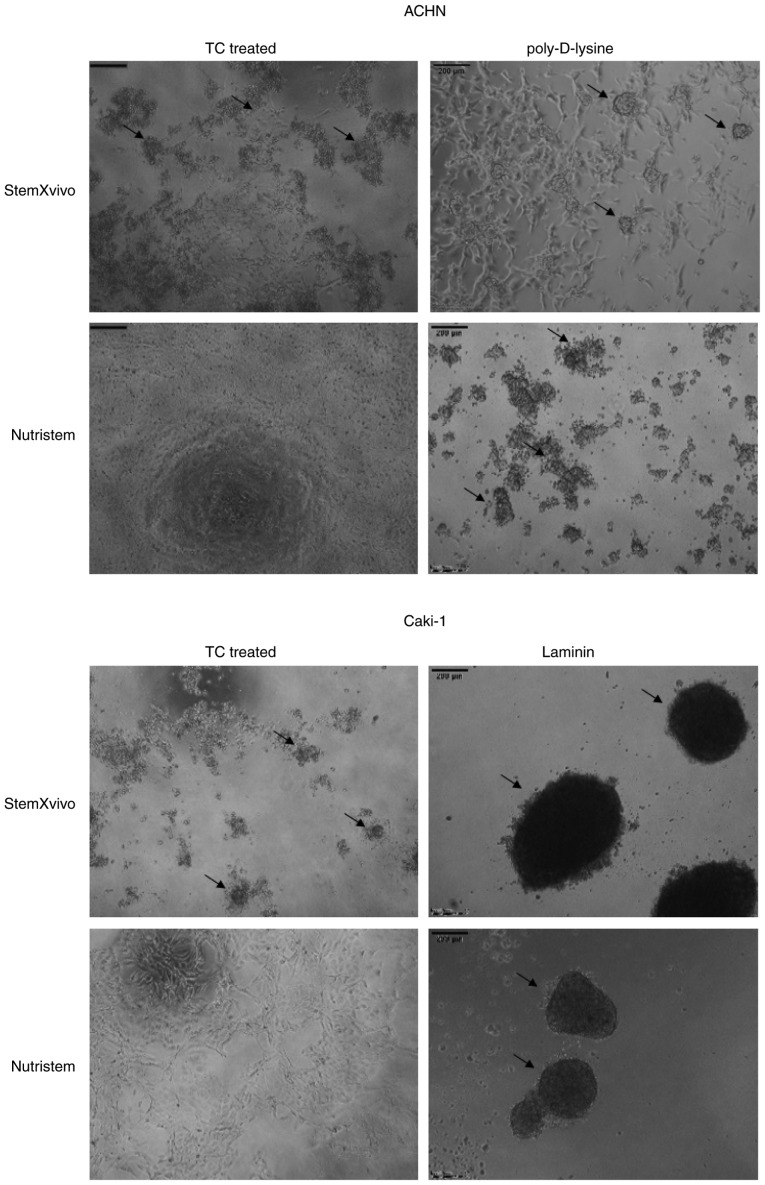
In our preliminary part of the study, we performed a screening to select the optimal culture conditions promoting formation of 3D spheroids by metastatic RCC cell lines, ACHN and Caki-1. We established that culture conditions of StemXVivo or Nutristem media on poly-D-lysine-coated plates for ACHN and laminin-coated plates for Caki-1 cells were the most efficient in promoting the 3D growth of cells [Fig. 1 and (50)].
Figure 1.
Morphology of 3D structures formed by Caki-1 and ACHN cells in the studied culture conditions after 6 days. Images were captured using an inverted microscope. TC, tissue culture.
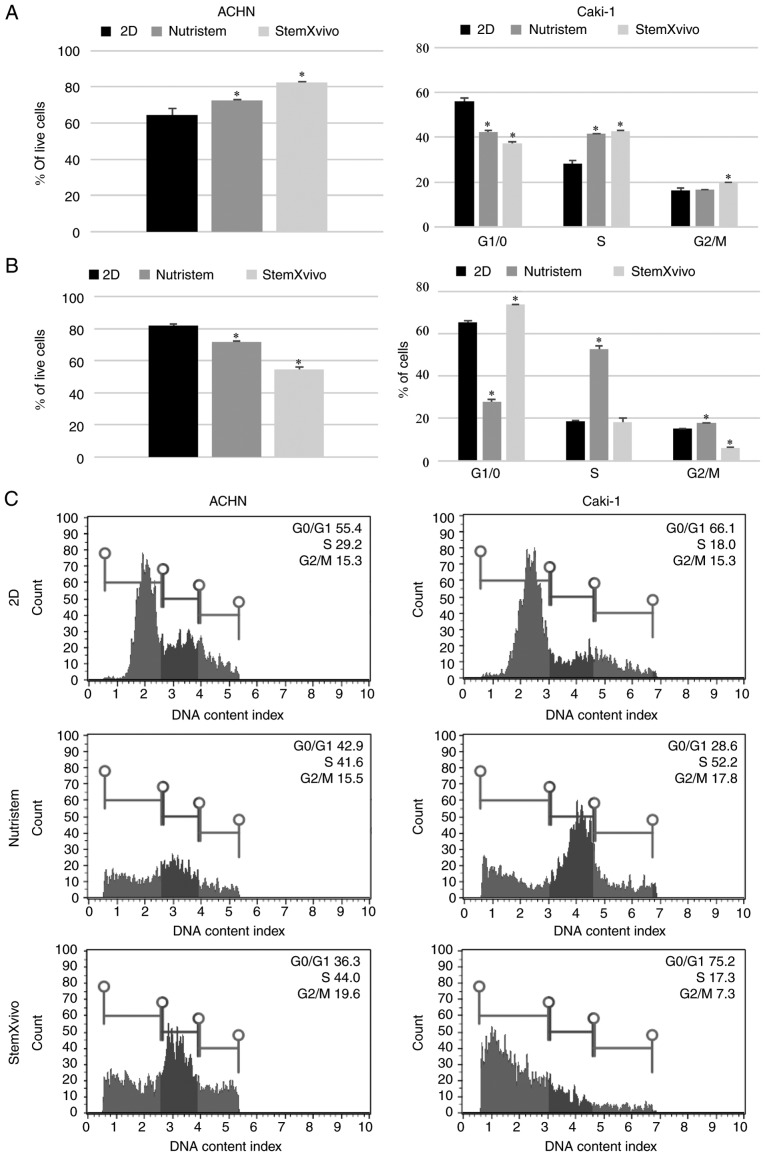
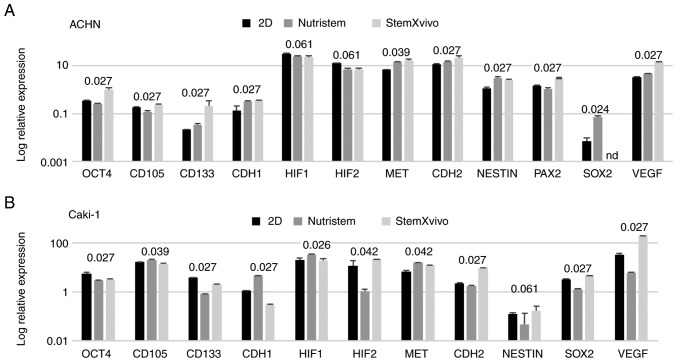
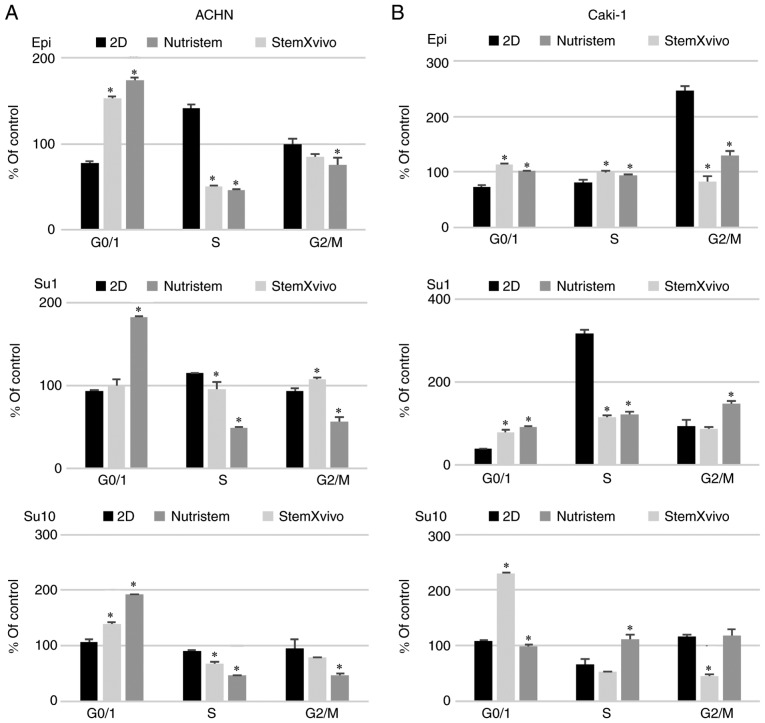
In optimal 3D spheroid-rich conditions (StemXVivo/poly-D-lysine combination) ACHN cells formed small spheroids with remaining adherent cells of an epithelial phenotype (Fig. 1). Such culture conditions significantly increased the viability of the cells and promoted their proliferation as defined by an increased percentage of cells in the S and G2 phases (Fig. 2A). The levels of expression of most stem-related genes tended to be increased in comparison to these levels in the 2D culture (P=0.081, Fig. 3A; SOX2 was not detected in this sample), but both CDH1 and CDH2 were overexpressed together with VEGF. In optimal 3D spheroid-rich conditions (StemXVivo/laminin combination), Caki-1 cells formed large dark structures (Fig. 1) which were later characterized by significantly reduced cell viability (Fig. 2B). Also a reduced percentage of proliferating cells (G2) was observed with a simultaneous G0/G1 phase shift (Fig. 2B). At the same time these cells exhibited the gene expression pattern characteristic of EMT, a decrease in CDH1 and increase in CDH2; however, most stem-related genes, apart from SOX2 and CD105, were downregulated in these cells in comparison to the 2D culture (Fig. 3B). In addition, the levels of VHL and NANOG gene expression were determined; however, they were not detected in the tested cell lines under these conditions.
Figure 2.
Effect of 3D structure formation on cell characteristics (viability and cell cycle distribution). (A) ACHN cells cultured on poly-D-lysine-coated surface in 3D media and standard monolayer. (B) Caki-1 cells cultured on laminin-coated surface in 3D media and standard monolayer. (C) Representative histograms of one technical replicate of cell cycle distribution in different culture variants. *P<0.05 vs. 2D culture, using Mann-Whitney U test; n=3 wells.
Figure 3.
Effect of 3D structure formation on gene expression (log relative expression) as determined using real-time PCR in (A) ACHN cells cultured on poly-D-lysine-coated surface in 3D media and standard monolayer or (B) Caki-1 cells cultured on laminin-coated surface in 3D media and standard monolayer. P-values using the Kruskal-Wallis test are presented over the graphs for each gene. P-values <0.05 are in bold print; n=2 wells. OCT4 or POU5F1, POU class 5 homeobox 1; CD105 or ENG, endoglin; CD133 or PROM1, prominin 1; CDH1, cadherin 1; HIF1, hypoxia-inducible factor 1-alpha; HIF2, hypoxia-inducible factor 2-alpha; MET, MET proto-oncogene, receptor tyrosine kinase; CDH2, cadherin 2; NESTIN, nestin; PAX2, paired box 2; SOX2, SRY-box 2; VEGF, vascular endothelial growth factor.
Growth factor deprivation does not promote a pluripotent phenotype of 3D RCC spheroids
Growth factor deprivation (Nutristem medium) on the poly-D-lysine surface promoted CDH1 overexpression in ACHN cells. These cells formed small, heterogeneous cell aggregates with high viability and characterized by accumulation in the S phase of the cell cycle (Figs. 1 and 2A). In the formed 3D structures, RCC stem-related genes (CD105, OCT4) displayed decreased expression while other genes (SOX2, CD133, NESTIN) were overexpressed in comparison to the 2D-cultured cells. c-MET, CDH2 and VEGF were overexpressed in these cells (Fig. 3A).
In contrast, Caki-1 growth in Nutristem medium upon attachment to laminin resulted in downregulation of the expression of various stem-related genes, CD133 and SOX2 (Fig. 3B), that were upregulated in ACHN. In optimal 3D spheroid-rich conditions (Nutristem/laminin), Caki-1 cells formed uniform spheroids with smooth edges (Fig. 1), but the viability of cells in spheroids was slightly reduced when compared to cells in the standard 2D monolayer culture (Fig. 2B). Under these conditions, HIF1 was overexpressed while VEGF expression was decreased (Fig. 3B), and an increased percentage of cells in the S and G2 phases (Fig. 2B) was noted.
3D growth affects RCC cell drug susceptibility
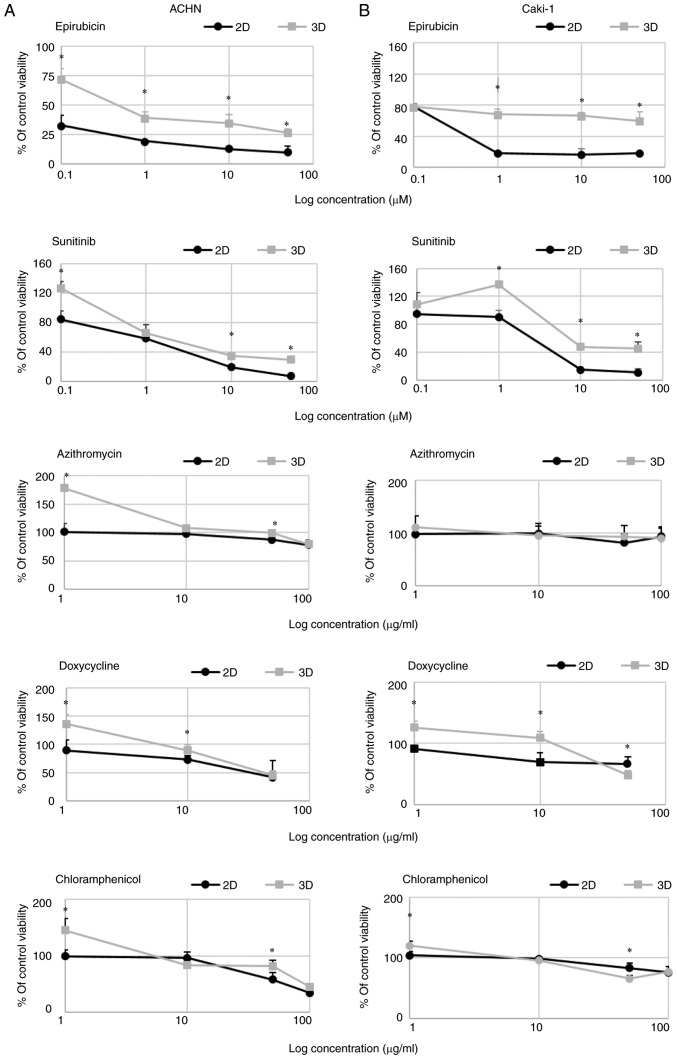
3D growth affected RCC cell susceptibility to TKIs and antibiotics in the tested model. Cells were more resistant to sunitinib and epirubicin in 3D than in 2D culture (Fig. 4). In 3D cell culture conditions, the IC50 values for sunitinib were >5.5- and 2.4-fold higher in comparison to the 2D culture for ACHN and Caki-1 cells, respectively. Moreover, low concentration of sunitinib caused a mild increase in 3D cell growth of the tested cells. The differences in the IC50 values were even higher for epirubicin; spheroids formed by Caki-1 cells were resistant to >80 times higher concentration of the drug, while in the case of ACHN cells the difference was 180-fold in comparison to the monolayer (Table IV).
Figure 4.
Dose-dependent changes in (A) ACHN and (B) Caki-1 cell viability (presented as % of control, untreated cells) in response to drug addition to 2D- (RPMI/FBS) or 3D-cultured cells (StemXvivo) as determined by Alamar Blue reduction. x-axis represents the log concentration of each tested drug. *P<0.05, 2D vs. 3D; using the Mann-Whitney U test; n=3 wells.
Table IV.
The calculated values of IC50 (µM) for epirubicin and sunitinib in the tested cell lines.
| Cell line | Drug | 2D | 3D |
|---|---|---|---|
| ACHN | Epirubicin | 0.005 | 0.9 |
| Sunitinib | 4.7 | 25.9 | |
| Caki-1 | Epirubicin | 0.9 | 75.1 |
| Sunitinib | 17.0 | 40.3 |
IC50, the half maximal inhibitory concentration.
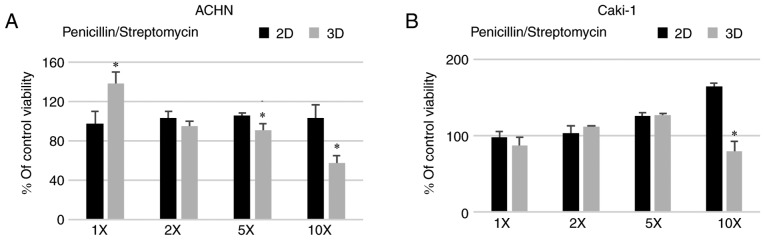
A standard concentration of penicillin/streptomycin (100 U/ml, 100 µg/ml, respectively) routinely used in cell culture did not affect the formation of 3D structures in the metastatic RCC cell lines (Fig. 5) with a similar observation in the case of the primary 786-O cell line (data not shown). On the contrary, penicillin/streptomycin enhanced 293 cell sphere formation (data not shown). However, when the antibiotic solution was added at a higher concentration, 5 or 10 times (5X and 10X) more than used in standard culture, the viability of the cells grown in 3D conditions was reduced for both ACHN and Caki-1 cell lines. Azithromycin had a non-significant effect on the growth of cells in 2D and 3D in the tested concentration range (Fig. 4) but surprisingly it increased the number of spheres formed by ACHN cells (Fig. 6C). RCC cells showed some susceptibility to chloramphenicol; however, it was not dependent on culture conditions. In both cell lines, 2D culture condition sensitized the cells to doxycycline (Fig. 4).
Figure 5.
Effect of different concentrations of penicillin/streptomycin on the viability (% of control, untreated cells) of (A) ACHN and (B) Caki-1 cells cultured in 2D (RPMI/FBS) or 3D (StemXvivo) media as determined by Alamar Blue reduction. *P<0.05, 2D vs. 3D; using Mann-Whitney U test; n=3 wells. 5X and 10X, 5 and 10 times more than used in standard culture.
Figure 6.
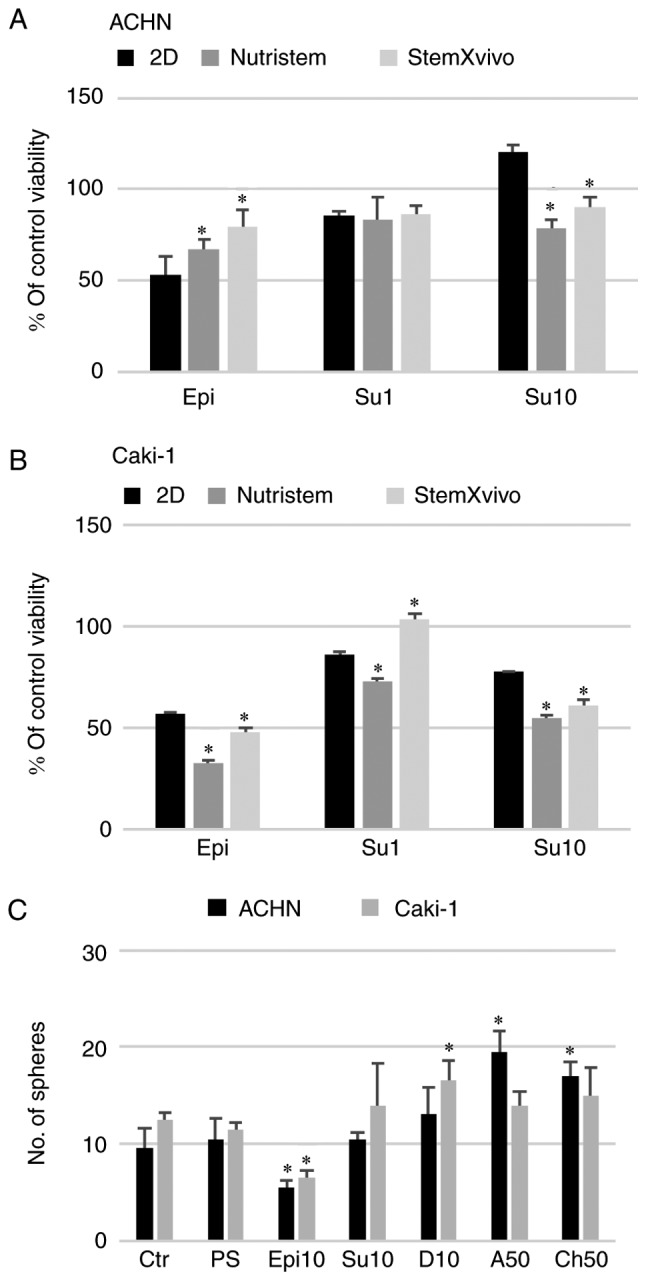
Effect of 3D structure formation on the cell reaction to in vitro drug treatment (viability). (A) ACHN cells cultured on poly-D-lysine-coated surface in 3D media and standard monolayer or (B) Caki-1 cells cultured on laminin-coated surface in 3D media and standard monolayer. *P<0.05 vs. 2D; using Mann-Whitney U test. y-axis represents % viability of the control, untreated cells. Epi, epirubicin at 1 µM; Su1, sunitinib at 1 µM; Su10, sunitinib at 10 µM. (C) Effect of the selected doses of the tested drugs on the sphere morphology and number. *P<0.05, 2D vs. 3D; using Mann-Whitney U test. Ctr, control; PS, Penicillin/Streptomycin; Epi10, epirubicin at 10 µM; Su10, sunitinib at 10 µM; D10, doxycycline at 10 µM; A50, azithromycin at 50 µM; Ch50, chloramphenicol at 50 µM. n=3 wells.
3D growth affects RCC cell susceptibility to chemotherapy and tyrosine kinase inhibitor
Sunitinib had a direct pro-apoptotic effect on RCC cells, although it was less potent than the control drug, epirubicin (Fig. 6). ACHN cells cultured in monolayer were sensitive to epirubicin (Epi, 1 µM) and low-dose sunitinib (Su1, 1 µM; Fig. 6A). However, a higher concentration of the drug (Su10, 10 µM) in this model promoted the survival of cells isolated from spheroids. A lower number of spheres was observed only after epirubicin addition (Fig. 6C). Moreover, in the StemXvivo ACHN cell culture, addition of sunitinib at a low concentration to formed 3D spheres was toxic and halted cells in the G0/G1 phase (Fig. 7A). 3D structures formed by ACHN cells also had enhanced resistance to epirubicin (Fig. 6A). These cells were also resistant to S phase arrest found in 2D conditions and accumulated in the G0/G1 phase (Fig. 7A) and had downregulated CSC-related genes in response to epirubicin (data not shown). In Caki-1 cells that had already compromised viability in the 3D structures (Fig. 2B), epirubicin further reduced the percentage of live cells; the addition of the drug to formed 3D structures had a strong cytotoxic effect. The percentage of live cells recovered from spheres after epirubicin treatment was lower than that noted in the monolayer culture (Fig. 6B) although in the Alamar Blue assay 3D cultures were more resistant to the drug (Fig. 4B). At the same time, there was no effect of Epi on the cell cycle; the cells were protected from a G2 arrest (observed in the monolayer culture), especially in Nutristem medium where simultaneously a slight increase in the G0/G1 contribution was observed (Fig. 7B). Caki-1 cells in the monolayer were sensitive to sunitinib in a dose-dependent manner (Fig. 6B) as observed previously (Fig. 4B) with S phase arrest at a low dose only. At the higher concentration in response to Su, the cells accumulated rather in the G2 phase, as it was observed for Epi treatment. In the case of the 3D culture, only the StemXvivo variant maintained an enhanced resistance to Su (at the lower tested concentration) observed in the initial experiment; simultaneously these cells accumulated in the G2 phase of the cell cycle. In growth factor deprivation medium (Nutristem medium), 10 µM sunitinib and epirubicin affected the cell cycle by retaining cells in the G0/G1 phase (Fig. 7B).
Figure 7.
Effect of 3D structure formation on cell reaction to in vitro drug treatment (cell cycle distribution): (A) ACHN cells cultured on poly-D-lysine-coated surface in 3D media and standard monolayer or (B) Caki-1 cells cultured on laminin-coated surface in 3D media and standard monolayer. *P<0.05 vs. 2D; using Mann-Whitney U test; n=2 wells. y-axis represents % of control, untreated cells in respective culture variant. Epi, epirubicin at 1 µM; Su1, sunitinib at 1 µM; Su10, sunitinib at 10 µM.
Discussion
In the present study, using bio-mimic 3D renal cell carcinoma (RCC) cell culture systems, we aimed to investigate the drug susceptibility of cells in a model more relevant to in vivo conditions (45) in terms of cell-cell interactions, cancer stem cell subpopulation presence and intratumoral hypoxia (51,52). We previously demonstrated that tyrosine kinase inhibitors (TKIs) are directly active against RCC stem cells (35). We also demonstrated that metastatic RCC cell lines (ACHN and Caki-1) had higher colony-forming ability in comparison to primary RCC cell lines (53), and that the ACHN cell line cultured under sphere formation induction conditions [b27 1X, epidermal growth factor (20 ng/ml) and basic fibroblast growth factor (25 ng/ml)] harbored a stem-like subpopulation (16). In the present study, we continued to describe specific factors influencing RCC and RCC-CSC 3D growth and verification of the altered drug susceptibility of RCC cells grown in 3D and molecular changes evoked by these growth conditions. We previously demonstrated that RCC-CSCs from primary tumors are targeted by TKIs (35) and here we described how metastatic tumor-derived cells behave under TKI treatment and how 3D growth alters drug susceptibility in RCC. With this we aimed to develop a system for future novel drug testing that would be more relevant for pre-clinical research than the standard 2D RPMI-based culture.
In the first step, optimal RCC cell 3D growth conditions and characteristics of resultant spheroids were described. In our model, StemXVivo or Nutristem media on poly-D-lysine-coated plates for ACHN and laminin-coated plates for Caki-1 were the most efficient conditions for promoting 3D growth of cells and these sets should be used in further studies. As epithelial mesenchymal transition (EMT) is an important step for cancer cells to acquire metastatic capability and malignant tumor progression, mesenchymal stem cell-inducing medium (StemXvivo) was used to activate cells. EMT is characterized by a phenotypic switch-acquisition of a fibroblast-like morphology, reducing cell polarity and cell-to-cell contacts (54). In accordance with this, we demonstrated reduced expression of the gene encoding E-cadherin (CDH1) epithelial marker and upregulation of the gene encoding N-cadherin (CDH2) mesenchymal marker in Caki-1 cells in mesenchymal medium on a laminin surface, which is the most important characteristic of cancer cells undergoing EMT. Moreover, anchorage-dependent cells cultured in low or serum-free media need cell-adhesive molecules for adequate cell attachment and spreading. These molecules are cell-specific and this was also confirmed for RCC. Optimal spheroid growth was shown for ACHN cells cultured on a poly-D-lysine-coated surface and Caki-1 cells cultured on laminin. Previously it was shown that association of laminin may serve as a stimulatory scaffold to support 3D growth and in melanoma stem cells it was found to upregulate the genes encoding Nestin and vascular endothelial growth factor receptor-1 (VEGFR-1). Laminin also promoted melanoma cell mitogenesis, growth and migration and as we report here this is also true for RCC cells (55). Moreover, under growth factor deprivation conditions (Nutristem medium) when attached to laminin the viability of cells in spheroids was reduced when compared to the standard 2D culture (Fig. 5B), suggesting the presence of necrotic areas inside these structures. In vivo, overexpression of HIF1 in these cells (Fig. 3B) may be related to reduced pO2 inside spheroids measured by electron paramagnetic resonance (EPR) as previously observed (56). In this previous study, the authors observed that the smallest spheroid had the smallest hypoxic zone. Hypoxia inside the tumor mass is widely observed in vivo (57–60), therefore the developed 3D model can indeed present a more biologically relevant alternative to standard monolayer cultures. Moreover, decreased expression of stem-related genes and the increase in the expression of the CDH1 gene encoding E-cadherin may indicate a low level of aggressiveness of the Nutristem/laminin model. In contrast, our results suggest that the StemXvivo/laminin culture variant of Caki-1 cells represents a potentially CSC-rich RCC culture. It was observed that individual cells were detaching from these spheres-cell shedding; this may suggest increased metastatic potential of these cells (61).
Although the presented culture conditions induced the formation of 3D structures by the tested cell lines, structures were heterogeneous with a limited number of CSCs within. Mostly, formation of the 3D structures coincided with an increase in the S phase of the cell cycle, but the expression of CSC-related genes was variable as defined by expression of stem transcription factors (OCT4, SOX2, NES) and RCC CSC putative markers (CD105, CD133).
Additionally, in one culture variant (Caki-1 cells on laminin surface and Nutristem medium) an increase in HIF1 expression occurred together with VEGF downregulation. In the other Caki-1 culture variant (StemXvivo medium), we observed the indication of EMT as CDH1 expression decreased with simultaneous upregulation of CDH2 while the expression of CSC-related genes, apart from NESTIN and SOX2, was decreased. This may be due to unaltered expression of the HIF1/2 genes which were shown to regulate the transcription factors responsible for cell pluripotency (62–64). We believe that the CSC gene expression pattern also changed as a consequence of long-term incubation while with sphere growth heterogeneity of cells increased as has been shown in other models (65–68). We also believe similarly that dynamic changes in pO2-mediated HIF expression are responsible for the observed results in prolonged 3D cultures (69,70).
Secondly, our research also aimed to verify the possibility to target CSCs in renal cancer by an antibiotic treatment that was reported to be effective in other cancers (41). Standard concentration of penicillin and streptomycin did not alter the spheroid forming ability of the tested RCC cells and was found to be safe for 3D experiments while it affects 3D growth at high concentrations. However, this may not apply to different cancers or even other, not assayed, RCC cell lines (41). Penicillin-streptomycin enhanced Caki-1 2D cell growth as observed previously for colon cancer cell lines (42), but its mechanism requires further analysis. The tested RCC cell lines were also insensitive to azithromycin, both the 2D and the sphere-forming population. In contrast, doxycycline was cytotoxic to RCC cells and spheroids with CSCs being more resistant than 2D cultured cells. Doxycycline targets mitochondria, due to homology of bacterial and mitochondrial ribosomes (71) and it may be that differential resistance to the drug characteristic for the 3D structures is a consequence of altered glycolytic metabolism (72). However, unresponsiveness to the other tested mitochondrial-targeting antibiotics remains to be explained. Tetracyclines including doxycycline also inhibit the activity of metalloproteinases (MMPs) which were shown to be important players in tumor progression (73), also in the case of RCC (74–76) and cells in spheroids are expected to express higher levels of MMPs (77). The mechanism of the cytotoxic effect of doxycycline against RCC cells needs to be elucidated.
In the final step, we analyzed the effects of cytotoxic and targeted drugs, epirubicin or sunitinib, on the RCC cells grown in spheroid cultures. Sunitinib was also tested as it was repeatedly reported that despite its antiangiogenic activity, this molecule also possesses a direct antitumor effect in vitro inducing apoptosis of RCC cells (44,78) in clinically relevant intratumoral concentrations (79). In our experiments, sunitinib exerted a visible anti-RCC activity at a concentration as low as 0.4 µM against cells cultured in the monolayer (Fig. 4). Cell proliferation was inhibited by sunitinib both in a mesenchymal phenotype (StemXvivo culture) and growth factor deprivation (Nutristem) conditions, which confirms its activity against CSCs (35). Higher concentrations of the drug were needed to effectively inhibit cells in 3D than those grown in 2D and the metastatic ACHN cell line was more susceptible (lower IC50 values). This is consistent with data reported for various in vitro cancer models (80–82). Moreover, increased resistance to cytotoxic drugs is suggested to result from the presence of CSCs in 3D cultures (83). Changes induced by the employed 3D culture conditions altered the RCC cell reaction to drugs. However, cells which were forming 3D structures were more resistant to the drug only when initially their viability was enhanced by the spheroid formation-promoting media. Since cell lines showed a consistent resistance to the drug in 3D spheroids (Fig. 4) we hypothesize that the drug resistance mechanism is activated in the CSC population (84). In accordance with this, ACHN spheroids in StemXVivo medium upon sunitinib treatment accumulated in the G0/G1 phase, which indicates that cell quiescence may be responsible for CSC drug resistance (85,86). On the contrary, a different mechanism seems to underlie Caki-1 resistance. Caki-1 spheroid-derived cells were found to shift to the G2/M cell cycle phase upon sunitinib treatment. We believe that this is a result of sunitinib inducing the cells to re-enter the cell cycle, from a population of G0/1 untreated cells, and as a result enhanced growth in 3D in response to the treatment.
The mechanism of epirubicin cellular toxicity was shown previously to be cell line dependent (87). Concordantly in our study, the Caki-1 cell line was more resistant to epirubicin than ACHN cells (Table IV). Furthermore, ACHN and Caki-1 cell growth in a monolayer was arrested by the drug in the S or G2 phase, respectively. The addition of epirubicin to 3D-cultured cells modified the cell cycle distribution. In particular, epirubicin-resistant ACHN cells accumulated in the G0/G1 phase which was previously associated with enhanced cell resistance (88). On the other hand, Caki-1 spheroid cells, which had reduced viability after epirubicin treatment in comparison to 2D culture, showed weak changes in the cell cycle with a small number of cells undergoing G2 arrest. This may be due to the lower contribution of spheroid-forming cells in the ACHN cell line, as opposed to Caki-1; there were remaining adherent cells in the culture (Fig. 1) which may have mediated a less pronounced effect on cell growth. This confirms previous research showing that cancer stem-like cells from tumor spheres are more resistant to epirubicin (89).
In the present study, we developed feasible and consistent methods for culturing RCC spheroids for two different cell lines using specific media and a surface coating stimulation method. We characterized in detail the necessary growth conditions for these cell lines as well as the growth viability over the duration of the spheroid growth. Our results revealed that the formation of spheres or 3D structures partially results from CSC phenotype induction upon epithelial-mesenchymal transition (EMT). We screened four types of 3D cultures that induced a different cell morphology and altered cell characteristics. Although 3D-grown cells tended to be more drug-resistant than those cultured in a standard monolayer, this phenomenon was dependent on the adopted protocol of cell viability measurement (Alamar Blue assay during culture or direct viability of cells isolated from spheres). Therefore, the results obtained from the 3D cultured cells need to be analyzed with caution in respect to cell specificity. Our model may also be of interest to understand issues related to hypoxia and growth necrosis in tumor spheroids. 3D spheroids are a useful model to determine the morphological growth characteristics of cell lines which are not found in routine 2D culture methods. 3D culture methods may provide a better alternative to study the process of epithelial-mesenchymal transition. Spheroids may be used as assays for stem cell activity and cancer cell self-renewal and RCC tumorigenesis and therefore represent a novel model for exploring RCC pathobiology. Due to the more complex architecture they may be more relevant to the in vivo tumor drug response.
Acknowledgements
The authors thank Mr. Paweł Krasowski for the administrative support of the research projects and Professor M. Rękas, Scientific Director of the Military Institute of Medicine for the continued support of the research.
Funding
The study was supported by the National Centre for Research and Development (NCBR, Poland) LIDER grant no. Lider/031/625/L-4/12/NCBR/2013. The cost of publication was supported by Military Institute of Medicine statutory scientific funding.
Availability of data and materials
Raw data is available upon E-mail request from KKB (kbrodaczewska@wim.mil.pl; ORCID: 0000-0003-2374-3600) and ZFB (lom@wim.mil.pl; ORCID: 0000-0002-1454-5437) or Science Department of the Military Institute of Medicine (Wojskowy Instytut Medyczny, Pion Nauki 01-141 Warszawa 44, ul. Szaserów 128, pionnauki@wim.mil.pl; nauka@wim.mil.pl; http://www.wim.mil.pl/nauka).
Authors' contributions
AMC designed the study, was responsible for acquisition of funding, developed the research conception, design and methodology, participated in data analysis and interpretation of data and manuscript drafting and revision. KKB, ZBF and KMO developed the research methods, participated in acquisition of data and manuscript writing including revisions. CS supervised revised it critically for important intellectual content; CP developed the concept and methods of the study, provided selected reagents. EB interpreted the data, edited and revised the manuscript. All authors approved the final manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Meyerrose TE, Herrbrich P, Hess DA, Nolta JA. Immune-deficient mouse models for analysis of human stem cells. BioTechniques. 2003;35:1262–1272. doi: 10.2144/03356ss06. [DOI] [PubMed] [Google Scholar]
- 2.Gedye C, Sirskyj D, Lobo NC, Meens J, Hyatt E, Robinette M, Fleshner N, Hamilton RJ, Kulkarni G, Zlotta A, et al. Cancer stem cells are underestimated by standard experimental methods in clear cell renal cell carcinoma. Sci Rep. 2016;6:25220. doi: 10.1038/srep25220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grotenhuis BA, Wijnhoven BP, van Lanschot JJ. Cancer stem cells and their potential implications for the treatment of solid tumors. J Surg Oncol. 2012;106:209–215. doi: 10.1002/jso.23069. [DOI] [PubMed] [Google Scholar]
- 4.Buczek M, Escudier B, Bartnik E, Szczylik C, Czarnecka A. Resistance to tyrosine kinase inhibitors in clear cell renal cell carcinoma: From the patient's bed to molecular mechanisms. Biochim Biophys Acta. 2014;1845:31–41. doi: 10.1016/j.bbcan.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Bielecka ZF, Czarnecka AM, Solarek W, Kornakiewicz A, Szczylik C. Mechanisms of acquired resistance to tyrosine kinase inhibitors in clear-cell renal cell carcinoma (ccRCC) Curr Signal Transduct Ther. 2014;8:218–228. doi: 10.2174/1574362409666140206223014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bussolati B, Bruno S, Grange C, Ferrando U, Camussi G. Identification of a tumor-initiating stem cell population in human renal carcinomas. FASEB J. 2008;22:3696–3705. doi: 10.1096/fj.08-102590. [DOI] [PubMed] [Google Scholar]
- 7.Czarnecka A, Matak D, Solarek W, Khan M, Szczylik C. Hypoxia response regulates clear cell renal cell carcinoma tumor initiating cells. BJU Int. 2013;112:1–17. [Google Scholar]
- 8.Debeb BG, Zhang X, Krishnamurthy S, Gao H, Cohen E, Li L, Rodriguez AA, Landis MD, Lucci A, Ueno NT, et al. Characterizing cancer cells with cancer stem cell-like features in 293T human embryonic kidney cells. Mol Cancer. 2010;9:180. doi: 10.1186/1476-4598-9-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gassenmaier M, Chen D, Buchner A, Henkel L, Schiemann M, Mack B, Schendel DJ, Zimmermann W, Pohla H. CXC chemokine receptor 4 is essential for maintenance of renal cell carcinoma-initiating cells and predicts metastasis. Stem Cells. 2013;31:1467–1476. doi: 10.1002/stem.1407. [DOI] [PubMed] [Google Scholar]
- 10.Ueda K, Ogasawara S, Akiba J, Nakayama M, Todoroki K, Ueda K, Sanada S, Suekane S, Noguchi M, Matsuoka K, Yano H. Aldehyde dehydrogenase 1 identifies cells with cancer stem cell-like properties in a human renal cell carcinoma cell line. PLoS One. 2013;8:e75463. doi: 10.1371/journal.pone.0075463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khan MI, Czarnecka AM, Lewicki S, Helbrecht I, Brodaczewska K, Koch I, Zdanowski R, Król M, Szczylik C. Comparative gene expression profiling of primary and metastatic renal cell carcinoma stem cell-like cancer cells. PLoS One. 2016;11:e0165718. doi: 10.1371/journal.pone.0165718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu J, Guan W, Liu P, Dai J, Tang K, Xiao H, Qian Y, Sharrow AC, Ye Z, Wu L, Xu H. Endoglin is essential for the maintenance of self-renewal and chemoresistance in renal cancer stem cells. Stem Cell Reports. 2017;9:464–477. doi: 10.1016/j.stemcr.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beaver CM, Ahmed A, Masters JR. Clonogenicity: Holoclones and meroclones contain stem cells. PLoS One. 2014;9:e89834. doi: 10.1371/journal.pone.0089834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rappa G, Mercapide J, Anzanello F, Prasmickaite L, Xi Y, Ju J, Fodstad O, Lorico A. Growth of cancer cell lines under stem cell-like conditions has the potential to unveil therapeutic targets. Exp Cell Res. 2008;314:2110–2122. doi: 10.1016/j.yexcr.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lichner Z, Saleh C, Subramaniam V, Seivwright A, Prud'homme GJ, Yousef GM. miR-17 inhibition enhances the formation of kidney cancer spheres with stem cell/tumor initiating cell properties. Oncotarget. 2015;6:5567–5581. doi: 10.18632/oncotarget.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matak D, Brodaczewska KK, Szczylik C, Koch I, Myszczyszyn A, Lipiec M, Lewicki S, Szymanski L, Zdanowski R, Czarnecka AM. Functional significance of CD105-positive cells in papillary renal cell carcinoma. BMC Cancer. 2017;17:21. doi: 10.1186/s12885-016-2985-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matak D, Szymanski L, Szczylik C, Sledziewski R, Lian F, Bartnik E, Sobocinska A, Czarnecka AM. Biology of renal tumour cancer stem cells applied in medicine. Contemp Oncol (Pozn) 2015;19:A44–A51. doi: 10.5114/wo.2014.47128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Czarnecka AM, Szczylik C. Renal cell carcinoma cancer stem cells as therapeutic targets. Curr Sign Trans Ther. 2013;8:203–209. doi: 10.2174/1574362409666140206222251. [DOI] [Google Scholar]
- 19.Peired AJ, Sisti A, Romagnani P. Renal cancer stem cells: Characterization and targeted therapies. Stem Cells Int. 2016;2016:8342625. doi: 10.1155/2016/8342625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bomken S, Fiser K, Heidenreich O, Vormoor J. Understanding the cancer stem cell. Br J Cancer. 2011;103:439–445. doi: 10.1038/sj.bjc.6605821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakano I, Chiocca EA. Finding drugs against CD133+ glioma subpopulations. J Neurosurg. 2011;114:648–650. doi: 10.3171/2009.11.JNS091668. [DOI] [PubMed] [Google Scholar]
- 22.Qiu W, Wang X, Leibowitz B, Liu H, Barker N, Okada H, Oue N, Yasui W, Clevers H, Schoen RE, et al. Chemoprevention by nonsteroidal anti-inflammatory drugs eliminates oncogenic intestinal stem cells via SMAC-dependent apoptosis. Proc Natl Acad Sci USA. 2010;107:20027–20032. doi: 10.1073/pnas.1010430107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prud'homme GJ, Glinka Y, Toulina A, Ace O, Subramaniam V, Jothy S. Breast cancer stem-like cells are inhibited by a non-toxic aryl hydrocarbon receptor agonist. PLoS One. 2010;5:e13831. doi: 10.1371/journal.pone.0013831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morrison R, Schleicher SM, Sun Y, Niermann KJ, Kim S, Spratt DE, Chung CH, Lu B. Targeting the mechanisms of resistance to chemotherapy and radiotherapy with the cancer stem cell hypothesis. J Oncol. 2011;2011:941876. doi: 10.1155/2011/941876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Escudier B, Eisen T, Stadler WM, Szczylik C, Oudard S, Siebels M, Negrier S, Chevreau C, Solska E, Desai AA, et al. Sorafenib in advanced clear-cell renal-cell carcinoma. N Engl J Med. 2007;356:125–134. doi: 10.1056/NEJMoa060655. [DOI] [PubMed] [Google Scholar]
- 26.Motzer RJ, Hutson TE, Tomczak P, Michaelson MD, Bukowski RM, Rixe O, Oudard S, Negrier S, Szczylik C, Kim ST, et al. Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. N Engl J Med. 2007;356:115–124. doi: 10.1056/NEJMoa065044. [DOI] [PubMed] [Google Scholar]
- 27.Sternberg CN, Davis ID, Mardiak J, Szczylik C, Lee E, Wagstaff J, Barrios CH, Salman P, Gladkov OA, Kavina A, et al. Pazopanib in locally advanced or metastatic renal cell carcinoma: Results of a randomized phase III trial. J Clin Oncol. 2011;28:1061–1068. doi: 10.1200/JCO.2009.23.9764. [DOI] [PubMed] [Google Scholar]
- 28.Yuan ZX, Mo J, Zhao G, Shu G, Fu HL, Zhao W. Targeting strategies for renal cell carcinoma: From renal cancer cells to renal cancer stem cells. Front Pharmacol. 2016;7:423. doi: 10.3389/fphar.2016.00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roskoski R., Jr Sunitinib: A VEGF and PDGF receptor protein kinase and angiogenesis inhibitor. Biochem Biophys Res Commun. 2007;356:323–328. doi: 10.1016/j.bbrc.2007.02.156. [DOI] [PubMed] [Google Scholar]
- 30.Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, et al. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 31.Huang D, Ding Y, Li Y, Luo WM, Zhang ZF, Snider J, Vandenbeldt K, Qian CN, the BT. Sunitinib acts primarily on tumor endothelium rather than tumor cells to inhibit the growth of renal cell carcinoma. Cancer Res. 2010;70:1053–1062. doi: 10.1158/0008-5472.CAN-09-3722. [DOI] [PubMed] [Google Scholar]
- 32.Brossa A, Grange C, Mancuso L, Annaratone L, Satolli MA, Mazzone M, Camussi G, Bussolati B. Sunitinib but not VEGF blockade inhibits cancer stem cell endothelial differentiation. Oncotarget. 2015;6:11295–11309. doi: 10.18632/oncotarget.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Potapova O, Laird AD, Nannini MA, Barone A, Li G, Moss KG, Cherrington JM, Mendel DB. Contribution of individual targets to the antitumor efficacy of the multitargeted receptor tyrosine kinase inhibitor SU11248. Mol Cancer Ther. 2006;5:1280–1289. doi: 10.1158/1535-7163.MCT-03-0156. [DOI] [PubMed] [Google Scholar]
- 34.Abrams TJ, Murray LJ, Pesenti E, Holway VW, Colombo T, Lee LB, Cherrington JM, Pryer NK. Preclinical evaluation of the tyrosine kinase inhibitor SU11248 as a single agent and in combination with ‘standard of care’ therapeutic agents for the treatment of breast cancer. Mol Cancer Ther. 2003;2:1011–1021. [PubMed] [Google Scholar]
- 35.Czarnecka AM, Solarek W, Kornakiewicz A, Szczylik C. Tyrosine kinase inhibitors target cancer stem cells in renal cell cancer. Oncol Rep. 2016;35:1433–1442. doi: 10.3892/or.2015.4514. [DOI] [PubMed] [Google Scholar]
- 36.Diaz R, Nguewa PA, Redrado M, Manrique I, Calvo A. Sunitinib reduces tumor hypoxia and angiogenesis, and radiosensitizes prostate cancer stem-like cells. Prostate. 2015;75:1137–1149. doi: 10.1002/pros.22980. [DOI] [PubMed] [Google Scholar]
- 37.Varna M, Gapihan G, Feugeas JP, Ratajczak P, Tan S, Ferreira I, Leboeuf C, Setterblad N, Duval A, Verine J, et al. Stem cells increase in numbers in perinecrotic areas in human renal cancer. Clin Cancer Res. 2015;21:916–924. doi: 10.1158/1078-0432.CCR-14-0666. [DOI] [PubMed] [Google Scholar]
- 38.Zhu Y, Liu H, Xu L, An H, Liu W, Liu Y, Lin Z, Xu J. p21-activated kinase 1 determines stem-like phenotype and sunitinib resistance via NF-kappaB/IL-6 activation in renal cell carcinoma. Cell Death Dis. 2015;6:e1637. doi: 10.1038/cddis.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chinchar E, Makey KL, Gibson J, Chen F, Cole SA, Megason GC, Vijayakumar S, Miele L, Gu JW. Sunitinib significantly suppresses the proliferation, migration, apoptosis resistance, tumor angiogenesis and growth of triple-negative breast cancers but increases breast cancer stem cells. Vasc Cell. 2014;6:12. doi: 10.1186/2045-824X-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conley SJ, Gheordunescu E, Kakarala P, Newman B, Korkaya H, Heath AN, Clouthier SG, Wicha MS. Antiangiogenic agents increase breast cancer stem cells via the generation of tumor hypoxia. Proc Natl Acad Sci USA. 2012;109:2784–2789. doi: 10.1073/pnas.1018866109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lamb R, Ozsvari B, Lisanti CL, Tanowitz HB, Howell A, Martinez-Outschoorn UE, Sotgia F, Lisanti MP. Antibiotics that target mitochondria effectively eradicate cancer stem cells, across multiple tumor types: Treating cancer like an infectious disease. Oncotarget. 2015;6:4569–4584. doi: 10.18632/oncotarget.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Relier S, Yazdani L, Ayad O, Choquet A, Bourgaux JF, Prudhomme M, Pannequin J, Macari F, David A. Antibiotics inhibit sphere-forming ability in suspension culture. Cancer Cell Int. 2016;16:6. doi: 10.1186/s12935-016-0277-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mays AN, Osheroff N, Xiao Y, Wiemels JL, Felix CA, Byl JA, Saravanamuttu K, Peniket A, Corser R, Chang C, et al. Evidence for direct involvement of epirubicin in the formation of chromosomal translocations in t(15;17) therapy-related acute promyelocytic leukemia. Blood. 2010;115:326–330. doi: 10.1182/blood-2009-07-235051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang XL, Zhang Y, Luo CL, Wu XH. Targeting renal cell carcinoma with gambogic acid in combination with sunitinib in vitro and in vivo. Asian Pac J Cancer Prev. 2012;13:6463–6468. doi: 10.7314/APJCP.2012.13.12.6463. [DOI] [PubMed] [Google Scholar]
- 45.Bielecka ZF, Maliszewska-Olejniczak K, Safir IJ, Szczylik C, Czarnecka AM. Three-dimensional cell culture model utilization in cancer stem cell research. Biol Rev Camb Philos Soc. 2017;92:1505–1520. doi: 10.1111/brv.12293. [DOI] [PubMed] [Google Scholar]
- 46.Brodaczewska KK, Szczylik C, Fiedorowicz M, Porta C, Czarnecka AM. Choosing the right cell line for renal cell cancer research. Mol Cancer. 2016;15:83. doi: 10.1186/s12943-016-0565-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim S, Alexander CM. Tumorsphere assay provides more accurate prediction of in vivo responses to chemotherapeutics. Biotechnol Lett. 2014;36:481–488. doi: 10.1007/s10529-013-1393-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 49.Wierzbicki PM, Klacz J, Rybarczyk A, Slebioda T, Stanislawowski M, Wronska A, Kowalczyk A, Matuszewski M, Kmiec Z. Identification of a suitable qPCR reference gene in metastatic clear cell renal cell carcinoma. Tumour Biol. 2014;35:12473–12487. doi: 10.1007/s13277-014-2566-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maliszewska-Olejniczak K, Brodaczewska KK, Bielecka ZF, Czarnecka AM. Three-dimensional cell culture model utilization in renal carcinoma cancer stem cell research. Methods Mol Biol. 2018;1817:47–66. doi: 10.1007/978-1-4939-8600-2_6. [DOI] [PubMed] [Google Scholar]
- 51.Kaminska K, Szczylik C, Bielecka ZF, Bartnik E, Porta C, Lian F, Czarnecka AM. The role of the cell-cell interactions in cancer progression. J Cell Mol Med. 2015;19:283–296. doi: 10.1111/jcmm.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Myszczyszyn A, Czarnecka AM, Matak D, Szymanski L, Lian F, Kornakiewicz A, Bartnik E, Kukwa W, Kieda C, Szczylik C. The role of hypoxia and cancer stem cells in renal cell carcinoma pathogenesis. Stem Cell Rev. 2015;11:919–943. doi: 10.1007/s12015-015-9611-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khan MI, Debski KJ, Dabrowski M, Czarnecka AM, Szczylik C. Gene set enrichment analysis and ingenuity pathway analysis of metastatic clear cell renal cell carcinoma cell line. Am J Physiol Renal Physiol. 2016;311:F424–F436. doi: 10.1152/ajprenal.00138.2016. [DOI] [PubMed] [Google Scholar]
- 54.Zhou N, Lu F, Liu C, Xu K, Huang J, Yu D, Bi L. IL-8 induces the epithelial-mesenchymal transition of renal cell carcinoma cells through the activation of AKT signaling. Oncol Lett. 2016;12:1915–1920. doi: 10.3892/ol.2016.4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Larson AR, Lee CW, Lezcano C, Zhan Q, Huang J, Fischer AH, Murphy GF. Melanoma spheroid formation involves laminin-associated vasculogenic mimicry. Am J Pathol. 2014;184:71–78. doi: 10.1016/j.ajpath.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Langan LM, Dodd NJ, Owen SF, Purcell WM, Jackson SK, Jha AN. Direct Measurements of Oxygen Gradients in Spheroid Culture System Using Electron Parametric Resonance Oximetry. PLoS One. 2016;11:e0149492. doi: 10.1371/journal.pone.0160795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng X, Wang X, Mao H, Wu W, Liu B, Jiang X. Hypoxia-specific ultrasensitive detection of tumours and cancer cells. Nat Commun. 2015;6:5834. doi: 10.1038/ncomms6834. [DOI] [PubMed] [Google Scholar]
- 58.Lal A, Peters H, St Croix B, Haroon ZA, Dewhirst MW, Strausberg RL, Kaanders JH, van der Kogel AJ, Riggins GJ. Transcriptional response to hypoxia in human tumors. J Natl Cancer Inst. 2019;93:1337–1343. doi: 10.1093/jnci/93.17.1337. [DOI] [PubMed] [Google Scholar]
- 59.McKenna1 DJ, Errington R, Pors K. Current challenges and opportunities in treating hypoxic prostate tumors. J Cancer Metastasis Treat. 2018;4:11. doi: 10.20517/2394-4722.2017.54. [DOI] [Google Scholar]
- 60.Ponte KF, Berro DH, Collet S, Constans JM, Emery E, Valable S, Guillamo JS. In vivo relationship between hypoxia and angiogenesis in human glioblastoma: A multimodal imaging study. J Nucl Med. 2017;58:1574–1579. doi: 10.2967/jnumed.116.188557. [DOI] [PubMed] [Google Scholar]
- 61.Gunther S, Ruhe C, Derikito MG, Bose G, Sauer H, Wartenberg M. Polyphenols prevent cell shedding from mouse mammary cancer spheroids and inhibit cancer cell invasion in confrontation cultures derived from embryonic stem cells. Cancer Lett. 2007;250:25–35. doi: 10.1016/j.canlet.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 62.Moreno-Manzano V, Rodriguez-Jimenez FJ, Acena-Bonilla JL, Fustero-Lardíes S, Erceg S, Dopazo J, Montaner D, Stojkovic M, Sánchez-Puelles JM. FM19G11, a new hypoxia-inducible factor (HIF) modulator, affects stem cell differentiation status. J Biol Chem. 2010;285:1333–1342. doi: 10.1074/jbc.M109.008326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Covello KL, Kehler J, Yu H, Gordan JD, Arsham AM, Hu CJ, Labosky PA, Simon MC, Keith B. HIF-2alpha regulates Oct-4: Effects of hypoxia on stem cell function, embryonic development, and tumor growth. Genes Dev. 2006;20:557–570. doi: 10.1101/gad.1399906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mathieu J, Zhang Z, Zhou W, Wang AJ, Heddleston JM, Pinna CM, Hubaud A, Stadler B, Choi M, Bar M, et al. HIF induces human embryonic stem cell markers in cancer cells. Cancer Res. 2011;71:4640–4652. doi: 10.1158/0008-5472.CAN-10-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Min SO, Lee SW, Bak SY, Kim KS. Ideal sphere-forming culture conditions to maintain pluripotency in a hepatocellular carcinoma cell lines. Cancer Cell Int. 2015;15:95. doi: 10.1186/s12935-015-0240-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gupta PB, Fillmore CM, Jiang G, Shapira SD, Tao K, Kuperwasser C, Lander ES. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146:633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 67.Chen YC, Cheng YH, Ingram P, Yoon E. Single cell proteolytic assays to investigate cancer clonal heterogeneity and cell dynamics using an efficient cell loading scheme. Sci Rep. 2016;6:27154. doi: 10.1038/srep27154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pece S, Tosoni D, Confalonieri S, Mazzarol G, Vecchi M, Ronzoni S, Bernard L, Viale G, Pelicci PG, Di Fiore PP. Biological and Molecular Heterogeneity of Breast Cancers Correlates with Their Cancer Stem Cell Content. Cell. 2010;140:62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 69.Nguyen LK, Cavadas MA, Scholz CC, Fitzpatrick SF, Bruning U, Cummins EP, Tambuwala MM, Manresa MC, Kholodenko BN, Taylor CT, Cheong A. A dynamic model of the hypoxia-inducible factor 1α (HIF-1α) network. J Cell Sci. 2013;126:1454–1463. doi: 10.1242/jcs.119974. [DOI] [PubMed] [Google Scholar]
- 70.Lafleur VN, Richard S, Richard DE. Transcriptional repression of hypoxia-inducible factor-1 (HIF-1) by the protein arginine methyltransferase PRMT1. Mol Biol Cell. 2014;25:925–935. doi: 10.1091/mbc.e13-07-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang X, Ryu D, Houtkooper RH, Auwerx J. Antibiotic use and abuse: a threat to mitochondria and chloroplasts with impact on research, health, and environment. Bioessays. 2015;37:1045–1053. doi: 10.1002/bies.201500071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sancho P, Barneda D, Heeschen C. Hallmarks of cancer stem cell metabolism. Br J Cancer. 2016;114:1305–1312. doi: 10.1038/bjc.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gialeli C, Theocharis AD, Karamanos NK. Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. Febs J. 2011;278:16–27. doi: 10.1111/j.1742-4658.2010.07919.x. [DOI] [PubMed] [Google Scholar]
- 74.Sumi T, Nakatani T, Yoshida H, Hyun Y, Yasui T, Matsumoto Y, Nakagawa E, Sugimura K, Kawashima H, Ishiko O. Expression of matrix metalloproteinases 7 and 2 in human renal cell carcinoma. Oncol Rep. 2003;10:567–570. [PubMed] [Google Scholar]
- 75.Lin YW, Lee LM, Lee WJ, Chu CY, Tan P, Yang YC, Chen WY, Yang SF, Hsiao M, Chien MH. Melatonin inhibits MMP-9 transactivation and renal cell carcinoma metastasis by suppressing Akt-MAPKs pathway and NF-κB DNA-binding activity. J Pineal Res. 2017;60:277–290. doi: 10.1111/jpi.12308. [DOI] [PubMed] [Google Scholar]
- 76.Roomi MW, Ivanov V, Kalinovsky T, Niedzwiecki A, Rath M. Modulation of human renal cell carcinoma 786-0 MMP-2 and MMP-9 activity by inhibitors and inducers in vitro. Med Oncol. 2006;23:245–250. doi: 10.1385/MO:23:2:245. [DOI] [PubMed] [Google Scholar]
- 77.Shrivastava S, Steele R, Sowadski M, Crawford SE, Varvares M, Ray RB. Identification of molecular signature of head and neck cancer stem-like cells. Sci Rep. 2015;5:7819. doi: 10.1038/srep07819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xin H, Zhang C, Herrmann A, Du Y, Figlin R, Yu H. Sunitinib inhibition of Stat3 induces renal cell carcinoma tumor cell apoptosis and reduces immunosuppressive cells. Cancer Res. 2009;69:2506–2513. doi: 10.1158/0008-5472.CAN-08-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gotink KJ, Broxterman HJ, Labots M, de Haas RR, Dekker H, Honeywell RJ, Rudek MA, Beerepoot LV, Musters RJ, Jansen G, et al. Lysosomal sequestration of sunitinib: A novel mechanism of drug resistance. Clin Cancer Res. 2011;17:7337–7346. doi: 10.1158/1078-0432.CCR-11-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu H, Wang H, Li C, Zhang T, Meng X, Zhang Y, Qian H. Spheres from cervical cancer cells display stemness and cancer drug resistance. Oncol Lett. 2016;12:2184–2188. doi: 10.3892/ol.2016.4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sun FF, Hu YH, Xiong LP, Tu XY, Zhao JH, Chen SS, Song J, Ye XQ. Enhanced expression of stem cell markers and drug resistance in sphere-forming non-small cell lung cancer cells. Int J Clin Exp Pathol. 2015;8:6287–6300. [PMC free article] [PubMed] [Google Scholar]
- 82.Chen SF, Chang YC, Nieh S, Liu CL, Yang CY, Lin YS. Nonadhesive culture system as a model of rapid sphere formation with cancer stem cell properties. PLoS One. 2012;7:e31864. doi: 10.1371/journal.pone.0031864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Harper LJ, Costea DE, Gammon L, Fazil B, Biddle A, Mackenzie IC. Normal and malignant epithelial cells with stem-like properties have an extended G2 cell cycle phase that is associated with apoptotic resistance. BMC Cancer. 2010;10:166. doi: 10.1186/1471-2407-10-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fernandez-Fuente G, Mollinedo P, Grande L, Vazquez-Barquero A, Fernandez-Luna JL. Culture dimensionality influences the resistance of glioblastoma stem-like cells to multikinase inhibitors. Mol Cancer Ther. 2014;13:1664–1672. doi: 10.1158/1535-7163.MCT-13-0854. [DOI] [PubMed] [Google Scholar]
- 85.Chen W, Dong J, Haiech J, Kilhoffer MC, Zeniou M. Cancer stem cell quiescence and plasticity as major challenges in cancer therapy. Stem Cells Int. 2016;2016:1740936. doi: 10.1155/2016/1740936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moore N, Lyle S. Quiescent, slow-cycling stem cell populations in cancer: A review of the evidence and discussion of significance. J Oncol. 2011;2011:396076. doi: 10.1155/2011/396076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hedenfalk IA, Baldetorp B, Borg A, Oredsson SM. Activated cell cycle checkpoints in epirubicin-treated breast cancer cells studied by BrdUrd-flow cytometry. Cytometry. 1997;29:321–327. doi: 10.1002/(SICI)1097-0320(19971201)29:4<321::AID-CYTO9>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 88.Grabarczyk P, Nahse V, Delin M, Przybylski G, Depke M, Hildebrandt P, Völker U, Schmidt CA. Increased expression of bcl11b leads to chemoresistance accompanied by G1 accumulation. PLoS One. 2010;5(pii):e12532. doi: 10.1371/journal.pone.0012532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang L, Guo H, Lin C, Yang L, Wang X. Enrichment and characterization of cancer stemlike cells from a cervical cancer cell line. Mol Med Rep. 2014;9:2117–2123. doi: 10.3892/mmr.2014.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw data is available upon E-mail request from KKB (kbrodaczewska@wim.mil.pl; ORCID: 0000-0003-2374-3600) and ZFB (lom@wim.mil.pl; ORCID: 0000-0002-1454-5437) or Science Department of the Military Institute of Medicine (Wojskowy Instytut Medyczny, Pion Nauki 01-141 Warszawa 44, ul. Szaserów 128, pionnauki@wim.mil.pl; nauka@wim.mil.pl; http://www.wim.mil.pl/nauka).