Abstract
The G4C2 hexanucleotide repeat expansion mutation in the C9orf72 gene is the most common genetic cause underlying both amyotrophic lateral sclerosis and frontotemporal dementia. Pathologically, these two neurodegenerative disorders are linked by the common presence of abnormal phosphorylated TDP-43 neuronal cytoplasmic inclusions. We compared the number and size of phosphorylated TDP-43 inclusions and their morphology in hippocampi from patients dying with sporadic versus C9orf72-related amyotrophic lateral sclerosis with pathologically defined frontotemporal lobar degeneration with phosphorylated TDP-43 inclusions, the pathological substrate of clinical frontotemporal dementia in patients with amyotrophic lateral sclerosis. In sporadic cases, there were numerous consolidated phosphorylated TDP-43 inclusions that were variable in size, whereas inclusions in C9orf72 amyotrophic lateral sclerosis/frontotemporal lobar degeneration were quantitatively smaller than those in sporadic cases. Also, C9orf72 amyotrophic lateral sclerosis/frontotemporal lobar degeneration homogenized brain contained soluble cytoplasmic TDP-43 that was largely absent in sporadic cases. To better understand these pathological differences, we modelled TDP-43 inclusion formation in fibroblasts derived from sporadic or C9orf72-related amyotrophic lateral sclerosis/frontotemporal dementia patients. We found that both sporadic and C9orf72 amyotrophic lateral sclerosis/frontotemporal dementia patient fibroblasts showed impairment in TDP-43 degradation by the proteasome, which may explain increased TDP-43 protein levels found in both sporadic and C9orf72 amyotrophic lateral sclerosis/frontotemporal lobar degeneration frontal cortex and hippocampus. Fibroblasts derived from sporadic patients, but not C9orf72 patients, demonstrated the ability to sequester cytoplasmic TDP-43 into aggresomes via microtubule-dependent mechanisms. TDP-43 aggresomes in vitro and TDP-43 neuronal inclusions in vivo were both tightly localized with autophagy markers and, therefore, were likely to function similarly as sites for autophagic degradation. The inability for C9orf72 fibroblasts to form TDP-43 aggresomes, together with the observations that TDP-43 protein was soluble in the cytoplasm and formed smaller inclusions in the C9orf72 brain compared with sporadic disease, suggests a loss of protein quality control response to sequester and degrade TDP-43 in C9orf72-related diseases.
Keywords: TDP-43, inclusion formation, aggresome, frontotemporal dementia, amyotrophic lateral sclerosis
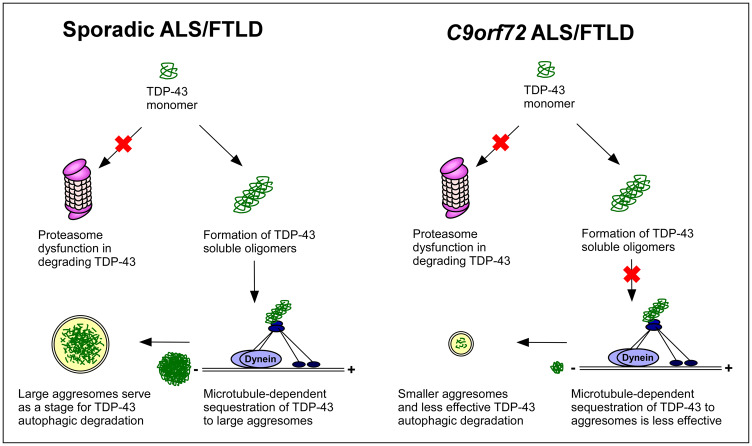
The mechanisms responsible for TDP-43 pathology in sporadic and C9orf72-associated amyotrophic lateral sclerosis/frontotemporal lobar degeneration remain largely unknown. Lee et al. report that C9orf72 patients demonstrate unique in vivo and in vitro features distinct from sporadic cases. These findings suggest a loss of protein quality control response to sequester and degrade TDP-43 in C9orf72 patients.
Graphical Abstract
Graphical Abstract.
Introduction
TDP-43 proteinopathies such as amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD) are progressive neurodegenerative diseases pathologically characterized by the presence of TAR DNA-binding protein 43 (TDP-43) neuronal cytoplasmic inclusions (Seelaar et al., 2007; Blokhuis et al., 2013). In addition to shared TDP-43 pathology, ALS and FTLD present as a clinical spectrum (Hinz and Geschwind, 2017). Approximately 15% of patients with clinical manifestation of FTLD, i.e. frontotemporal dementia (FTD), is diagnosed with ALS, and up to 30% of ALS patients meet the diagnostic criteria for FTD (Hinz and Geschwind, 2017). These shared pathologies and overlapping clinical presentations, together with the discovery of the G4C2 hexanucleotide repeat expansion (HRE) mutation in the C9orf72 gene as the most common genetic cause of ALS and/or FTLD (ALS/FTLD; Renton et al., 2011; Rademakers, 2012), suggest common pathogenic mechanisms for ALS and FTLD. Patients with C9orf72-related ALS/FTD are clinically indistinguishable from those with sporadic ALS/FTD (Sha et al., 2012). Pathologically, C9orf72-associated disease is characterized by the pathognomonic finding of cerebellar neuronal inclusions that are positive by immunohistochemistry for p62 (sequestosome-1), but negative for phosphorylated TDP-43 (pTDP-43; Pikkarainen et al., 2010; Al-Sarraj et al., 2011; Bigio et al., 2013).
TDP-43 is essential for regulating transcriptional events (Sephton et al., 2012) and its expression level in neurons is tightly regulated (Polymenidou et al., 2011). Previous studies reported that expression of exogenous human TDP-43 protein in transgenic mice causes neurodegeneration (Wils et al., 2010; Igaz et al., 2011), suggesting that excess TDP-43 could be toxic to the central nervous system. TDP-43 is prone to spontaneous aggregation into oligomers that are cytotoxic (Johnson et al., 2009; Takalo et al., 2013), which supports a role for the overabundance of TDP-43 to cause neuronal injury. However, it remains unknown whether an increase of TDP-43 protein level is linked to ALS and FTLD pathogenesis because the protein level of TDP-43 in the brains of ALS and FTLD patients has not been established. Although dysfunction in protein quality control systems that target excess proteins for degradation, such as the ubiquitin-proteasome system (UPS; Ciechanover, 1994) and the autophagy pathway (Klionsky et al., 2012), have been proposed for ALS and FTLD pathogenesis (Shahheydari et al., 2017), it is unclear whether these pathways are affected in sporadic and C9orf72-associated ALS and FTLD patients.
The mechanisms responsible for TDP-43 pathology in sporadic and C9orf72-associated ALS and FTLD remain largely unknown. Differences in p62 inclusions between sporadic and C9orf72-associated ALS and FTLD in the cerebellum prompted us to determine whether TDP-43 inclusion body formation is different between sporadic and C9orf72-associated ALS and FTLD, which may provide valuable insights into the pathogenesis of these diseases. Studies of TDP-43 inclusions in the FTLD brain is difficult because the concomitant diagnosis of ALS can affect FTLD pathology (Mackenzie et al., 2006; Cairns et al., 2007; Mackenzie et al., 2011a). An advantage to focusing on ALS patients with FTD and FTLD is that this is a more homogeneous group of subjects to investigate the differences in TDP-43 pathology between sporadic and C9orf72-associated diseases. In addition to examining TDP-43 pathology in vivo, we also performed complimentary in vitro experiments using patient-derived cells to investigate differences in the underlying mechanisms of TDP-43 inclusion body formation and protein control pathways between sporadic and C9orf72 patients. Overall, our findings demonstrate that C9orf72 patients are characterized by unique in vivo and in vitro features in TDP-43 inclusion body formation that are distinct from sporadic cases, which may have clinical and pathophysiological implications.
Materials and methods
Patient tissue samples and generation of patient-derived fibroblasts
For pathological studies, post-mortem brain tissues were obtained from the brain bank maintained by the Emory Alzheimer Disease Research Center under proper Institutional Review Board protocols. All patients were diagnosed with ALS and were cared for at the Emory ALS Center (Umoh et al., 2016). The clinical diagnosis of FTD, for selection of cases for fibroblast generation, was made based on established clinical diagnostic criteria (Rascovsky et al., 2011) by an experienced neurologist (J.D.G.). The neuropathological diagnosis of FTLD with TDP-43 pathology was made by experienced neuropathologists (M.G. and J.D.G.) based on published criteria (Cairns et al., 2007; Mackenzie et al., 2011b). The presence of a C9orf72 HRE mutation was assessed from DNA extracted from the post-mortem brains of ALS/FTLD patients using the published PCR method as described previously (DeJesus-Hernandez et al., 2011; Umoh et al., 2016). Clinical and pathological information from all individual cases, including disease status, age, sex and post-mortem interval are provided in Supplementary Table 1 and summarized in Supplementary Table 2. All patients with ALS/FTLD had a clinical diagnosis of ALS with behavioural variant FTD.
For the generation of fibroblast lines, patients diagnosed clinically as ALS/FTD or ALS underwent skin biopsy after informed consent was obtained from the patient and the patient’s legal power of attorney. Samples were de-identified and processed as described previously to obtain fibroblasts (Konrad et al., 2017). Fibroblasts were cultured in Dulbecco’s modified Eagle medium supplemented with 10% foetal bovine serum. Cultured fibroblasts were studied at passages ranging between 7 and 15. Experiments with patient-derived fibroblasts were approved by the Institutional Review Board at Emory University (IRB00064365).
Immunohistochemistry of brain tissue
Paraffin-embedded sections from frontal cortex, hippocampus, occipital cortex and cerebellum (8 μm thickness) were de-paraffinized by incubation at 60°C for 30 min and rehydrated by immersion in graded ethanol solutions. Antigen retrieval was by microwaving in 10 mM citrate buffer (pH 6.0) for 5 min followed by allowing slides to cool to room temperature for 30 min. Endogenous peroxidase activity was eliminated by incubating slides with hydrogen peroxide block solution (Fisher) for 10 min at room temperature followed by rinsing in phosphate buffered saline. Non-specific binding was reduced by blocking in ultraVision Block (Fisher) for 5 min at room temperature. Sections were then incubated overnight with primary antibodies (refer to Supplementary Table 3 for a full list of antibodies used in this study) diluted in 1% BSA in phosphate buffered saline for 30 min in room temperature or incubated without primary antibody as a negative control. Sections were rinsed in phosphate buffered saline and incubated in labelled ultraVision LP detection system horseradish peroxidase-polymer secondary antibody (Fisher) for 15 min at room temperature. Slides were imaged for analysis using an Aperio Digital Pathology Slide Scanner (Leica Biosystems). For immunofluorescence, slides were processed as described above except that fluorescent secondary antibodies (Jackson ImmunoResearch Laboratories) were used instead of biotinylated antibodies. Tissue sections were analysed using immunofluorescence confocal microscopy as described previously (Lee et al., 2011, 2012).
Quantification of inclusions in the hippocampus
Hippocampal sections were chosen for quantification analyses because this brain region was routinely examined in previous reports of C9orf72-associated FTLD cases and is known to have abundant TDP-43 inclusions in FTLD cases (Al-Sarraj et al., 2011; Bigio et al., 2013). For the quantification of TDP-43 and p62 neuronal cytoplasmic inclusions, sections stained with an antibody against pTDP-43 (Seilhean et al., 2009; Brettschneider et al., 2014) or p62 were analysed. The dentate gyrus in each image was manually annotated as the region of interest using custom JavaScript web-based applications. Images then underwent colour normalization using the Reinhard method (Reinhard et al., 2001) and colour deconvolution into two separate channels to discriminate the neuronal nuclei stained with haematoxylin from pTDP-43 or p62 inclusions visualized by 3,3′-diaminobenzidine chromogen. For each channel, Laplacian-of-Gaussian filtering as applied by the HistomicsTK Python toolkit (GitHub, https://github.com/DigitalSlideArchive/HistomicsTK) was used, and the Al-Kofahi method (Al-Kofahi et al., 2010) was used to detect individual nuclei and inclusions. To prevent background staining from being counted as an inclusion, we calculated the Euclidean distance from the middle point of each pTDP-43 or p62 marking to the middle point of every nucleus and excluded any immunostaining that was not near any nucleus as determined by a pre-defined threshold. Using these methods, the inclusion sizes, the total number of hippocampal inclusions, the total number of hippocampal neuronal nuclei, the number of inclusions per non-overlapping low-power field, the number of neuronal nuclei per non-overlapping low-power field and the area of each inclusion were measured. The median inclusion sizes and the percentages of neurons with pTDP-43 inclusions and/or p62 inclusions were calculated for the entire hippocampus from each case. In addition, the percentages of hippocampal neurons with pTDP-43 inclusions from at least 20 low-power fields were determined and averaged for each case. Statistical analyses of the inclusion sizes and the percentages of neurons with inclusions were performed using individual cases as the experimental unit.
For quantification of double immunofluorescence staining, hippocampal sections stained with antibodies against p62 and the C-terminal of TDP-43 were subjected to confocal microscopic analyses. The C-terminal TDP-43 antibody, in contrast to the pTDP-43 antibody, reacts with all species of TDP-43 protein. Low-power field images were examined in a blinded manner and scored for the number of TDP-43-positive/p62-positive (defined as p62-positive inclusion that co-localized with TDP-43) and TDP-43-negative/p62-positive (defined as p62-positive inclusion that did not co-localize with TDP-43) inclusions. The total number of hippocampal cells in each field was determined by 4′,6-diamidino-2-phenylindole (DAPI) staining. The percentages of hippocampal cells with each type of inclusion were determined and averaged from at least 10 low-power fields per case. Statistical analyses of the percentages of hippocampal cells with inclusions were performed using individual cases as the experimental unit.
Fractionation of human frontal cortex tissue
Frozen frontal cortex tissue (∼0.1 mg) was randomly selected from three to five patients per group by our co-author (S.A.) who was blinded to group (three control patients, three C9orf72 ALS/FTLD and five sporadic ALS/FTLD cases total). The tissue was thawed in 1 ml of ice-cold homogenization buffer (10 mM HEPES/KOH, pH 7.4, 1 mM EDTA) containing 250 mM sucrose, and homogenates were centrifuged at 10 000 g for 10 min to pellet the non-soluble nuclei/unbroken cells/inclusion bodies fraction and to obtain the supernatant as the soluble cytosol fraction as previously described (Shaiken and Opekun, 2014). Equal volumes of the total, nuclear/unbroken cells/inclusion bodies and cytosol fractions were analysed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and by sequential immunoblotting for TDP-43, the nuclear marker NeuN (Aber et al., 2003; Misztal et al., 2017), the cytosolic marker glyceraldehyde 3-phosphate dehydrogenase (GADPH) and actin. Soluble cytoplasmic TDP-43 protein levels from non-over-exposed images obtained using the Bio-Rad ChemiDoc MP imaging system were quantified (Giles et al., 2008; Lee et al., 2011) using NIH ImageJ and were normalized to total TDP-43 protein level. To further control for protein quantities in each fraction, soluble cytoplasmic and total TDP-43 protein levels were normalized to the cytoplasmic and total GADPH protein levels, respectively, followed by normalization of soluble TDP-43 protein level to total TDP-43 protein level.
Quantification of TDP-43 level in brain tissues
Frozen brain tissue from the frontal cortex, hippocampus, occipital cortex and cerebellum were homogenized with a Teflon-glass homogenizer in 10 volumes of 1% SDS, followed by a combination of boiling and sonification. An equal amount of total proteins from each lysate was subjected to immunoblot analyses. TDP-43 protein levels from non-over-exposed images obtained using the Bio-Rad ChemiDoc MP imaging system were quantified as described (Giles et al., 2008; Lee et al., 2011) using NIH ImageJ and were normalized to actin protein level. Actin was chosen as an appropriate loading control since its protein level is not significantly altered between control, C9orf72 ALS/FTLD, sporadic ALS/FTLD and sporadic ALS tissues from various brain regions when normalized to the corresponding total protein as determined by Ponceau S staining (data not shown).
Treatment of fibroblasts with proteasome and autophagy inhibitors
Fibroblasts from two control patients, three C9orf72 patients (one with clinical ALS/FTD and two with clinical ALS) and three sporadic patients (one with clinical ALS/FTD and two with clinical ALS) were incubated for 24 h at 37°C with proteasome inhibitor MG132 (20 μM, Sigma), autophagy inhibitor bafilomycin A1 (50 nM, Sigma), or vehicle (0.1% DMSO) as described previously (Giles et al., 2008; Lee et al., 2011). For immunoblot analysis, fibroblasts were lysed with 1% SDS, and an equal amount of total protein from each lysate was analysed by SDS-PAGE. The protein levels of TDP-43 were quantified using NIH ImageJ (Giles et al., 2008; Lee et al., 2011) and then normalized to the corresponding actin levels. Quantification of LC3-II/LC3-I ratio was performed as described previously (Klionsky et al., 2012).
Immunocytochemistry and quantification of co-localization and signal intensity
Fibroblasts were fixed in 4% paraformaldehyde and processed for immunofluorescence confocal microscopy as described previously (Olzmann et al., 2007; Lee et al., 2012). The percentage of co-localization for each cell was determined by the percentage of the immunostaining by the first primary antibody [anti-pTDP-43 (Seilhean et al., 2009; Sabatelli et al., 2015), anti-TDP-43 or anti-p62] that was co-localized with the immunostaining by the second primary antibody (anti-p62 or anti-TDP-43) or with DAPI staining. Co-localization was quantified on unprocessed images of cells double-labelled for TDP-43, p62 and/or DAPI as previously described (Webber et al., 2008; Lee et al., 2011). Single cells were selected by manually tracing the cell outlines. The background was subtracted and the percentage of the first primary antibody overlapping with the second primary antibody or DAPI staining was determined for each cell (Webber et al., 2008; Lee et al., 2011). The signal intensities of pTDP-43 and p62 immunostaining were quantified on unprocessed images of cells stained with either pTDP-43 or p62 as previously described (Lee et al., 2012). Single cells were selected and with background subtracted as above, and the signal intensity for either pTDP-43 or p62 was measured with NIS Elements (Nikon) and normalized to DAPI signal intensity level for each cell (Kreiling et al., 2011; Serebryannyy et al., 2016). For statistical analyses of signal intensity and co-localization, results from at least 30 randomly selected cells were averaged per patient, and analyses were performed using individual cases as the experimental unit. Fibroblasts from two control patients, three C9orf72 patients (one with clinical ALS/FTD and two with clinical ALS) and three sporadic disease patients (one with clinical ALS/FTD and two with clinical ALS) were analysed.
Statistical analysis
Data were subjected to statistical analyses by Student’s t-tests, one-way ANOVA or two-way ANOVA using the SigmaPlot software (Systat Software, Inc.) Results were expressed as mean ± standard error of the mean (SEM). A P-value of <0.05 was considered statistically significant. The experimental units, the specific statistical tests, the test statistics, degrees of freedom as subscripts to the test statistic and the exact P-values were reported in Supplementary Table 4.
Data availability
The data supporting the findings of this study are available from the corresponding authors upon reasonable request.
Results
Sporadic ALS/FTLD brain has more frequent and larger pTDP-43 inclusions compared with C9orf72 brain
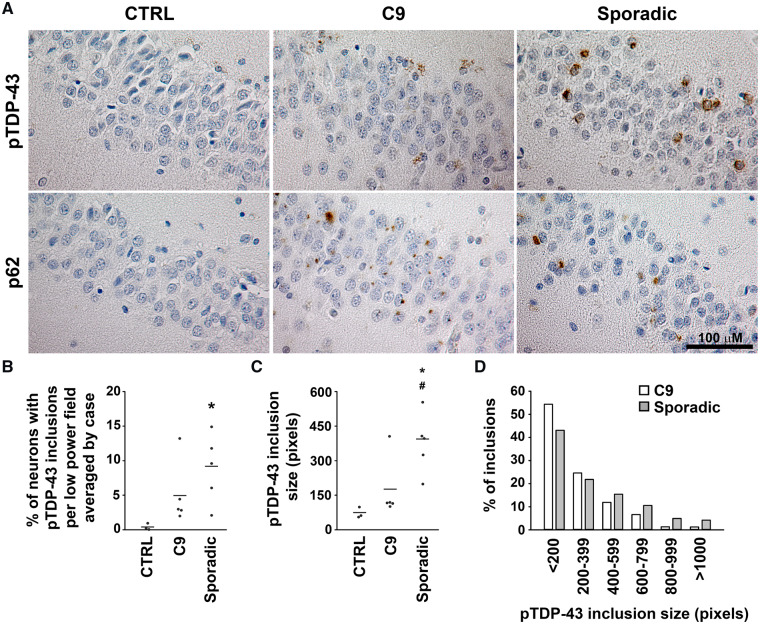
Using a phospho-TDP-43-specific antibody, we performed immunohistochemistry on C9orf72 ALS/FTLD brain and confirmed the pathognomonic findings of p62-positive inclusions in the absence of pTDP-43 inclusions in the cerebellum (Supplementary Fig. 1). In contrast, sporadic ALS/FTLD brains showed complete absence of pTDP-43 and p62 inclusions in the cerebellum (Supplementary Fig. 1). We further compared sporadic and C9orf72 disease pathology in the hippocampus with quantification analyses and found that sporadic ALS/FTLD brain contains numerous round and circumferential pTDP-43 inclusions throughout the hippocampal dentate gyrus as previously reported (Tan et al., 2017), whereas hippocampal pTDP-43 inclusions were notably less obvious in C9orf72 patients (Fig. 1A). Quantitative analyses showed that sporadic ALS/FTLD cases demonstrated a trend to having more overall pTDP-43 inclusions in the hippocampus compared with C9orf72 patients, though this did not meet statistical significance (Supplementary Fig. 2A). However, pTDP-43 inclusions were more frequently seen in sporadic ALS/FTLD when quantifying the density of pTDP-43 inclusions under low-power fields for each case and analysed using each case as the experimental unit (Fig. 1B). In contrast to pTDP-43 inclusions, p62 inclusions were significantly more numerous in the hippocampal dentate gyrus of C9orf72 patients compared with sporadic patients (Fig. 1A, Supplementary Fig. 2B). Another notable finding was that pTDP-43 inclusions in the hippocampus of C9orf72 patients were quantitatively smaller than those seen in sporadic cases (Fig. 1A, C and D). Both round and circumferential pTDP-43 inclusions were also found in the frontal cortex of sporadic ALS/FTLD cases but were less obvious in C9orf72 ALS/FTLD frontal cortex (Supplementary Fig. 3). These findings were not influenced by TDP-43 inclusion subtypes (Mackenzie et al., 2006; Cairns et al., 2007; Mackenzie et al., 2011a). The results illustrated in Fig. 1B and C were unchanged when only cases with TDP-43 type B inclusions were present in the quantification analyses (data not shown). Compared with the dentate gyrus and frontal cortex, pTDP-43 inclusions were rarely found in the occipital cortex and absent in the cerebellum in sporadic and C9orf72 ALS/FTLD (Supplementary Figs. 1 and 3).
Figure 1.
pTDP-43 inclusions are more frequent and larger in sporadic compared with C9orf72 ALS/FTLD hippocampus. (A) Paraffin-embedded sections from the dentate gyrus of the hippocampus were subjected to immunohistochemistry with anti-pTDP-43 antibody or anti-p62 antibody and counterstained with haematoxylin. (B) The percentages of neurons with pTDP-43 inclusions in low-power fields were determined and averaged for each case and analysed using each case as the experimental unit as described in the Materials and Methods section. (C) The sizes of pTDP-43 inclusions were determined for each case, and the median inclusion sizes for these cases were compared among control, C9orf72 and sporadic ALS/FTLD hippocampi as described in the Materials and Methods section. The mean for each experimental group was represented by a horizontal line. *P < 0.05 compared with control and #P < 0.05 compared with C9orf72 ALS/FTLD based on one-way ANOVA and post-hoc Tukey’s test. (D) Histogram showing the percentages of pTDP-43 inclusions of different sizes in C9orf72 and sporadic ALS/FTLD hippocampi. C9, C9orf72; CTRL, control; pTDP-43, phosphorylated TDP-43.
Cytosolic TDP-43 is soluble in C9orf72 ALS/FTLD brain but forms inclusions in sporadic cases
To further define TDP-43 pathology in sporadic and C9orf72 ALS/FTLD brains, we chose a well-characterized antibody against the C-terminal of TDP-43 that detects both phosphorylated and non-phosphorylated forms of full-length TDP-43 and its C-terminal fragments (Wils et al., 2010). We found that the frontal cortex and hippocampus (Fig. 2A) of sporadic ALS/FTLD brain contained abundant TDP-43 cytoplasmic neuronal inclusions. These inclusions had the same morphologies as those detected using the anti-pTDP-43 antibody. In contrast to sporadic ALS/FTLD, the C-terminal TDP-43 antibody detected fewer neurons with TDP-43 cytoplasmic inclusions in C9orf72 ALS/FTLD brains (Fig. 2A). In addition to consolidated TDP-43 inclusions, the C-terminal TDP-43 antibody identified a subpopulation of neurons in C9orf72 ALS/FTLD with diffuse cytoplasmic staining in the frontal cortex (Fig. 2A, Supplementary Table 1). This diffuse immunostaining by the C-terminal TDP-43 antibody was distinct from the inclusion morphology identified by pTDP-43 antibody in C9orf72 ALS/FTLD brains (Fig. 1A), and was not always associated with complete loss of nuclear TDP-43 expression (i.e. nuclear clearing) as typically reported with TDP-43 cytoplasmic inclusions (Neumann et al., 2006). In contrast, neurons with TDP-43 cytoplasmic inclusions did show nuclear clearing (Fig. 2A).
Figure 2.
TDP-43 is soluble in the cytoplasm of C9orf72 ALS/FTLD brain but not sporadic ALS/FTLD brain. (A) Paraffin-embedded sections from the frontal cortex and dentate gyrus of the hippocampus were subjected to immunohistochemistry with anti-C-terminal TDP-43 antibody and counterstained with haematoxylin. Black arrows indicate examples of TDP-43 neuronal cytoplasmic inclusions. Black arrow heads indicate examples of neurons with diffuse cytosolic TDP-43 staining. (B) Total lysates from the frontal cortex were separated into nuclear/inclusion fraction and soluble fraction. Aliquots representing an equal percentage of each fraction were subjected to immunoblot analysis with antibodies against C-terminal of TDP-43, GADPH and NeuN. The proportion of TDP-43 protein in the soluble fraction was higher in C9orf72 tissue (C) and also after normalization to GADPH (D). The mean for each experimental group was represented by a horizontal line. *P < 0.05 compared with control and #P < 0.05 compared with C9orf72 ALS/FTLD based on one-way ANOVA and post-hoc Tukey’s test. C9, C9orf72; CTRL, control; GADPH, glyceraldehyde 3-phosphate dehydrogenase; HMW TDP-43, higher molecular weight species identified by the C-terminal anti-TDP-43 antibody.
To determine whether the diffuse cytoplasmic TDP-43 staining seen in C9orf72 brain represents mis-localization of soluble, cytoplasmic TDP-43, we performed subcellular fractionation of frontal cortex from C9orf72 and sporadic ALS/FTLD patients. We isolated a fraction consisting of soluble cytosolic proteins and another fraction containing the nucleus, unbroken cells and inclusion bodies (Fig. 2B, Supplementary Fig. 4). Our results showed that C9orf72 frontal cortex contained a soluble cytosolic fraction of TDP-43 that was absent in control and minimally present in sporadic ALS/FTLD frontal cortices (Fig. 2B–D), suggesting that the diffuse cytosolic staining seen by immunohistochemistry in C9orf72 likely represents soluble cytoplasmic TDP-43.
C9orf72 ALS/FTLD is characterized by TDP-43 negative/p62-positive inclusions
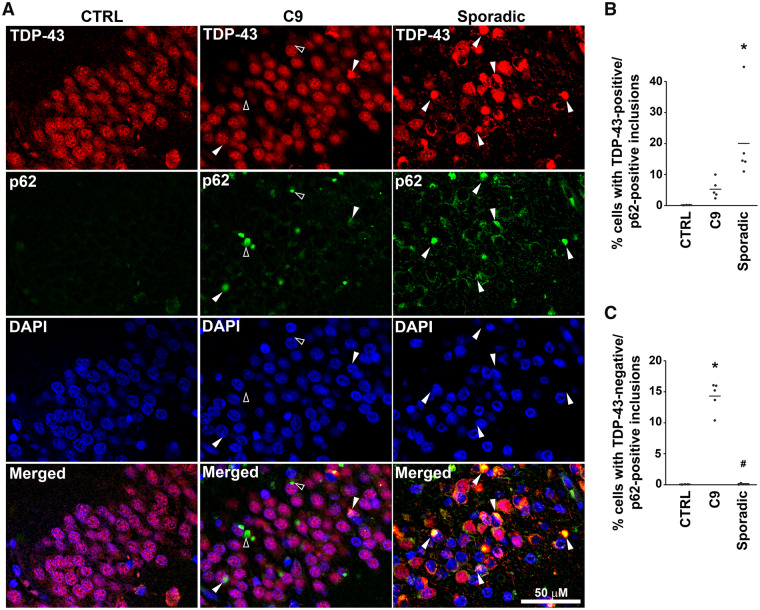
We addressed the occurrence of co-aggregation of TDP-43 and p62 using double fluorescence immunohistochemistry. In sporadic ALS/FTLD brains, the overwhelming majority of hippocampal dentate gyrus neurons containing TDP-43 inclusions were positive for p62 (Fig. 3A and B). TDP-43-positive/p62-positive neurons were also seen in the C9orf72 hippocampus (Fig. 3A), but were much less abundant than in sporadic ALS/FTLD (Fig. 3A and B). There were numerous inclusions positive for p62 but negative for TDP-43 in C9orf72 hippocampus, reminiscent of pathology in the cerebellum, but this pattern of staining was virtually absent in sporadic ALS/FTLD (Fig. 3A and C). These findings in the C9orf72 hippocampus, together with a complete lack of TDP-43 inclusions and an abundance of p62 inclusions in the cerebellum (Supplementary Fig. 1), indicate that TDP-43-negative/p62-positive inclusions are a pathological feature that distinguishes C9orf72 ALS/FTLD from sporadic ALS/FTLD in hippocampus as well as in the cerebellum.
Figure 3.
C9orf72 ALS/FTLD hippocampus showed more TDP-43-negative/p62-positive inclusions compared with sporadic cases. (A) Paraffin-embedded sections from the hippocampus were subjected to double immunofluorescence histochemistry with anti-C-terminal TDP-43 (red) and p62 (green) antibodies. White arrow heads indicate examples of neurons with TDP-43-positive/p62-positive inclusions. Open arrow heads indicate examples of neurons with TDP-43-negative/p62-positive inclusions. Nuclei were visualized by DAPI stain (blue). The percentages of cells with TDP-43-positive/p62-positive (B) and TDP-43-negative/p62-positive (C) inclusions were determined as described in the Materials and Methods section and analysed using each case as the experimental unit. The mean for each experimental group was represented by a horizontal line. *P < 0.05 compared with control and #P < 0.05 compared with C9orf72 ALS/FTLD based on one-way ANOVA and post-hoc Tukey’s test. C9, C9orf72; CTRL, control; sporadic, sporadic ALS/FTLD.
TDP-43 inclusions in sporadic and C9orf72 ALS/FTLD are autophagy substrates
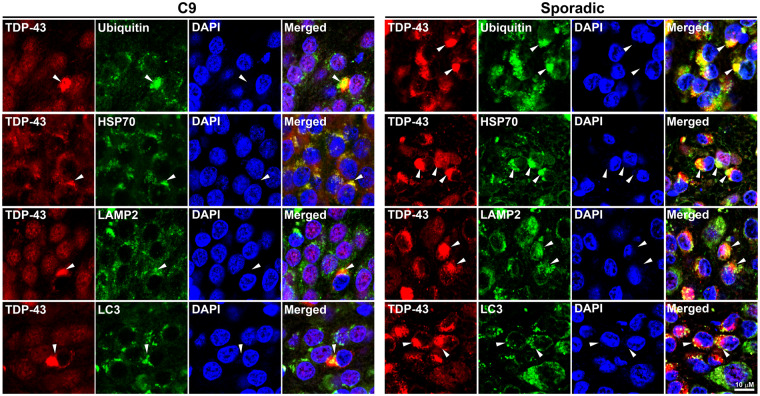
Additional double immunofluorescence analyses using the C-terminal TDP-43 antibody demonstrated that TDP-43 inclusions of various sizes in both sporadic and C9orf72 ALS/FTLD hippocampi co-localized with ubiquitin (Fig. 4) which represents the deposition of ubiquitinated proteins, including TDP-43 (Neumann et al., 2006; Richter-Landsberg and Leyk, 2013). In addition, the chaperone heat shock protein 70 kDa (HSP70), which is thought to protect neurons from toxicity of misfolded proteins (Turturici et al., 2011), co-localized with TDP-43 inclusions in both sporadic and C9orf72 ALS/FTLD hippocampus (Fig. 4, Supplementary Fig. 5). These TDP-43 inclusions also co-localized with and/or were surrounded by the autophagosome marker LC3 and the lysosome marker lysosome-associated membrane protein 2 (LAMP2; Fig. 4). Hippocampi from control patients did not demonstrate co-localization between TDP-43 and these markers (Supplementary Fig. 5). Thus, TDP-43 inclusions in sporadic and C9orf72 ALS/FTLD share similar characteristics and are likely substrates for autophagy.
Figure 4.
TDP-43 inclusions in C9orf72 and sporadic ALS/FTLD hippocampus co-localized with ubiquitin and aggresome markers. Paraffin-embedded hippocampal sections from C9orf72 or sporadic ALS/FTLD patients were subjected to double immunofluorescence histochemistry with antibodies against TDP-43 (red) and ubiquitin, HSP70, LAMP2 or LC3 (green). White arrowheads indicate examples of neurons with TDP-43 inclusions. Nuclei were visualized by DAPI stain (blue). C9, C9orf72; HSP70, heat shock protein 70 kDa; LAMP2, lysosomal-associated membrane protein 2.
Sporadic and C9orf72 ALS/FTLD patients have elevated TDP-43 protein levels in the frontal cortex and hippocampus
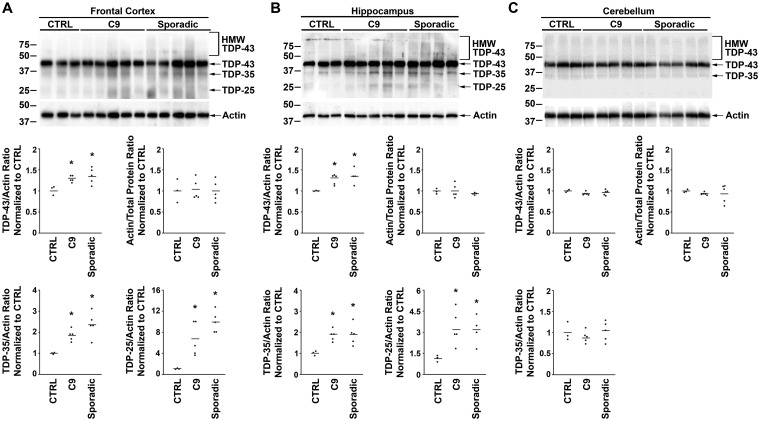
Whether TDP-43 protein levels are altered in sporadic and C9orf72 ALS/FTLD brains has not been established. Immunoblotting of post-mortem frontal cortex and hippocampus demonstrated elevated TDP-43 levels in both sporadic and C9orf72 ALS/FTLD patients as compared with controls (Fig. 5A and B, Supplementary Fig. 6), consistent with results from our recent proteomic analysis of frontal cortex tissues (Umoh et al., 2018). The actin loading control did not show differences between sporadic and C9orf72 ALS/FTLD patients and controls (Fig. 5A and B). TDP-43 protein levels did not differ between sporadic and C9orf72 cases (Fig. 5A and B). Elevated levels of TDP-43 protein were not observed in clinically unaffected regions of the brain such as the cerebellum (Fig. 5C, Supplementary Fig. 6). Increased TDP-43 protein levels were also not seen in the frontal cortex and hippocampus of sporadic ALS patients without clinical FTD or pathological FTLD diagnoses (Supplementary Fig. 7).
Figure 5.
C9orf72 and sporadic ALS/FTLD have increased TDP-43 protein levels in the frontal cortex and hippocampus. Equal amounts of total proteins from the indicated human patient frontal cortex (A), hippocampus (B) and cerebellum (C) were subjected to immunoblot analysis with anti-C-terminal TDP-43 antibody and anti-actin antibodies. The protein levels of full-length TDP-43 and the C-terminal fragments TDP-35 and TDP-25 were quantified and normalized to actin, and the protein levels of actin were quantified and normalized to total protein. The mean for each experimental group was represented by a horizontal line. Quantification of TDP-25 protein levels in the cerebellum was not provided due to absence of this species in the immunoblot. *P < 0.05 compared with control based on one-way ANOVA and post-hoc Tukey’s test. C9, C9orf72; CTRL, control; HMW TDP-43, higher molecular weight species identified by the C-terminal anti-TDP-43 antibody.
Western blot analysis of sporadic and C9orf72 ALS/FTLD also revealed the presence of lower molecular weight TDP-43 species in the frontal cortex (Fig. 5A) and hippocampus (Fig. 5B) that were significantly less prominent in controls and likely represent C-terminal fragments of TDP-43. Amongst these fragments, the protein bands at 35 kDa and at 25 kDa correspond to previously reported TDP-43 C-terminal fragments termed TDP-35 and TDP-25, respectively (Neumann et al., 2006; Igaz et al., 2008). TDP-25 is of particular interest as it was recently implicated to be cytotoxic (Yang et al., 2010; Gregory et al., 2012). These results suggest that excess TDP-43 in the frontal cortex and hippocampus is associated with the increased presence of potentially toxic C-terminal fragments that could contribute to the pathogenesis of sporadic and C9orf72 ALS/FTLD.
Sporadic and C9orf72 ALS/FTD fibroblasts demonstrate impairment in proteasomal degradation of TDP-43
The finding of elevated TDP-43 protein levels in both sporadic and C9orf72 ALS/FTLD brains and the smaller TDP-43 inclusion size in C9orf72 ALS/FTLD brains prompted us to further investigate these observations using an in vitro cellular model. Patient-derived fibroblasts from clinically diagnosed sporadic and C9orf72 ALS/FTD patients show abnormally increased TDP-43 protein levels as compared with controls (Sabatelli et al., 2015), which made this an attractive cellular model for studying how excess TDP-43 protein is processed in patient-derived cells. Note that the sporadic ALS/FTD fibroblasts were generated from living FTD patients who also had ALS, since the pathology of sporadic FTD in ALS patients is exclusively FTLD–TDP regardless of the clinical FTD subtype (Neumann et al., 2006; Neumann et al., 2007).
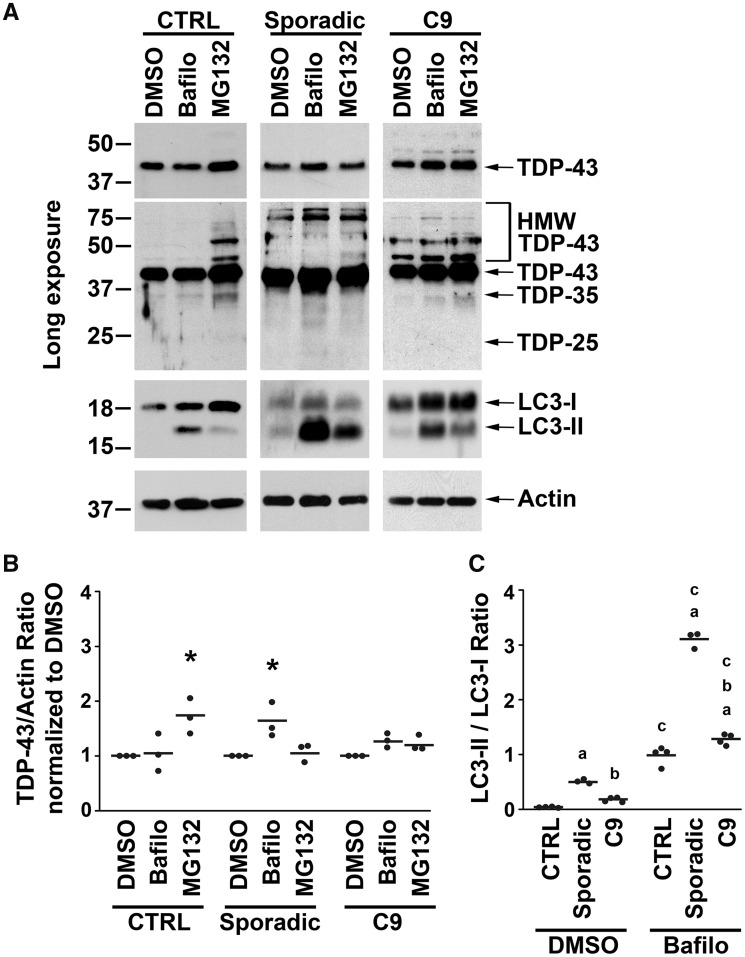
Our finding of excess TDP-43 protein in the frontal cortex and hippocampus of sporadic and C9orf72 ALS/FTLD patients (Fig. 5) raised the possibility that TDP-43 turnover via the UPS may be impaired in these patients. To test this possibility, we first assessed the effects of proteasome inhibition on steady-state protein levels of TDP-43 in patient-derived fibroblasts (Fig. 6A, Supplementary Fig. 8A). Proteasome inhibition by MG132 had no effect on TDP-43 protein levels in sporadic and C9orf72 ALS/FTD and ALS fibroblasts, suggesting that TDP-43 protein is not degraded by the UPS in fibroblasts from these patients (Fig. 6A and B). In contrast, proteasome inhibition significantly increased TDP-43 protein levels in control fibroblasts, indicating that TDP-43 is normally degraded by the UPS (Fig. 6A and B). In addition, HMW TDP-43 species were seen in vehicle-treated sporadic and C9orf72 disease fibroblasts but were only found in control fibroblasts after proteasome inhibition with MG132 (Fig. 6A), indicating that HMW species likely represent accumulation of ubiquitinated TDP-43. Proteasome inhibition by MG132 also had no effect on poly-ubiquitinated protein levels in sporadic and C9orf72 disease fibroblasts, suggesting the inability of the proteasome pathway to degrade poly-ubiquitinated proteins in these fibroblasts (Supplementary Fig. 8B and C). Together, these findings suggest that TDP-43 degradation via the proteasome pathway is impaired in both sporadic and C9orf72 disease fibroblasts which could be responsible for the accumulation of TDP-43 protein in ALS/FTLD brains.
Figure 6.
Sporadic and C9orf72 disease fibroblasts showed proteasome impairment and autophagic degradation of TDP-43. (A) Patient-derived fibroblasts were treated for 24 h with 20 μM MG132, 50 nM of bafilomycin A1 (Bafilo) or vehicle control (DMSO). Equal amounts of total proteins from the indicated fibroblasts were subjected to immunoblot analysis with anti-C-terminal TDP-43, anti-actin and anti-LC3 antibodies. (B) The protein levels of full-length TDP-43 from control, sporadic and C9orf72 ALS/FTD were quantified and normalized to vehicle treatment and to actin. The effects of MG132 and bafilomycin A1 on full-length TDP-43 protein level among control, sporadic and C9orf72 ALS/FTD fibroblasts were shown. The mean for each experimental group was represented by a horizontal line.*P < 0.05 compared with the corresponding vehicle treatment group based on two-way ANOVA and post-hoc Tukey’s test. (C) The ratios of LC3-II and LC3-I protein levels with vehicle treatment were compared between control, sporadic and C9orf72 ALS/FTD fibroblasts. The mean for each experimental group was represented by a horizontal line. aP < 0.05 when compared with the corresponding control fibroblasts, bP < 0.05 when compared with the corresponding sporadic ALS/FTD fibroblasts, cP < 0.05 when compared with the corresponding vehicle-treated fibroblasts based on two-way ANOVA and post-hoc Tukey’s test. C9, C9orf72; CTRL, control; bafilo, bafilomycin A1; HMW TDP-43, higher molecular weight species identified by the C-terminal anti-TDP-43 antibody.
Sporadic ALS/FTD fibroblasts degrade TDP-43 via autophagy more effectively than C9orf72 fibroblasts
When the UPS fails, proteins such as TDP-43 have the potential to form oligomers which are small protein aggregates (Ross and Poirier, 2005; Johnson et al., 2009). Aggregated proteins are cleared by the autophagy pathway where autophagosomes engulf these aggregates and then fuse with lysosomes to eliminate them via enzymatic degradation (Klionsky et al., 2012). Autophagy activation causes the conversion of LC3-I to LC3-II, and LC3-II is subsequently degraded by autophagy (Klionsky et al., 2012). We first examined whether there was a difference in autophagy activation by studying LC3 conversion in these fibroblasts. Our results show that LC3-II/LC3-I ratio is low in control fibroblasts and is significantly higher in both sporadic and C9orf72 disease fibroblasts (Fig. 6A and C, Supplementary Fig. 8). To see whether this increase in LC3-II/LC3-I ratio in sporadic and C9orf72 ALS/FTD and ALS fibroblasts resulted from autophagy activation or from impairment of autophagic degradation and subsequent build-up of LC3-II, we treated fibroblasts with the autophagy inhibitor bafilomycin A1. Bafilomycin A1 treatment resulted in further increase in the LC3-II/LC3-I ratio in sporadic and C9orf72 disease fibroblasts, indicating that autophagy degradation of LC3-II was intact in these cells (Fig. 6A and C). Therefore, it is likely that the increased LC3-II/LC3-I ratio in sporadic and C9orf72 disease fibroblasts compared with control fibroblasts reflects an activation of autophagy in sporadic and C9orf72-associated disease.
We examined the effects of bafilomycin A1 on TDP-43 protein levels and found that it raised TDP-43 protein levels in sporadic disease fibroblasts (Fig. 6A and B). A trend towards an increase in TDP-43 protein levels in C9orf72 cells did not reach statistical significance (Fig. 6A and B). These findings suggest that although UPS is impaired in both sporadic and C9orf72 disease fibroblasts, the autophagy pathway is responsible for degrading TDP-43 in these fibroblasts and further implies that autophagic degradation of TDP-43 may be more robust in sporadic cases compared with C9orf72 cases.
Proteasome inhibition causes TDP-43 aggresome formation in sporadic ALS/FTD but not in C9orf72 fibroblasts
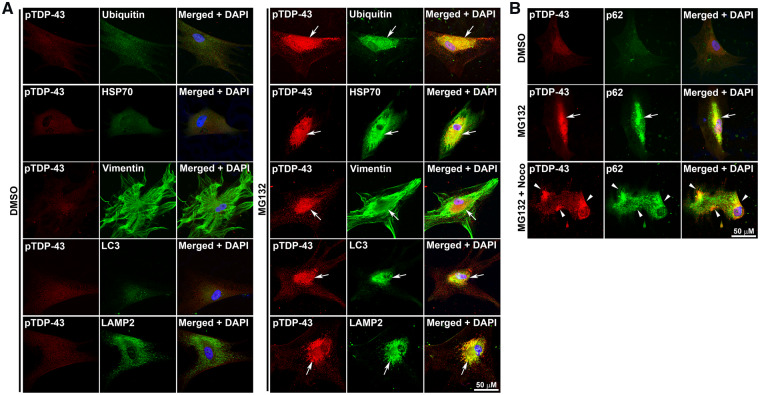
The pTDP-43 inclusions were not seen in vehicle-treated sporadic ALS/FTD fibroblasts (Fig. 7A), but proteasome inhibition with MG132 resulted in pTDP-43 accumulation in inclusion bodies (Fig. 7A). These findings are consistent with prior studies demonstrating the induction of TDP-43 phoshorylation by MG132 in cells (Nonaka et al., 2009; van Eersel et al., 2011). The perinuclear location of these inclusions suggested that they might be aggresomes. Aggresome formation is a cellular response that occurs when the capacity of the proteasome is exceeded by the production of misfolded and/or toxic proteins (Johnston et al., 1998). Aggresomes can also be induced by proteasome inhibition (Richter-Landsberg and Leyk, 2013). Cells target toxic proteins to perinuclear aggresomes via microtubule-dependent retrograde transport and sequester these proteins by encapsulating them with vimentin intermediate filaments or neurofilaments (Johnston et al., 1998). The pTDP-43 inclusion bodies in MG132-treated fibroblasts were caged by vimentin and also were enriched with ubiquitin, HSP70, p62, LC3, LAMP2, defining them as aggresomes (Fig. 7A and B). Further proof that these pTDP-43 inclusions were aggresomes comes from the demonstration that they did not co-localize with the Golgi apparatus marker GM130 nor the mitochondrial marker TIM23 but juxtaposed the endoplasmic reticulum marker KDEL (Supplementary Fig. 9), all of which are known features of aggresomes (Kopito and Sitia, 2000; Walker et al., 2013). Treatment with nocodazole, a microtubule-depolymerizing drug, abolished the pTDP-43 inclusions providing further confirmation that these were bona fide aggresomes (Fig. 7B).
Figure 7.
Proteasome impairment induced the formation of pTDP-43 aggresomes in sporadic ALS/FTD fibroblasts. (A) Fibroblasts from sporadic ALS/FTD were treated for 24 h with 20 μM MG132 or vehicle control (DMSO) and then immunostained with pTDP-43 (red) and ubiquitin, HSP70, vimentin, LC3 or LAMP2 (green) antibodies. (B) Formation of pTDP-43 aggresome was microtubule-dependent. Fibroblasts from sporadic ALS/FTD were treated for 24 h with vehicle control, 20 μM MG132, or 20 μM MG132 plus 2 μg/ml nocodazole (Noco) for 24 h and were immunostained with anti-pTDP-43 (red) and anti-p62 (green) antibodies. Nuclei were visualized by DAPI stain (blue). Aggresomes were indicated by white arrows. pTDP-43 microaggregates were indicated by white arrow heads. HSP70, heat shock protein 70 kDa; LAMP2, lysosomal-associated membrane protein 2; noco, nocodazole; pTDP-43, phosphorylated TDP-43.
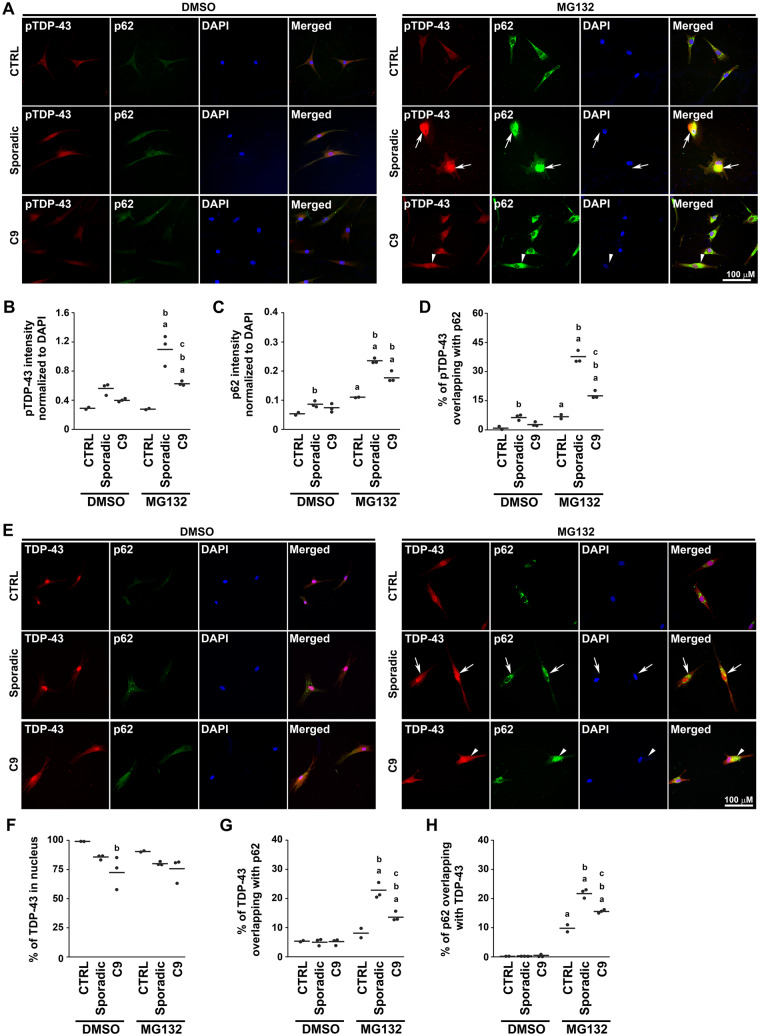
Our finding of the reduced abundance of TDP-43 inclusions and the presence of soluble cytoplasmic TDP-43 in C9orf72 ALS/FTLD brains (Figs. 1 and 2) suggested that the C9orf72 mutation may inhibit the formation of insoluble TDP-43 inclusions. To test this possibility in vitro, we examined patient-derived fibroblasts under the condition of proteasome inhibition with MG132. Treatment with MG132 resulted in increased pTDP-43 levels and the formation of large pTDP-43 positive inclusions in sporadic ALS/FTD and ALS fibroblasts but to a lesser degree in C9orf72 fibroblasts (Fig. 8A and B). However, p62 levels were similar in sporadic and C9orf72 fibroblasts (Fig. 8A and C). As these aggresomes were both pTDP-43 and p62-positive (Fig. 7A and B), we measured the formation of pTDP-43 aggresomes by examining pTDP-43 co-localization with p62. We found a reduced pTDP-43 co-localization with p62-positive aggresomes in C9orf72 fibroblasts compared with sporadic disease fibroblasts (Fig. 8A and D), indicating that C9orf72 fibroblasts were impaired in their ability to form pTDP-43 aggresomes. The reduced pTDP-43 inclusion formation and lack of pTDP-43/p62 co-localization in C9orf72 patient fibroblasts were consistent with our observations in C9orf72 and sporadic ALS/FTLD brains (Figs. 1 and 3).
Figure 8.
Targeting of TDP-43 to aggresomes was impaired in C9orf72 fibroblasts. (A) Human patient-derived fibroblasts were treated for 24 h with 20 μM MG132 or vehicle control (DMSO) and then immunostained with phospho-TDP-43 (pTDP-43, red) and p62 (green) antibodies. Examples of pTDP-43-positive/p62-positive aggresomes were indicated by white arrows. Examples of areas where pTDP-43 had some co-localization with p62 were indicated by white arrow heads. Nuclei were visualized by DAPI stain (blue). (B, C) The intensity of pTDP-43 (B) and p62 (C) immunostaining in patient fibroblasts was normalized to DAPI intensity and quantified as described in Materials and Methods section. (D) The percentages of pTDP-43 co-localized with p62 in patient fibroblasts were quantified as described in the Materials and Methods section. (E) Human patient-derived fibroblasts were treated for 24 h with 20 μM MG132 or vehicle control (DMSO) and then immunostained with C-terminal TDP-43 (TDP-43, red) and p62 (green) antibodies. Examples of TDP-43-positive aggresomes were indicated by white arrows. Examples of areas where TDP-43 had some co-localization with p62 were indicated by white arrow heads. Nuclei were visualized by DAPI stain (blue). The percentages of TDP-43 co-localized with nucleus as visualized by DAPI stain (F), TDP-43 co-localized with p62 (G) and p62 co-localized with TDP-43 (H) were quantified as described in the Materials and Methods section. All statistical analyses in this figure were carried out using each case as the experimental unit as described in the Materials and Methods section. The mean for each experimental group was represented by a horizontal line. aP < 0.05 when compared with the corresponding vehicle-treated fibroblasts, bP < 0.05 when compared with the corresponding control fibroblasts, cP < 0.05 when compared with the corresponding sporadic disease fibroblasts based on two-way ANOVA and post-hoc Tukey’s test. C9, C9orf72; CTRL, control; pTDP-43, phosphorylated TDP-43.
Using the C-terminal TDP-43 antibody to examine other forms of TDP-43, we found that TDP-43 immunostaining overlapped with the nuclear staining marker DAPI in vehicle-treated control fibroblasts (Fig. 8E and F). However, in vehicle-treated C9orf72 fibroblasts, TDP-43 immunostaining was found in the cytoplasm with reduced nuclear TDP-43 signal as compared with fibroblasts from controls (Fig. 8E and F). A similar pattern of TDP-43 staining was seen in vehicle-treated sporadic disease fibroblasts but was not statistically significant (Fig. 8E and F). Proteasome inhibition again resulted in formation of TDP-43-positive/p62-positive aggresomes in sporadic disease fibroblasts that were reduced in C9orf72 fibroblasts (Fig. 8E, G, and H, Supplementary Fig. 10). Rather than being sequestered to the aggresome, TDP-43 accumulated as diffuse cytoplasmic staining in C9orf72 fibroblasts (Fig. 8E and F, Supplementary Fig. 10).
Discussion
The current study identified several major pathological differences in TDP-43 inclusion formation between sporadic and C9orf72-related disease patients. We confirmed in our pathological samples the previously reported TDP-43-negative/p62-positive inclusions in the cerebellum and hippocampus that define C9orf72 ALS/FTLD neuropathology (Al-Sarraj et al., 2011; Troakes et al., 2012). In these cases, the hippocampi from sporadic ALS/FTLD contained numerous and easily identified circumferential and round pTDP-43 inclusions, whereas pTDP-43 inclusions were proportionally fewer and were morphologically smaller in C9orf72 ALS/FTLD hippocampi. These findings are consistent with two recently published reports on C9orf72 cases without robust TDP-43 pathology (Vatsavayai et al., 2016; Nana et al., 2019). Another important finding was the demonstration of soluble cytoplasmic TDP-43 in C9orf72 ALS/FTLD that was largely absent in sporadic ALS/FTLD. Both sporadic and C9orf72 brains had similarly elevated TDP-43 protein levels in the frontal cortex and hippocampus; therefore, the differences in TDP-43 inclusion body formation between sporadic and C9orf72-related diseases could not be attributed to a difference in TDP-43 protein abundance. The lesser presence of TDP-43 inclusions in C9orf72 neurons cannot be attributed to a generalized reduced ability to form cytoplasmic inclusion bodies, as p62 inclusions are more abundant in the C9orf72 hippocampus than sporadic cases. The presence of cytosolic TDP-43 protein in C9orf72 ALS/FTLD neurons is not surprising given recent studies indicating that the nucleocytoplasmic transport of TDP-43 is impaired in Drosophila and neuronal culture models of C9orf72-related disease (Zhang et al., 2015; Khosravi et al., 2017). We suggest that the presence of soluble TDP-43 protein in the cytoplasm of C9orf72 ALS/FTLD neurons, together with the smaller sizes and proportionally fewer TDP-43 inclusions in these neurons, might implicate an impairment of cytoplasmic TDP-43 inclusion body formation in C9orf72 patients in vivo. Supporting this conclusion are the experiments in patient-derived fibroblasts, which revealed that the induction of microtubule-dependent TDP-43 inclusion body formation by proteasome inhibition was deficient in C9orf72 fibroblasts compared with sporadic disease fibroblasts. Taken together, these data from ALS/FTLD brains and patient-derived fibroblasts demonstrate that turnover of TDP-43 protein is handled differently in C9orf72-related diseases, and suggests that there is an impairment of TDP-43 inclusion body formation in C9orf72 patients compared with sporadic disease patients.
The abnormal HRE in C9orf72 is transcribed bidirectionally to form nuclear RNA foci (DeJesus-Hernandez et al., 2011; Wojciechowska and Krzyzosiak, 2011) and is translated in an AUG-independent manner from both sense and antisense C9orf72 transcripts into five distinct dipeptide protein repeats (Mann et al., 2013; Mori et al., 2013). These distinct dipeptide protein repeats form neuronal cytoplasmic inclusions that stain positive for p62 (Mann et al., 2013; Mori et al., 2013). The fact that C9orf72 ALS/FTLD was uniquely characterized by TDP-43-negative/p62-positive inclusions, together with the presence of distinct dipeptide protein repeats-positive/p62-positive inclusions, suggest a possibility that distinct dipeptide protein repeats might compete with TDP-43 to become the major constituent of inclusion bodies in C9orf72 patients. It is also possible that RNA foci could block the formation of TDP-43 inclusions via unknown mechanisms. In addition, the abnormal HRE in C9orf72 causes haploinsufficiency in the C9orf72 protein which could affect protein quality control systems (Burk and Pasterkamp, 2019) to disrupt TDP-43 inclusion body formation. Another possible reason for impaired TDP-43 inclusion body formation could be that the C9orf72 mutation, via yet-to-be-identified mechanisms, inhibits the phosphorylation of TDP-43 to block the targeting of TDP-43 into inclusion bodies (Brady et al., 2011). This hypothesis is supported by our findings that the diffuse cytoplasmic soluble TDP-43 in C9orf72 patient-derived fibroblasts was not labelled by the anti-phospho-TDP-43 antibody and thus represents non-phosphorylated forms of TDP-43. Why the presence of diffuse cytosolic TDP-43 was not always associated with TDP-43 nuclear clearing, which is typically seen in neurons with TDP-43 cytoplasmic inclusions, is unknown, but may have to do with different degree of nucleocytoplasmic transport dysfunction in these C9orf72 neurons. Additional studies are needed to investigate the possible mechanisms underlying how the C9orf72 mutation inhibits the targeting of TDP-43 into inclusion bodies and dysregulates nucleocytoplasmic transport of TDP-43, which could have important implications in understanding the pathogenesis of C9orf72-related TDP-43 proteinopathies such as FTD and ALS.
Our observation of excess TDP-43 protein and its toxic C-terminal fragments in the frontal cortex and hippocampus of sporadic and C9orf72 ALS/FTLD, together with previous studies showing that an overabundance of TDP-43 protein causes neurotoxicity and ALS/FTLD phenotype and pathology in transgenic mice (Wils et al., 2010; Igaz et al., 2011), provides strong evidence to suggest that aberrantly increased TDP-43 protein levels may contribute to sporadic and C9orf72 ALS/FTLD pathogenesis, possibly via a toxic gain-of-function mechanism. In support of this mechanistic hypothesis is that elevated TDP-43 protein levels in the frontal cortex and hippocampus were seen only in the patients with FTD and pathological FTLD, but TDP-43 protein levels in non-demented, sporadic ALS patients were the same as controls. In addition, our previous work using quantitative proteomics also demonstrated elevated levels of TDP-43 in the frontal cortex of FTD patients as compared with controls (Umoh et al., 2018).
Our in vitro studies demonstrated that TDP-43 degradation by the UPS is impaired in sporadic and C9orf72 ALS/FTD fibroblasts, which might explain the abnormal accumulation of TDP-43 protein in both patient-derived ALS/FTD fibroblasts (Sabatelli et al., 2015) and ALS/FTLD brains. The selectively increased TDP-43 protein levels in clinically affected brain regions in ALS/FTLD (frontal cortex and hippocampus), but not in relatively unaffected areas (cerebellum), raises the possibility that the dysfunction in proteasome degradation of TDP-43 correlates with clinical disease activity. Why elevation of TDP-43 protein levels associated with sporadic and C9orf72 ALS/FTD is found in fibroblasts without known skin manifestations in these patients remains unknown. One possibility is that certain brain regions are more vulnerable to TDP-43 toxicity, which is supported by a transgenic mouse model expressing human TDP-43 in all tissues but only demonstrating central nervous system manifestation and pathology without causing disease in other organs (Swarup et al., 2011). In the setting of proteasome inhibition, sporadic ALS/FTD fibroblasts formed TDP-43 aggresomes that were tightly associated with autophagosomes and lysosomes. This finding is consistent with numerous reports that suggest aggresomes are staging areas for the degradation of misfolded proteins by autophagy (Iwata et al., 2005a, b; Mehrpour et al., 2010; Lee et al., 2011) and may explain why aggresome formation is not associated with increased protein levels in cells (Zaarur et al., 2008). Several studies have reported that aggresome formation correlates with cell survival (Kawaguchi et al., 2003; Taylor et al., 2003; Shiarli et al., 2006). TDP-43 aggresomes in fibroblasts resembled the TDP-43 cytoplasmic neuronal inclusions in ALS/FTLD brains in that they were both perinuclear in location and that they co-localized with ubiquitin, p62 and HSP70. These inclusions also co-localized with autophagosomes and lysosomes and, therefore, likely served as a location for degradation of excess TDP-43 protein by autophagy.
TDP-43 aggresome formation in response to proteasome inhibition was impaired in C9orf72 fibroblasts compared with sporadic disease fibroblasts. The inability to consolidate TDP-43 into aggresomes likely resulted in inefficient autophagic degradation of excess TDP-43, which suggested an impairment of protein quality control responses and increased cellular toxicity in C9orf72-related diseases. Despite a conundrum in the neurodegenerative disease literature regarding whether soluble protein oligomers or insoluble protein inclusions are the culprits for neurotoxicity, the general consensus is that the sequestration of soluble aggregated toxic proteins including TDP-43 by active microtubule-dependent mechanisms into insoluble aggregates is beneficial by preventing toxic, misfolded proteins from saturating chaperones and the proteasome and facilitating their clearance via autophagy (Ross and Poirier, 2004; Cohen et al., 2006; Scotter et al., 2014; Sontag et al., 2014; Sweeney et al., 2017; Soto and Pritzkow, 2018). Supporting this view, several studies have reported that aggresome formation correlates with cell survival (Kawaguchi et al., 2003; Taylor et al., 2003; Shiarli et al., 2006). Our study of mutant SOD1 proteins also showed that soluble mutant proteins were demonstrable only in pathologically affected spinal cord tissues, further suggesting that soluble protein species are responsible for ALS pathogenesis (Brotherton et al., 2012). However, cells are known to be dynamic creatures that can form insoluble proteins from soluble proteins (Bagola and Sommer, 2008) and, therefore, it is virtually impossible to purely isolate and test which protein species mediate cytotoxicity.
The demonstration of aggresome formation in the current work is a cell culture phenomenon that does not necessarily represent the inclusion bodies found in brains. It is also unclear whether the impairment of TDP-43 inclusion body formation in C9orf72 patients has clinical significance given that there are no major phenotypic differences between C9orf72 and sporadic ALS/FTLD patients. Furthermore, impairment of protein quality control systems in these patients could affect not only TDP-43 but also other potentially toxic proteins to contribute to disease pathogenesis. Future studies are needed to determine the clinical and pathophysiological significances of these findings.
Supplementary material
Supplementary material is available at BrainCommunications online.
Supplementary Material
Acknowledgements
We thank the Emory University Alzheimer Disease Research Center (ADRC) for providing central nervous system tissue samples and the patient family members who contributed those tissues. We also thank Emory University School of Medicine Neurology Residency Training Program for providing 6 months of protected research time for S.M.L.
Funding
This work was supported in part by National Institutes of Health grants (AG025688 to J.D.G. and M.G., NS055077 to M.G., NS092343 to L.L., NS093550 to L.S.C. and K08-NS087121 to C.M.H.) and by research funding from the Amyotrophic Lateral Sclerosis Association and the Muscular Dystrophy Association.
Competing interests
The authors report no competing interests.
Glossary
Abbreviations
- ALS =
amyotrophic lateral sclerosis
- C9 =
C9orf72
- FTD =
frontotemporal dementia
- FTLD =
frontotemporal lobar degeneration
- HRE =
hexanucleotide repeat expansion
- pTDP-43 =
phosphorylated TAR DNA-binding protein 43
- TDP-43 =
TAR DNA-binding protein 43
- UPS =
ubiquitin-proteasome system
References
- Aber KM, Nori P, MacDonald SM, Bibat G, Jarrar MH, Kaufmann WE.. Methyl-CpG-binding protein 2 is localized in the postsynaptic compartment: an immunochemical study of subcellular fractions. Neuroscience 2003; 116: 77–80. [DOI] [PubMed] [Google Scholar]
- Al-Kofahi Y, Lassoued W, Lee W, Roysam B.. Improved automatic detection and segmentation of cell nuclei in histopathology images. IEEE Trans Biomed Eng 2010; 57: 841–52. [DOI] [PubMed] [Google Scholar]
- Al-Sarraj S, King A, Troakes C, Smith B, Maekawa S, Bodi I, et al. p62 positive, TDP-43 negative, neuronal cytoplasmic and intranuclear inclusions in the cerebellum and hippocampus define the pathology of C9orf72-linked FTLD and MND/ALS. Acta Neuropathol 2011; 122: 691–702. [DOI] [PubMed] [Google Scholar]
- Bagola K, Sommer T.. Protein quality control: on IPODs and other JUNQ. Curr Biol 2008; 18: R1019–21. [DOI] [PubMed] [Google Scholar]
- Bigio EH, Weintraub S, Rademakers R, Baker M, Ahmadian SS, Rademaker A, et al. Frontotemporal lobar degeneration with TDP-43 proteinopathy and chromosome 9p repeat expansion in C9ORF72: clinicopathologic correlation. Neuropathology 2013; 33: 122–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokhuis AM, Groen EJ, Koppers M, van den Berg LH, Pasterkamp RJ.. Protein aggregation in amyotrophic lateral sclerosis. Acta Neuropathol 2013; 125: 777–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady OA, Meng P, Zheng Y, Mao Y, Hu F.. Regulation of TDP-43 aggregation by phosphorylation and p62/SQSTM1. J Neurochem 2011; 116: 248–59. [DOI] [PubMed] [Google Scholar]
- Brettschneider J, Arai K, Del Tredici K, Toledo JB, Robinson JL, Lee EB, et al. TDP-43 pathology and neuronal loss in amyotrophic lateral sclerosis spinal cord. Acta Neuropathol 2014; 128: 423–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotherton TE, Li Y, Cooper D, Gearing M, Julien JP, Rothstein JD, et al. Localization of a toxic form of superoxide dismutase 1 protein to pathologically affected tissues in familial ALS. Proc Natl Acad Sci USA 2012; 109: 5505–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk K, Pasterkamp RJ.. Disrupted neuronal trafficking in amyotrophic lateral sclerosis. Acta Neuropathol 2019; 137: 859–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns NJ, Bigio EH, Mackenzie IR, Neumann M, Lee VM, Hatanpaa KJ, et al. Neuropathologic diagnostic and nosologic criteria for frontotemporal lobar degeneration: consensus of the Consortium for Frontotemporal Lobar Degeneration. Acta Neuropathol 2007; 114: 5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell 1994; 79: 13–21. [DOI] [PubMed] [Google Scholar]
- Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A.. Opposing activities protect against age-onset proteotoxicity. Science 2006; 313: 1604–10. [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 2011; 72: 245–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles LM, Chen J, Li L, Chin LS.. Dystonia-associated mutations cause premature degradation of torsinA protein and cell-type-specific mislocalization to the nuclear envelope. Hum Mol Genet 2008; 17: 2712–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory JM, Barros TP, Meehan S, Dobson CM, Luheshi LM.. The aggregation and neurotoxicity of TDP-43 and its ALS-associated 25 kDa fragment are differentially affected by molecular chaperones in Drosophila. PLoS One 2012; 7: e31899.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz FI, Geschwind DH.. Molecular genetics of neurodegenerative dementias. Cold Spring Harb Perspect Biol 2017; 9: a023705.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaz LM, Kwong LK, Lee EB, Chen-Plotkin A, Swanson E, Unger T, et al. Dysregulation of the ALS-associated gene TDP-43 leads to neuronal death and degeneration in mice. J Clin Invest 2011; 121: 726–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaz LM, Kwong LK, Xu Y, Truax AC, Uryu K, Neumann M, et al. Enrichment of C-terminal fragments in TAR DNA-binding protein-43 cytoplasmic inclusions in brain but not in spinal cord of frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Am J Pathol 2008; 173: 182–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata A, Christianson JC, Bucci M, Ellerby LM, Nukina N, Forno LS, et al. Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation. Proc Natl Acad Sci USA 2005a; 102: 13135–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata A, Riley BE, Johnston JA, Kopito RR.. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem 2005b; 280: 40282–92. [DOI] [PubMed] [Google Scholar]
- Johnson BS, Snead D, Lee JJ, McCaffery JM, Shorter J, Gitler AD.. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J Biol Chem 2009; 284: 20329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JA, Ward CL, Kopito RR.. Aggresomes: a cellular response to misfolded proteins. J Cell Biol 1998; 143: 1883–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP.. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 2003; 115: 727–38. [DOI] [PubMed] [Google Scholar]
- Khosravi B, Hartmann H, May S, Mohl C, Ederle H, Michaelsen M, et al. Cytoplasmic poly-GA aggregates impair nuclear import of TDP-43 in C9orf72 ALS/FTLD. Hum Mol Genet 2017; 26: 790–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8: 445–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad C, Kawamata H, Bredvik KG, Arreguin AJ, Cajamarca SA, Hupf JC, et al. Fibroblast bioenergetics to classify amyotrophic lateral sclerosis patients. Mol Neurodegener 2017; 12: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopito RR, Sitia R.. Aggresomes and Russell bodies. Symptoms of cellular indigestion? EMBO Rep 2000; 1: 225–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiling JA, Tamamori-Adachi M, Sexton AN, Jeyapalan JC, Munoz-Najar U, Peterson AL, et al. Age-associated increase in heterochromatic marks in murine and primate tissues. Aging Cell 2011; 10: 292–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Chin LS, Li L.. Charcot-Marie-Tooth disease-linked protein SIMPLE functions with the ESCRT machinery in endosomal trafficking. J Cell Biol 2012; 199: 799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Olzmann JA, Chin LS, Li L.. Mutations associated with Charcot-Marie-Tooth disease cause SIMPLE protein mislocalization and degradation by the proteasome and aggresome-autophagy pathways. J Cell Sci 2011; 124 (Pt 19): 3319–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Baborie A, Pickering-Brown S, Du Plessis D, Jaros E, Perry RH, et al. Heterogeneity of ubiquitin pathology in frontotemporal lobar degeneration: classification and relation to clinical phenotype. Acta Neuropathol 2006; 112: 539–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Munoz DG, Kusaka H, Yokota O, Ishihara K, Roeber S, et al. Distinct pathological subtypes of FTLD-FUS. Acta Neuropathol 2011a; 121: 207–18. [DOI] [PubMed] [Google Scholar]
- Mackenzie IR, Neumann M, Baborie A, Sampathu DM, Du Plessis D, Jaros E, et al. A harmonized classification system for FTLD-TDP pathology. Acta Neuropathol 2011b; 122: 111–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann DM, Rollinson S, Robinson A, Bennion Callister J, Thompson JC, Snowden JS, et al. Dipeptide repeat proteins are present in the p62 positive inclusions in patients with frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9ORF72. Acta Neuropathol Commun 2013; 1: 68.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrpour M, Esclatine A, Beau I, Codogno P.. Overview of macroautophagy regulation in mammalian cells. Cell Res 2010; 20: 748–62. [DOI] [PubMed] [Google Scholar]
- Misztal K, Brozko N, Nagalski A, Szewczyk LM, Krolak M, Brzozowska K, et al. TCF7L2 mediates the cellular and behavioral response to chronic lithium treatment in animal models. Neuropharmacology 2017; 113: 490–501. [DOI] [PubMed] [Google Scholar]
- Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 2013; 339: 1335–8. [DOI] [PubMed] [Google Scholar]
- Nana AL, Sidhu M, Gaus SE, Hwang JL, Li L, Park Y, et al. Neurons selectively targeted in frontotemporal dementia reveal early stage TDP-43 pathobiology. Acta Neuropathol 2019; 137: 27–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Kwong LK, Sampathu DM, Trojanowski JQ, Lee VM.. TDP-43 proteinopathy in frontotemporal lobar degeneration and amyotrophic lateral sclerosis: protein misfolding diseases without amyloidosis. Arch Neurol 2007; 64: 1388–94. [DOI] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 2006; 314: 130–3. [DOI] [PubMed] [Google Scholar]
- Nonaka T, Arai T, Buratti E, Baralle FE, Akiyama H, Hasegawa M.. Phosphorylated and ubiquitinated TDP-43 pathological inclusions in ALS and FTLD-U are recapitulated in SH-SY5Y cells. FEBS Lett 2009; 583: 394–400. [DOI] [PubMed] [Google Scholar]
- Olzmann JA, Li L, Chudaev MV, Chen J, Perez FA, Palmiter RD, et al. Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J Cell Biol 2007; 178: 1025–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikkarainen M, Hartikainen P, Alafuzoff I.. Ubiquitinated p62-positive, TDP-43-negative inclusions in cerebellum in frontotemporal lobar degeneration with TAR DNA binding protein 43. Neuropathology 2010; 30: 197–9. [DOI] [PubMed] [Google Scholar]
- Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci 2011; 14: 459–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademakers R. C9orf72 repeat expansions in patients with ALS and FTD. Lancet Neurol 2012; 11: 297–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rascovsky K, Hodges JR, Knopman D, Mendez MF, Kramer JH, Neuhaus J, et al. Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain 2011; 134: 2456–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard E, Adhikhmin M, Gooch B, Shirley P.. Color transfer between images. IEEE Comput Grap Appl 2001; 21: 34–41. [Google Scholar]
- Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 2011; 72: 257–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter-Landsberg C, Leyk J.. Inclusion body formation, macroautophagy, and the role of HDAC6 in neurodegeneration. Acta Neuropathol 2013; 126: 793–807. [DOI] [PubMed] [Google Scholar]
- Ross CA, Poirier MA.. Protein aggregation and neurodegenerative disease. Nat Med 2004; 10 (Suppl): S10–7. [DOI] [PubMed] [Google Scholar]
- Ross CA, Poirier MA.. Opinion: what is the role of protein aggregation in neurodegeneration? Nat Rev Mol Cell Biol 2005; 6: 891–8. [DOI] [PubMed] [Google Scholar]
- Sabatelli M, Zollino M, Conte A, D, Grande A, Marangi G, Lucchini M, et al. Primary fibroblasts cultures reveal TDP-43 abnormalities in amyotrophic lateral sclerosis patients with and without SOD1 mutations. Neurobiol Aging 2015; 36: 2005.e5–e13. [DOI] [PubMed] [Google Scholar]
- Scotter EL, Vance C, Nishimura AL, Lee YB, Chen HJ, Urwin H, et al. Differential roles of the ubiquitin proteasome system and autophagy in the clearance of soluble and aggregated TDP-43 species. J Cell Sci 2014; 127 (Pt 6): 1263–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelaar H, Schelhaas HJ, Azmani A, Kusters B, Rosso S, Majoor-Krakauer D, et al. TDP-43 pathology in familial frontotemporal dementia and motor neuron disease without Progranulin mutations. Brain 2007; 130 (Pt 5): 1375–85. [DOI] [PubMed] [Google Scholar]
- Seilhean D, Cazeneuve C, Thuries V, Russaouen O, Millecamps S, Salachas F, et al. Accumulation of TDP-43 and alpha-actin in an amyotrophic lateral sclerosis patient with the K17I ANG mutation. Acta Neuropathol 2009; 118: 561–73. [DOI] [PubMed] [Google Scholar]
- Sephton CF, Cenik B, Cenik BK, Herz J, Yu G.. TDP-43 in central nervous system development and function: clues to TDP-43-associated neurodegeneration. Biol Chem 2012; 393: 589–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serebryannyy LA, Cruz CM, de Lanerolle P.. A role for nuclear actin in HDAC 1 and 2 regulation. Sci Rep 2016; 6: 28460.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha SJ, Takada LT, Rankin KP, Yokoyama JS, Rutherford NJ, Fong JC, et al. Frontotemporal dementia due to C9ORF72 mutations: clinical and imaging features. Neurology 2012; 79: 1002–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahheydari H, Ragagnin A, Walker AK, Toth RP, Vidal M, Jagaraj CJ, et al. Protein quality control and the amyotrophic lateral sclerosis/frontotemporal dementia continuum. Front Mol Neurosci 2017; 10: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaiken TE, Opekun AR.. Dissecting the cell to nucleus, perinucleus and cytosol. Sci Rep 2014; 4: 4923.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiarli AM, Jennings R, Shi J, Bailey K, Davidson Y, Tian J, et al. Comparison of extent of tau pathology in patients with frontotemporal dementia with Parkinsonism linked to chromosome 17 (FTDP-17), frontotemporal lobar degeneration with Pick bodies and early onset Alzheimer’s disease. Neuropathol Appl Neurobiol 2006; 32: 374–87. [DOI] [PubMed] [Google Scholar]
- Sontag EM, Vonk WIM, Frydman J.. Sorting out the trash: the spatial nature of eukaryotic protein quality control. Curr Opin Cell Biol 2014; 26: 139–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto C, Pritzkow S.. Protein misfolding, aggregation, and conformational strains in neurodegenerative diseases. Nat Neurosci 2018; 21: 1332–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup V, Phaneuf D, Bareil C, Robertson J, Rouleau GA, Kriz J, et al. Pathological hallmarks of amyotrophic lateral sclerosis/frontotemporal lobar degeneration in transgenic mice produced with TDP-43 genomic fragments. Brain 2011; 134: 2610–26. [DOI] [PubMed] [Google Scholar]
- Sweeney P, Park H, Baumann M, Dunlop J, Frydman J, Kopito R, et al. Protein misfolding in neurodegenerative diseases: implications and strategies. Transl Neurodegener 2017; 6: 6.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takalo M, Salminen A, Soininen H, Hiltunen M, Haapasalo A.. Protein aggregation and degradation mechanisms in neurodegenerative diseases. Am J Neurodegener Dis 2013; 2: 1–14. [PMC free article] [PubMed] [Google Scholar]
- Tan RH, Yang Y, Kim WS, Dobson-Stone C, Kwok JB, Kiernan MC, et al. Distinct TDP-43 inclusion morphologies in frontotemporal lobar degeneration with and without amyotrophic lateral sclerosis. Acta Neuropathol Commun 2017; 5: 76.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JP, Tanaka F, Robitschek J, Sandoval CM, Taye A, Markovic-Plese S, et al. Aggresomes protect cells by enhancing the degradation of toxic polyglutamine-containing protein. Hum Mol Genet 2003; 12: 749–57. [DOI] [PubMed] [Google Scholar]
- Troakes C, Maekawa S, Wijesekera L, Rogelj B, Siklos L, Bell C, et al. An MND/ALS phenotype associated with C9orf72 repeat expansion: abundant p62-positive, TDP-43-negative inclusions in cerebral cortex, hippocampus and cerebellum but without associated cognitive decline. Neuropathology 2012; 32: 505–14. [DOI] [PubMed] [Google Scholar]
- Turturici G, Sconzo G, Geraci F.. Hsp70 and its molecular role in nervous system diseases. Biochem Res Int 2011; 2011: 618127.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umoh ME, Dammer EB, Dai J, Duong DM, Lah JJ, Levey AI, et al. A proteomic network approach across the ALS-FTD disease spectrum resolves clinical phenotypes and genetic vulnerability in human brain. EMBO Mol Med 2018; 10: 48–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umoh ME, Fournier C, Li Y, Polak M, Shaw L, Landers JE, et al. Comparative analysis of C9orf72 and sporadic disease in an ALS clinic population. Neurology 2016; 87: 1024–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eersel J, Ke YD, Gladbach A, Bi M, Gotz J, Kril JJ, et al. Cytoplasmic accumulation and aggregation of TDP-43 upon proteasome inhibition in cultured neurons. PLoS One 2011; 6: e22850.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatsavayai SC, Yoon SJ, Gardner RC, Gendron TF, Vargas JN, Trujillo A, et al. Timing and significance of pathological features in C9orf72 expansion-associated frontotemporal dementia. Brain 2016; 139 (Pt 12): 3202–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AK, Soo KY, Sundaramoorthy V, Parakh S, Ma Y, Farg MA, et al. ALS-associated TDP-43 induces endoplasmic reticulum stress, which drives cytoplasmic TDP-43 accumulation and stress granule formation. PLoS One 2013; 8: e81170.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber E, Li L, Chin LS.. Hypertonia-associated protein Trak1 is a novel regulator of endosome-to-lysosome trafficking. J Mol Biol 2008; 382: 638–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wils H, Kleinberger G, Janssens J, Pereson S, Joris G, Cuijt I, et al. TDP-43 transgenic mice develop spastic paralysis and neuronal inclusions characteristic of ALS and frontotemporal lobar degeneration. Proc Natl Acad Sci USA 2010; 107: 3858–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciechowska M, Krzyzosiak WJ.. Cellular toxicity of expanded RNA repeats: focus on RNA foci. Hum Mol Genet 2011; 20: 3811–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Tan W, Whittle C, Qiu L, Cao L, Akbarian S, et al. The C-terminal TDP-43 fragments have a high aggregation propensity and harm neurons by a dominant-negative mechanism. PLoS One 2010; 5: e15878.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaarur N, Meriin AB, Gabai VL, Sherman MY.. Triggering aggresome formation. Dissecting aggresome-targeting and aggregation signals in synphilin 1. J Biol Chem 2008; 283: 27575–84. [DOI] [PubMed] [Google Scholar]
- Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 2015; 525: 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available from the corresponding authors upon reasonable request.