Keywords: circRNAs, endothelial cells, RNA sequencing, cerebral ischemia reperfusion injury, microRNAs, neural regeneration
Abstract
Circular RNAs (circRNAs) are generated by head-to-tail splicing and are ubiquitously expressed in all multicellular organisms. Their important biological functions are increasingly recognized. Cerebral ischemia reperfusion injury-induced brain microvascular endothelial cell dysfunction is an initial stage of blood-brain barrier disruption. The expression profile and potential function of circRNAs in brain microvascular endothelial cells is unknown. Rat brain microvascular endothelial cells were extracted and cultured in glucose-free medium for 4 hours with 5% CO2 and 95% N2, and the medium was then replaced with complete growth medium for 6 hours. The RNA in these cells was then extracted. The circRNA was identified by Find_circ and CIRI2 software. Functional and pathway enrichment analysis of genes that were common to differentially expressed mRNAs and circRNA host genes was performed by the Database for Annotation, Visualization and Integrated Discovery Functional Annotation Tool. Miranda software was used to predict microRNAs that were potentially sponged by circRNAs. Furthermore, cytoscape depicted the circR-NA-microRNA interaction network. The results showed that there were 1288 circRNAs in normal and oxygen-glucose deprived/recovered primary brain microvascular endothelial cells. There are 211 upregulated and 326 downregulated differentially expressed circRNAs. The host genes of these differentially expressed circRNAs overlapped with those of differentially expressed mRNAs. The shared genes were further studied by functional enrichment analyses, which revealed that circRNAs may contribute to calcium ion function and the cyclic guanosine 3′,5′-monophosphate (CAMP) dependent protein kinase (PKα) signaling pathway. Next, quantitative reverse transcription polymerase chain reaction assays were performed to detect circRNA levels transcribed from the overlapping host genes. Eight out of the ten circRNAs with the highest fold-change identified by sequencing were successfully verified. Subsequently, the circRNA-microRNA interaction networks of these eight circRNAs were explored by bioinformatic analysis. These results demonstrate that altered circRNAs may be important in the pathogenesis of cerebral ischemia reperfusion injury and consequently may also be potential therapeutic targets for cerebral ischemia diseases. All animal experiments were approved by the Chongqing Medical University Committee on Animal Research, China (approval No. CQMU20180086) on March 22, 2018.
Chinese Library Classification No. R446; R743; R741
Introduction
The blood-brain barrier consists of brain microvascular endothelial cells (BMECs), astrocytes, microglial cells, and pericytes and the microvasculature in the brain delivers oxygen and nutrients to neurocytes. Under physiological con-ditions, the blood-brain barrier selectively allows molecules to pass through the barrier to protect the central nervous system. In particular, BMECs can express special transport proteins to carry glucose, amino acids, and other factors. BMECs also secrete neurotrophins and enzymes to nourish neural cells and degrade harmful molecules. Thus, BMECs contribute to revascularization and neurological recovery after ischemic injury (Yu et al., 2015; Toth and Nielsen, 2018).
Circular RNAs (circRNAs) have recently been described as novel regulatory noncoding RNAs (Barrett et al., 2015). circRNAs are transcribed from the exons and introns of genes and form covalently closed head-to-tail (or backspliced) circularized transcripts (Hansen et al., 2013; Barrett et al., 2015). circRNAs can act as competing endogenous RNA or microRNA (miRNA) sponges. circRNAs play important roles in stroke and endothelial function. For example, circRNA-ZNF609 adsorbs miR-615-5p, which withstands oxidative stress and promotes vascular endothelial cell mi-gration (Liu et al., 2017b). Furthermore, circRNA Hectd1 acts as a sponge that inhibits miR-142 and contributes to ischemic stroke via astrocyte activation (Han et al., 2018). Therefore, we suggest that circRNAs are worthy of further exploration.
BMECs contribute significantly to integrity and function of the brain vasculature. Oxygen and nutrient deprivation may induce BMEC dysfunction and increased blood-brain barrier permeability (Yu et al., 2015). However, how oxy-gen-glucose shortages affect circRNAs in BMECs is unknown. Here, we used RNA sequencing to measure the global changes of circRNAs in BMECs subjected to oxygen-glucose deprivation (OGD)/recovery (OGD/R) treatment. Differentially expressed (DE) circRNAs as well as potential mechanisms were explored.
Materials and Methods
Isolation and cultivation of primary BMECs
All animal methods were approved by the Chongqing Medical University Committee on Animal Research, China (approval No. CQMU20180086) on March 22, 2018. Adult male Sprague-Dawley rats aged 6 weeks and weighing 160–200 g were housed in a specific-pathogen-free animal room of the Animal Breeding Center of Chongqing Medical University, China [license No. SYXK (Yu) 2017-0023]. Eight rats were anesthetized by inhalation of isoflurane (2% in oxygen) (RWD Life Science, Shenzhen, China), euthanized, and their brains collected. Tissue was homogenized and then centrifuged at 720 × g for 5 minutes at 4°C. The supernatant was discarded, and the pellet resuspended in phosphate-buffered saline. This was then layered over 15 mL 18% dextran and centrifuged at 4500 × g for 20 minutes at 4°C. The pellet was resuspended in 10 mL phosphate-buffered saline (containing 0.1% bovine serum albumin). Ten milliliters of the suspension were added to 100 µL collagenase (100 mg/mL), 40 µL DNase I (10 mg/mL) and 100 µL N-alpha-tosyl-L-lysine chloromethyl ketone hydrochloride (14.7 µg/mL) and digested for 1 hour at 37°C. After centrifugation at 1000 × g for 5 minutes, the pellet was resuspended in 2 mL phosphate-buffered saline containing 0.1% bovine serum albumin and 100 µL biotin-labeled anti-CD31 antibody (DSB-X Biotin Protein Labeling Kit, Thermo Fisher, Waltham, MA, USA). After incubation for 10 minutes at 4°C, BMECs were isolated with a Dynabeads FlowComp Flexi Kit (Thermo Fisher). Finally, bead-free cells were cultured in ordinary medium. The medium comprised DMEM basic medium (Thermo Fisher), 10% fetal bovine serum, 20 mg/L endothelial cell growth supplement (ECGS), 2 mM L-glutamine and 100 mg/L heparin sodium (Plácido et al., 2017; Sawaguchi et al., 2017). The endothelial cell markers, factor VIII and CD31 (Thermo Fisher), were used to identify BMECs by immunofluorescence (Bachetti and Morbidelli, 2000).
OGD/R treatment
Cells at 90–100% confluency were digested with trypsin and re-seeded in new dishes. The passage dilution was 1:4. Passage 6 BMECs were subjected to OGD/R treatment. At 48 hours after seeding, the medium was replaced by glu-cose-free medium, which was prebubbled with 95% N2 and 5% CO2 for 1 hour. BMECs were then cultured in an incubator flushed with 5% CO2 and 95% N2. The chamber was sealed and kept at 37°C for 4 hours with an oxygen concentration of < 0.2%. Upon OGD termination, the cells were placed back into a normal incubator (5% CO2, 95% air at 37°C), and the medium was replaced with complete growth medium. Control BMECs were not exposed to OGD. The cells were harvested for further analyses after 6 hours.
RNA extraction and sequencing
A total of 5 µg of RNA per sample was prepared using TRIzol reagent according to the manufacturer’s recommenda-tions (Thermo Fisher). A Nanodrop 2000 (Thermo Fisher), an Agilent Bioanalyzer 2100 (Agilent, USA) and agarose gel electrophoresis were used to determine RNA quality. Samples with an optical density (OD)260/280 between 1.7–2.0, OD260/230 > 2, 28S/18S > 1.8 and RNA integrity number > 9 were considered reliable. Five samples of equal mass in each group were mixed. An Epicentre Ribozero ribosomal RNA Removal Kit (Epicentre, Madison, WI, USA) was used to remove ribosomal RNA, and RNase R (Epicentre) was used to digest linear RNA. Subsequently, sequencing libraries were generated using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB, Ipswich, MA, USA) following the manufacturer’s recommendations. After cluster generation, the libraries were sequenced on an Illumina Hiseq 4000 platform, and 150-bp paired-end reads were generated.
For mRNA sequencing, ribosomal RNA was removed from 3 µg of total RNA. Sequencing libraries were generated directly without linear RNA digestion. Sequencing and DE circRNA/mRNA analysis were completed by Novogene, Beijing, China.
Bioinformatic analysis
Find_circ (GitHub, San Francisco, CA, USA) and CIRI2 (GitHub) software were used to identify circRNAs (Memczak et al., 2013; Gao et al., 2018). Raw data were normalized by standardized TPM (transcripts per million clean tags). The DESeq R package (1.10.1) (Bioconductor, Riverside, CA, USA) calculated differences in the expression of circRNAs.
For mRNA analysis, bowtie2 (v2.2.8) (Johns Hopkins University, Baltimore, MD, USA) and HISAT2 (v2.0.4) (Johns Hopkins University) mapped data to the reference genome. Cuffdiff (v2.1.1) (GenePattern, CA, USA) was used to calculate FPKMs (expected number of Fragments Per Kilobase of transcript sequence per Million base pairs sequenced) of mRNA. Next, the FPKM value was used to calculate mRNA differential expression.
The intersecting genes of DE circRNA host genes and DE mRNAs were subjected to functional and pathway enrichment analysis by the Database for Annotation, Visualization and Integrated Discovery (DAVID) Functional Annotation Tool (DAVID Bioinformatic Team, Frederick, USA) (Huang da et al., 2009). Miranda software (cBio-MSKCC, New York, USA) was used to predict the miRNAs that were potentially sponged by circRNAs (John et al., 2004). Cytoscape (NIGMS, Bethesda, MD, USA) depicted the circRNA-miRNA interaction network (Shannon et al., 2003).
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
As described previously (Shang et al., 2016), RNA was extracted using TRIzol reagent (Thermo Fisher). Two mi-crograms of RNA were reverse-transcribed to cDNA using a RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher). qRT-PCR was performed with 20 µL SYBR Green reaction mix containing specific primers (Sangon Biotech, Shanghai, China) using a standard protocol The primers are listed in Additional Table 1. Each group had five experimental replicates; and each experimental replicate was performed three times. The Ct value of target circRNAs was normalized to the geometrical average of glyceraldehyde-3-phosphate dehydrogenase, β-actin and U6.
Additional Table 1.
The list of CircRNAs primers
| ID | Primers sequence | Length (bp) |
|---|---|---|
| novel_circ_0000173 | F: 5’-AAG GAC GTG CTG TCG GTG GAG AGT-3’ R: 5’-GCA GCA AGC GGC AAT TAA CAG AGA CC-3’ | 103 |
| novel_circ_0004586 | F: 5’-CTC CTG CCC TGC TGT GTC CCA TAT CT-3’ R: 5’-CCA CCC ACC ACT GAG AAC AAG CCA AA-3’ | 268 |
| novel_circ_0003342 | F: 5’-TGC TGA GGT GTG GGT GCT GTC A-3’ R: 5’-ACA GTG GTG TGA AGG CTC CAG C-3’ | 157 |
| novel_circ_0004860 | F: 5’-TGG TCT TCT TTG TTC AGG GCC TTG TT-3’ R: 5’-TGG TGG TCA CCC AGT TTG GGT TAT AC-3’ | 185 |
| novel_circ_0002085 | F: 5’-GGT CTT CCT CCT GAT GTG AGC TGA AC-3’ R: 5’-ACA ACA AAG GCA GAG ACT GGC GTT T-3’ | 216 |
| novel_circ_0003760 | F: 5’-ACC TAA AGG AAG CAA GGA CCC GAC AA-3’ R: 5’-CCT TCT TCA GGA CCT TGA GGC AGT AC-3’ | 105 |
| novel_circ_0002086 | F: 5’-GGT TTA TTT CTG TTG GCT TCA GTG CAG T-3’ R: 5’-CCC AGG ATA CAT TGA CCC GGA TTC TTT G-3’ | 116 |
| novel_circ_0000456 | F: 5’-GCT GAC AGG ACC CGA CGA TGG CTT T-3’ R: 5’-CAA CGC ATT CTG CTG ATG CAC CTC CA-3’ | 108 |
| novel_circ_0001606 | F: 5’-TGG TGG TCG TCC GCA TGG TGT T-3’ R: 5’-AAA GAT CTG CTG CCC GGT GAA CCC-3’ | 109 |
| novel_circ_0002023 | F: 5’-CCG TGT GGC TCC GCA AGT GAA TCT T-3’ R: 5’-CAG CAA CTC CAG TGA CCG TGC CAA G-3’ | 116 |
F: Forward primer; R: reversed primer.
Statistical analysis
For sequencing, a model based on negative binomial distribution was used. The resulting P values were adjusted to q values using Benjamini and Hochberg’s approach for controlling the false discovery rate. In DAVID enrichment anal-ysis, Fisher’s exact test was used to determine whether the proportions of those falling into each category differed by group. P value was adopted to measure the gene enrichment in annotation terms. In the bubble chart, the number of genes that fall into each category divided by the total number of this category was the RichFactor. Bubble chart analysis was performed using OmicShare tools (Genedenovo, Guangzhou, China). qRT-PCR analysis was performed using Student’s t-test with a two-tailed P value (GraphPad Software Company, San Diego, CA, USA). All summary statistics of the results are presented as the mean ± standard error of the mean (SEM). P < 0.05 was considered statistically significant.
Results
Identification of differentially expressed circRNAs
At least 10G of clean sequencing data were obtained from each sample for further analysis. The intersection of the Find_circ and CIRI2 software results identified 1195 circRNAs in the control group and 1109 circRNAs in the OGD/R group (Memczak et al., 2013; Gao et al., 2018), with 1016 circRNAs found in both groups (Figure 1A). The detailed information of all 1288 identified circRNAs is shown in Additional Table 2 (2MB, pdf) , including chromosome, length, strand, host genes and expression level. Most circRNAs were transcribed from exons and circRNAs were uniformly distributed across chromosomes. We selected 10 representative chromosomes to show the distribution of circRNAs (Figure 1B).
Figure 1.
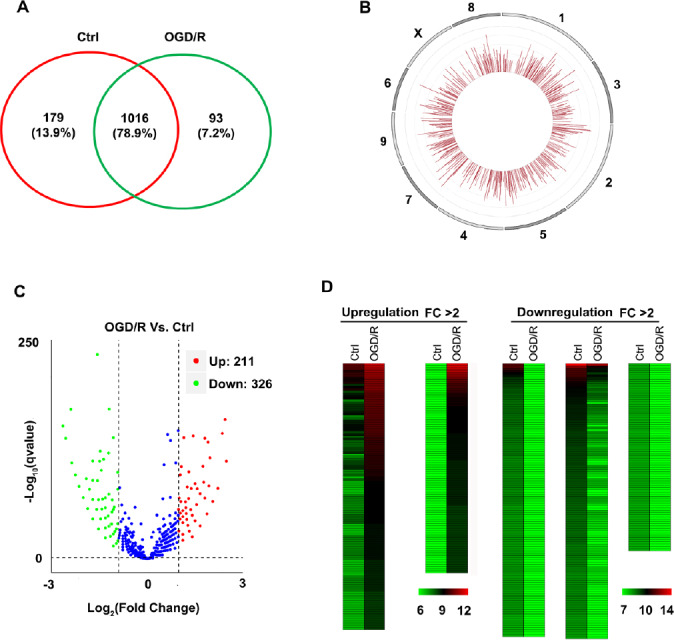
Profile of circular RNAs (circRNAs) in brain microvascular endothelial cells in control (Ctrl) and oxygen glucose deprivation/recovery (OGD/R) groups.
(A) RNA sequencing identified 1195 circRNAs in the Ctrl group and 1109 circRNAs in the OGD/R group. Among these circRNAs, 1016 circRNAs existed in both groups. (B) The circRNAs were uniformly distributed across chromosomes. Ten representative chromosomes are shown. (C) Volcano plot showing the global change in circRNAs. A q value (ordinate) < 0.01 and fold change (abscissa) > 2 defined the differentially expressed circRNAs. The blue dots represent the unchanged circRNAs. (D) Heatmaps show 211 upregulated and 326 downregulated circRNAs in primary brain microvascular endothelial cells after OGD/R treatment. FC: Fold change.
Next, we performed a global analysis of the DE circRNAs in the OGD/R and control BMECs. Standardized TPM values were applied to compare gene expression between two groups (Zhou et al., 2010). The fold change (FC) in the expression of each circRNA was calculated as the log2 ratio using normalized TPM values (Audic and Claverie, 1997). Subsequently, the resulting q values for all genes were corrected for multiple tests using a DEGseq adjustment (Wang et al., 2010). A Volcano Plot was used to show the filter and distribution of DE circRNAs (Figure 1C). Finally, the circRNAs with FC > 2 and q values < 0.01 were identified as DE circRNAs. As shown in Figure 1D, there were 211 upregulated and 326 downregulated circRNAs in OGD/R-treated cells. The detailed information is shown in Additional Table 3. These results demonstrate that OGD/R dramatically altered circRNA expression profiles in vascular endothelial cells. Ischemia-reperfusion-induced endothelial dysfunction has been attributed to angiogenesis, oxidative stress, and inflammation (Guo et al., 2018; Pang et al., 2018; Zhu et al., 2018). Whether the OGD/R-induced circRNA alterations participate in endothelial dysfunction by affecting these responses remains to be determined in future studies.
Additional Table 3.
The list of differentially expressed circRNAs
| ID | OGD/R | Ctrl | log2.Fold_c | p.value | q.value | Expression change |
|---|---|---|---|---|---|---|
| novel_circ_0000007 | 979.7346 | 474.0868 | 1.0472 | 1.63E-44 | 5.22E-45 | up |
| novel_circ_0000012 | 1110.366 | 474.0868 | 1.2278 | 1.55E-63 | 7.84E-64 | up |
| novel_circ_0000062 | 2612.625 | 0 | 12.351 | 0 | 0 | up |
| novel_circ_0000083 | 1045.05 | 355.5651 | 1.5554 | 9.69E-84 | 1.22E-83 | up |
| novel_circ_0000104 | 457.2095 | 118.5217 | 1.9477 | 2.70E-50 | 9.10E-51 | up |
| novel_circ_0000123 | 522.5251 | 177.7826 | 1.5554 | 9.16E-43 | 2.85E-43 | up |
| novel_circ_0000147 | 326.5782 | 0 | 9.3513 | 6.42E-61 | 2.91E-61 | up |
| novel_circ_0000153 | 326.5782 | 0 | 9.3513 | 6.42E-61 | 2.91E-61 | up |
| novel_circ_0000167 | 391.8938 | 0 | 9.6143 | 3.13E-70 | 2.21E-70 | up |
| novel_circ_0000170 | 522.5251 | 177.7826 | 1.5554 | 9.16E-43 | 2.85E-43 | up |
| novel_circ_0000211 | 391.8938 | 0 | 9.6143 | 3.13E-70 | 2.21E-70 | up |
| novel_circ_0000217 | 457.2095 | 177.7826 | 1.3627 | 1.98E-31 | 5.23E-32 | up |
| novel_circ_0000228 | 457.2095 | 0 | 9.8367 | 3.80E-79 | 3.69E-79 | up |
| novel_circ_0000238 | 326.5782 | 0 | 9.3513 | 6.42E-61 | 2.91E-61 | up |
| novel_circ_0000243 | 2286.047 | 0 | 12.159 | 0 | 0 | up |
| novel_circ_0000257 | 326.5782 | 0 | 9.3513 | 6.42E-61 | 2.91E-61 | up |
| novel_circ_0000261 | 979.7346 | 177.7826 | 2.4623 | 0 | 0 | up |
| novel_circ_0000301 | 391.8938 | 118.5217 | 1.7253 | 3.16E-37 | 9.25E-38 | up |
| novel_circ_0000327 | 914.4189 | 414.826 | 1.1403 | 2.97E-47 | 9.81E-48 | up |
| novel_circ_0000328 | 391.8938 | 0 | 9.6143 | 3.13E-70 | 2.21E-70 | up |
| novel_circ_0000335 | 457.2095 | 0 | 9.8367 | 3.80E-79 | 3.69E-79 | up |
| novel_circ_0000353 | 326.5782 | 0 | 9.3513 | 6.42E-61 | 2.91E-61 | up |
| novel_circ_0000360 | 391.8938 | 177.7826 | 1.1403 | 3.37E-21 | 7.14E-22 | up |
| novel_circ_0000368 | 326.5782 | 118.5217 | 1.4623 | 3.32E-25 | 7.63E-26 | up |
| novel_circ_0000371 | 326.5782 | 0 | 9.3513 | 6.42E-61 | 2.91E-61 | up |
| novel_circ_0000377 | 522.5251 | 0 | 10.029 | 9.88E-88 | 1.52E-87 | up |
| novel_circ_0000379 | 457.2095 | 118.5217 | 1.9477 | 2.70E-50 | 9.10E-51 | up |
| novel_circ_0000396 | 587.8407 | 0 | 10.199 | 4.93E-96 | 9.65E-96 | up |
| novel_circ_0000407 | 457.2095 | 0 | 9.8367 | 3.80E-79 | 3.69E-79 | up |
| novel_circ_0000431 | 522.5251 | 0 | 10.029 | 9.88E-88 | 1.52E-87 | up |
| novel_circ_0000446 | 457.2095 | 177.7826 | 1.3627 | 1.98E-31 | 5.23E-32 | up |
| novel_circ_0000447 | 522.5251 | 177.7826 | 1.5554 | 9.16E-43 | 2.85E-43 | up |
| novel_circ_0000456 | 522.5251 | 118.5217 | 2.1403 | 3.83E-64 | 2.40E-64 | up |
| novel_circ_0000486 | 783.7876 | 296.3043 | 1.4034 | 7.08E-55 | 2.46E-55 | up |
| novel_circ_0000533 | 326.5782 | 0 | 9.3513 | 6.42E-61 | 2.91E-61 | up |
| novel_circ_0000619 | 391.8938 | 118.5217 | 1.7253 | 3.16E-37 | 9.25E-38 | up |
| novel_circ_0000665 | 457.2095 | 0 | 9.8367 | 3.80E-79 | 3.69E-79 | up |
| novel_circ_0000667 | 587.8407 | 0 | 10.199 | 4.93E-96 | 9.65E-96 | up |
| novel_circ_0000691 | 1632.891 | 0 | 11.673 | 0 | 0 | up |
| novel_circ_0000714 | 326.5782 | 118.5217 | 1.4623 | 3.32E-25 | 7.63E-26 | up |
| novel_circ_0000766 | 718.472 | 296.3043 | 1.2779 | 3.42E-44 | 1.09E-44 | up |
| novel_circ_0000776 | 979.7346 | 474.0868 | 1.0472 | 1.63E-44 | 5.22E-45 | up |
| novel_circ_0000784 | 783.7876 | 0 | 10.614 | 0 | 0 | up |
| novel_circ_0000791 | 1045.05 | 355.5651 | 1.5554 | 9.69E-84 | 1.22E-83 | up |
| novel_circ_0000814 | 457.2095 | 177.7826 | 1.3627 | 1.98E-31 | 5.23E-32 | up |
| novel_circ_0000863 | 391.8938 | 177.7826 | 1.1403 | 3.37E-21 | 7.14E-22 | up |
| novel_circ_0000883 | 522.5251 | 237.0434 | 1.1403 | 1.00E-27 | 2.48E-28 | up |
| novel_circ_0000908 | 326.5782 | 0 | 9.3513 | 6.42E-61 | 2.91E-61 | up |
| novel_circ_0000988 | 457.2095 | 0 | 9.8367 | 3.80E-79 | 3.69E-79 | up |
| novel_circ_0001002 | 326.5782 | 118.5217 | 1.4623 | 3.32E-25 | 7.63E-26 | up |
| novel_circ_0001012 | 326.5782 | 118.5217 | 1.4623 | 3.32E-25 | 7.63E-26 | up |
| novel_circ_0001025 | 391.8938 | 177.7826 | 1.1403 | 3.37E-21 | 7.14E-22 | up |
| novel_circ_0001031 | 653.1564 | 0 | 10.351 | 0 | 0 | up |
| novel_circ_0001041 | 457.2095 | 177.7826 | 1.3627 | 1.98E-31 | 5.23E-32 | up |
| novel_circ_0001051 | 2090.1 | 1007.434 | 1.0529 | 1.01E-93 | 1.86E-93 | up |
| novel_circ_0001053 | 783.7876 | 0 | 10.614 | 0 | 0 | up |
| novel_circ_0001073 | 391.8938 | 0 | 9.6143 | 3.13E-70 | 2.21E-70 | up |
| novel_circ_0001090 | 718.472 | 355.5651 | 1.0148 | 1.35E-31 | 3.64E-32 | up |
| novel_circ_0001092 | 3004.519 | 0 | 12.553 | 0 | 0 | up |
| novel_circ_0001121 | 391.8938 | 0 | 9.6143 | 3.13E-70 | 2.21E-70 | up |
| novel_circ_0001156 | 326.5782 | 0 | 9.3513 | 6.42E-61 | 2.91E-61 | up |
| novel_circ_0001158 | 326.5782 | 118.5217 | 1.4623 | 3.32E-25 | 7.63E-26 | up |
| novel_circ_0001167 | 457.2095 | 0 | 9.8367 | 3.80E-79 | 3.69E-79 | up |
| novel_circ_0001221 | 391.8938 | 0 | 9.6143 | 3.13E-70 | 2.21E-70 | up |
| novel_circ_0001225 | 391.8938 | 177.7826 | 1.1403 | 3.37E-21 | 7.14E-22 | up |
| novel_circ_0001235 | 391.8938 | 0 | 9.6143 | 3.13E-70 | 2.21E-70 | up |
| novel_circ_0001237 | 1306.313 | 533.3477 | 1.2924 | 3.40E-80 | 4.09E-80 | up |
| novel_circ_0001275 | 457.2095 | 0 | 9.8367 | 3.80E-79 | 3.69E-79 | up |
| novel_circ_0001300 | 522.5251 | 237.0434 | 1.1403 | 1.00E-27 | 2.48E-28 | up |
| novel_circ_0001327 | 587.8407 | 237.0434 | 1.3103 | 8.75E-38 | 2.61E-38 | up |
| novel_circ_0001339 | 391.8938 | 118.5217 | 1.7253 | 3.16E-37 | 9.25E-38 | up |
| novel_circ_0001425 | 457.2095 | 118.5217 | 1.9477 | 2.70E-50 | 9.10E-51 | up |
| novel_circ_0001428 | 457.2095 | 0 | 9.8367 | 3.80E-79 | 3.69E-79 | up |
| novel_circ_0001498 | 326.5782 | 0 | 9.3513 | 6.42E-61 | 2.91E-61 | up |
| novel_circ_0001503 | 979.7346 | 414.826 | 1.2399 | 4.47E-57 | 1.98E-57 | up |
| novel_circ_0001517 | 391.8938 | 118.5217 | 1.7253 | 3.16E-37 | 9.25E-38 | up |
| novel_circ_0001523 | 522.5251 | 177.7826 | 1.5554 | 9.16E-43 | 2.85E-43 | up |
| novel_circ_0001541 | 326.5782 | 0 | 9.3513 | 6.42E-61 | 2.91E-61 | up |
| novel_circ_0001551 | 1371.628 | 651.8694 | 1.0732 | 6.21E-64 | 3.54E-64 | up |
| novel_circ_0001589 | 326.5782 | 118.5217 | 1.4623 | 3.32E-25 | 7.63E-26 | up |
| novel_circ_0001606 | 522.5251 | 118.5217 | 2.1403 | 3.83E-64 | 2.40E-64 | up |
| novel_circ_0001624 | 1045.05 | 474.0868 | 1.1403 | 9.39E-54 | 3.26E-54 | up |
| novel_circ_0001670 | 391.8938 | 0 | 9.6143 | 3.13E-70 | 2.21E-70 | up |
| novel_circ_0001676 | 391.8938 | 0 | 9.6143 | 3.13E-70 | 2.21E-70 | up |
| novel_circ_0001687 | 457.2095 | 177.7826 | 1.3627 | 1.98E-31 | 5.23E-32 | up |
| novel_circ_0001688 | 326.5782 | 0 | 9.3513 | 6.42E-61 | 2.91E-61 | up |
| novel_circ_0001696 | 457.2095 | 118.5217 | 1.9477 | 2.70E-50 | 9.10E-51 | up |
| novel_circ_0001736 | 1110.366 | 0 | 11.117 | 0 | 0 | up |
| novel_circ_0001737 | 326.5782 | 0 | 9.3513 | 6.42E-61 | 2.91E-61 | up |
| novel_circ_0001739 | 522.5251 | 0 | 10.029 | 9.88E-88 | 1.52E-87 | up |
| novel_circ_0001744 | 457.2095 | 0 | 9.8367 | 3.80E-79 | 3.69E-79 | up |
| novel_circ_0001764 | 783.7876 | 355.5651 | 1.1403 | 9.47E-41 | 2.91E-41 | up |
| novel_circ_0001811 | 326.5782 | 118.5217 | 1.4623 | 3.32E-25 | 7.63E-26 | up |
| novel_circ_0001814 | 391.8938 | 0 | 9.6143 | 3.13E-70 | 2.21E-70 | up |
| novel_circ_0001841 | 1567.575 | 770.3911 | 1.0249 | 6.51E-68 | 4.43E-68 | up |
| novel_circ_0001850 | 326.5782 | 0 | 9.3513 | 6.42E-61 | 2.91E-61 | up |
| novel_circ_0001894 | 522.5251 | 0 | 10.029 | 9.88E-88 | 1.52E-87 | up |
| novel_circ_0001898 | 391.8938 | 177.7826 | 1.1403 | 3.37E-21 | 7.14E-22 | up |
| novel_circ_0001910 | 326.5782 | 118.5217 | 1.4623 | 3.32E-25 | 7.63E-26 | up |
| novel_circ_0001943 | 326.5782 | 118.5217 | 1.4623 | 3.32E-25 | 7.63E-26 | up |
| novel_circ_0001962 | 326.5782 | 118.5217 | 1.4623 | 3.32E-25 | 7.63E-26 | up |
| novel_circ_0001996 | 326.5782 | 0 | 9.3513 | 6.42E-61 | 2.91E-61 | up |
| novel_circ_0002023 | 522.5251 | 118.5217 | 2.1403 | 3.83E-64 | 2.40E-64 | up |
| novel_circ_0002025 | 522.5251 | 0 | 10.029 | 9.88E-88 | 1.52E-87 | up |
| novel_circ_0002053 | 326.5782 | 118.5217 | 1.4623 | 3.32E-25 | 7.63E-26 | up |
| novel_circ_0002067 | 653.1564 | 177.7826 | 1.8773 | 7.23E-68 | 4.86E-68 | up |
| novel_circ_0002086 | 587.8407 | 118.5217 | 2.3103 | 1.37E-78 | 1.32E-78 | up |
| novel_circ_0002090 | 522.5251 | 0 | 10.029 | 9.88E-88 | 1.52E-87 | up |
| novel_circ_0002105 | 522.5251 | 177.7826 | 1.5554 | 9.16E-43 | 2.85E-43 | up |
| novel_circ_0002193 | 457.2095 | 177.7826 | 1.3627 | 1.98E-31 | 5.23E-32 | up |
| novel_circ_0002208 | 718.472 | 177.7826 | 2.0148 | 2.44E-81 | 3.04E-81 | up |
| novel_circ_0002217 | 391.8938 | 0 | 9.6143 | 3.13E-70 | 2.21E-70 | up |
| novel_circ_0002226 | 1175.681 | 296.3043 | 1.9883 | 0 | 0 | up |
| novel_circ_0002230 | 522.5251 | 118.5217 | 2.1403 | 3.83E-64 | 2.40E-64 | up |
| novel_circ_0002240 | 2743.257 | 0 | 12.422 | 0 | 0 | up |
| novel_circ_0002241 | 522.5251 | 0 | 10.029 | 9.88E-88 | 1.52E-87 | up |
| novel_circ_0002242 | 1306.313 | 0 | 11.351 | 0 | 0 | up |
| novel_circ_0002255 | 522.5251 | 177.7826 | 1.5554 | 9.16E-43 | 2.85E-43 | up |
| novel_circ_0002289 | 326.5782 | 0 | 9.3513 | 6.42E-61 | 2.91E-61 | up |
| novel_circ_0002327 | 326.5782 | 0 | 9.3513 | 6.42E-61 | 2.91E-61 | up |
| novel_circ_0002336 | 653.1564 | 237.0434 | 1.4623 | 1.00E-48 | 3.37E-49 | up |
| novel_circ_0002346 | 391.8938 | 177.7826 | 1.1403 | 3.37E-21 | 7.14E-22 | up |
| novel_circ_0002397 | 457.2095 | 0 | 9.8367 | 3.80E-79 | 3.69E-79 | up |
| novel_circ_0002409 | 522.5251 | 237.0434 | 1.1403 | 1.00E-27 | 2.48E-28 | up |
| novel_circ_0002433 | 849.1033 | 0 | 10.73 | 0 | 0 | up |
| novel_circ_0002493 | 718.472 | 296.3043 | 1.2779 | 3.42E-44 | 1.09E-44 | up |
| novel_circ_0002513 | 1045.05 | 296.3043 | 1.8184 | 0 | 0 | up |
| novel_circ_0002520 | 1306.313 | 355.5651 | 1.8773 | 0 | 0 | up |
| novel_circ_0002540 | 326.5782 | 0 | 9.3513 | 6.42E-61 | 2.91E-61 | up |
| novel_circ_0002559 | 326.5782 | 0 | 9.3513 | 6.42E-61 | 2.91E-61 | up |
| novel_circ_0002570 | 1567.575 | 770.3911 | 1.0249 | 6.51E-68 | 4.43E-68 | up |
| novel_circ_0002599 | 849.1033 | 0 | 10.73 | 0 | 0 | up |
| novel_circ_0002643 | 391.8938 | 0 | 9.6143 | 3.13E-70 | 2.21E-70 | up |
| novel_circ_0002649 | 457.2095 | 177.7826 | 1.3627 | 1.98E-31 | 5.23E-32 | up |
| novel_circ_0002667 | 391.8938 | 0 | 9.6143 | 3.13E-70 | 2.21E-70 | up |
| novel_circ_0002679 | 326.5782 | 0 | 9.3513 | 6.42E-61 | 2.91E-61 | up |
| novel_circ_0002715 | 849.1033 | 237.0434 | 1.8408 | 1.69E-85 | 2.19E-85 | up |
| novel_circ_0002740 | 326.5782 | 0 | 9.3513 | 6.42E-61 | 2.91E-61 | up |
| novel_circ_0002743 | 522.5251 | 118.5217 | 2.1403 | 3.83E-64 | 2.40E-64 | up |
| novel_circ_0002812 | 2155.416 | 1007.434 | 1.0973 | 0 | 0 | up |
| novel_circ_0002853 | 522.5251 | 0 | 10.029 | 9.88E-88 | 1.52E-87 | up |
| novel_circ_0002859 | 979.7346 | 237.0434 | 2.0472 | 0 | 0 | up |
| novel_circ_0002877 | 326.5782 | 0 | 9.3513 | 6.42E-61 | 2.91E-61 | up |
| novel_circ_0002881 | 587.8407 | 0 | 10.199 | 4.93E-96 | 9.65E-96 | up |
| novel_circ_0002894 | 849.1033 | 355.5651 | 1.2558 | 1.26E-50 | 4.30E-51 | up |
| novel_circ_0002903 | 522.5251 | 177.7826 | 1.5554 | 9.16E-43 | 2.85E-43 | up |
| novel_circ_0002958 | 326.5782 | 118.5217 | 1.4623 | 3.32E-25 | 7.63E-26 | up |
| novel_circ_0002986 | 979.7346 | 414.826 | 1.2399 | 4.47E-57 | 1.98E-57 | up |
| novel_circ_0002991 | 326.5782 | 0 | 9.3513 | 6.42E-61 | 2.91E-61 | up |
| novel_circ_0002999 | 326.5782 | 118.5217 | 1.4623 | 3.32E-25 | 7.63E-26 | up |
| novel_circ_0003074 | 457.2095 | 177.7826 | 1.3627 | 1.98E-31 | 5.23E-32 | up |
| novel_circ_0003096 | 1110.366 | 533.3477 | 1.0579 | 5.53E-51 | 1.89E-51 | up |
| novel_circ_0003103 | 326.5782 | 0 | 9.3513 | 6.42E-61 | 2.91E-61 | up |
| novel_circ_0003115 | 653.1564 | 0 | 10.351 | 0 | 0 | up |
| novel_circ_0003129 | 391.8938 | 0 | 9.6143 | 3.13E-70 | 2.21E-70 | up |
| novel_circ_0003154 | 326.5782 | 0 | 9.3513 | 6.42E-61 | 2.91E-61 | up |
| novel_circ_0003162 | 783.7876 | 237.0434 | 1.7253 | 1.10E-72 | 1.01E-72 | up |
| novel_circ_0003222 | 391.8938 | 118.5217 | 1.7253 | 3.16E-37 | 9.25E-38 | up |
| novel_circ_0003226 | 1828.838 | 651.8694 | 1.4883 | 0 | 0 | up |
| novel_circ_0003243 | 653.1564 | 177.7826 | 1.8773 | 7.23E-68 | 4.86E-68 | up |
| novel_circ_0003259 | 391.8938 | 118.5217 | 1.7253 | 3.16E-37 | 9.25E-38 | up |
| novel_circ_0003270 | 783.7876 | 237.0434 | 1.7253 | 1.10E-72 | 1.01E-72 | up |
| novel_circ_0003274 | 391.8938 | 0 | 9.6143 | 3.13E-70 | 2.21E-70 | up |
| novel_circ_0003280 | 522.5251 | 177.7826 | 1.5554 | 9.16E-43 | 2.85E-43 | up |
| novel_circ_0003377 | 1045.05 | 355.5651 | 1.5554 | 9.69E-84 | 1.22E-83 | up |
| novel_circ_0003426 | 1436.944 | 0 | 11.489 | 0 | 0 | up |
| novel_circ_0003437 | 1110.366 | 414.826 | 1.4205 | 2.57E-78 | 2.46E-78 | up |
| novel_circ_0003459 | 326.5782 | 0 | 9.3513 | 6.42E-61 | 2.91E-61 | up |
| novel_circ_0003471 | 1502.26 | 592.6085 | 1.342 | 4.26E-97 | 9.01E-97 | up |
| novel_circ_0003487 | 522.5251 | 0 | 10.029 | 9.88E-88 | 1.52E-87 | up |
| novel_circ_0003492 | 457.2095 | 0 | 9.8367 | 3.80E-79 | 3.69E-79 | up |
| novel_circ_0003520 | 457.2095 | 177.7826 | 1.3627 | 1.98E-31 | 5.23E-32 | up |
| novel_circ_0003528 | 522.5251 | 177.7826 | 1.5554 | 9.16E-43 | 2.85E-43 | up |
| novel_circ_0003530 | 326.5782 | 0 | 9.3513 | 6.42E-61 | 2.91E-61 | up |
| novel_circ_0003535 | 326.5782 | 0 | 9.3513 | 6.42E-61 | 2.91E-61 | up |
| novel_circ_0003591 | 979.7346 | 0 | 10.936 | 0 | 0 | up |
| novel_circ_0003621 | 326.5782 | 118.5217 | 1.4623 | 3.32E-25 | 7.63E-26 | up |
| novel_circ_0003630 | 457.2095 | 0 | 9.8367 | 3.80E-79 | 3.69E-79 | up |
| novel_circ_0003686 | 522.5251 | 118.5217 | 2.1403 | 3.83E-64 | 2.40E-64 | up |
| novel_circ_0003730 | 391.8938 | 0 | 9.6143 | 3.13E-70 | 2.21E-70 | up |
| novel_circ_0003754 | 326.5782 | 0 | 9.3513 | 6.42E-61 | 2.91E-61 | up |
| novel_circ_0003760 | 718.472 | 118.5217 | 2.5998 | 0 | 0 | up |
| novel_circ_0003787 | 457.2095 | 177.7826 | 1.3627 | 1.98E-31 | 5.23E-32 | up |
| novel_circ_0003829 | 522.5251 | 0 | 10.029 | 9.88E-88 | 1.52E-87 | up |
| novel_circ_0003844 | 391.8938 | 0 | 9.6143 | 3.13E-70 | 2.21E-70 | up |
| novel_circ_0003856 | 783.7876 | 296.3043 | 1.4034 | 7.08E-55 | 2.46E-55 | up |
| novel_circ_0003868 | 522.5251 | 0 | 10.029 | 9.88E-88 | 1.52E-87 | up |
| novel_circ_0004003 | 979.7346 | 474.0868 | 1.0472 | 1.63E-44 | 5.22E-45 | up |
| novel_circ_0004015 | 783.7876 | 237.0434 | 1.7253 | 1.10E-72 | 1.01E-72 | up |
| novel_circ_0004057 | 391.8938 | 0 | 9.6143 | 3.13E-70 | 2.21E-70 | up |
| novel_circ_0004100 | 457.2095 | 118.5217 | 1.9477 | 2.70E-50 | 9.10E-51 | up |
| novel_circ_0004120 | 326.5782 | 0 | 9.3513 | 6.42E-61 | 2.91E-61 | up |
| novel_circ_0004238 | 1502.26 | 592.6085 | 1.342 | 4.26E-97 | 9.01E-97 | up |
| novel_circ_0004333 | 326.5782 | 118.5217 | 1.4623 | 3.32E-25 | 7.63E-26 | up |
| novel_circ_0004399 | 718.472 | 355.5651 | 1.0148 | 1.35E-31 | 3.64E-32 | up |
| novel_circ_0004468 | 2547.31 | 1125.956 | 1.1778 | 0 | 0 | up |
| novel_circ_0004527 | 391.8938 | 0 | 9.6143 | 3.13E-70 | 2.21E-70 | up |
| novel_circ_0004605 | 1045.05 | 414.826 | 1.333 | 1.89E-67 | 1.26E-67 | up |
| novel_circ_0004616 | 522.5251 | 0 | 10.029 | 9.88E-88 | 1.52E-87 | up |
| novel_circ_0004652 | 1045.05 | 177.7826 | 2.5554 | 0 | 0 | up |
| novel_circ_0004711 | 522.5251 | 237.0434 | 1.1403 | 1.00E-27 | 2.48E-28 | up |
| novel_circ_0004761 | 391.8938 | 0 | 9.6143 | 3.13E-70 | 2.21E-70 | up |
| novel_circ_0004762 | 391.8938 | 118.5217 | 1.7253 | 3.16E-37 | 9.25E-38 | up |
| novel_circ_0004783 | 914.4189 | 296.3043 | 1.6258 | 4.70E-78 | 4.47E-78 | up |
| novel_circ_0004796 | 1306.313 | 651.8694 | 1.0028 | 4.73E-55 | 1.66E-55 | up |
| novel_circ_0004890 | 326.5782 | 118.5217 | 1.4623 | 3.32E-25 | 7.63E-26 | up |
| novel_circ_0004913 | 587.8407 | 0 | 10.199 | 4.93E-96 | 9.65E-96 | up |
| novel_circ_0004933 | 718.472 | 237.0434 | 1.5998 | 2.14E-60 | 9.66E-61 | up |
| novel_circ_0004934 | 457.2095 | 177.7826 | 1.3627 | 1.98E-31 | 5.23E-32 | up |
| novel_circ_0004961 | 326.5782 | 118.5217 | 1.4623 | 3.32E-25 | 7.63E-26 | up |
| novel_circ_0005050 | 391.8938 | 177.7826 | 1.1403 | 3.37E-21 | 7.14E-22 | up |
| novel_circ_0000010 | 0 | 414.826 | -9.6964 | 4.73E-72 | 3.76E-72 | down |
| novel_circ_0000032 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0000045 | 130.6313 | 296.3043 | -1.1816 | 5.87E-15 | 1.05E-15 | down |
| novel_circ_0000068 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0000070 | 0 | 533.3477 | -10.059 | 1.54E-87 | 2.07E-87 | down |
| novel_circ_0000077 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0000101 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0000113 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0000141 | 0 | 414.826 | -9.6964 | 4.73E-72 | 3.76E-72 | down |
| novel_circ_0000158 | 0 | 414.826 | -9.6964 | 4.73E-72 | 3.76E-72 | down |
| novel_circ_0000173 | 130.6313 | 948.1736 | -2.8597 | 0 | 0 | down |
| novel_circ_0000178 | 130.6313 | 474.0868 | -1.8597 | 1.11E-44 | 3.61E-45 | down |
| novel_circ_0000182 | 0 | 414.826 | -9.6964 | 4.73E-72 | 3.76E-72 | down |
| novel_circ_0000204 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0000210 | 130.6313 | 414.826 | -1.667 | 8.02E-34 | 2.21E-34 | down |
| novel_circ_0000224 | 261.2625 | 592.6085 | -1.1816 | 2.42E-28 | 6.10E-29 | down |
| novel_circ_0000245 | 326.5782 | 829.6519 | -1.3451 | 7.94E-48 | 2.63E-48 | down |
| novel_circ_0000282 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0000297 | 130.6313 | 296.3043 | -1.1816 | 5.87E-15 | 1.05E-15 | down |
| novel_circ_0000305 | 457.2095 | 1066.695 | -1.2222 | 1.97E-52 | 6.79E-53 | down |
| novel_circ_0000306 | 130.6313 | 592.6085 | -2.1816 | 2.83E-68 | 1.96E-68 | down |
| novel_circ_0000333 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0000342 | 195.9469 | 533.3477 | -1.4446 | 6.55E-35 | 1.87E-35 | down |
| novel_circ_0000404 | 130.6313 | 414.826 | -1.667 | 8.02E-34 | 2.21E-34 | down |
| novel_circ_0000406 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0000408 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0000414 | 522.5251 | 1066.695 | -1.0296 | 1.22E-39 | 3.72E-40 | down |
| novel_circ_0000428 | 261.2625 | 651.8694 | -1.3191 | 7.92E-37 | 2.30E-37 | down |
| novel_circ_0000434 | 0 | 533.3477 | -10.059 | 1.54E-87 | 2.07E-87 | down |
| novel_circ_0000442 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0000470 | 522.5251 | 1185.217 | -1.1816 | 5.50E-55 | 1.92E-55 | down |
| novel_circ_0000471 | 0 | 1837.086 | -11.843 | 0 | 0 | down |
| novel_circ_0000475 | 130.6313 | 533.3477 | -2.0296 | 3.29E-56 | 1.42E-56 | down |
| novel_circ_0000480 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0000482 | 0 | 592.6085 | -10.211 | 6.25E-95 | 1.17E-94 | down |
| novel_circ_0000494 | 457.2095 | 1422.26 | -1.6373 | 0 | 0 | down |
| novel_circ_0000501 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0000507 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0000525 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0000601 | 130.6313 | 296.3043 | -1.1816 | 5.87E-15 | 1.05E-15 | down |
| novel_circ_0000604 | 130.6313 | 355.5651 | -1.4446 | 7.99E-24 | 1.76E-24 | down |
| novel_circ_0000638 | 1045.05 | 2133.391 | -1.0296 | 1.54E-77 | 1.45E-77 | down |
| novel_circ_0000640 | 0 | 414.826 | -9.6964 | 4.73E-72 | 3.76E-72 | down |
| novel_circ_0000642 | 195.9469 | 474.0868 | -1.2747 | 7.44E-26 | 1.78E-26 | down |
| novel_circ_0000643 | 1306.313 | 2666.738 | -1.0296 | 1.76E-96 | 3.66E-96 | down |
| novel_circ_0000673 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0000686 | 587.8407 | 1363 | -1.2133 | 1.01E-65 | 6.50E-66 | down |
| novel_circ_0000707 | 130.6313 | 651.8694 | -2.3191 | 8.94E-81 | 1.08E-80 | down |
| novel_circ_0000719 | 849.1033 | 2785.26 | -1.7138 | 0 | 0 | down |
| novel_circ_0000721 | 130.6313 | 296.3043 | -1.1816 | 5.87E-15 | 1.05E-15 | down |
| novel_circ_0000757 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0000781 | 0 | 533.3477 | -10.059 | 1.54E-87 | 2.07E-87 | down |
| novel_circ_0000812 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0000815 | 130.6313 | 355.5651 | -1.4446 | 7.99E-24 | 1.76E-24 | down |
| novel_circ_0000836 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0000850 | 0 | 474.0868 | -9.889 | 6.37E-80 | 6.85E-80 | down |
| novel_circ_0000871 | 130.6313 | 414.826 | -1.667 | 8.02E-34 | 2.21E-34 | down |
| novel_circ_0000884 | 130.6313 | 296.3043 | -1.1816 | 5.87E-15 | 1.05E-15 | down |
| novel_circ_0000926 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0000936 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0000964 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0000976 | 0 | 474.0868 | -9.889 | 6.37E-80 | 6.85E-80 | down |
| novel_circ_0000991 | 849.1033 | 2133.391 | -1.3291 | 0 | 0 | down |
| novel_circ_0001048 | 261.2625 | 829.6519 | -1.667 | 6.75E-66 | 4.38E-66 | down |
| novel_circ_0001049 | 457.2095 | 1363 | -1.5759 | 1.37E-98 | 2.99E-98 | down |
| novel_circ_0001082 | 0 | 474.0868 | -9.889 | 6.37E-80 | 6.85E-80 | down |
| novel_circ_0001089 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0001096 | 0 | 414.826 | -9.6964 | 4.73E-72 | 3.76E-72 | down |
| novel_circ_0001099 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0001114 | 130.6313 | 296.3043 | -1.1816 | 5.87E-15 | 1.05E-15 | down |
| novel_circ_0001115 | 0 | 592.6085 | -10.211 | 6.25E-95 | 1.17E-94 | down |
| novel_circ_0001138 | 0 | 474.0868 | -9.889 | 6.37E-80 | 6.85E-80 | down |
| novel_circ_0001163 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0001170 | 0 | 533.3477 | -10.059 | 1.54E-87 | 2.07E-87 | down |
| novel_circ_0001183 | 0 | 414.826 | -9.6964 | 4.73E-72 | 3.76E-72 | down |
| novel_circ_0001250 | 261.2625 | 592.6085 | -1.1816 | 2.42E-28 | 6.10E-29 | down |
| novel_circ_0001260 | 326.5782 | 770.3911 | -1.2382 | 3.85E-39 | 1.16E-39 | down |
| novel_circ_0001274 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0001279 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0001280 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0001283 | 195.9469 | 533.3477 | -1.4446 | 6.55E-35 | 1.87E-35 | down |
| novel_circ_0001290 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0001310 | 261.2625 | 948.1736 | -1.8597 | 1.47E-87 | 2.07E-87 | down |
| novel_circ_0001312 | 326.5782 | 948.1736 | -1.5377 | 1.11E-66 | 7.34E-67 | down |
| novel_circ_0001316 | 326.5782 | 829.6519 | -1.3451 | 7.94E-48 | 2.63E-48 | down |
| novel_circ_0001345 | 0 | 1244.478 | -11.281 | 0 | 0 | down |
| novel_circ_0001348 | 261.2625 | 651.8694 | -1.3191 | 7.92E-37 | 2.30E-37 | down |
| novel_circ_0001367 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0001384 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0001390 | 0 | 474.0868 | -9.889 | 6.37E-80 | 6.85E-80 | down |
| novel_circ_0001412 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0001413 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0001429 | 0 | 414.826 | -9.6964 | 4.73E-72 | 3.76E-72 | down |
| novel_circ_0001443 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0001446 | 0 | 414.826 | -9.6964 | 4.73E-72 | 3.76E-72 | down |
| novel_circ_0001490 | 130.6313 | 296.3043 | -1.1816 | 5.87E-15 | 1.05E-15 | down |
| novel_circ_0001506 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0001524 | 261.2625 | 770.3911 | -1.5601 | 1.09E-55 | 4.70E-56 | down |
| novel_circ_0001588 | 130.6313 | 296.3043 | -1.1816 | 5.87E-15 | 1.05E-15 | down |
| novel_circ_0001621 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0001626 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0001630 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0001643 | 130.6313 | 414.826 | -1.667 | 8.02E-34 | 2.21E-34 | down |
| novel_circ_0001653 | 130.6313 | 414.826 | -1.667 | 8.02E-34 | 2.21E-34 | down |
| novel_circ_0001675 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0001690 | 195.9469 | 414.826 | -1.082 | 9.99E-18 | 1.94E-18 | down |
| novel_circ_0001726 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0001732 | 0 | 3200.086 | -12.644 | 0 | 0 | down |
| novel_circ_0001768 | 195.9469 | 533.3477 | -1.4446 | 6.55E-35 | 1.87E-35 | down |
| novel_circ_0001777 | 130.6313 | 355.5651 | -1.4446 | 7.99E-24 | 1.76E-24 | down |
| novel_circ_0001779 | 195.9469 | 474.0868 | -1.2747 | 7.44E-26 | 1.78E-26 | down |
| novel_circ_0001790 | 0 | 1422.26 | -11.474 | 0 | 0 | down |
| novel_circ_0001804 | 195.9469 | 592.6085 | -1.5966 | 1.01E-44 | 3.30E-45 | down |
| novel_circ_0001866 | 130.6313 | 474.0868 | -1.8597 | 1.11E-44 | 3.61E-45 | down |
| novel_circ_0001871 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0001902 | 195.9469 | 711.1302 | -1.8597 | 3.96E-66 | 2.60E-66 | down |
| novel_circ_0001904 | 326.5782 | 829.6519 | -1.3451 | 7.94E-48 | 2.63E-48 | down |
| novel_circ_0001920 | 0 | 770.3911 | -10.589 | 0 | 0 | down |
| novel_circ_0001926 | 130.6313 | 414.826 | -1.667 | 8.02E-34 | 2.21E-34 | down |
| novel_circ_0001938 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0001953 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0001963 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0001969 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0001972 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0001986 | 261.2625 | 592.6085 | -1.1816 | 2.42E-28 | 6.10E-29 | down |
| novel_circ_0002027 | 0 | 474.0868 | -9.889 | 6.37E-80 | 6.85E-80 | down |
| novel_circ_0002037 | 195.9469 | 414.826 | -1.082 | 9.99E-18 | 1.94E-18 | down |
| novel_circ_0002039 | 522.5251 | 1303.739 | -1.3191 | 6.65E-72 | 5.25E-72 | down |
| novel_circ_0002062 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0002085 | 130.6313 | 711.1302 | -2.4446 | 1.24E-93 | 2.25E-93 | down |
| novel_circ_0002118 | 261.2625 | 533.3477 | -1.0296 | 1.23E-20 | 2.56E-21 | down |
| novel_circ_0002122 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0002127 | 130.6313 | 296.3043 | -1.1816 | 5.87E-15 | 1.05E-15 | down |
| novel_circ_0002141 | 130.6313 | 296.3043 | -1.1816 | 5.87E-15 | 1.05E-15 | down |
| novel_circ_0002145 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0002155 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0002177 | 0 | 474.0868 | -9.889 | 6.37E-80 | 6.85E-80 | down |
| novel_circ_0002182 | 195.9469 | 414.826 | -1.082 | 9.99E-18 | 1.94E-18 | down |
| novel_circ_0002185 | 326.5782 | 711.1302 | -1.1227 | 4.86E-31 | 1.27E-31 | down |
| novel_circ_0002191 | 130.6313 | 414.826 | -1.667 | 8.02E-34 | 2.21E-34 | down |
| novel_circ_0002239 | 0 | 414.826 | -9.6964 | 4.73E-72 | 3.76E-72 | down |
| novel_circ_0002246 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0002254 | 0 | 414.826 | -9.6964 | 4.73E-72 | 3.76E-72 | down |
| novel_circ_0002278 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0002374 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0002385 | 0 | 414.826 | -9.6964 | 4.73E-72 | 3.76E-72 | down |
| novel_circ_0002418 | 261.2625 | 651.8694 | -1.3191 | 7.92E-37 | 2.30E-37 | down |
| novel_circ_0002437 | 195.9469 | 770.3911 | -1.9751 | 1.52E-77 | 1.43E-77 | down |
| novel_circ_0002448 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0002452 | 326.5782 | 711.1302 | -1.1227 | 4.86E-31 | 1.27E-31 | down |
| novel_circ_0002457 | 0 | 651.8694 | -10.348 | 0 | 0 | down |
| novel_circ_0002463 | 6727.511 | 15170.78 | -1.1732 | 0 | 0 | down |
| novel_circ_0002469 | 391.8938 | 1066.695 | -1.4446 | 4.51E-68 | 3.09E-68 | down |
| novel_circ_0002477 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0002486 | 0 | 414.826 | -9.6964 | 4.73E-72 | 3.76E-72 | down |
| novel_circ_0002562 | 0 | 651.8694 | -10.348 | 0 | 0 | down |
| novel_circ_0002600 | 1763.522 | 4859.39 | -1.4623 | 0 | 0 | down |
| novel_circ_0002618 | 0 | 2311.173 | -12.174 | 0 | 0 | down |
| novel_circ_0002663 | 0 | 414.826 | -9.6964 | 4.73E-72 | 3.76E-72 | down |
| novel_circ_0002670 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0002672 | 130.6313 | 296.3043 | -1.1816 | 5.87E-15 | 1.05E-15 | down |
| novel_circ_0002692 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0002741 | 326.5782 | 770.3911 | -1.2382 | 3.85E-39 | 1.16E-39 | down |
| novel_circ_0002744 | 326.5782 | 770.3911 | -1.2382 | 3.85E-39 | 1.16E-39 | down |
| novel_circ_0002746 | 0 | 414.826 | -9.6964 | 4.73E-72 | 3.76E-72 | down |
| novel_circ_0002748 | 195.9469 | 533.3477 | -1.4446 | 6.55E-35 | 1.87E-35 | down |
| novel_circ_0002772 | 195.9469 | 711.1302 | -1.8597 | 3.96E-66 | 2.60E-66 | down |
| novel_circ_0002778 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0002781 | 195.9469 | 474.0868 | -1.2747 | 7.44E-26 | 1.78E-26 | down |
| novel_circ_0002791 | 0 | 533.3477 | -10.059 | 1.54E-87 | 2.07E-87 | down |
| novel_circ_0002807 | 0 | 414.826 | -9.6964 | 4.73E-72 | 3.76E-72 | down |
| novel_circ_0002818 | 0 | 414.826 | -9.6964 | 4.73E-72 | 3.76E-72 | down |
| novel_circ_0002819 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0002821 | 195.9469 | 1185.217 | -2.5966 | 0 | 0 | down |
| novel_circ_0002824 | 130.6313 | 414.826 | -1.667 | 8.02E-34 | 2.21E-34 | down |
| novel_circ_0002837 | 130.6313 | 296.3043 | -1.1816 | 5.87E-15 | 1.05E-15 | down |
| novel_circ_0002847 | 457.2095 | 1066.695 | -1.2222 | 1.97E-52 | 6.79E-53 | down |
| novel_circ_0002849 | 195.9469 | 533.3477 | -1.4446 | 6.55E-35 | 1.87E-35 | down |
| novel_circ_0002856 | 0 | 533.3477 | -10.059 | 1.54E-87 | 2.07E-87 | down |
| novel_circ_0002875 | 130.6313 | 592.6085 | -2.1816 | 2.83E-68 | 1.96E-68 | down |
| novel_circ_0002901 | 130.6313 | 533.3477 | -2.0296 | 3.29E-56 | 1.42E-56 | down |
| novel_circ_0002915 | 0 | 474.0868 | -9.889 | 6.37E-80 | 6.85E-80 | down |
| novel_circ_0002928 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0002933 | 0 | 414.826 | -9.6964 | 4.73E-72 | 3.76E-72 | down |
| novel_circ_0002939 | 0 | 414.826 | -9.6964 | 4.73E-72 | 3.76E-72 | down |
| novel_circ_0002952 | 326.5782 | 770.3911 | -1.2382 | 3.85E-39 | 1.16E-39 | down |
| novel_circ_0003011 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0003044 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0003068 | 587.8407 | 1659.304 | -1.4971 | 0 | 0 | down |
| novel_circ_0003086 | 0 | 592.6085 | -10.211 | 6.25E-95 | 1.17E-94 | down |
| novel_circ_0003088 | 195.9469 | 474.0868 | -1.2747 | 7.44E-26 | 1.78E-26 | down |
| novel_circ_0003118 | 130.6313 | 296.3043 | -1.1816 | 5.87E-15 | 1.05E-15 | down |
| novel_circ_0003119 | 0 | 414.826 | -9.6964 | 4.73E-72 | 3.76E-72 | down |
| novel_circ_0003126 | 195.9469 | 414.826 | -1.082 | 9.99E-18 | 1.94E-18 | down |
| novel_circ_0003130 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0003138 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0003153 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0003160 | 130.6313 | 296.3043 | -1.1816 | 5.87E-15 | 1.05E-15 | down |
| novel_circ_0003173 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0003201 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0003206 | 130.6313 | 296.3043 | -1.1816 | 5.87E-15 | 1.05E-15 | down |
| novel_circ_0003211 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0003214 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0003229 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0003254 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0003256 | 130.6313 | 296.3043 | -1.1816 | 5.87E-15 | 1.05E-15 | down |
| novel_circ_0003312 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0003316 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0003319 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0003327 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0003342 | 130.6313 | 770.3911 | -2.5601 | 0 | 0 | down |
| novel_circ_0003355 | 195.9469 | 651.8694 | -1.7341 | 3.67E-55 | 1.29E-55 | down |
| novel_circ_0003364 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0003402 | 326.5782 | 1185.217 | -1.8597 | 0 | 0 | down |
| novel_circ_0003432 | 0 | 888.9128 | -10.796 | 0 | 0 | down |
| novel_circ_0003438 | 130.6313 | 533.3477 | -2.0296 | 3.29E-56 | 1.42E-56 | down |
| novel_circ_0003441 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0003453 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0003462 | 0 | 533.3477 | -10.059 | 1.54E-87 | 2.07E-87 | down |
| novel_circ_0003468 | 130.6313 | 355.5651 | -1.4446 | 7.99E-24 | 1.76E-24 | down |
| novel_circ_0003499 | 195.9469 | 414.826 | -1.082 | 9.99E-18 | 1.94E-18 | down |
| novel_circ_0003501 | 130.6313 | 651.8694 | -2.3191 | 8.94E-81 | 1.08E-80 | down |
| novel_circ_0003509 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0003532 | 261.2625 | 829.6519 | -1.667 | 6.75E-66 | 4.38E-66 | down |
| novel_circ_0003553 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0003563 | 1306.313 | 2963.043 | -1.1816 | 0 | 0 | down |
| novel_circ_0003564 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0003586 | 261.2625 | 533.3477 | -1.0296 | 1.23E-20 | 2.56E-21 | down |
| novel_circ_0003587 | 261.2625 | 533.3477 | -1.0296 | 1.23E-20 | 2.56E-21 | down |
| novel_circ_0003608 | 1240.997 | 3081.564 | -1.3122 | 0 | 0 | down |
| novel_circ_0003620 | 130.6313 | 533.3477 | -2.0296 | 3.29E-56 | 1.42E-56 | down |
| novel_circ_0003632 | 0 | 3081.564 | -12.589 | 0 | 0 | down |
| novel_circ_0003645 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0003646 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0003649 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0003659 | 522.5251 | 1600.043 | -1.6145 | 0 | 0 | down |
| novel_circ_0003661 | 195.9469 | 829.6519 | -2.082 | 2.40E-89 | 4.32E-89 | down |
| novel_circ_0003680 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0003695 | 130.6313 | 474.0868 | -1.8597 | 1.11E-44 | 3.61E-45 | down |
| novel_circ_0003702 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0003746 | 0 | 474.0868 | -9.889 | 6.37E-80 | 6.85E-80 | down |
| novel_circ_0003748 | 0 | 414.826 | -9.6964 | 4.73E-72 | 3.76E-72 | down |
| novel_circ_0003750 | 130.6313 | 355.5651 | -1.4446 | 7.99E-24 | 1.76E-24 | down |
| novel_circ_0003764 | 326.5782 | 1066.695 | -1.7076 | 3.60E-87 | 4.72E-87 | down |
| novel_circ_0003800 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0003806 | 261.2625 | 533.3477 | -1.0296 | 1.23E-20 | 2.56E-21 | down |
| novel_circ_0003816 | 0 | 770.3911 | -10.589 | 0 | 0 | down |
| novel_circ_0003825 | 130.6313 | 355.5651 | -1.4446 | 7.99E-24 | 1.76E-24 | down |
| novel_circ_0003832 | 130.6313 | 296.3043 | -1.1816 | 5.87E-15 | 1.05E-15 | down |
| novel_circ_0003849 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0003862 | 326.5782 | 888.9128 | -1.4446 | 5.01E-57 | 2.21E-57 | down |
| novel_circ_0003869 | 130.6313 | 414.826 | -1.667 | 8.02E-34 | 2.21E-34 | down |
| novel_circ_0003907 | 130.6313 | 296.3043 | -1.1816 | 5.87E-15 | 1.05E-15 | down |
| novel_circ_0003940 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0003947 | 130.6313 | 296.3043 | -1.1816 | 5.87E-15 | 1.05E-15 | down |
| novel_circ_0003952 | 0 | 533.3477 | -10.059 | 1.54E-87 | 2.07E-87 | down |
| novel_circ_0003970 | 130.6313 | 355.5651 | -1.4446 | 7.99E-24 | 1.76E-24 | down |
| novel_circ_0003973 | 130.6313 | 414.826 | -1.667 | 8.02E-34 | 2.21E-34 | down |
| novel_circ_0003977 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0004011 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0004013 | 130.6313 | 355.5651 | -1.4446 | 7.99E-24 | 1.76E-24 | down |
| novel_circ_0004021 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0004024 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0004061 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0004062 | 0 | 533.3477 | -10.059 | 1.54E-87 | 2.07E-87 | down |
| novel_circ_0004067 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0004105 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0004118 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0004121 | 130.6313 | 296.3043 | -1.1816 | 5.87E-15 | 1.05E-15 | down |
| novel_circ_0004129 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0004164 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0004180 | 0 | 829.6519 | -10.696 | 0 | 0 | down |
| novel_circ_0004188 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0004193 | 326.5782 | 1066.695 | -1.7076 | 3.60E-87 | 4.72E-87 | down |
| novel_circ_0004198 | 587.8407 | 1303.739 | -1.1492 | 1.22E-57 | 5.47E-58 | down |
| novel_circ_0004220 | 195.9469 | 414.826 | -1.082 | 9.99E-18 | 1.94E-18 | down |
| novel_circ_0004229 | 0 | 533.3477 | -10.059 | 1.54E-87 | 2.07E-87 | down |
| novel_circ_0004249 | 130.6313 | 296.3043 | -1.1816 | 5.87E-15 | 1.05E-15 | down |
| novel_circ_0004311 | 261.2625 | 651.8694 | -1.3191 | 7.92E-37 | 2.30E-37 | down |
| novel_circ_0004330 | 130.6313 | 414.826 | -1.667 | 8.02E-34 | 2.21E-34 | down |
| novel_circ_0004335 | 130.6313 | 296.3043 | -1.1816 | 5.87E-15 | 1.05E-15 | down |
| novel_circ_0004340 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0004354 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0004384 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0004444 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0004454 | 130.6313 | 355.5651 | -1.4446 | 7.99E-24 | 1.76E-24 | down |
| novel_circ_0004457 | 0 | 533.3477 | -10.059 | 1.54E-87 | 2.07E-87 | down |
| novel_circ_0004500 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0004522 | 0 | 474.0868 | -9.889 | 6.37E-80 | 6.85E-80 | down |
| novel_circ_0004542 | 130.6313 | 414.826 | -1.667 | 8.02E-34 | 2.21E-34 | down |
| novel_circ_0004547 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0004586 | 130.6313 | 888.9128 | -2.7665 | 0 | 0 | down |
| novel_circ_0004590 | 195.9469 | 414.826 | -1.082 | 9.99E-18 | 1.94E-18 | down |
| novel_circ_0004593 | 0 | 474.0868 | -9.889 | 6.37E-80 | 6.85E-80 | down |
| novel_circ_0004601 | 0 | 474.0868 | -9.889 | 6.37E-80 | 6.85E-80 | down |
| novel_circ_0004608 | 130.6313 | 533.3477 | -2.0296 | 3.29E-56 | 1.42E-56 | down |
| novel_circ_0004628 | 0 | 888.9128 | -10.796 | 0 | 0 | down |
| novel_circ_0004653 | 195.9469 | 474.0868 | -1.2747 | 7.44E-26 | 1.78E-26 | down |
| novel_circ_0004661 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0004706 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0004707 | 0 | 414.826 | -9.6964 | 4.73E-72 | 3.76E-72 | down |
| novel_circ_0004728 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0004736 | 195.9469 | 474.0868 | -1.2747 | 7.44E-26 | 1.78E-26 | down |
| novel_circ_0004743 | 130.6313 | 355.5651 | -1.4446 | 7.99E-24 | 1.76E-24 | down |
| novel_circ_0004758 | 195.9469 | 474.0868 | -1.2747 | 7.44E-26 | 1.78E-26 | down |
| novel_circ_0004820 | 130.6313 | 296.3043 | -1.1816 | 5.87E-15 | 1.05E-15 | down |
| novel_circ_0004824 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0004833 | 195.9469 | 414.826 | -1.082 | 9.99E-18 | 1.94E-18 | down |
| novel_circ_0004846 | 261.2625 | 711.1302 | -1.4446 | 5.66E-46 | 1.86E-46 | down |
| novel_circ_0004855 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0004860 | 130.6313 | 770.3911 | -2.5601 | 0 | 0 | down |
| novel_circ_0004861 | 391.8938 | 1066.695 | -1.4446 | 4.51E-68 | 3.09E-68 | down |
| novel_circ_0004866 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0004935 | 130.6313 | 296.3043 | -1.1816 | 5.87E-15 | 1.05E-15 | down |
| novel_circ_0004941 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0004943 | 130.6313 | 355.5651 | -1.4446 | 7.99E-24 | 1.76E-24 | down |
| novel_circ_0004944 | 130.6313 | 296.3043 | -1.1816 | 5.87E-15 | 1.05E-15 | down |
| novel_circ_0004969 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0004989 | 130.6313 | 533.3477 | -2.0296 | 3.29E-56 | 1.42E-56 | down |
| novel_circ_0004997 | 0 | 355.5651 | -9.474 | 6.97E-64 | 3.54E-64 | down |
| novel_circ_0005014 | 0 | 296.3043 | -9.2109 | 2.33E-55 | 8.23E-56 | down |
| novel_circ_0005016 | 391.8938 | 829.6519 | -1.082 | 7.57E-34 | 2.14E-34 | down |
| novel_circ_0005018 | 130.6313 | 651.8694 | -2.3191 | 8.94E-81 | 1.08E-80 | down |
| novel_circ_0005035 | 0 | 533.3477 | -10.059 | 1.54E-87 | 2.07E-87 | down |
| novel_circ_0005045 | 130.6313 | 296.3043 | -1.1816 | 5.87E-15 | 1.05E-15 | down |
| novel_circ_0005052 | 0 | 474.0868 | -9.889 | 6.37E-80 | 6.85E-80 | down |
OGD/R,oxygen-glucose deprivation/recovery; Ctrl, control
Functional enrichment of the overlapping host genes for DE circRNAs and mRNAs
circRNAs are synthesized via backsplicing and are generated from mRNA precursors. Several circRNAs can also reg-ulate the expression of their host genes (Barrett et al., 2015). Therefore, characterizing the function of these mRNAs may enhance our understanding of the features of circRNAs. We analyzed the genes that colocalized on chromosomes with these DE circRNAs and found that the exonic and intronic circRNAs originated from 416 host genes (Figure 2B).
Figure 2.
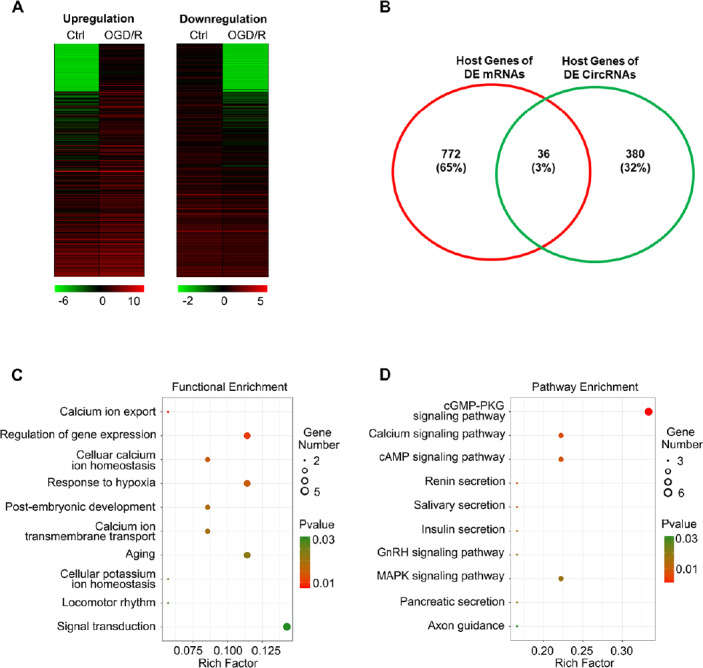
Functional annotation clustering of differentially expressed (DE) circular RNAs (circRNAs) combined with DE mRNAs.
(A) Heatmaps show mRNAs with FC > 2 in the control (Ctrl) and oxygen glucose deprivation/recovery (OGD/R) groups. There were 448 upregulated and 439 downregulated mRNAs. (B) The intersection of DE mRNA host genes and DE circRNA host genes. A total of 36 genes were identified in the intersection. (C, D) The functional (C) and signaling (D) pathway enrichment analyses of these genes, respectively. The P value indicates significance for enrichment. P < 0.05 was considered significant. The RichFactor is calculated as the number of genes that fall into each category divided by the total number in that category. The size of the circle reflects the absolute number of genes that fall into each category. FC: Fold change.
Next, RNA sequencing was used to explore the mRNA profile of BMECs. After quality trimming of raw reads, 13G high-quality data remained. We mapped the clean reads to the Ensembl human genome database. The proportion of total reads in the OGD/R and control transcriptome libraries that mapped to the genome ranged from 92.9% to 93.2%. Cuffdiff calculated the FPKM of each transcript and the FC of DE mRNAs. We set the threshold as FC > 2.0 and P < 0.05 and identified 808 DE mRNAs, including 448 upregulated mRNAs and 439 downregulated mRNAs (Figure 2A and B).
To predict the potential function of the DE circRNAs, software was used to identify the host genes of the DE circRNAs and DE mRNAs. Figure 2B shows the intersection of host genes for DE circRNAs and DE mRNA. A total of 36 genes were identified to transcribe both DE mRNA and circRNA and were then subjected to functional and pathway enrichment analysis. In Figure 2C and D, the color of the bubble distinguishes the P value, and the size represents the number of genes that are enriched. More detailed information is shown in Additional Table 4. We identified calcium ion export, cellular calcium ion homeostasis and calcium ion transmembrane transport to be significantly enriched functional terms and the calcium signaling pathway as a top term in pathway enrichment analysis.
Additional Table 4.
The list of functional and signal pathway enrichment
| Term | Genes |
|---|---|
| Functional enrichment list | |
| GO:1901660~calcium ion export | Slc8a1, Atp2b1 |
| GO:0010468~regulation of gene expression | ATPBS, Thoc2, Rc3h2, Bcl11b |
| GO:0006874~cellular calcium ion homeostasis | ATPBS, Slc8a1, Atp2b1 |
| GO:0001666~response to hypoxia | ATPBS, Slc8a1, Kcnma1, Flt1 |
| GO:0009791~post-embryonic development | Slc8a1, Rc3h2, Bcl11b |
| GO:0070588~calcium ion transmembrane transport | Slc8a1, Atp2b1, Cacna1d |
| GO:0007568~aging | Kcnma1, Atp2b1, Flt1, Cacna1d |
| GO:0030007~cellular potassium ion homeostasis | ATPBS, Kcnma1 |
| GO:0045475~locomotor rhythm | Mapk10, Kcnma1 |
| GO:0007165~signal transduction | Ptprm, Srgap2, Magi3, Bcl11b, Cap2 |
| GO:0006883~cellular sodium ion homeostasis | ATPBS, Slc8a1 |
| GO:0070509~calcium ion import | Slc8a1, Cacna1d |
| GO:0002028~regulation of sodium ion transport | Slc8a1, Wnk1 |
| GO:0001782~B cell homeostasis | Sos2, Rc3h2 |
| GO:0001824~blastocyst development | ATPBS, Thoc2 |
| GO:0050769~positive regulation of neurogenesis | Spen, Man2a1 |
| GO:0001569~patterning of blood vessels | Flt1, Sema5a |
| GO:0051924~regulation of calcium ion transport | Slc8a1, Cacna1d |
| GO:0006468~protein phosphorylation | Mapk10, Flt1, Nek4, Wnk1 |
| GO:0045909~positive regulation of vasodilation | Ptprm, Kcnma1 |
| GO:0007628~adult walking behavior | Kcnma1, Oxr1 |
| GO:0060048~cardiac muscle contraction | ATPBS, Slc8a1 |
| GO:0048286~lung alveolus development | Man2a1, Rc3h2 |
| GO:0010923~negative regulation of phosphatase activity | Tmem132d, Wnk1 |
| Pathway enrichment list | |
| rno04022:cGMP-PKG signaling pathway | ATPBS, Ppp3cc, Slc8a1, Kcnma1, Atp2b1, Cacna1d |
| rno04020:Calcium signaling pathway | Ppp3cc, Slc8a1, Atp2b1, Cacna1d |
| rno04024:cAMP signaling pathway | ATPBS, Mapk10, Atp2b1, Cacna1d |
| rno04924:Renin secretion | Ppp3cc, Kcnma1, Cacna1d |
| rno04970:Salivary secretion | ATPBS, Kcnma1, Atp2b1 |
| rno04911:Insulin secretion | ATPBS, Kcnma1, Cacna1d |
| rno04912:GnRH signaling pathway | Mapk10, Sos2, Cacna1d |
| rno04010:MAPK signaling pathway | Mapk10, Sos2, Ppp3cc, Cacna1d |
| rno04972:Pancreatic secretion | ATPBS, Kcnma1, Atp2b1 |
| rno04360:Axon guidance | Srgap2, Ppp3cc, Sema5a |
| rno04728:Dopaminergic synapse | Mapk10, Ppp3cc, Cacna1d |
| rno04261:Adrenergic signaling in cardiomyocytes | ATPBS, Atp2b1, Cacna1d |
| rno04510:Focal adhesion | Mapk10, Sos2, Flt1 |
| rno04973:Carbohydrate digestion and absorption | ATPBS, Cacna1d |
| rno04014:Ras signaling pathway | Mapk10, Sos2, Flt1 |
Validation of the DE circRNAs
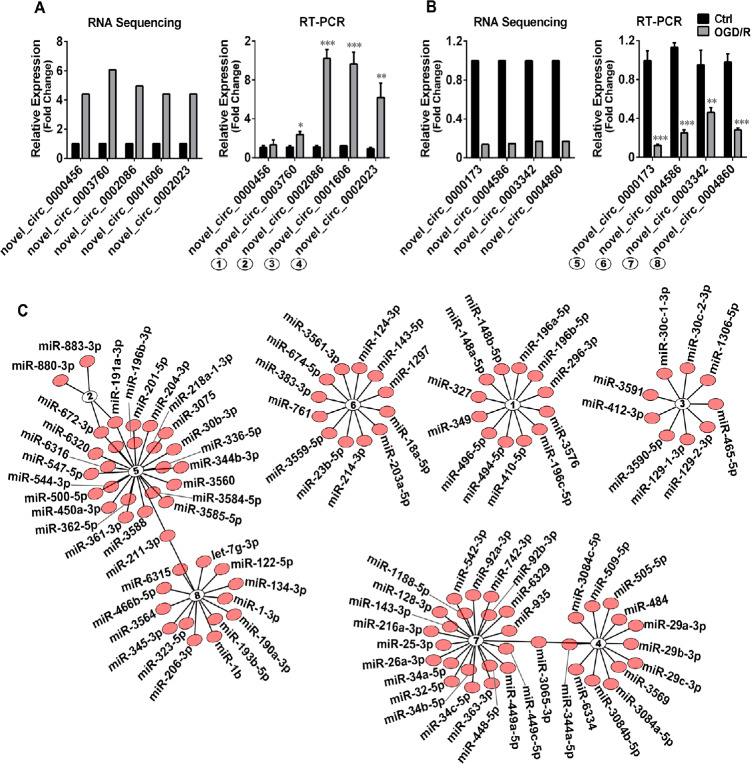
Next, we continued to investigate the circRNAs transcribed from the overlapping host genes (Figure 2B). We adopted qRT-PCR to verify changes in the expression of circRNAs. According to the FC value, we selected the five most up-regulated or downregulated circRNAs for expression validation. As shown in Figure 3A and B, the quantification of circRNA expression was well correlated with the qRT-PCR results. Nine out of ten circRNA candidates identified in the samples were successfully amplified by qRT-PCR using the RevertAid First Strand cDNA Synthesis Kit and SYBR Green PCR Kits (Thermo Fisher). Additionally, the PCR results of eight circRNAs were consistent with sequencing, demonstrating the high reliability of the high-throughput RNA sequencing of circRNA.
Figure 3.
The microRNA (miRNA)-circular RNA (circRNA) interaction network map.
(A, B) The five most upregulated or downregulated circRNAs, the host genes of which were identified in both the differentially expressed (DE) mRNA and DE circRNA analyses, were selected for verification. One circRNA was not successfully amplified by quantitative reverse transcription-polymerase chain reaction (RT-PCR). The quantification of eight circRNAs was consistent with the RNA sequencing results. (C) The miRNA-circRNA interaction network map of these eight circRNAs created by bioinformatic analysis. ①novel_circ_0003760; ②novel_circ_0002086; ③novel_circ_0001606; ④;novel_circ_0002023; ⑤novel_circ_0000173; ⑥novel_circ_0004586; ⑦nov-el_circ_0003342; ⑧novel_circ_0004860. Red dots indicate miRNAs.
circRNA-miRNA network analysis
Recent studies have shown that some circRNAs act as miRNA sponges (Hansen et al., 2013). To determine the possible miRNA targets of OGD/R-induced DE circRNAs, Miranda software was employed to analyze the binding sites of DE circRNAs and miRNAs. Figure 3C shows the circRNA-miRNA interaction networks of the eight circRNAs verified by qRT-PCR. A detailed list of the predicted circRNA-miRNA interactions is provided in Additional Table 5. In cerebral ischemia, some miRNAs are sponged by circRNAs. For instance, circRNA DLGAP4 functions as an endogenous miR-143 sponge to inhibit miR-143 activity, resulting in the inhibition of endothelial-mesenchymal transition by regulating tight junction protein and mesenchymal cell marker expression (Bai et al., 2018). Our results also show that miR-143 may interact with novel_circ_0003342 (Figure 3C).
Additional Table 5.
The list of the predicted circRNA-miRNA interactions
| miRNA | CircRNAs |
|---|---|
| rno-let-7g-3p | novel_circ_0004860 |
| rno-miR-1188-5p | novel_circ_0003342 |
| rno-miR-122-5p | novel_circ_0004860 |
| rno-miR-124-3p | novel_circ_0004586 |
| rno-miR-128-3p | novel_circ_0003342 |
| rno-miR-129-1-3p | novel_circ_0001606 |
| rno-miR-129-2-3p | novel_circ_0001606 |
| rno-miR-1297 | novel_circ_0004586 |
| rno-miR-1306-5p | novel_circ_0001606 |
| rno-miR-134-3p | novel_circ_0004860 |
| rno-miR-1-3p | novel_circ_0004860 |
| rno-miR-143-3p | novel_circ_0003342 |
| rno-miR-143-5p | novel_circ_0004586 |
| rno-miR-148a-5p | novel_circ_0003760 |
| rno-miR-148b-5p | novel_circ_0003760 |
| rno-miR-17-2-3p | novel_circ_0000173 |
| rno-miR-17-2-3p | novel_circ_0002086 |
| rno-miR-18a-5p | novel_circ_0004586 |
| rno-miR-190a-3p | novel_circ_0004860 |
| rno-miR-191a-3p | novel_circ_0000173 |
| rno-miR-191a-3p | novel_circ_0002086 |
| rno-miR-193b-5p | novel_circ_0004860 |
| rno-miR-196a-5p | novel_circ_0003760 |
| rno-miR-196b-3p | novel_circ_0000173 |
| rno-miR-196b-5p | novel_circ_0003760 |
| rno-miR-196c-5p | novel_circ_0003760 |
| rno-miR-1b | novel_circ_0004860 |
| rno-miR-201-5p | novel_circ_0000173 |
| rno-miR-203a-5p | novel_circ_0004586 |
| rno-miR-204-3p | novel_circ_0000173 |
| rno-miR-206-3p | novel_circ_0004860 |
| rno-miR-211-3p | novel_circ_0000173 |
| rno-miR-211-3p | novel_circ_0004860 |
| rno-miR-214-3p | novel_circ_0004586 |
| rno-miR-216a-3p | novel_circ_0003342 |
| rno-miR-218a-1-3p | novel_circ_0000173 |
| rno-miR-23b-5p | novel_circ_0004586 |
| rno-miR-25-3p | novel_circ_0003342 |
| rno-miR-26a-3p | novel_circ_0003342 |
| rno-miR-296-3p | novel_circ_0003760 |
| rno-miR-29a-3p | novel_circ_0002023 |
| rno-miR-29b-3p | novel_circ_0002023 |
| rno-miR-29c-3p | novel_circ_0002023 |
| rno-miR-3065-3p | novel_circ_0002023 |
| rno-miR-3065-3p | novel_circ_0003342 |
| rno-miR-3075 | novel_circ_0000173 |
| rno-miR-3084a-5p | novel_circ_0002023 |
| rno-miR-3084b-5p | novel_circ_0002023 |
| rno-miR-3084c-5p | novel_circ_0002023 |
| rno-miR-30b-3p | novel_circ_0000173 |
| rno-miR-30c-1-3p | novel_circ_0001606 |
| rno-miR-30c-2-3p | novel_circ_0001606 |
| rno-miR-323-5p | novel_circ_0004860 |
| rno-miR-32-5p | novel_circ_0003342 |
| rno-miR-327 | novel_circ_0003760 |
| rno-miR-336-5p | novel_circ_0000173 |
| rno-miR-344a-5p | novel_circ_0002023 |
| rno-miR-344b-3p | novel_circ_0000173 |
| rno-miR-345-3p | novel_circ_0004860 |
| rno-miR-349 | novel_circ_0003760 |
| rno-miR-34a-5p | novel_circ_0003342 |
| rno-miR-34b-5p | novel_circ_0003342 |
| rno-miR-34c-5p | novel_circ_0003342 |
| rno-miR-3559-5p | novel_circ_0004586 |
| rno-miR-3560 | novel_circ_0000173 |
| rno-miR-3561-3p | novel_circ_0004586 |
| rno-miR-3564 | novel_circ_0004860 |
| rno-miR-3569 | novel_circ_0002023 |
| rno-miR-3576 | novel_circ_0003760 |
| rno-miR-3584-5p | novel_circ_0000173 |
| rno-miR-3585-5p | novel_circ_0000173 |
| rno-miR-3588 | novel_circ_0000173 |
| rno-miR-3590-5p | novel_circ_0001606 |
| rno-miR-3591 | novel_circ_0001606 |
| rno-miR-361-3p | novel_circ_0000173 |
| rno-miR-362-5p | novel_circ_0000173 |
| rno-miR-363-3p | novel_circ_0003342 |
| rno-miR-383-3p | novel_circ_0004586 |
| rno-miR-410-5p | novel_circ_0003760 |
| rno-miR-412-3p | novel_circ_0001606 |
| rno-miR-448-5p | novel_circ_0003342 |
| rno-miR-449a-5p | novel_circ_0003342 |
| rno-miR-449c-5p | novel_circ_0003342 |
| rno-miR-450a-3p | novel_circ_0000173 |
| rno-miR-465-5p | novel_circ_0001606 |
| rno-miR-466b-5p | novel_circ_0004860 |
| rno-miR-484 | novel_circ_0002023 |
| rno-miR-494-5p | novel_circ_0003760 |
| rno-miR-496-5p | novel_circ_0003760 |
| rno-miR-500-5p | novel_circ_0000173 |
| rno-miR-505-5p | novel_circ_0002023 |
| rno-miR-509-5p | novel_circ_0002023 |
| rno-miR-542-3p | novel_circ_0003342 |
| rno-miR-544-3p | novel_circ_0000173 |
| rno-miR-547-5p | novel_circ_0000173 |
| rno-miR-6315 | novel_circ_0004860 |
| rno-miR-6316 | novel_circ_0000173 |
| rno-miR-6320 | novel_circ_0000173 |
| rno-miR-6329 | novel_circ_0003342 |
| rno-miR-6334 | novel_circ_0002023 |
| rno-miR-672-3p | novel_circ_0000173 |
| rno-miR-674-5p | novel_circ_0004586 |
| rno-miR-742-3p | novel_circ_0003342 |
| rno-miR-761 | novel_circ_0004586 |
| rno-miR-880-3p | novel_circ_0002086 |
| rno-miR-883-3p | novel_circ_0002086 |
| rno-miR-92a-3p | novel_circ_0003342 |
| rno-miR-92b-3p | novel_circ_0003342 |
| rno-miR-935 | novel_circ_0003342 |
Discussion
Lin et al. (2016) explored the circRNA profile of mouse hippocampal HT22 cells in an OGD/R model and identified several DE circRNAs that may be involved in apoptosis, metabolism and immunoreaction. A further study also identified 1027 DE circRNAs in a middle cerebral artery occlusion model (Liu et al., 2017a). However, none of the circRNAs that we identified in BMECs overlapped with those identified in neural cells or tissues in previous papers. In view of the specificity of circRNA expression, endothelial cells and neural cells likely have different expression profiles in hypoxic-ischemic conditions. These newly identified circRNAs might also contribute to the features and functions of BMECs.
We analyzed the intersection of DE circRNA and DE mRNA host genes. The results indicate the circRNAs are closely related to calcium ions. Calcium signaling plays an important role in the regulation of vascular endothelial cell function. For example, increased calcium binds to calmodulin and interacts with related proteins to release vasodilators, such as nitric oxide and prostacyclin (Yamamoto et al., 2000). Differences in the amplitude and duration of intracellular calcium oscillations contribute to the differential activation of various transcription factors, leading to regulated gene expression (Chen et al., 2019). This is consistent with the high score for “regulation of gene expression” in our results. Additionally, “response to hypoxia” had a high enrichment score. Therefore, we suggest that circRNAs transcribed from the 36 genes identified may play important roles in calcium ion regulation during cerebral ischemia-reperfusion injury.
Surprisingly, the most significant term in the pathway results was cyclic guanosine 3′,5′-monophosphate (cGMP)-cGMP-dependent protein kinase (PKG) signaling. The first mechanism proposed for cGMP-dependent relaxation of smooth muscle was the reduction of free intracellular cytosolic calcium concentration (Johnson and Lincoln, 1985). Several sites of action have been proposed to account for cGMP-dependent regulation of cytosolic calcium, and these have been reviewed (Lincoln et al., 2001). Nitric oxide-cGMP signaling was recognized by the 1998 Nobel Prize in Physiology and Medicine (Arnold et al., 1977). Nitric oxide diffuses across vascular smooth muscle cell membranes and activates the enzyme-soluble guanylate cyclase, which catalyzes the conversion of guanosine tri-phosphate into cGMP (Denninger and Marletta, 1999). cGMP activates PKG, which promotes multiple phosphorylation of targets, lowering cellular calcium concentrations and promoting vascular relaxation (Surks et al., 1999). The cGMP-PKG pathway also decreases calcium release, which inhibits caspase-3 activation and apoptosis. Therefore, identification of this term in our pathway enrichment analysis is consistent with the identification of calcium ion terms in the functional enrichment analysis.
circRNA was first identified in 1976 by Sanger et al. (1976). circRNAs are not susceptible to degradation by RNA exonucleases because of their covalently closed circular structure that lacks accessible ends (Altesha et al., 2019). These characteristics give circRNAs significantly longer half-lives than linear RNAs. BMEC-released circRNAs can be easily detected in plasma; for example circHectd1 is significantly increased in the plasma of model stroke mice (Han et al., 2018). circRNAs are, therefore, potential candidates for diagnostic and prognostic biomarkers of disease.
miRNAs negatively regulate gene expression by partial base pairing with the untranslated region of its target mRNA. But interaction of the miRNA seed region with the mRNA is not unidirectional. Transcribed pseudogenes, long noncoding RNAs and circRNAs compete for the same pool of miRNAs, thereby regulating miRNA activity. This phenomenon of regulating other RNA transcripts by competing for shared miRNAs is performed by RNAs termed competing endogenous RNAs. The binding and holding of miRNAs by circRNAs has been termed the “sponging effect”, which results in the increased expression of miRNA target mRNAs (Altesha et al., 2019). In the present study, we obtained several circRNA-miRNA networks by bioinformatics. Some of the miRNAs have been reported in cerebral ischemia reperfusion injury. For example, miR-26a can promote endothelial lumen formation and cell proliferation in BMECs via the phosphatidylinositol 3′-kinase/Akt and mitogen-activated protein kinase/extracellular signal-regulated kinase pathway (Liang et al., 2018). In addition, overexpression of miR-544 ameliorated the inflammation and apoptotic responses in brain tissue after ischemia reperfusion by down-regulating the expression of interleukin-1 receptor-associated kinase 4 (Fang et al., 2018). Moreover, miR-124, one the most abundant miRNAs in the central nervous system, is a potential partner of novel_circ_0004586. miR-124 plays multiple functions in brain ischemia, such as in inflammation, glycolysis and cell damage (Zhu et al., 2014; Hamzei Taj et al., 2016; Caruso et al., 2017). Future research should explore the regulatory mechanisms of circRNAs and functional miRNAs.
The most significant result of the present study is the association of circRNAs with calcium ion-related pathways. Calcium overload is well known to be induced by brain ischemia and oxygen and glucose deprivation and to induce dysfunctional adenosine triphosphate and cell damage. Targeting calcium ion-related proteins, such as sodium-calcium exchanger, voltage-sensitive calcium channels, transient receptor potential channels, and N-methyl-D-aspartate recep-tors has been confirmed to be an effective treatment (Khananshvili, 2013; Kumar et al., 2014). cGMP and PKGs are widely involved in the physiological processes of the vascular system. This pathway stimulates endothelial cell prolif-eration and inhibits vascular smooth muscle cell proliferation. Dysfunction of the cGMP-PKG signaling pathway at any step of the cascade has been implicated in numerous vascular diseases, ranging from cerebral ischemia to atherosclerosis and angiogenesis (Zhang et al., 2003; Tsai and Kass, 2009). In future studies we will, therefore, explore the mechanism between circRNAs and calcium ion-related pathways.
In the present study, we used rat BMECs to detect DE circRNAs and mRNAs after OGD/R injury. Bioinformatics predicted the functions of circRNAs and indicated downstream pathways. We believe this will be benefit treatment strategies for cerebral ischemia reperfusion injury. However, one contentious question is whether animal samples and in vitro experimental results can be used to predict responses in human. circRNAs are poorly conserved among species (Chen and Yang, 2015) and all biological functions are determined by the genes of the individual. Every species has a unique genetic code for the biological activities associated with that species (Shanks et al., 2009). Currently, nine out of ten experimental drugs fail in clinical studies because we cannot accurately predict how they will behave in people based on laboratory and animal studies (Shanks et al., 2009). Therefore, we will explore the circRNAs from the present study that are homologous with human orthologs.
In summary, a number of previously unrecorded circRNAs have been revealed to be differentially expressed in primary BMECs after OGD/R treatment. Furthermore, calcium ion and cGMP-PKG signaling pathways may be important regulatory targets of circRNAs. Altered circRNAs may be important in the pathogenesis of cerebral ische-mia-reperfusion injury and, consequently, may be potential therapeutic targets for cerebral ischemia diseases.
Additional files:
Additional Table 1: The list of CircRNAs primers.
Additional Table 2 (2MB, pdf) : Identified 1288 circRNAs by RNA sequencing.
Identified 1288 circRNAs by RNA sequencing
Additional Table 3: The list of differentially expressed circRNAs.
Additional Table 4: The list of functional and signal pathway enrichment.
Additional Table 5: The list of the predicted circRNA-miRNA interactions.
Footnotes
Conflicts of interest: The authors declare no conflicts of interest.
Financial support: This work was supported by the National Natural Science Foundation for Young Scientists of China, No. 81601058 (to WL); and Basic Research and Frontier Science Exploration Foundation of Yuzhong District, Chongqing, China, No. 20180106 (to FFS). The funding bodies played no role in the study design, in the collection, analysis and interpretation of data, in the writing of the paper, and in the decision to submit the paper for publication.
Institutional review board statement: All animal methods were approved and guided by the Chongqing Medical University Committee on Animal Research (approval No. CQMU20180086) on March 22, 2018.
Copyright license agreement: The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement: Datasets analyzed during the current study are available from the corresponding author on reasonable request.
Plagiarism check: Checked twice by iThenticate.
Peer review: Externally peer reviewed.
Open peer reviewer: Fei Ding, Nantong University, China.
Funding: This work was supported by the National Natural Science Foundation for Young Scientists of China, No. 81601058 (to WL); and Basic Research and Frontier Science Exploration Foundation of Yuzhong District, Chongqing, China, No. 20180106 (to FFS).
P-Reviewer: Ding F; C-Editor: Zhao M; S-Editors: Yu J, Li CH; L-Editors: Yu J, Song LP; T-Editor: Jia Y
References
- 1.Altesha MA, Ni T, Khan A, Liu K, Zheng X. Circular RNA in cardiovascular disease. J Cell Physiol. 2019;234:5588–5600. doi: 10.1002/jcp.27384. [DOI] [PubMed] [Google Scholar]
- 2.Arnold WP, Mittal CK, Katsuki S, Murad F. Nitric oxide activates guanylate cyclase and increases guanosine 3′:5′-cyclic monophos-phate levels in various tissue preparations. Proc Natl Acad Sci U S A. 1977;74:3203–3207. doi: 10.1073/pnas.74.8.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- 4.Bachetti T, Morbidelli L. Endothelial cells in culture: a model for studying vascular functions. Pharmacol Res. 2000;42:9–19. doi: 10.1006/phrs.1999.0655. [DOI] [PubMed] [Google Scholar]
- 5.Bai Y, Zhang Y, Han B, Yang L, Chen X, Huang R, Wu F, Chao J, Liu P, Hu G, Zhang JH, Yao H. Circular RNA DLGAP4 ameliorates ischemic stroke outcomes by targeting miR-143 to regulate endothelial-mesenchymal transition associated with blood-brain barrier integrity. J Neurosci. 2018;38:32–50. doi: 10.1523/JNEUROSCI.1348-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife. 2015;4:e07540. doi: 10.7554/eLife.07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caruso P, Dunmore BJ, Schlosser K, Schoors S, Dos Santos C, Perez-Iratxeta C, Lavoie JR, Zhang H, Long L, Flockton AR, Frid MG, Upton PD, D’Alessandro A, Hadinnapola C, Kiskin FN, Taha M, Hurst LA, Ormiston ML, Hata A, Stenmark KR, et al. Identification of microRNA-124 as a major regulator of enhanced endothelial cell glycolysis in pulmonary arterial hypertension via PTBP1 (polypyrimidine tract binding protein) and pyruvate kinase M2. Circulation. 2017;136:2451–2467. doi: 10.1161/CIRCULATIONAHA.117.028034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen ZZ, Yuan WM, Xiang C, Zeng DP, Liu B, Qin KR. A microfluidic device with spatiotemporal wall shear stress and ATP signals to investigate the intracellular calcium dynamics in vascular endothelial cells. Biomech Model Mechanobiol. 2019;18:189–202. doi: 10.1007/s10237-018-1076-x. [DOI] [PubMed] [Google Scholar]
- 10.Denninger JW, Marletta MA. Guanylate cyclase and the .NO/ cGMP signaling pathway. Biochim Biophys Acta. 1999;1411:334–350. doi: 10.1016/s0005-2728(99)00024-9. [DOI] [PubMed] [Google Scholar]
- 11.Fang R, Zhao NN, Zeng KX, Wen Q, Xiao P, Luo X, Liu XW, Wang YL. MicroRNA-544 inhibits inflammatory response and cell apoptosis after cerebral ischemia reperfusion by targeting IRAK4. Eur Rev Med Pharmacol Sci. 2018;22:5605–5613. doi: 10.26355/eurrev_201809_15825. [DOI] [PubMed] [Google Scholar]
- 12.Gao Y, Zhang J, Zhao F. Circular RNA identification based on multiple seed matching. Brief Bioinform. 2018;19:803–810. doi: 10.1093/bib/bbx014. [DOI] [PubMed] [Google Scholar]
- 13.Guo ZN, Jin H, Sun H, Zhao Y, Liu J, Ma H, Sun X, Yang Y. Antioxidant melatonin: potential functions in improving cerebral autoregulation after subarachnoid hemorrhage. Front Physiol. 2018;9:1146. doi: 10.3389/fphys.2018.01146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamzei Taj S, Kho W, Riou A, Wiedermann D, Hoehn M. MiRNA-124 induces neuroprotection and functional improvement after focal cerebral ischemia. Biomaterials. 2016;91:151–165. doi: 10.1016/j.biomaterials.2016.03.025. [DOI] [PubMed] [Google Scholar]
- 15.Han B, Zhang Y, Zhang Y, Bai Y, Chen X, Huang R, Wu F, Leng S, Chao J, Zhang JH, Hu G, Yao H. Novel insight into circular RNA HECTD1 in astrocyte activation via autophagy by targeting MIR142-TIPARP: implications for cerebral ischemic stroke. Autophagy. 2018;14:1164–1184. doi: 10.1080/15548627.2018.1458173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 17.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 18.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson RM, Lincoln TM. Effects of nitroprusside, glyceryl trinitrate, and 8-bromo cyclic GMP on phosphorylase a formation and myosin light chain phosphorylation in rat aorta. Mol Pharmacol. 1985;27:333–342. [PubMed] [Google Scholar]
- 20.Khananshvili D. The SLC8 gene family of sodium-calcium exchangers (NCX) - structure, function, and regulation in health and disease. Mol Aspects Med. 2013;34:220–235. doi: 10.1016/j.mam.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 21.Kumar VS, Gopalakrishnan A, Naziroglu M, Rajanikant GK. Calcium ion--the key player in cerebral ischemia. Curr Med Chem. 2014;21:2065–2075. doi: 10.2174/0929867321666131228204246. [DOI] [PubMed] [Google Scholar]
- 22.Liang Z, Chi YJ, Lin GQ, Luo SH, Jiang QY, Chen YK. MiRNA-26a promotes angiogenesis in a rat model of cerebral infarction via PI3K/AKT and MAPK/ERK pathway. Eur Rev Med Pharmacol Sci. 2018;22:3485–3492. doi: 10.26355/eurrev_201806_15175. [DOI] [PubMed] [Google Scholar]
- 23.Lin SP, Ye S, Long Y, Fan Y, Mao HF, Chen MT, Ma QJ. Circular RNA expression alterations are involved in OGD/R-induced neuron injury. Biochem Biophys Res Commun. 2016;471:52–56. doi: 10.1016/j.bbrc.2016.01.183. [DOI] [PubMed] [Google Scholar]
- 24.Lincoln TM, Dey N, Sellak H. Invited review: cGMP-dependent protein kinase signaling mechanisms in smooth muscle: from the regulation of tone to gene expression. J Appl Physiol (1985) 2001;91:1421–1430. doi: 10.1152/jappl.2001.91.3.1421. [DOI] [PubMed] [Google Scholar]
- 25.Liu C, Zhang C, Yang J, Geng X, Du H, Ji X, Zhao H. Screening circular RNA expression patterns following focal cerebral ischemia in mice. Oncotarget. 2017a;8:86535–86547. doi: 10.18632/oncotarget.21238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu C, Yao MD, Li CP, Shan K, Yang H, Wang JJ, Liu B, Li XM, Yao J, Jiang Q, Yan B. Silencing of circular RNA-ZNF609 ameliorates vascular endothelial dysfunction. Theranostics. 2017b;7:2863–2877. doi: 10.7150/thno.19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 28.Pang D, Wang L, Dong J, Lai X, Huang Q, Milner R, Li L. Integrin alpha5beta1-Ang1/Tie2 receptor cross-talk regulates brain endo-thelial cell responses following cerebral ischemia. Exp Mol Med. 2018;50:117. doi: 10.1038/s12276-018-0145-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plácido AI, Pereira CM, Correira SC, Carvalho C, Oliveira CR, Moreira PI. Phosphatase 2A inhibition affects endoplasmic reticulum and mitochondria homeostasis via cytoskeletal alterations in brain endothelial cells. Mol Neurobiol. 2017;54:154–168. doi: 10.1007/s12035-015-9640-1. [DOI] [PubMed] [Google Scholar]
- 30.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sawaguchi S, Varshney S, Ogawa M, Sakaidani Y, Yagi H, Takeshita K, Murohara T, Kato K, Sundaram S, Stanley P, Okajima T. O-GlcNAc on NOTCH1 EGF repeats regulates ligand-induced Notch signaling and vascular development in mammals. Elife. 2017;6:e24419. doi: 10.7554/eLife.24419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shang FF, Xia QJ, Liu W, Xia L, Qian BJ, You L, He M, Yang JL, Wang TH. miR-434-3p and DNA hypomethylation co-regulate eIF5A1 to increase AChRs and to improve plasticity in SCT rat skeletal muscle. Sci Rep. 2016;6:22884. doi: 10.1038/srep22884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shanks N, Greek R, Greek J. Are animal models predictive for humans? Philos Ethics Humanit Med. 2009;4:2. doi: 10.1186/1747-5341-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environ-ment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Surks HK, Mochizuki N, Kasai Y, Georgescu SP, Tang KM, Ito M, Lincoln TM, Mendelsohn ME. Regulation of myosin phosphatase by a specific interaction with cGMP-dependent protein kinase Ialpha. Science. 1999;286:1583–1587. doi: 10.1126/science.286.5444.1583. [DOI] [PubMed] [Google Scholar]
- 36.Toth AE, Nielsen MS. Analysis of the trafficking system in blood-brain barrier models by high content screening microscopy. Neural Regen Res. 2018;13:1883–1884. doi: 10.4103/1673-5374.239435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsai EJ, Kass DA. Cyclic GMP signaling in cardiovascular pathophysiology and therapeutics. Pharmacol Ther. 2009;122:216–238. doi: 10.1016/j.pharmthera.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto K, Korenaga R, Kamiya A, Ando J. Fluid shear stress activates Ca(2+) influx into human endothelial cells via P2X4 puri-noceptors. Circ Res. 2000;87:385–391. doi: 10.1161/01.res.87.5.385. [DOI] [PubMed] [Google Scholar]
- 40.Yu QJ, Tao H, Wang X, Li MC. Targeting brain microvascular endothelial cells: a therapeutic approach to neuroprotection against stroke. Neural Regen Res. 2015;10:1882–1891. doi: 10.4103/1673-5374.170324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang R, Wang L, Zhang L, Chen J, Zhu Z, Zhang Z, Chopp M. Nitric oxide enhances angiogenesis via the synthesis of vascular endothelial growth factor and cGMP after stroke in the rat. Circ Res. 2003;92:308–313. doi: 10.1161/01.res.0000056757.93432.8c. [DOI] [PubMed] [Google Scholar]
- 42.Zhou L, Chen J, Li Z, Li X, Hu X, Huang Y, Zhao X, Liang C, Wang Y, Sun L, Shi M, Xu X, Shen F, Chen M, Han Z, Peng Z, Zhai Q, Chen J, Zhang Z, Yang R, et al. Integrated profiling of microRNAs and mRNAs: microRNAs located on Xq27.3 associate with clear cell renal cell carcinoma. PLoS One. 2010;5:e15224. doi: 10.1371/journal.pone.0015224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu F, Liu JL, Li JP, Xiao F, Zhang ZX, Zhang L. MicroRNA-124 (miR-124) regulates Ku70 expression and is correlated with neuronal death induced by ischemia/reperfusion. J Mol Neurosci. 2014;52:148–155. doi: 10.1007/s12031-013-0155-9. [DOI] [PubMed] [Google Scholar]
- 44.Zhu S, Gao X, Huang K, Gu Y, Hu Y, Wu Y, Ji Z, Wang Q, Pan S. Glibenclamide enhances the therapeutic benefits of early hypothermia after severe stroke in rats. Aging Dis. 2018;9:685–695. doi: 10.14336/AD.2017.0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Identified 1288 circRNAs by RNA sequencing