Keywords: nerve regeneration, peripheral nerves, peripheral nerve injuries, Wallerian degeneration, tibial nerve, common peroneal nerve, RNA sequencing, proteomic; real-time quantitative polymerase chain reaction, neural regeneration
Abstract
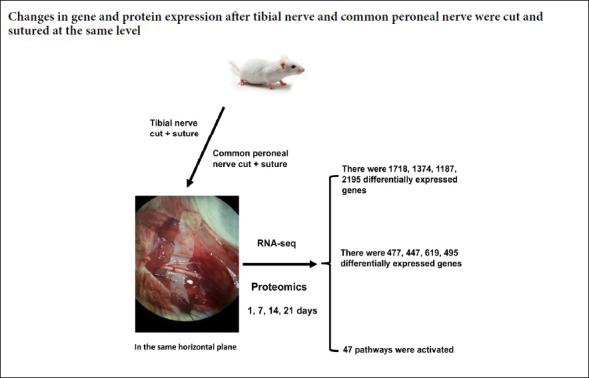
Wallerian degeneration and nerve regeneration after injury are complex processes involving many genes, proteins and cytokines. After different peripheral nerve injuries the regeneration rate can differ. Whether this is caused by differential expression of genes and proteins during Wallerian degeneration remains unclear. The right tibial nerve and the common peroneal nerve of the same rat were exposed and completely cut through and then sutured in the same horizontal plane. On days 1, 7, 14, and 21 after surgery, 1–2 cm of nerve tissue distal to the suture site was dissected out from the tibial and common peroneal nerves. The differences in gene and protein expression during Wallerian degeneration of the injured nerves were then studied by RNA sequencing and proteomic techniques. In the tibial and common peroneal nerves, there were 1718, 1374, 1187, and 2195 differentially expressed genes, and 477, 447, 619, and 495 differentially expressed proteins on days 1, 7, 14, and 21 after surgery, respectively. Forty-seven pathways were activated during Wallerian degeneration. Three genes showing significant differential expression by RNA sequencing (Hoxd4, Lpcat4 and Tbx1) were assayed by real-time quantitative polymerase chain reaction. RNA sequencing and real-time quantitative polymerase chain reaction results were consistent. Our findings showed that expression of genes and proteins in injured tibial and the common peroneal nerves were significantly different during Wallerian degeneration at different time points. This suggests that the biological processes during Wallerian degeneration are different in different peripheral nerves after injury. The procedure was approved by the Animal Experimental Ethics Committee of the Second Military Medical University, China (approval No. CZ20160218) on February 18, 2016.
Chinese Library Classification No. R446; R364; Q2
Introduction
Peripheral nerve injury is commonly encountered in the clinic. Treatment of peripheral nerve injury is difficult and, in general, damaged function often cannot be fully recovered, which can cause impairment of sensory and motor functions (Cattin et al., 2015; Trehan et al., 2016; Burtt et al., 2017; Panagopoulos et al., 2017). Therefore, the promotion of nerve regeneration and functional recovery has been intensively investigated (Bozkurk et al., 2008; Sadeghian et al., 2010). In a preliminary study, we found that different peripheral nerves have substantially different functional recovery after injury (Zhang et al., 2016). The tibial nerve recovers better than the common peroneal nerve after damage in the same plane. This phenomenon has been reported previously (Gousheh et al., 2008; Murovic, 2009; George et al., 2014). Kim et al. (2003) found that the degree of functional recovery after ulnar nerve injury was much lower than that after injury to the median and radial nerves. Roganovic et al. (2006) compared the repair of 393 different peripheral nerve injuries. They found a large difference in regeneration potential depending on the peripheral nerve injured.
Peripheral nerve regeneration is a complex and coordinated process and there are currently two theories on its mechanism: the theory of nerve chemotaxis regeneration and the contact-oriented theory (Mackinnon et al., 1989). The chemotaxis theory is generally widely accepted (Shubayev et al., 2006; Corriden et al., 2012; Yuan et al., 2013). Its basic premise is that during nerve regeneration the axons of the regenerating nerve are directed to grow by the release of chemical substances from the distal nerve or target tissue (Jung et al., 2009). A series of cellular and molecular changes (Wallerian degeneration) occur at the distal nerve end after nerve injury, including clearance of axons and myelin sheaths and proliferation of Schwann cells. Mature Schwann cells are transformed into an undifferentiated state. These undifferentiated Schwann cells form Büngner bands in the basement membrane and produce a variety of neuro-chemokines and trophic factors that guide and promote nerve regeneration (Guertin et al., 2005; Waetzig et al., 2005; Maness et al., 2007; Navarro et al., 2007; Parkinson et al., 2008; Kim et al., 2009). Therefore, the distal nerve plays a crucial role in chemotaxis regeneration.
Wallerian degeneration and nerve regeneration after nerve injury involve the mutual regulation of several genes, proteins and cytokines in a complex network involving many related molecular mechanisms, such as apoptosis and regeneration (Raivich et al., 2004; Yoo et al., 2010; Hamby et al., 2012; Chang et al., 2013; Merianda et al., 2013; Jiang et al., 2014), and are not determined by a single or even a few genes and proteins (Zhou et al., 2012; Christie et al., 2013; Merianda et al., 2013; Chan et al., 2014). Yao et al. (2013) found that during Wallerian degeneration and regeneration after sciatic nerve injury in rats, there were 6076 differentially expressed genes with 23 expression trends, and 93 differentially expressed proteins. They also identified 108 signaling pathways that participated in formation of the Wallerian degeneration signal conditioning network. Bosse et al. (2012) found that nerve regeneration capacity gradually declines with age, which is mainly because of decreased secretion of endogenous neurotrophic factors after nerve injury. Differences in chemical substances released by distal nerves or target tissues during Wallerian degeneration and regeneration, and in chemokines and nutritional factors affecting the precise growth of axons after different peripheral nerve injuries, ultimately result in differences in functional recovery. However, these differences have not been characterized. In recent years, the development of gene and protein chip technology has provided effective tools to systematically and macroscopically study the expression and function of genes and proteins (Kogenaru et al., 2012; Cheng et al., 2017; Pan et al., 2017). This technology generates large amounts of information enabling the study of gene expression and functional changes in whole organisms (Li et al., 2013).
There are several classes of up- or down-regulated genes during Wallerian degeneration and regeneration after sciatic nerve injury in rats (Allodi et al., 2012; Yao et al., 2013; Gordon et al., 2014). However, differences in the biological changes that occur during Wallerian degeneration after injury to different peripheral nerves have not been studied. In this study, the same injury to tibial and common peroneal nerves was established in rats. High-throughput RNA sequencing and proteomic techniques were used to study the Wallerian degeneration of the distal nerves. Differential gene and protein expression analysis, functional analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) signal pathway analysis were also performed. The molecular mechanism of Wallerian degeneration and nerve regeneration after peripheral nerve injury was examined by observing whether there was a difference in gene expression and protein function during Wallerian degeneration after tibial nerve and common peroneal nerve injury.
Materials and Methods
Animal preparation and surgical procedures
Sixteen male Sprague-Dawley rats aged 8 weeks and weighing 200 ± 20 g were provided by the Shanghai JieSiJie Experimental Animal Co., Ltd., Shanghai, China (license No. SCXK (Hu) 2013-0006). The rats were maintained in cages at 22°C with 14 hours of light and 10 hours of darkness per day. The experimental procedures were approved by the Animal Ethics Committee of the Second Military Medical University, China (approval No. CZ20160218) on February 18, 2016.
The rats were equally and randomly divided into four groups according to survival time (1, 7, 14, and 21 days). Identical tibial and common peroneal nerve injury was surgically induced in each rat. The experimental rats were anesthetized by intraperitoneal injection with 2.5% pentobarbital sodium (30 mg/kg), and fixed in the prone position. A 2 cm incision was made in a posterior medial position to expose the tibial nerve and the common peroneal nerve, and the two were simultaneously cut 1 cm below the piriformis with microsurgical scissors. Accurate end-to-end suturing was immediately performed with 11-0 nylon. Rats were anesthetized again on days 1, 7, 14, and 21 after surgery, and the distal nerve stumps of the tibial and common peroneal nerves (approximately 1 cm in length) were obtained. Samples were collected into liquid nitrogen, and stored at −80°C until use.
If a rat has no autonomic activity after recovery, the knee, ankle and toe joints are difficult to bend or they bend and are limp. In these cases, the model can be considered to have been successfully established.
RNA sequencing
Total RNA was isolated from the distal nerve stumps using TRIzol reagent (Invitrogen, Grand Island, NY, USA) according to manufacturer’s instructions at 1, 7, 14, and 21 days after surgery. Synthesis of double-stranded cDNA, purification of double-stranded cDNA, and construction of a two-terminal sequencing library were performed according to the Illumina operating manual (Illumina, San Diego, CA, USA). The constructed library was analyzed using the Agilent 2100 Bio analyzer and the ABI Step One Plus Real-time PCR System (Applied Biosystems, Waltham, MA, USA), and sequenced on an Illumina HiSeq™ 2000 sequencer to obtain raw reads. Unigene transcripts were obtained after processing the reads using the SOAP denovo program.
Real-time quantitative polymerase chain reaction
Nerve tissue samples were placed in RNA Stabilization Reagent (Qiagen, Valencia, CA, USA). The total RNA fraction was extracted using an RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. Reverse transcription using a reverse transcription kit (PrimeScript™ RT reagent Kit, TaKaRa, Shiga, Japan), and the resulting cDNA was subjected to real-time fluorescent quantitative PCR amplification. The relative expression values of each mRNA were calculated using comparative CT analysis and were normalized using glyceraldehyde phosphate dehydrogenase (Gapdh) mRNA for each data point. All reactions were run in triplicate. The primers used in RT-PCR assays are shown in Table 1.
Table 1.
Primer sequences of Hoxd4, Lpcat4, and Tbx1 genes
| Gene | Sequences (5′–3′) | Product size (bp) |
|---|---|---|
| GAPDH | Sense: AGA ACG CCG CAT CTT CTT GT | 281 |
| Anti-sense: CTT GCC GTG GGT AGAGTC AT | ||
| Hoxd4 | Sense: CAG CAA GTC CTA GAA CTG GAA AAG | 168 |
| Anti-sense: CTT GGT GTT GGG CAG TTT GT | ||
| Lpcat4 | Sense: GCC CTG GCG TAC TCA AAG T | 142 |
| Anti-sense: ATG ACT CGC TGG ACG TTG TT | ||
| Tbx1 | Sense: GGA AAT GAA GGC GCT ATG G | 93 |
| Anti-sense: ACT TGG AAG GTG GGG AAC A |
Proteomics
Distal nerve stump samples were homogenized in radioimmunoprecipitation assay buffer (RIPA), quantified using a bicinchoninic acid protein quantification kit (Pierce™ BCA protein Assay kit, Thermo Fisher Scientific, Waltham, MA, USA), and subjected to proteolysis by NH4HCO3 (Sigma A6141), iodoacetamide (Sigma), dithiothreitol (PlusOne), and trypsin. The peptides were separated and loaded into a mass spectrometer (Thermo Scientific Q Exactive, Shanghai, China) for analysis. The parameters were as follows: Primary mass spectrometry parameters: Resolution: 70,000, AGCtarget: 3e6, MaximumIT: 40 ms, Scanrange: 350–1800 m/z; Secondary mass spectrometry parameters: Resolution: 17,500, AGCtarget: 1e5, MaximumIT: 60 ms, TopN: 20, NCE/steppedNCE: 27.
Bioinformatic analysis
Randomized variation models were used to analyze differentially expressed genes and proteins, and significant trend models were clustered to obtain differentially expressed genes and proteins. We performed GO enrichment analysis on differentially expressed genes (DESeq software; Ander et al., 2010) and proteins (OmicsBean software; http://www.omicsbean.cn; Ashburner et al., 2000; Conesa et al., 2005) and described their functions. In addition, the KEGG Database was used to determine the significant pathways of differentially expressed genes and proteins (Kanehisa et al., 2004; Xie et al., 2011). The default screening discrepancies were P < 0.05 and difference multiples were > 2.
Statistical analysis
Data are expressed as the mean ± SD. SPSS 17.0 statistical software (SPSS, Chicago, IL, USA) was used to analyze data. Paired t-tests or repeated measures analysis of variance were used for comparisons across different time points in each group. The variance between tibial nerve and common peroneal nerve data was compared using one-way analysis of variance followed by Dunnett’s post hoc test, with a test level of α = 0.05.
Results
Differential gene expression
Two criteria were adopted to compare and analyze whether the same gene in two samples was differentially expressed. One was FoldChange, which is the fold change in expression level of the same gene in two samples. The other was P-value or FDR (adjusted P-value). The FDR value was calculated by calculating the P-value for each gene. The FDR error control method was used to perform multi-hypothesis test correction on the P-value. The default screening difference was P < 0.05 and the difference multiple was > 2. On days 1, 7, 14, and 21 after surgery, there were 1718, 1374, 1187, and 2195 genes, respectively, with significantly different expression levels between the tibial and common peroneal nerves (Table 2).
Table 2.
Differential gene expression in the tibial and common peroneal nerves
| Time post-surgery (day) | Up_diff | Down_diff | Total_diff |
|---|---|---|---|
| 1 | 922 | 796 | 1718 |
| 7 | 670 | 704 | 1374 |
| 14 | 532 | 655 | 1187 |
| 21 | 493 | 1702 | 2195 |
Up_diff: Significant difference in up-regulated gene quantity; Down_diff: significant difference in down-regulated gene quantity; Total_diff: significant difference in total number of genes.
Functional analysis of differentially expressed genes
GO enrichment analysis of differentially expressed genes
GO enrichment analysis according to three GO categories, biological process, molecular functions, and cellular component, was performed on the differentially expressed genes. Functions were described and statistical classification was performed.
The GO enrichment analysis results showed that in the early stage of Wallerian degeneration, the differentially expressed genes were mainly concentrated in biological processes, such as apoptosis, release of inflammatory mediators, and cellular inflammatory response. At the late stage of Wallerian degeneration, differentially expressed genes were enriched in cytokine release, reconstruction of muscle and blood vessels, and neurotransmitter release (Figures 1–3).
Figure 1.
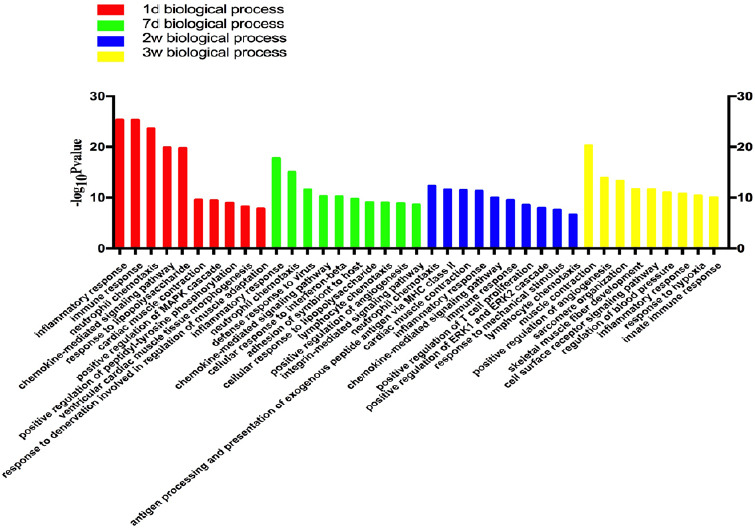
Statistics of differentially expressed gene counts in biological processes from RNA sequencing data.
On days 1 and 7 after tibial nerve and common peroneal nerve injury, the differentially expressed genes in biological processes were mainly concentrated in inflammatory and immune responses. On days 14 and 21 after surgery, the differentially expressed genes were concentrated in vascular production and muscle fiber development. d: Day(s); w: weeks.
Figure 3.
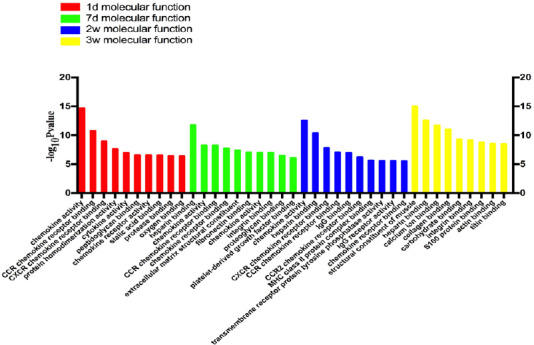
Statistics of differentially expressed gene counts in molecular function from RNA sequencing data.
During tibial nerve and common peroneal nerve injury, the molecular functions of differentially expressed genes mainly involved CCR chemokine receptor binding, CXCR, and chemokine activity. d: Day(s); w: weeks.
Figure 2.
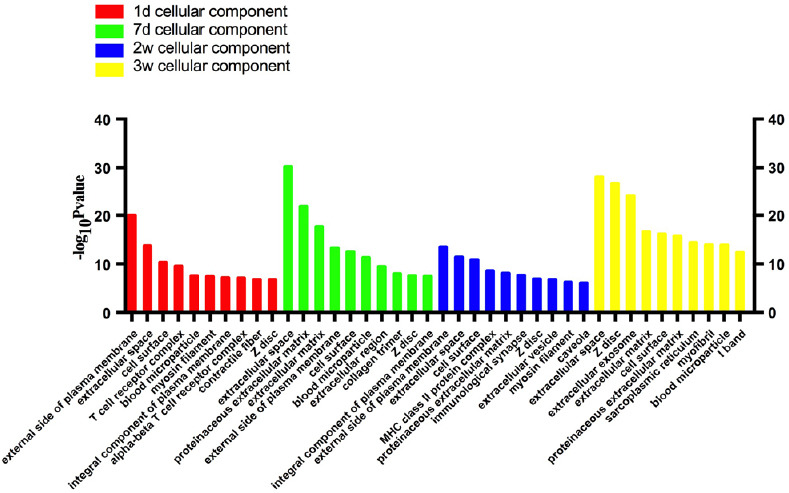
Statistics of differentially expressed gene counts in cellular components from RNA sequencing data.
During tibial nerve and common peroneal nerve injury, the cellular components involved were mainly located in the extracellular space, such as on the external side of the plasma membrane and in the proteinaceous and extracellular matrix. d: Day(s); w: weeks.
KEGG enrichment analysis of differentially expressed genes
KEGG enrichment analysis indicated that 47 signaling pathways were activated after tibial nerve and common peroneal nerve injury. In the early stage of Wallerian degeneration, the main pathways activated included the peroxisome proliferator-activated receptor (PPAR) signaling pathway, the PI3K-Akt signaling pathway, the chemokine signaling pathway, and cytokine-cytokine receptor interaction. In the late stage of Wallerian degeneration, the main pathways activated involved cell adhesion molecules, calcium ion transport, the ErbB signaling pathway, the tumor necrosis factor signaling pathway, the 5′-adenosine monophosphate-activated protein kinase (AMPK) signaling pathway, the mitogen-activated protein kinase (MAPK) signaling pathway, the PPAR signaling pathway, and the Wnt signaling pathway (Figures 4–7).
Figure 4.
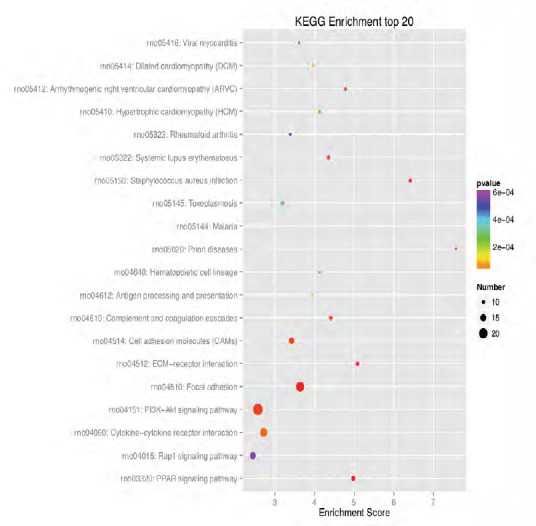
KEGG enrichment analysis of the 20 most highly differentially expressed genes between tibial and common peroneal nerves 1 day after surgery.
The signaling pathways involved include the PPAR, PI3K-Akt, and Rap1 signaling pathways. The X-axis is the enrichment score. The larger the bubble, the larger the number of differentially expressed genes. The color of the bubble changes in the order red-blue-green-yellow, with increasing P-value enrichment. KEGG: Kyoto encyclopedia of genes and genomes; PPAR: peroxisome proliferator-activated receptor; PI3K: phosphoinositide 3-kinase.
Figure 7.
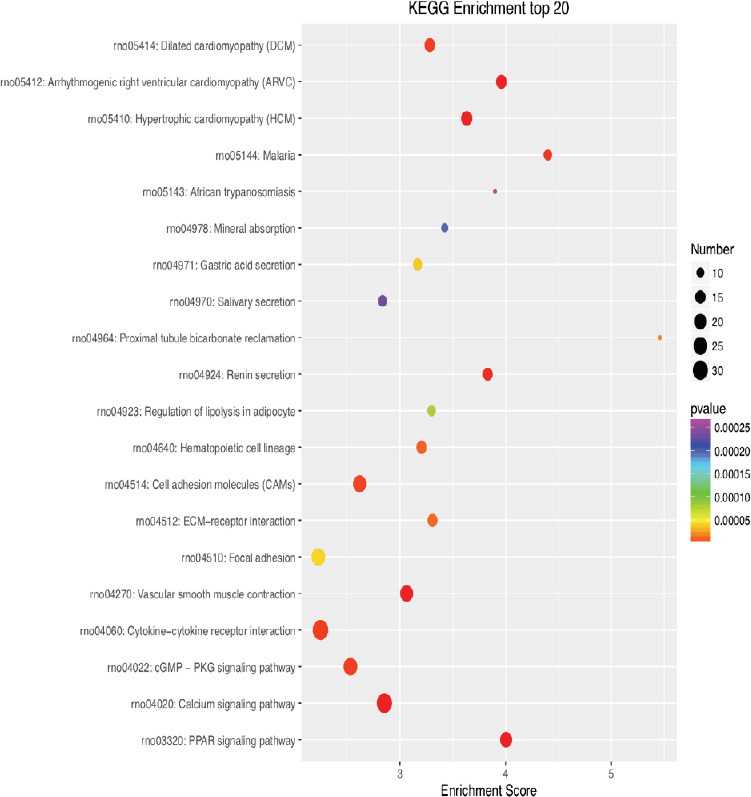
KEGG enrichment analysis of the 20 most highly differentially expressed genes between tibial and common peroneal nerves 21 days after surgery.
The signaling pathways involved include the PPAR, calcium, and cGMP-PKG pathways. The X-axis is the enrichment score. The larger the bubble, the larger the number of differentially expressed genes. The color of the bubble changes in the order red-blue-green-yellow, with increasing P-value enrichment. KEGG: Kyoto encyclopedia of genes and genomes; PPAR: peroxisome proliferator-activated receptor; cGMP: cyclic guanosine monophosphate; PKG: protein kinase G.
Figure 5.
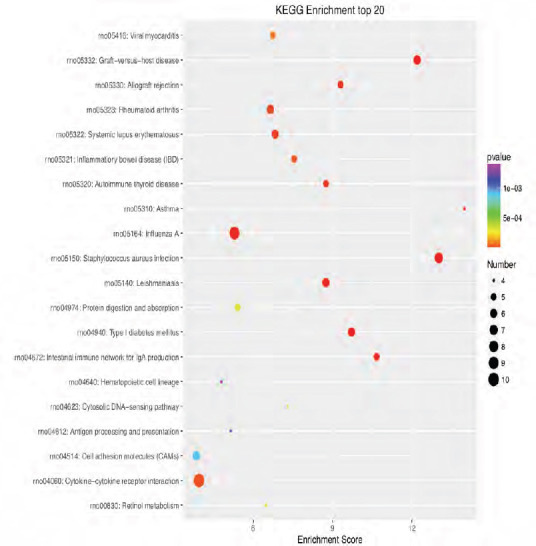
KEGG enrichment analysis of the 20 most highly differentially expressed genes between tibial and common peroneal nerves 7 days after surgery.
The signaling pathways involved include the Chemokine, B cell receptor, Toll-like receptor, and nuclear factor-kappa B signaling pathways. The X-axis is the enrichment score. The larger the bubble, the larger the number of differentially expressed genes. The color of the bubble changes in the order red-blue-green-yellow, with increasing in P-value enrichment. KEGG: Kyoto encyclopedia of genes and genomes.
Figure 6.
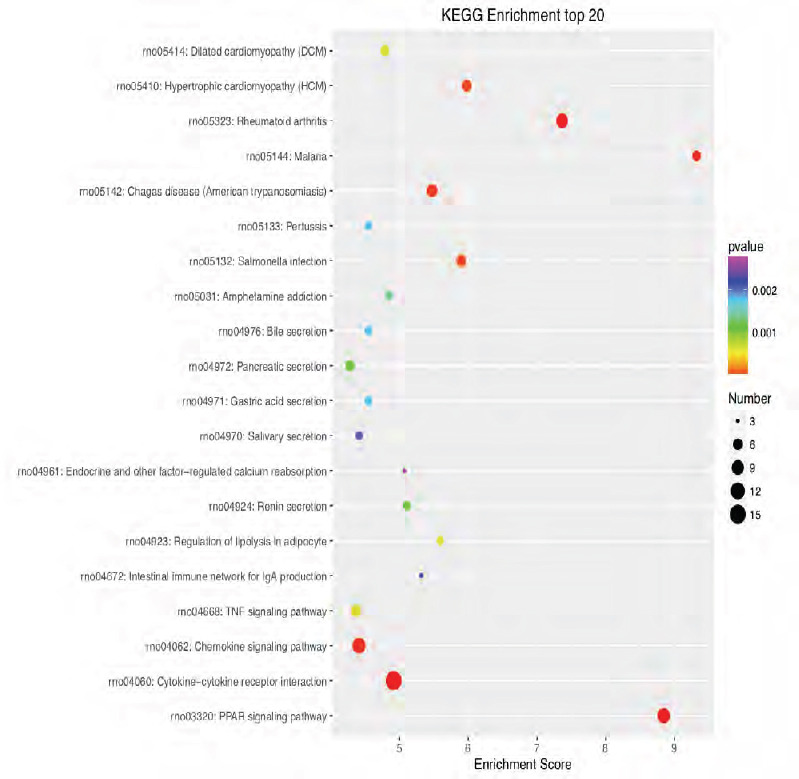
KEGG enrichment analysis of the 20 most highly differentially expressed genes between tibial and common peroneal nerves 14 days after surgery.
The signaling pathways involved include the PPAR, chemokine, and tumor necrosis factor pathways. The X-axis is the enrichment score. The larger the bubble, the larger the number of differentially expressed genes. The color of the bubble changes in the order red-blue-green-yellow, with increasing P-value enrichment. KEGG: Kyoto encyclopedia of genes and genomes; PPAR: peroxisome proliferator-activated receptor.
Differentially expressed proteins
FoldChange and FDR were adopted to compare and analyze whether the same protein in two samples was differentially expressed. The default screening difference was P < 0.05 and the difference fold was > 2.
At 1, 7, 14, and 21 days after surgery, the numbers of proteins showing significant differences in expression between the tibial and common peroneal nerves were 477, 447, 619, and 495, respectively (Table 3).
Table 3.
Statistics of proteins that were differentially expressed in the tibial and common peroneal nerves (FC > 2 or < 0.5)
| Time post-surgery (days) | Up_diff | Down_diff | Total_diff |
|---|---|---|---|
| 1 | 310 | 167 | 477 |
| 7 | 177 | 220 | 447 |
| 14 | 254 | 365 | 619 |
| 21 | 261 | 234 | 495 |
Up_diff: Significant difference in up-regulated protein quantity; Down_diff: significant difference in down-regulated protein quantity; Total_diff: significant difference total number of proteins; FC: Fold Change.
Functional analysis of differentially expressed proteins
GO enrichment analysis of differentially expressed proteins
Biological process: In this analysis, 8893 biological processes were co-enriched, of which 2480 were statistically significant (P < 0.05; Figure 8).
Figure 8.
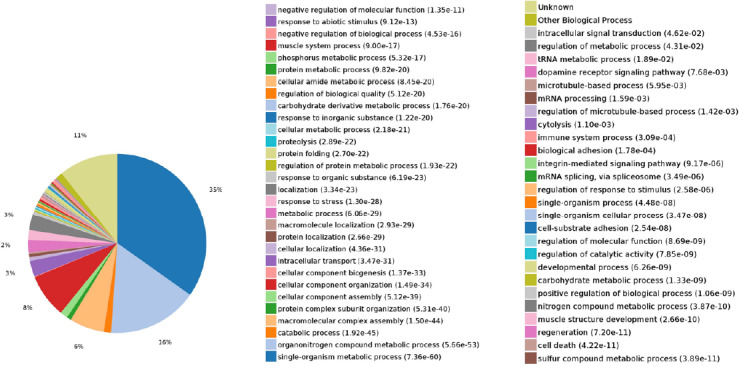
Statistics of differentially expressed proteins in biological processes.
The expressed proteins were mainly involved in biological processes such as the single-organism metabolic process, organonitrogen compound metabolism, macromolecular complex assembly, cellular component organization, intracellular transport, and localization.
Cellular component: In this analysis, 1152 cellular components were co-enriched, of which 489 were statistically significant (P < 0.05; Figure 9).
Figure 9.

Statistics of differentially expressed proteins in cellular components.
The expressed proteins were mainly involved in cellular components, such as extracellular organelles, cytoplasm, extracellular region part, macromolecular complex, and cytosol.
Molecular function: In this analysis, 2111 molecular functions were co-enriched, of which 557 were statistically significant (P < 0.05; Figure 10).
Figure 10.
Statistics of differentially expressed proteins in molecular functions.
The expressed proteins were mainly involved in molecular functions, such as protein binding, catalytic activity, RNA binding, poly(A) RNA binding, and enzyme binding.
KEGG enrichment analysis of differentially expressed proteins
As shown in Figure 11, the enriched KEGG pathways mainly included proximal tubule bicarbonate reclamation, ribosomes, gap junctions, and metabolic pathways.
Figure 11.
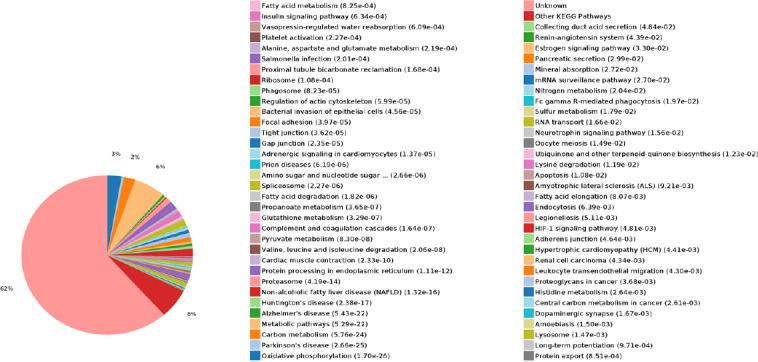
KEGG classification of protein expression.
The KEGG pathways mainly included proximal tubule bicarbonate reclamation, ribosomes, gap junctions, and metabolic pathways. KEGG: Kyoto encyclopedia of genes and genomes.
Real-time quantitative PCR assay
To validate our data, three genes with significant differences in expression at various time points between the tibial and common peroneal nerves were selected for PCR validation: Tbx1, Hoxd4, and Lpcat4. Tbx1 is a member of the T-box transcription factor family and plays an important role in vertebrate embryonic development (Papaioannou et al., 1998). Hoxd4 is closely related to embryonic development and plays an important role in the development of the nervous system. Lpcat4 has lysophospholipid acyltransferase activity and is involved in phospholipid metabolism. The RT-PCR results showed significant differences in the expression of these genes between the tibial and common peroneal nerves at various time points except on the first day after surgery. The changes observed were mostly consistent with the RNA sequencing results (Figure 12).
Figure 12.

Real-time quantitative polymerase chain reaction analysis of differential gene expression in the distal tibial nerve and common peroneal nerve stumps of rats at 1, 7, 14, 21 days post-surgery.
The relative expression of each mRNA was calculated using the comparative Ct method and normalized using Gapdh for each data point. *P < 0.05, ****P < 0.0001, vs. tibial nerve (n = 16; mean ± SD, one-way analysis of variance followed by Dunnett’s post hoc test). d: Day(s).
Discussion
Although the molecular mechanisms of Wallerian degeneration have been extensively studied, the exact mechanism of degeneration and regeneration of distal nerves after injury remains unclear (Loreto et al., 2015; Vargas et al., 2015; Chang et al., 2016). The recovery of function after peripheral nerve injury depends on whether the broken end achieves a bridging repair, the distance between the injured nerve and the target organ, the speed of nerve regeneration, and whether the regenerated axonal can accurately grow into the original target organ (Skouras et al., 2011). Highly developed microsurgery enables the bridging of nerve ends (Jiang et al., 2006; Ray et al., 2010; Faroni et al., 2015). We designed the injury of the tibial nerve and the common peroneal nerve to be in the same plane, and there was no obvious difference in the distance to the corresponding target muscle, so regeneration distance was not the decisive factor. Kang et al. (2013) confirmed that the nerve regeneration rate of aged rats is remarkably slower than that of young rats. However, there was no difference in regeneration rate after injury between nerves. Our early studies found that recovery of the tibial nerve was markedly better than that of the common peroneal nerve after injury. Therefore, we inflicted injury on the two nerves in the same horizontal plane on the same thigh of the same mouse. We used RNA sequencing and proteomics to explore whether there were differences in gene and protein expression in Wallerian degeneration between the two nerves.
Our study showed that the biological processes occurring after injury to the tibial and common peroneal nerves are not the same, indicating differences in the Wallerian degeneration processes between the two nerves. We tested both nerves and identified differentially expressed genes. In the early stages of Wallerian degeneration, the differentially expressed genes were mainly involved apoptosis, release of inflammatory mediators, and cellular inflammatory response. In the late stage of Wallerian degeneration, the differentially expressed genes were mainly involved in cytokine release, muscle and vascular remodeling, and neurotransmission. KEGG enrichment analysis showed that 47 different signaling pathways were activated during Wallerian degeneration after injury of the tibial and common peroneal nerves. These were mainly PPAR, PI3K-Akt, and chemokine signaling pathways, and cytokine-cytokine receptor interaction in early Wallerian degeneration. In the late stage, the signaling pathways were mainly ErbB, tumor necrosis factor, AMPK, MAPK, PPAR, and Wnt.
To determine whether there was any difference between tibial and common peroneal nerves after injury we performed a proteomics assay. We identified a significant difference in protein synthesis between the tibial and common peroneal nerves after injury. The differentially expressed proteins were mainly involved in processes such as organonitrogen compound metabolism, macromolecular complex assembly, cellular component organization, intracellular transport, localization, catalytic activity, ion binding, blinding, cation binding, and metal ion binding.
Finally, we screened for genes expressed in both nerves at various time points, and we selected three genes with significant differential expression for qRT-PCR confirmation. The RT-PCR results showed significant differences in the expression of these genes between the tibial and common peroneal nerves except on the first day after surgery. These results were consistent with the results of RNA sequencing. Differential expression of these genes indicated differences in the Wallerian degeneration between the tibial nerve and common peroneal nerve, which indicates differences in neural regeneration.
In summary, Wallerian degeneration differs depending on the peripheral nerve injured. Our study adds data to the Wallerian degeneration theory, and provides a basis for further exploration of the molecular mechanisms underlying differences in regeneration potential among different peripheral nerves.
Additional file: Open peer review report 1 (98.7KB, pdf) .
Acknowledgments:
We are very grateful to Professor Chun-Lin Hou of Shanghai Changzheng Hospital, China for his help in clinical and experimental studies
Footnotes
Conflicts of interest: The authors declare that there are no conflicts of interest associated with this manuscript.
Financial support: This study was funded by the National Natural Science Foundation of China, No. 81572146 (to HDL); the Program of Outstanding Medical Talent of Shanghai Municipal Health Bureau, China, No. 2017BR034 (to HDL); the Shuguang Program of Shanghai Education Development Foundation and Shanghai Municipal Education Commission, China, No. 15SG34 (to HDL). The funding sources had no role in study conception and design, data analysis or interpretation, paper writing or deciding to submit this paper for publication.
Institutional review board statement: The procedure was approved by the Animal Experimental Ethics Committee of the Second Military Medical University, China (approval No. CZ20160218) on February 18, 2016. The experimental procedure followed the United States National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publication No. 85-23, revised 1985).
Copyright license agreement: The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement: Datasets analyzed during the current study are available from the corresponding author on reasonable request.
Plagiarism check: Checked twice by iThenticate.
Peer review: Externally peer reviewed.
Open peer reviewer: Ben Christensen, University of Utah, USA.
Funding: This study was funded by the National Natural Science Foundation of China, No. 81572146 (to HDL); the Program of Outstanding Medical Talent of Shanghai Municipal Health Bureau, China, No. 2017BR034 (to HDL); the Shuguang Program of Shanghai Education Development Foundation and Shanghai Municipal Education Commission, China, No. 15SG34 (to HDL).
P-Reviewer: Christensen B; C-Editor: Zhao M; S-Editors: Wang J, Li CH; L-Editors: Allen J, Norman C, Qiu Y, Song LP; T-Editor: Jia Y
References
- 1.Allodi I, Udina E, Navarro X. Specificity of peripheral nerve regeneration: interactions at the axon level. Prog Neurobiol. 2012;98:16–37. doi: 10.1016/j.pneurobio.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Ander S, Huber W. Differential expression analysis for sequence count data. Genome Biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosse F. Extrinsic cellular and molecular mediators of peripheral axonal regeneration. Cell Tissue Res. 2012;349:5–14. doi: 10.1007/s00441-012-1389-5. [DOI] [PubMed] [Google Scholar]
- 5.Bozkurt A, Tholl S, Wehner S, Tank J, Cortese M, O’Dey Dm, Deumens R, Lassner F, Schügner F, Gröger A, Smeets R, Brook G, Pallua N. Evaluation of functional nerve recovery with Visual-SSI-a novel computerized approach for the assessment of the static sciatic index (SSI) Neurosci Methods. 2008;170:117–122. doi: 10.1016/j.jneumeth.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 6.Burtt KE, Badash I, Wu B. Assessing surgical methods for treatment of cubital tunnel syndrome - which is the best? Clin Trials Orthop Disord. 2017;2:123–124. [Google Scholar]
- 7.Cattin AL, Burden JJ, Van Emmenis L, Mackenzie FE, Hoving JJ, Garcia Calavia N, Guo Y, McLaughlin M, Rosenberg LH, Quereda V, Jamecna D, Napoli I, Parrinello S, Enver T, Ruhrberg C, Lloyd AC. Macrophage-induced blood vessels guide Schwann cell mediated regeneration of peripheral nerves. Cell. 2015;162:1127–1139. doi: 10.1016/j.cell.2015.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan KM, Gordon T, Zochodne DW, Power HA. Improving peripheral nerve regeneration: from molecular mechanisms to potential therapeutic targets. Exp Neurol. 2014;261:826–835. doi: 10.1016/j.expneurol.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 9.Chang B, Quan Q, Lu S. Molecular mechanisms in the initiation phase of Wallerian degeneration. Eur J Neurosci. 2016;44:2040–2048. doi: 10.1111/ejn.13250. [DOI] [PubMed] [Google Scholar]
- 10.Chang LW, Viader A, Varghese N, Payton JE, Milbrandt J, Nagarajan R. An integrated approach to characterize transcription factor and microRNA regulatory networks involved in Schwann cell response to peripheral nerve injury. BMC Genomics. 2013;14:84. doi: 10.1186/1471-2164-14-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng Q, Wang YX, Yu J, Yi S. Critical signaling pathways during Wallerian degeneration of peripheral nerve. Neural Regen Res. 2017;12:995–1002. doi: 10.4103/1673-5374.208596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christie KJ, Zochodne D. Peripheral axon regrowth: new molecular approaches. Neuroscience. 2013;240:310–324. doi: 10.1016/j.neuroscience.2013.02.059. [DOI] [PubMed] [Google Scholar]
- 13.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 14.Corriden R, Insel PA. New insights regarding the regulation of chemotaxis by nucleotides, adenosine, and their receptors. Purinergic Signal. 2012;8:587–598. doi: 10.1007/s11302-012-9311-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faroni A, Mobasseri SA, Kingham PJ, Reid AJ. Peripheral nerve regeneration: experimental strategies and future perspectives. Adv Drug Deliv Rev 82-83. 2015:160–167. doi: 10.1016/j.addr.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 16.George SC, Boyce DE. An evidence-based structured review to assess the results of common peroneal nerve repair. Plast Reconstr Surg. 2014;134:302e–311e. doi: 10.1097/PRS.0000000000000318. [DOI] [PubMed] [Google Scholar]
- 17.Gordon T. Neurotrophic factor expression in denervated motor and sensory Schwann cells: relevance to specificity of peripheral nerve regeneration. Exp Neurol. 2014;254:99–108. doi: 10.1016/j.expneurol.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 18.Gousheh J, Arasteh E, Beikpour H. Therapeutic results of sciatic nerve repair in Iran-Iraq war casualties. Plast Reconstr Surg. 2008;121:878–886. doi: 10.1097/01.prs.0000299286.67932.88. [DOI] [PubMed] [Google Scholar]
- 19.Guertin AD, Zhang DP, Mak KS. Microanatomy of axon/glial signaling during Wallerian degeneration. J Neurosci. 2005;25:3478–3487. doi: 10.1523/JNEUROSCI.3766-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamby ME, Copola G, Ao Y, Geschwind DH, Khakh BS, Sofroniew MV. Inflammatory mediators alter the astrocyte transcriptome and Calcium signaling elicited by multiple G-protein-coupled receptors. J Neurosci. 2012;32:14489–14510. doi: 10.1523/JNEUROSCI.1256-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang B, Zhang P, Zhang D, Fu Z, Yin X, Zhang H. Study on small gap sleeve bridging peripheral nerve injury. Artif Cells Blood Substit Immobil Biotechnol. 2006;34:55–74. doi: 10.1080/10731190500430149. [DOI] [PubMed] [Google Scholar]
- 22.Jiang N, Li H, Sun Y, Yin D, Zhao Q, Cui S, Yao D. Differential gene expression in proximal and distal nerve segments of rats with sciatic nerve injury during Wallerian degeneration. Neural Regen Res. 2014;9:1186–94. doi: 10.4103/1673-5374.135325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jung H, Bhangoo S, Banisadr G, Freitag C, Ren D, White FA, Miller RJ. Visualization of chemokine receptor activation in transgenic mice reveals peripheral activation of CCR2 receptors in states of neuropathic pain. J Neurosci. 2009;29:8051–8062. doi: 10.1523/JNEUROSCI.0485-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang H, Lichtman JW. Motor axon regeneration and muscle reinnervation in young adult and aged animals. J Neurosci. 2013;33:19480–19491. doi: 10.1523/JNEUROSCI.4067-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim D, Lee S, Lee SJ. Toll-like receptors in peripheral nerve injury and neuropathic pain. Curr Top Microbiol Immunol. 2009;336:169–186. doi: 10.1007/978-3-642-00549-7_10. [DOI] [PubMed] [Google Scholar]
- 27.Kim DH, Han K, Tiel RL, Murovic JA, Kline DG. Surgical outcomes of 654 ulnar nerve lesions. J Neurosurg. 2003;98:993–1004. doi: 10.3171/jns.2003.98.5.0993. [DOI] [PubMed] [Google Scholar]
- 28.Kogenaru S, Qing Y, Guo Y, Wang N. RNA-seq and microarray complement each other in transcriptome profiling. BMC Genomics. 2012;13:629. doi: 10.1186/1471-2164-13-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li M, Guo W, Zhang P, Li H, Gu X, Yao D. Signal flow and pathways in response to early wallerian degeneration after rat sciatic nerve injury. Neurosci Lett. 2013;536:56–63. doi: 10.1016/j.neulet.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 30.Loreto A, Di Stefano M, Gering M, Conforti L. Wallerian degeneration is executed by an NMN-SARM1-Dependent Late Ca(2+) influx but only modestly influenced by mitochondria. Cell Rep. 2015;13:2539–2552. doi: 10.1016/j.celrep.2015.11.032. [DOI] [PubMed] [Google Scholar]
- 31.Mackinnon SE. New directions in peripheral nerve surgery. Ann Plast Surg. 1989;22:257–273. doi: 10.1097/00000637-198903000-00013. [DOI] [PubMed] [Google Scholar]
- 32.Maness PF, Schachner M. Neural recognition molecules of the immunoglobulin super-family: signaling transducers of zxon guidance and neuronal migration. Nat Neurosci. 2007;10:19–26. doi: 10.1038/nn1827. [DOI] [PubMed] [Google Scholar]
- 33.Merianda T, Twiss J. Peripheral nerve axons contain machinery for cotranslational secretion of axonally-generated proteins. Neurosci Bull. 2013;29:493–500. doi: 10.1007/s12264-013-1360-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murovic JA. Lower-extremity peripheral nerve injuries: a Louisiana State University Health Sciences Center literature review with comparison of the operative outcomes of 806 Louisiana State University Health Sciences Center sciatic, common peroneal, and tibial nerve lesions. Neurosurgery. 2009;65:A18–A23. doi: 10.1227/01.NEU.0000339123.74649.BE. [DOI] [PubMed] [Google Scholar]
- 35.Navarro X, Vivó M, Valero Cabré A. Neural plasticity after peripheral nerve injury and regeneration. Prog Neurobiol. 2007;82:163–201. doi: 10.1016/j.pneurobio.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 36.Pan B, Liu Y, Yan JY, Wang Y, Yao X, Zhou HX, Lu L, Kong XH, Feng SQ. Gene expression analysis at multiple time-points identifies key genes for nerve regeneration. Muscle Nerve. 2017;55:373–383. doi: 10.1002/mus.25225. [DOI] [PubMed] [Google Scholar]
- 37.Panagopoulos GN, Megaloikonomos PD, Mavrogenis AF. The present and future for peripheral nerve regeneration. Orthopedics. 2017;40:e141–156. doi: 10.3928/01477447-20161019-01. [DOI] [PubMed] [Google Scholar]
- 38.Papaioannou VE, Silver LM. The T-box gene family. Bioessays. 1998;20:9–19. doi: 10.1002/(SICI)1521-1878(199801)20:1<9::AID-BIES4>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 39.Parkinson DB, Bhaskaran A, Arthur-Farraj P. C-Jun is a negative regulator of myelination. J Cell Biol. 2008;181:625–637. doi: 10.1083/jcb.200803013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raivich G, Bohatschek M, Da Costa C, Iwata O, Galiano M, Hristova M, Nateri AS, Makwana M, Riera-Sans L, Wolfer DP, Lipp HP, Aguzzi A, Wagner EP, Behrens A. The AP-1transcription factor c-Jun is required for efifcient axonal regeneration. Neuron. 2004;43:57–67. doi: 10.1016/j.neuron.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 41.Ray WZ, Mackinnon SE. Management of nerve gaps: autografts, allografts, nerve transfers, and end-to-side neurorrhaphy. Exp Neurol. 2010;223:77–85. doi: 10.1016/j.expneurol.2009.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roganovic Z, Pavlicevic G. Difference in recovery potential of peripheral nerves after graft repairs. Neurosurgery. 2006;59:621–633. doi: 10.1227/01.NEU.0000228869.48866.BD. [DOI] [PubMed] [Google Scholar]
- 43.Sadeghian H, Wolfe GI. Therapy update in nerve, neuromuscular junction and myopathic disorders. Curr Opin Neurol. 2010;23:496–501. doi: 10.1097/WCO.0b013e32833d7367. [DOI] [PubMed] [Google Scholar]
- 44.Shubayev VI, Angert M, Dolkas J, Campana WM, Palenscar K, Myers RR. TNF alpha induced MMP-9 promotes macrophage recruitment into injured peripheral nerve. Mol Cell Neurosci. 2006;3:407–415. doi: 10.1016/j.mcn.2005.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skouras E, Ozsoy U, Sarikcioglu L, Angelov D. Intrinsic and therapeutic factors determining the recovery of motor function after peripheral nerve transection. Ann Anat. 2011;193:286–303. doi: 10.1016/j.aanat.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 46.Trehan SK, Model Z, Lee SK. Nerve repair and nerve grafting. Hand Clin. 2016;32:119–125. doi: 10.1016/j.hcl.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 47.Vargas ME, Yamagishi Y. Live imaging of calcium dynamics during axon degeneration reveals two functionally distinct phases of calcium influx. J Neurosci. 2015;35:15026–15038. doi: 10.1523/JNEUROSCI.2484-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waetzig V, Herdegen T. MEKK1controls neurite regrowth after experimental injury by balancing ERK1/2 and JNK2 signaling. Mol Cell Neurosci. 2005;30:67–78. doi: 10.1016/j.mcn.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 49.Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li C, Wei L. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yao D, Li M, Shen D, Ding F, Lu S, Zhao Q, Gu X. Expression changes and bioinformatic analysis of Wallerian degeneration after sciatic nerve injury in rat. Neurosci Bull. 2013;29:321–332. doi: 10.1007/s12264-013-1340-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoo S, van Niekerk EA, Merianda TT, Twiss JL. Dynamics of axonal mRNA transport and implications for peripheral nerve regeneration. Exp Neurol. 2010;223:19–27. doi: 10.1016/j.expneurol.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuan J, Chan S, Mortimer D, Nguyen H, Goodhill GJ. Optimality and saturation in axonal chemotaxis. Neural Computation. 2013;25:833–853. doi: 10.1162/NECO_a_00426. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Q, Chen H, Liu G, Zong H, Lin H, Hou C. Comparison of healing results between tibial nerve and common peroneal nerve after sciatic nerve injury repair in rhesus monkey. Zhongguo Xiufu Chongjian Waike Zazhi. 2016;30:608–611. doi: 10.7507/1002-1892.20160123. [DOI] [PubMed] [Google Scholar]
- 54.Zhou S, Shen D, Wang Y, Gong L, Tang X, Yu B, Gu X, Ding F. micro RNA-222 targeting PTEN promotes neurite outgrowth from adult dorsal root ganglion neurons following sciatic nerve transection. PLoS One. 2012;7:e44768. doi: 10.1371/journal.pone.0044768. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



