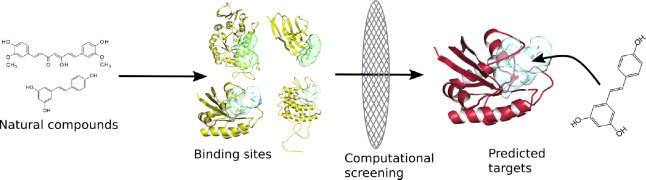
The development of new drugs is traditionally focused on a single target protein. Only recently has it been recognized that drugs, i.e., small molecules, usually bind to several target proteins and thus have a wider spectrum of different effects (Konc, 2019). Natural substances are very interesting in this respect, as more than 32% of all drugs on the market today are derived from natural sources. However, natural substances are largely unexplored as their exact mechanisms of action are often unknown (Baran, 2018). In order to determine their actions, we can resort to computational methods. Computational screening enables the identification of potential target proteins, the explanation of observed effects and the prediction of novel effects of natural compounds. Computer algorithms enable targeted investigations, such as focused experimental tests on a specific target protein or a family of potential target proteins. Recently, we used computational screening to identify new potential target proteins for two natural compounds, curcumin and resveratrol, components of turmeric and grape, respectively (Furlan et al., 2018; Kores et al., 2019). These two compounds have been shown to have anti-neuroinflammatory activity and therapeutic effects in the treatment of neurodegenerative diseases. We identified new potential target proteins of these two compounds, the modulation of which with resveratrol and curcumin might help to explain their therapeutic effects in neurodegenerative diseases, in particular in the Alzheimer’s disease. We have discovered some already known target proteins and also new potential target proteins involved in neurodegeneration. Consisting of two complementary methods, inverse molecular docking and comparison of protein binding sites, our computational approach is general and can be applied to any natural or synthetic compound (Figure 1). Our results confirm the usefulness of this approach for the determination of new target proteins of natural compounds and for the investigation of their potential effects in the treatment of neurological diseases.
Figure 1.
Computational screening for the detection of new target proteins of natural compounds.
Inverse molecular docking: Inverse molecular docking is an approach that tests the potential of a compound to bind multiple protein structures (Furlan et al., 2018). The docking process consists of fitting three-dimensional conformations of a compound into a protein structure with the goal of finding the best possible geometric fit of a compound in a protein structure that maximizes the number of its attractive interactions. After docking, the scoring follows, i.e., the assessment of the binding affinity of the docked compounds to a protein. The binding affinity can be approximated in various ways, e.g., as the sum of hydrogen bonds or van der Waals interactions formed between a compound and a protein. We have developed an inverse docking method (Furlan et al., 2018), which enables fast and efficient docking of compounds into tens of thousands of protein structures. These are derived from the ProBiS database (Konc and Janežič, 2017) of predicted binding site structures for the entire Protein Data Bank (PDB) (https://rcsb.org). The end result of inverse molecular docking is a list of potential target proteins for the search compound, ranked by their binding energies from the most preferred to the least preferred.
The successful realization of inverse molecular docking requires a database of protein structures that enables efficient and automated docking of compounds to thousands of proteins. Since small molecules most often bind to protein binding sites, predicting binding sites for each protein provides significant acceleration. To this end, we have developed the ProBiS database (Konc and Janežič, 2017), which contains more than 200,000 binding sites for currently > 150,000 protein structures determined by X-ray crystallography, nuclear magnetic resonance or electron microscopy available in the PDB and a subset of human binding sites, which currently contains more than 38,000 predicted binding sites. The prediction of binding sites allows us to concentrate docking only on binding sites, thus limiting the search space and increasing the efficiency of the inverse molecular docking. This enables a rapid scan of tens of thousands of protein structures using the inverse molecular docking method, which was previously impossible.
Binding site comparison: Another method that enables the prediction of new target proteins is the binding site comparison. Here, it is assumed that similar binding sites very probably bind similar or identical ligands (Konc, 2019). A limitation of this procedure is that a natural compound for which we wish to detect its target proteins must be co-crystallized as a ligand in at least one binding site in the PDB. New target proteins are predicted by structurally comparing this binding site with other proteins (or binding sites) from the PDB. Proteins containing detected binding sites similar to the query binding site are deemed as new potential targets for the investigated natural compound. We have developed the ProBiS algorithm, which allows a local structural comparison of protein binding sites or complete protein structures without reference to known binding sites (Konc and Janežič, 2017). The ProBiS algorithm compares the local geometry of the binding sites and their physico-chemical properties. The result of each pairwise comparison is the three-dimensional superposition of the compared binding sites and the estimation of the degree of their similarity, expressed as z-score. The ProBiS algorithm also enables the detection of similar binding sites in proteins whose amino acid sequences and global folds differ. Based on this algorithm, we have developed the ProBiS web server (http://probis.cmm.ki.si) and the ProBiS-CHARMMing web server (https://probis.nih.gov), both of which enable the comparison of a protein with the entire PDB (Konc and Janežič, 2017). The result of this computation is a list of potential target proteins for the investigated compound, ranked by the degree of their binding site similarity to the known binding site for this compound.
Identification of potential target proteins of resveratrol: We identified new potential target proteins for resveratrol using the inverse molecular docking and the binding site comparison methods (Kores et al., 2019). One of the target proteins of resveratrol identified by us is lysine-specific histone demethylase 1A (LSD1). This flavoenzyme selectively demethylates lysine in histones and plays an important role in cell growth, differentiation, cancer and neuronal diseases. Recent results underline the role of LSD1 as a central regulator of the autophagy pathway, which supports cellular homeostasis and is a protective mechanism against neurodegenerative diseases (Ambrosio and Majello, 2018). The inhibition of LSD1 has already been investigated as an approach for the treatment of neurodegenerative diseases and resveratrol has been shown to inhibit this enzyme in vitro (Abdulla et al., 2013). Further, we found that resveratrol potentially binds to mitofusin-1, which is expressed in hypothalamic proopiomelanocortin neurons that are important for regulating glucose sensing (Ramírez et al., 2017). We also predicted resveratrol binding to dynamin-1, an enzyme with GTP-ase activity expressed in the brain and involved as a microtubule-associated protein in vesicular transport processes such as endocytosis. Dynamin-1 has been linked to neurodegenerative diseases and inhibitors of this enzyme could find various applications in neurological and other diseases. The expression of this protein is known to be down-regulated by resveratrol (Navarro et al., 2017), but a direct interaction with resveratrol was previously not known.
Identification of potential target proteins of curcumin: In another study, we predicted target proteins for curcumin (Furlan et al., 2018). It has been recognized that a curcumin diet improves the cognitive function of patients with neurodegenerative diseases. Among the various reported effects of curcumin are reduced formation of β-amyloid plaques, inhibition of neuron degradation, anti-inflammatory and antioxidant effects, and reduced formation of microglia (Mishra and Palanivelu, 2008). However, it remains largely unexplored which target proteins are responsible for the observed effects of curcumin. Using the inverse molecular docking method, we have identified the binding potential of curcumin to cAMP-specific 3′,5′-cyclic phosphodiesterase 4D (PDE4D), which belongs to the family of cyclic nucleotide phosphodiesterase 4. This enzyme is mainly distributed in inflammatory and immune cells and is, among other things, a potential target for the treatment of cognitive impairment in dementia and the inhibition of neuroinflammatory and apoptotic processes in Alzheimer’s disease. Inhibition of the enzyme PDE4D leads to an increase in intracellular cyclic adenosine-3′,5′ monophosphate and cyclic guanosine-3′,5′ monophosphate concentrations, which may improve cognitive abilities and lead to inhibition of neuroinflammatory and apoptotic pathways. Our finding that curcumin binds to PDE4D is consistent with a study that attributes the cancer-preventive and therapeutic effects of curcumin to the inhibition of PDE4D (Abusnina et al., 2015). The predicted structure of the curcumin complex with PDE4D allows the determination of interactions with specific amino acid residues at the active site of this enzyme. In contrast to the previous study (Abusnina et al., 2015), our results provide a detailed mechanistic view of the curcumin binding to PDE4D. In addition, we have detected the potential binding of curcumin to the mitochondrial enzyme 17β-hydroxysteroid dehydrogenase type 10 (17β-HSD10). This enzyme is found in various brain regions and is essential for the maintenance of neurosteroid homeostasis. It interacts with amyloid-β and is associated with neuronal dysfunction in Alzheimer’s disease. The brain of people with Alzheimer’s disease and the mouse model of this disease show abnormally elevated values of 17β-HSD10 (Yang et al., 2014), which lead to a worsening of the structure, function and dynamics of mitochondria and can be the basis for the pathogenesis of synaptic and neuronal defects characteristic of Alzheimer’s disease. The positioning of 17β-HSD10 as a potential target of curcumin may open a new therapeutic approach for the treatment of this disease and is a novel finding as a link between curcumin and this enzyme has not yet been established.
Discussion and future perspectives: Inverse molecular docking and binding site comparison are promising new methods to identify potential target proteins for natural compounds. We found that both resveratrol and curcumin potentially bind to proteins that play an important role in cognitive disorders associated with Alzheimer’s disease. The main value of the computational methods presented is that they enable the identification of new target proteins of therapeutically interesting natural products, which should reduce the number of experiments needed to determine their molecular mechanisms of action.
This work was supported by the Slovenian Research Agency Research Project L7-8269 (to JK).
Footnotes
Copyright license agreement: The Copyright License Agreement has been signed by the author before publication.
Plagiarism check: Checked twice by iThenticate.
Peer review: Externally peer reviewed.
Open peer reviewer: T. Bucky Jones, Midwestern University, USA.
P-Reviewer: Jones TB; C-Editors: Zhao M, Li JY; T-Editor: Jia Y
References
- 1.Abdulla A, Zhao X, Yang F. Natural polyphenols inhibit lysine-specific demethylase-1 in vitro. J Biochem Pharmacol Res. 2013;1:56–63. [PMC free article] [PubMed] [Google Scholar]
- 2.Abusnina A, Keravis T, Zhou Q, Justiniano H, Lobstein A, Lugnier C. Tumour growth inhibition and anti-angiogenic effects using curcumin correspond to combined PDE2 and PDE4 inhibition. Thromb Haemost. 2015;114:319–328. doi: 10.1160/TH14-05-0454. [DOI] [PubMed] [Google Scholar]
- 3.Ambrosio S, Majello B. Targeting histone demethylase LSD1/KDM1a in neurodegenerative diseases. J Exp Neurosci. 2018;12:1–3. doi: 10.1177/1179069518765743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baran PS. Natural product total synthesis: as exciting as ever and here to stay. J Am Chem Soc. 2018;140:4751–4755. doi: 10.1021/jacs.8b02266. [DOI] [PubMed] [Google Scholar]
- 5.Furlan V, Konc J, Bren U. Inverse molecular docking as a novel approach to study anticarcinogenic and anti-neuroinflammatory effects of curcumin. Molecules. 2018;23:3351. doi: 10.3390/molecules23123351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Konc J. Binding site comparisons for target-centered drug discovery. Expert Opin Drug Discovery. 2019;14:445–454. doi: 10.1080/17460441.2019.1588883. [DOI] [PubMed] [Google Scholar]
- 7.Konc J, Janežič D. ProBiS tools (algorithm, database, and web servers) for predicting and modeling of biologically interesting proteins. Prog Biophys Mol Biol. 2017;128:24–32. doi: 10.1016/j.pbiomolbio.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 8.Kores K, Lešnik S, Bren U, Janežič D, Konc J. Discovery of novel potential human targets of resveratrol by inverse molecular docking. J Chem Inf Model. 2019;59:2467–2478. doi: 10.1021/acs.jcim.8b00981. [DOI] [PubMed] [Google Scholar]
- 9.Mishra S, Palanivelu K. The effect of curcumin (turmeric) on Alzheimer’s disease: An overview. Ann Indian Acad Neurol. 2008;11:13–19. doi: 10.4103/0972-2327.40220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Navarro G, Martínez-Pinilla E, Sánchez-Melgar A, Ortiz R, Noé V, Martín M, Ciudad C, Franco R. A genomics approach identifies selective effects of trans-resveratrol in cerebral cortex neuron and glia gene expression. PLoS One. 2017;12:e0176067. doi: 10.1371/journal.pone.0176067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramírez S, Gómez-Valadés AG, Schneeberger M, Varela L, Haddad-Tóvolli R, Altirriba J, Noguera E, Drougard A, Flores-Martínez A, Imbernón M, Chivite I, Pozo M, Vidal-Itriago A, Garcia A, Cervantes S, Gasa R, Nogueiras R, Gama-Pérez P, Garcia-Roves PM, Cano DA, et al. Mitochondrial dynamics mediated by mitofusin 1 is required for POMC neuron glucose-sensing and insulin release control. Cell Metab. 2017;25:1390–1399. doi: 10.1016/j.cmet.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 12.Yang SY, He XY, Isaacs C, Dobkin C, Miller D, Philipp M. Roles of 17β-hydroxysteroid dehydrogenase type 10 in neurodegenerative disorders. J Steroid Biochem Mol Biol. 2014;143:460–472. doi: 10.1016/j.jsbmb.2014.07.001. [DOI] [PubMed] [Google Scholar]