Abstract
The ability to introduce targeted edits in the genome of model organisms is revolutionizing the field of genetics. State-of-the-art methods for precision genome editing use RNA-guided endonucleases to create double-strand breaks (DSBs) and DNA templates containing the edits to repair the DSBs. Following this strategy, we have developed a protocol to create precise edits in the C. elegans genome. The protocol takes advantage of two innovations to improve editing efficiency: direct injection of CRISPR-Cas9 ribonucleo-protein complexes and use of linear DNAs with short homology arms as repair templates. The protocol requires no cloning or selection, and can be used to generate base and gene-size edits in just 4 days. Point mutations, insertions, deletions and gene replacements can all be created using the same experimental pipeline.
Keywords: Genome editing, CRISPR, C. elegans, Cas9 RNP, Linear repair templates, Short homology, Gene conversion, Homology-dependent repair (HDR)
1. Introduction
The ability to introduce precise changes in genomic DNA (“precision genome editing”) is revolutionizing our ability to decode the genome [1]. Current methods for precision genome editing in eukaryotic cells rely on endogenous DNA repair pathways to introduce custom-designed alterations in the genome [2]. First, a double-strand break (DSB) is generated in or near the target sequence. This first step has been greatly facilitated by the discovery of RNA-guided endonucleases, such as CRISPR-Cas9, that can be programmed with a short RNA (“guide RNA”) to cleave a specific DNA sequence [3]. In a second step, the DSB is repaired using the cell’s endogenous homology-dependent repair (HDR) mechanisms and a user-provided DNA template, which contains the desired edit flanked by sequences homologous to the target (“homology arms”). HDR, however, can be an inefficient process especially for large (GFP-sized) knock-in edits, which require long homology arms and/or selection markers to isolate the rare recombinants. We have found that, in C. elegans, HDR efficiency is enhanced when using linear DNAs as repair templates [4,5]. Linear DNAs engage in a gene conversion mechanism that only requires short (~35 nt) stretches of homology. The gene conversion process most likely involves DNA synthesis templated by the linear DNAs and primed by 3′ ends at the DSB [5]. Based on these observations, we developed a simple protocol for editing the C. elegans genome. Unlike other editing protocols available to worm breeders [6–9], our protocol does not require selection or a specific genetic background and uses the same experimental pipeline to create base changes, insertions, deletions and gene replacements. In principle, the protocol may also be adapted for use in other nematode species [10].
1.1. Protocol overview
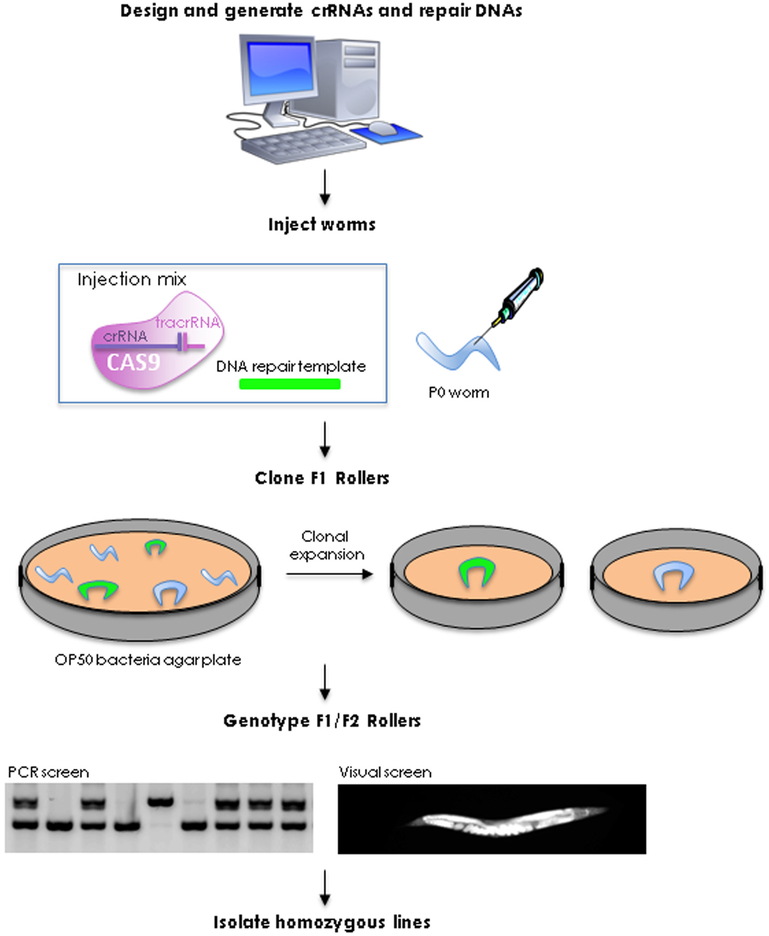
Each editing project requires two custom-made reagents: the guide RNA(s) that will direct the RNA-guided endonuclease (Cas9) to the target sequence, and the repair template(s) containing the edit. Guide RNAs and repair templates are chemically synthesized (by a commercial source) as RNA and DNA single-stranded oligonucleotides, respectively. For large edits such as GFP insertion, the repair template is synthesized as a double-stranded DNA fragment by PCR amplification. On the day of injection, the RNA-guided endonuclease is assembled in vitro using recombinant protein [typically Streptococcus pyogenes Cas9 (spy-Cas9)], the custom-ordered guide RNA (crRNA), and a universal structural RNA (tracrRNA). The endonuclease/RNA complex is injected with the DNA repair templates in the gonad of adult hermaphrodites. To facilitate the identification of successfully injected hermaphrodites, the injection mix also contains reagents (guide RNA and DNA repair oligonucleotide) designed to edit the dpy-10 gene [11]. Edited worms are recovered as early as four days later by PCR screening or visual inspection of broods that segregate dpy-10 edits (“Roller” worms) (Fig. 1).
Fig. 1.
Protocol overview. See Section 1.1 for details.
1.2. Advantages and challenges of the protocol
Two features distinguish our protocol from other editing protocols available for C. elegans: direct injection of the endonuclease/ RNA complexes [12] and linear DNA templates with short homology arms [4]. These innovations provide distinct advantages and challenges. First, because plasmids are not needed to express the endonuclease and guide RNA or for construction of the repair templates, the entire protocol is cloning-free and requires only reagents that can be outsourced (Cas9 protein, crRNAs and oligonucleotide templates) or synthesized in the lab using simple methods (Cas9 protein and PCR templates). Second, because editing frequency is generally high (5–50% of Roller F1 progeny depending on the guide RNA), the protocol does not require selection and therefore does not require a specific genetic background. Editing can be performed on strains that have been edited previously and/or carry other useful mutations or markers. The co-conversion strategy [11] requires a strain background that permits expression of the dpy-10 Roller phenotype, but is only used to enrich for edits and can be omitted or altered if necessary (see Section 3.3 for an alternative co-conversion marker). Third, because edits are identified in the heterozygous state in the first generation after injection, edits that cause dominant sterility or maternal-effect lethality can be recovered [13]. Although these edits cannot be propagated, they can be analyzed directly in the first generation after injection and re-generated when needed for further analyses.
The use of linear templates, however, also imposes certain limitations. First, the repair mechanism engaged by linear DNAs (a gene conversion mechanism related to synthesis-dependent strand annealing [14]) is a local process with conversion tracts that rarely extends beyond 30 nt away from the DSB [4,5,11]. Therefore, linear templates are best suited for edits within 30 nt of an endonuclease recognition site. When using spyCas9, this means that the edit needs to be within 30 nt of a GG dinucleotide, the PAM sequence recognized by Cas9 (Fig. 2). The second limitation is that gene conversion is very sensitive to the molarity of the DNA template [5], possibly because of the low processivity of repair DNA polymerases, which leads to frequent switching from one template to another [15]. High template copy number decreases the chance of aberrant template switching events between non-homologous sequences. For best results, we recommend using repair templates at ~0.5 pmol/μl or higher in the injection mix. This requirement is easy to achieve when using ssODNs or GFP-sized PCR fragments, but is more difficult for edits that require larger templates (>1 kb).
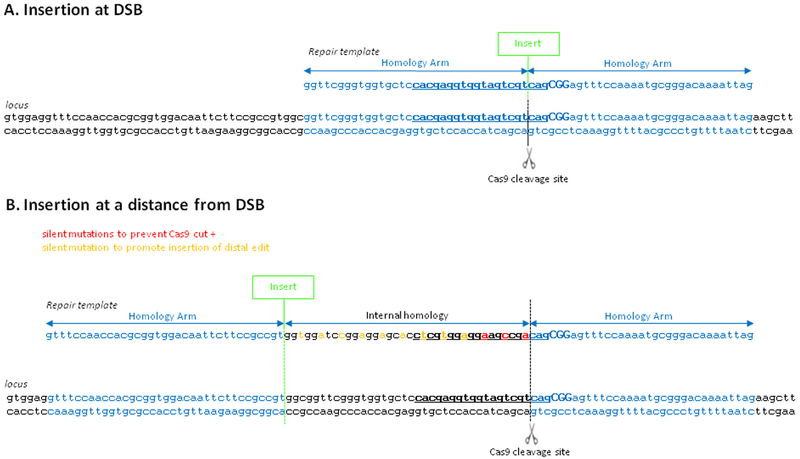
Fig. 2.
Template design. Sequence of locus and repair templates are shown for two insertional edits, one right at the Cas9-induced double-strand break (DSB) (A) and one away from the DSB (B). Homology arms are in blue, inserts in green, crRNA sequence is bolded and underlined, PAM is in capital letters. In (A), the insert disrupts the Cas9 targeting sequence and so no recoding is needed to prevent recutting. In (B), the insert is away from the Cas9 targeting sequence and so silent mutations are required to prevent re-cutting (red) and to prevent premature template switching (orange) during copying of the template. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
For edits with large inserts or that are situated 30 nt or more from a Cas9 targeting sequence, we recommend protocols that use plasmid templates with long homology arms (e.g. [7]). These types of templates appear to participate in a different repair mechanism that is less efficient but generates longer conversion tracts. Plasmid templates can also accommodate selection markers which make them ideal for low-efficiency editing projects or in situations where PCR screening strategies are not possible or desired.
2. Protocol
2.1. Choice of crRNA
The spyCas9 recognition sequence consists of 20 bases complementary to the guide RNA (crRNA) followed by the sequence NGG (PAM sequence) [16]. In many cases, there will be several potential Cas9 targeting sites near the desired edit (be sure to check both strands!). Because HDR efficiency decreases with increasing distance from the DSB, the most important consideration when choosing a crRNA is that the DSB (−3 nt relative to the PAM) be as close as possible to the desired edit and no further than 30 nt away [4,5] (Fig. 2). Another consideration is that not all crRNAs perform with equal efficiency: crRNAs with a G or a GG in the position immediately preceding the PAM and with 50–70% GC content are thought to be most efficient. In contrast, crRNAs with a C preceding the PAM are less efficient [17] [18] [19]. crRNA efficiency is difficult to predict, yet can significantly affect editing efficiency [4]. Finally, it is useful to check whether the crRNA targets other sites in the C. elegans genome using the CRISPR MIT website (http://crispr.mit.edu/). Two or more mismatches in the first 15 nt upstream the PAM are sufficient to prevent Cas9 cleavage reducing the chances of off-target mutations [20]. Also keep in mind that unwanted mutations in predicted off-target loci can be removed after editing by outcrossing.
2.2. Designing the repair template
At a minimum, the repair template should contain the desired edit and two 35 nt homology arms. In cases where the edit is to be inserted right at the DSB, both homology arms will correspond to sequences directly flanking the DSB (Fig. 2). In cases where the edit is inserted at a distance from the DSB, one homology arm will correspond to sequences directly flanking the DSB and the other will flank the edit. The sequence between the edit and the DSB is not considered part of the homology arm. This sequence creates an internal homology on the repair template which can lead to premature switching from the repair template back to the chromo-some without copying the edit [5]. To mitigate this problem and raise the frequency of repair events that introduce the edit, we recommend recoding this sequence, for example by introducing silent mutations that will interrupt the homology (Fig. 2).
The repair template should also contain mutations to prevent re-cutting by Cas9 after editing. In some cases, the edit itself (or the recoding described above) will provide that protection. If not, additional mutations should be incorporated in the template. These mutations can be base changes, insertions/deletions that disrupt the PAM or crRNA sequence (Fig. 2). Avoid changing the NGG PAM to NGA, since Cas9 can also weakly recognize NGA [20]. When disrupting the crRNA sequence, we recommend incorporating the mismatch on the same side of the DSB as the edit: one mismatch within 3 nt upstream of the PAM or 2 or more mismatches 4–20 nt upstream of the PAM. Mutations closest to the PAM are the most effective [20].
Ultimately, successful editing will depend on a robust screening strategy that will allow precise edits to be distinguished from other imprecise repair events. The choice of screening strategy should be considered when designing the repair template. Insertions and deletions greater than 20 nt are easy to identify by gel electrophoresis of PCR fragments, provided the amplified fragment is not too large relative to the edit (<0.5 kb for a ~20 nt edit). Small edits, however, can be more difficult to identify. For those edits, we recommend including a novel restriction enzyme site in the repair template. Webcutter 2.0 can be used to design a restriction site using degenerate nucleotide code (http://rna.lundberg.gu.se/cutter2/). Be sure to place the restriction site distal to the edit relative to the DSB to avoid false positives.
2.3. Editing strategies
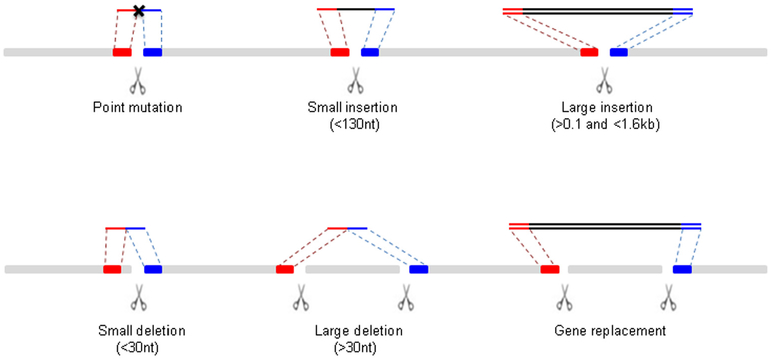
Using different combinations of crRNAs and repair templates, it is possible to generate a wide variety of edits. Here we describe strategies for the most common types of edits (Fig. 3).
Fig. 3.
Editing strategies. Schematic representation of chromosome (grey), DSB (scissors) and DNA repair templates above chromosome (ssODNs: single line or PCR fragments: two lines). Homology arms are highlighted in red (left arm) and blue (right arm). Inserts are in black. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.4. Point mutation
Single base changes can be used to introduce nonsense or mis-sense mutations in a coding sequence or to mutate a splice site or a transcription factor binding site. For those types of edits, we recommend using a single-stranded DNA oligonucleotide (ssODN) template, that includes a new restriction site to facilitate screening. When introducing a stop codon, you can use a NheI site which includes a TAG codon. If the goal is to knock out gene activity, we recommend also adding an extra nucleotide to create a frame shift after the premature stop codon.
2.5. Small insertion (up to 130 bp)
This strategy can be used to introduce small antigenic tags into an ORF. Use one crRNA and one ssODN with two 35 nt homology arms. Currently, custom ssODNs can be ordered up to 200 nt which limits the size of the insert to 130 nt (+2 × 35 nt homology arms). If your insert is larger than 130 nt, consider using two overlapping ssODNs (see Section 2.10).
2.6. Large insertion (0.1–1.6 kb)
This strategy is useful to tag ORFs with GFP or other large tags. Use one crRNA and one PCR fragment with 35 bp homology arms flanking the insertion. To synthesize the PCR fragment, order two custom primers, each containing one homology arm and ~20 nt to amplify the insert.
2.7. Small deletion (up to 30 bp)
Use one crRNA near one end of the deletion and an ssODN repair template containing one homology arm that extends to the DSB and a second homology arm at a distance up to 30 nt from the DSB.
2.8. Large deletion (gene size)
Use two crRNAs located at each extremity of the genomic region to be deleted and an ssODN containing homology arms that flank each DSB. The ssODN can also include a restriction site at the junction to distinguish precise deletions with the desired junction from imprecise deletions.
2.9. Gene replacement
Gene replacement can be performed in one or two experiments depending on the type of replacement. If the goal is to replace one sequence with an unrelated sequence (e.g. replace an ORF with GFP for example), the replacement can be completed in one editing experiment. Use two crRNAs flanking the sequence to be replaced and a PCR fragment containing the insert flanked by two 35 bp homology arms. For this approach to succeed, it is very important that the insert does not contain any sequence homologous to the sequence to replace (outside of the homology arms flanking the insert). If the goal is to replace an ORF with a related sequence, we recommend performing two editing experiments sequentially. First, delete the ORF and insert a novel Cas9 targeting sequence not present in C. elegans genome at the junction (using a ssODN repair template). Next, in a second editing experiment, use the new Cas9 targeting sequence as an insertion site for the modified ORF (using a PCR repair template). This approach can be used to insert multiple mutations at once in an ORF or to recode an entire ORF.
2.10. Recombineering
Overlapping repair templates can be used to overcome the 200 nt size limit of ssODNs or to assemble sequences that cannot be amplified by PCR from a unique DNA template. ssODNs can also be combined with PCR fragments to provide homology arms in trans. In all cases, the overlaps should be 35 nt long. Refer to [5] for the best recombineering strategies. Note that recombineering reduces the frequency of correct edits [5], so this strategy is best used with high efficiency crRNAs. Custom-made synthetic double-stranded DNAs (gBlocks) are another good alternative for knock-in edits that are too large to fit in a standard-size ssODN.
3. Assembling experimental reagents
3.1. Cas9 reagents
The Cas9 RNP consists of Cas9 protein and two RNAs: the custom guide RNA (crRNA) and a universal structural RNA (tracrRNA) which links the crRNA to Cas9 protein (Fig. 1). Cas9 protein can be purchased from a commercial vendor. Alternatively, Cas9 protein can be made in lab using a streamlined bacterial purification protocol [12]. The crRNA and tracrRNA can be purchased as RNA oligonucleotides from IDT or Dharmacon. (IDT and Dharmacon crRNAs and tracrRNA are not cross-compatible, only use RNAs from the same manufacturer). Reconstitute the dried RNAs by spinning down the tubes and adding 5 mM Tris-HCl pH7.5 for a final concentration of 0.6 mM and 0.17 mM for crRNA and tracrRNA respectively, and mix by pipetting. We recommend using sterile solutions (filtered or autoclaved). DEPC treatment is not recommended.
3.2. Repair templates
3.2.1. ssODNs
ssODNs can be purchased from a variety of commercial vendors. Design ssODN with the requisite homology arms (~35 nt) and desired edit (<130 nt) (Figs. 2 and 3). If necessary, also include mutations to prevent recutting by Cas9 after editing (Fig. 2). Spin the ssODNs and reconstitute in water to 25 μM. Please note that salt free purification of ssODNs is sufficient to achieve efficient genome editing but PAGE purification and modified ssODNs have been reported to improve editing efficiency in C. elegans [21] and mammalians cells [22].
3.2.2. PCR templates
Design primers with the desired homology arms (~35 nt) and sequences (18–20 nt) to amplify the insert. If necessary, also include mutations to prevent recutting by Cas9 after editing. Where possible, limit the size of the primers to less than 65 nt to avoid primer contamination after PCR purification.
-
To generate the PCR repair template, use the following reaction mix: Mix 0.8 μl of template plasmid (prepared using standard miniprep procedure), 2 μl of forward and reverse primers (100 μM stock), 195.2 μl of H2O, 200 μl of Phusion Master Mix 2X (NEB, #M0531L). Split the mix equally into 8 PCR tubes and perform the PCR reaction as follow. (We use 8 PCR reactions to ensure sufficient yield and to avoid propagation of PCR errors).
98 °C for 2 min
98 °C for 30 s, 61.5 °C for 30 s, 72 °C for variable time depending on amplicon size (45 s is sufficient for ~0.7–1 kb inserts).
Repeat 29 times
72 °C for 10 min
Hold at 4 °C
Most PCR reactions perform well with an annealing temperature of 61.5 °C. However if the PCR reaction yield is low or contaminated with other bands, we recommend performing a gradient PCR with annealing temperatures ranging from 60 to 72 °C.
Do a second (nested) PCR step if the PCR primers are >65 nt to remove unincorporated PCR primers and improve yield: use forward and reverse primers (18–20 nt) that correspond to the extreme ends of the template made in the first PCR. Run the second PCR as before using 0.8 μl of the 1st round purified PCR amplicon.
Pool the PCRs (up to 8) in one tube and purify them in one Qiagen minelute column (#28006), elution with 10 ml of H2O. The expected yield ranges from 1 to 1.5 μg/ml for fragments <1 kb.
3.3. Co-conversion reagents
In order to identify the broods with the highest HDR edit frequencies, we use the co-conversion method [11]. Arribere et al., 2014 have found that worms edited at one locus have a higher probability to be edited at a second locus. The most straightforward co-conversion method uses a crRNA and repair ssODN designed to introduce a missense mutation at the dpy-10 locus. This mutation causes a dominant Roller phenotype that is easily spotted among the adult progeny of injected hermaphrodites. Worms that are edited at the dpy-10 locus have a higher probability to also be edited at the locus of interest. This is also true (although to a lesser extent) for worms that themselves are not edited at the dpy-10 locus but are siblings of dpy-10 edited worms [11].
dpy-10 crRNA: GCUACCAUAGGCACCACGAG
dpy-10 ssODN: CACTTGAACTTCAATACGGCAAGATGAGAATGACT GGAAACCGTACCGCATGCGGTGCCTATGGTAGCGGAGCTTCACAT GGCTTCAGACCAACAGCCTAT
If it is not possible or desirable to modify dpy-10, an alternative approach is to tag the gtbp-1 gene with GFP (or another fluorescent protein) [12]. gtbp-1::GFP worms express a strong green fluorescent signal that is easy to screen using a standard fluorescent microscope.
gtbp-1 crRNA: CCACGAGGUGGUAUGCGCAG
gtbp-1 eGFP repair template: amplify by PCR using the plasmid pAP682 and the following primers (5′–3′, lower case indicating the homology arm sequence)
Forward primer: ggttcgggtggtgctccacgaggtggtatgcgcGTGAGTAA AGGAGAAGAAC
Reverse primer: cttctaattttgtcccgcattttggaaaccgctCTTGTACAG CTCGTCCATGCC
gtbp-1 mCherry repair template: amplify by PCR using the plasmid pAP582 and the following primers (5′–3′, lower case indicating the homology arm sequence)
Forward primer: ggttcgggtggtg ctccacgaggtggtatgcgcGTCTCAA AGGGTGAAGAAGATAAC
Reverse primer: cttctaattttgtcccgcattttggaaaccgctCTTATACAATT CATCCATGCC
4. Experimental protocol for editing the C. elegans genome
4.1. Assembling the injection mix
-
1To Cas9 protein (5 μl, 10 μg/μl, 20 mM HEPES pH7.5, 500 mM KCl, 20% Glycerol), add in the following order (after each addition mix by pipetting)
- KCl (1 M): 0.5 μl
- HEPES pH 7.4 (200 mM): 0.75 μl
- tracrRNA (0.17 mM): 5 μl
- dpy-10 crRNA (0.6 mM): 0.4 μl
- Targeted gene crRNA (0.6 mM): 1 μl
- dpy-10 ssODN (16 μM): 0.55 μl
- Repair template. For ssODN, use 0.4 μl of 25 μM ssODN stock (10 pmol total). For PCR repair template, use 10 pmol total (determine the DNA quantity needed using this equivalence: 1 μg of 1 bp dsDNA = 1624 pmol).
- H2O: to reach a final volume of injection mix of 20 μl
Carefully monitor the injection mix when adding each component. If the mix shows sign of precipitation (whitening), start again using a 3 M KCl solution instead of a 1 M solution.
For large deletion or gene replacement strategies that use two DSBs, use 0.75 ml of crRNA for the first DSB and 0.75 μl of crRNA for the second DSB and 6.7 μl of tracrRNA.
To edit two loci at once (in addition to dpy-10), use 0.75 μl of crRNA to target the first locus and 0.75 μl of crRNA to target the second locus, and 6.7 μl of tracrRNA. Use 10 pmol of repair template for each locus.
For edits linked to the dpy-10 on LGII, we recommend using only 0.28 μl of dpy-10 crRNA to maximize the number of edits among non-Roller worms.
-
2
Incubate the injection mix at 37 °C for 10–15 min. Immediately load the injection needles and proceed with the injections.
4.2. Injections, worm recovery and handling
We recommend “Transformation and microinjection” by Tom Evans (http://www.wormbook.org/chapters/www_transformationmicroinjection/transformationmicroinjection.html) for a detailed protocol of C. elegans microinjection. We outline here only basic steps.
-
Load the injection needle with the injection mix.
Prepare loading and injection needles a few minutes ahead of loading in order to let them cool down before use. Use a loading needle with a wide opening to allow the injection mix to flow freely into the injection needle by capillary action to avoid any bubbles.
-
Inject both arms of young adult hermaphrodites
Use young adult hermaphrodites with a single row of embryos in their uteri. We recommend using synchronized worms:~3 days before injection, plate ~8000 embryos at 20 °C on large NEP plates seeded with NA22 bacteria. Use plates that are not fully covered with bacteria to ensure that some worms will be off the bacteria and thus easier to pick for injection. To ensure young adults of the correct stage on injection day, prepare several plates and incubate them at different temperatures.
After injection, recover the worms in recovery buffer (5 mM HEPES pH 7.2, 3 mM CaCl2, 3 mM MgCl2, 66 mM NaCl, 2.4 mM KCl, 4% Glucose(w/v). After 30 min, add every 5–10 min, 5 μl/5 μl/10 μl/10 μl/20 μl/20 μl/40 μl of 1X M9 (420 mM Na2HPO4, 220 mM KH2PO4, 860 mM NaCl, 10 mM MgSO4).
Clone out the injected hermaphrodites (P0s) onto small NGM plates (1 P0 per plate, at 20 °C). Be sure not to transfer any mineral oil from the injection pad onto the plates with the worms. Use fresh NGM plates with a thin layer of OP50 bacteria at the center to facilitate screening for Rollers. After 20–24 h, transfers the P0s to a second plate (again one P0 per plate) and let the P0s continue laying embryos.
4–7 days after cloning the P0s, examine their F1 broods for worms exhibiting the Roller phenotype. You should expect ~30–70% of P0s to segregate Roller worms. Determine the number of Roller F1s per P0 and select the three P0s broods that give the most Rollers. These are your “jackpot broods”. Clone out the F1 Roller worms from jackpot broods onto small NGM plates (one F1 Roller per plate, at 20 °C). We recommend cloning ~50 Roller F1 worms.
You can also clone non-Roller F1s (from jackpot broods) if your edit is linked to dpy-10 (LGII) and you wish to recover the edit in a non-Roller background.
4.3. Screening for edits
The F1 Roller progenies (or F2s) can be screened for the presence of the desired HDR edits in two ways: 1) PCR amplification of the locus or 2) visual screen using microscopy.
4.3.1. PCR screening
PCR screening can be performed on single F1s or, more easily, on their progeny (F2 cohorts).
Worm lysis: Place 20 μL of worm lysis buffer (10 mM Tris pH 8.3, 50 mM KCl, 2.5 mM MgCl2, 0.45% NP-40, 0.45% Tween-20, 0.1 μg/μL Proteinase K) into individual PCR tubes. Place one F1 worm, or 10–15 F2 worms, into the lysis buffer. Lysis worms at 60 °C for 1 h, followed by 95 °C for 15 min and store at −20 °C.
To genotype the targeted locus, design forward and reverse primers approximately 250 bp upstream and downstream the edited site. Use “NCBI/Primer-BLAST” website for help in selecting primers (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). It is critical to test the genotyping primers on wild-type lysates using a gradient of annealing temperature to determine optimal PCR conditions. If there are many non-specific bands in the reaction, design new genotyping primers. We recommend Phusion taq polymerase for genotyping.
Genotyping PCR reaction mixture: Make a PCR master mix. For each 25 μl PCR reaction, use 0.125 μl of forward and reverse primers (100 μM stock), 10.25 μl of H2O and 12.5 μl of Phusion 2X mix. Distribute the master mix into PCR strip tubes and add 2 μl of worm lysis.
Analyze PCR products by agarose gel electrophoresis by either size shift (edits 20 nt or greater) or restriction enzyme digestion (edits <20 nt).
Screening by size shift: use a 2.5% agarose gel for edits between 20 and 200 bp and a 1.25% agarose gel for edits > to 200 bp.
Screening by Restriction Enzyme (RE): Use 5 μl of the PCR product and add 1 μl of the desired RE (approximately 20 units), 1 μl of appropriate 10X buffer and 3 μl of H2O. Allow the digestion to proceed overnight. Analyse the digests on a 2.5% gel. Most restriction enzymes work well in the Phusion taq PCR reaction, but we recommend testing enzyme efficiency before use for genotyping.
4.3.2. Visual examination of fluorescent protein insertion
If the desired edit will result in a known expression pattern, you can visually screen cloned F1 Rollers (after allowing them to lay eggs overnight) by placing them in an M9 drop containing levami-sole (1 mM) (Sigma-Aldrich, L9756) on a 3 or 12-well microscope slide (with coverslip) using the 10X objective of a fluorescent microscope.
If the fluorescence signal is strong enough, you can also omit the coverslip and recover the fluorescent F1s worms after examination. Wash the positive worms in a drop of M9 without levami-sole before transferring them on a OP50 plate.
4.3.3. Strain establishment
Identify homozygous edited animals in the F2 or later generations by genotyping. Only clone non-Roller worms if you want to isolate your edit in a dpy-10(+) background. For fluorescent protein edits, examine the segregation of the fluorescent signal in F3 worms derived from singled-out F2s to identify homozygous lines. Once homozygous lines are established, verify the edits by sequencing. Use sequencing primers that anneal inside the geno-typing PCR product. Sequence the entire sequence present in the rescue template and at least 30 nt upstream and downstream. Note that, even when in frame, tagged genes may not be fully functional – check the homozygous edited lines for brood size and viability at different temperatures. When using guide RNAs with potential off-target sites, check the integrity of the off-target loci by sequencing and outcross any undesired mutations. It is good practice to generate at least two independent lines for each edit (derived from a different F1).
5. Suggestion for antigenic tags, fluorescent proteins, Cas9 targeting sequence
5.1. Peptide tags
We have successfully used Ollas and 3xFlag tags to label germline-expressed genes
Ollas sequence: tccggattcgccaacgagctcggaccacgtctcatgggaaag (contains SacI site). The Ollas tag can be detected using the anti-Ollas antibody (Novus Biologicals, NBP1–06713) at 1/200 dilution for Immuno-Fluorescence and 1/1000 for Western-Blot.
3xFlag sequence: gactacaaagaccatgacggtgattataaagatcatgatatcgattacaaggatgacgatgacaag (contains EcoRV and ClaI sites). The 3xFlag tag can be detected using the anti-Flag antibody (F1804, Sigma-Aldrich) at 1/500 dilution for Immuno-Fluorescence and the anti-Flag HRP conjugate antibody at a 1/1000 dilution for Western-Blot (2044-S, Cell Signaling Technology).
5.2. Fluorescent proteins
Many fluorescent proteins are available for microscopy or for more specific applications such as as FRET. Refer to [23] for a systematic comparison of the performance of different fluorescent tags in C. elegans. We have generated a set of plasmids coding for fluorescent proteins optimized for C. elegans expression (available through Addgene) (Table S1, File S1).
5.3. Cas9 targeting sequence
For some multi-step editing projects (such as gene replacements), it is useful to include a new Cas9 targeting sequence in the repair template for use in a second editing step. We successfully used this Cas9 targeting sequence (PAM in bold): GGGAAGTTTGTCCAGAGCAG(AGG).
6. Trouble shooting
Precipitate forms during preparation of injection mix. Solution: Increase the concentration of KCl in injection mixture.
-
Injection needle clogs during injection
Solution: Spin the injection mixture for 10 min at 13,000 rpm.
-
No F1 Rollers
Solution: Increase the amount of dpy-10 crRNA in injection mixture. Careful, however, using too much dpy-10 reagents will result in a higher proportion of Roller worms that do not contain the edit at the locus of interest.
-
Genotyping PCR gives extra bands
Solution: Optimize PCR with different polymerase and vary oligo annealing temperatures. If problems continue to persist consider designing new screening PCR primers.
- No desired edit or low editing efficiency
- crRNA doesn’t cut efficiently. Design new crRNA. You can test crRNA activity in vitro [16] but we do not yet know how in vitro and in vivo activities compare. One way to determine crRNA efficiency in vivo is to use the crRNA in combination with an ssODN repair template designed to insert a restriction site right at the DSB. Insertion of a restriction site is efficient and easy to screen for.
- Edit is too far from Cas9 cleavage site. Try a Cas9 targeting sequence closer to the edit.
- Not enough template. Make sure template was present at high enough molarity (0.5 pmol/μL) in injection mix.
- Screening method cannot identify edit. When screening for an insertion, make sure that the size difference is visible on gel, and that the smaller wild-type product does not out-compete synthesis of the larger insertion product.
Supplementary Material
Acknowledgments
We thank members of the Seydoux lab for their contributions during the development of this protocol.
Funding
This work was supported by National Institutes of Health (NIH) [Grant No. R01HD37047]. G.S. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.ymeth.2017.03.023.
References
- [1].Doudna JA, Charpentier E, Genome editing. The new frontier of genome engineering with CRISPR-Cas9, Science 346 (6213) (2014) 1258096. [DOI] [PubMed] [Google Scholar]
- [2].Jasin M, Haber JE, The democratization of gene editing: Insights from site-specific cleavage and double-strand break repair, DNA Repair 44 (2016) 6–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Jiang W, Marraffini LA, CRISPR-Cas: new tools for genetic manipulations from bacterial immunity systems, Annu. Rev. Microbiol 69 (2015) 209–228. [DOI] [PubMed] [Google Scholar]
- [4].Paix A, Wang Y, Smith HE, Lee CY, Calidas D, Lu T, Smith J, Schmidt H, Krause MW, Seydoux G, Scalable and versatile genome editing using linear DNAs with microhomology to Cas9 Sites in Caenorhabditis elegans, Genetics 198 (4) (2014) 1347–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Paix A, Schmidt H, Seydoux G, Cas9-assisted recombineering in C. elegans: genome editing using in vivo assembly of linear DNAs, Nucleic Acids Res (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Xu S, The application of CRISPR-Cas9 genome editing in Caenorhabditis elegans, J. Genet. Genomics 42 (8) (2015) 413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Dickinson DJ, Goldstein B, CRISPR-based methods for Caenorhabditis elegans genome engineering, Genetics 202 (3) (2016) 885–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kim HM, Colaiacovo MP, CRISPR-Cas9-guided genome engineering in C. elegans, Curr. Protoc. Mol. Biol 115 (2016) 3171–31718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Schwartz ML, Jorgensen EM, SapTrap, a toolkit for high-throughput CRISPR/ Cas9 gene modification in Caenorhabditis elegans, Genetics (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lo TW, Pickle CS, Lin S, Ralston EJ, Gurling M, Schartner CM, Bian Q, Doudna JA, Meyer BJ, Precise and heritable genome editing in evolutionarily diverse nematodes using TALENs and CRISPR/Cas9 to engineer insertions and deletions, Genetics 195 (2) (2013) 331–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Arribere JA, Bell RT, Fu BX, Artiles KL, Hartman PS, Fire AZ, Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans, Genetics 198 (3) (2014) 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Paix A, Folkmann A, Rasoloson D, Seydoux G, High efficiency, homology-directed genome editing in Caenorhabditis elegans using CRISPR-Cas9 ribonucleoprotein complexes, Genetics 201 (1) (2015) 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Smith J, Calidas D, Schmidt H, Lu T, Rasoloson D, Seydoux G, Spatial patterning of P granules by RNA-induced phase separation of the intrinsically-disordered protein MEG-3, eLife 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Morrical SW, DNA-pairing and annealing processes in homologous recombination and homology-directed repair, Cold Spring Harb. Perspect. Biol 7 (2) (2015) a016444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].McVey M, Khodaverdian VY, Meyer D, Cerqueira PG, Heyer WD, Eukaryotic DNA polymerases in homologous recombination, Annu. Rev. Genet 50 (2016) 393–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E, A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity, Science 337 (6096) (2012) 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, Root DE, Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation, Nat. Biotechnol 32 (12) (2014) 1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gagnon JA, Valen E, Thyme SB, Huang P, Akhmetova L, Pauli A, Montague TG, Zimmerman S, Richter C, Schier AF, Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs, PLoS One 9 (5) (2014) e98186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Farboud B, Meyer BJ, Dramatic enhancement of genome editing by CRISPR/ Cas9 through improved guide RNA design, Genetics 199 (4) (2015) 959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F, DNA targeting specificity of RNA-guided Cas9 nucleases, Nat. Biotechnol 31 (9) (2013) 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ward JD, Rapid and precise engineering of the Caenorhabditis elegans genome with lethal mutation co-conversion and inactivation of NHEJ repair, Genetics 199 (2) (2015) 363–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Renaud JB, Boix C, Charpentier M, De Cian A, Cochennec J, Duvernois-Berthet E, Perrouault L, Tesson L, Edouard J, Thinard R, Cherifi Y, Menoret S, Fontaniere S, de Croze N, Fraichard A, Sohm F, Anegon I, Concordet JP, Giovannangeli C, Improved genome editing efficiency and flexibility using modified oligonucleotides with TALEN and CRISPR-Cas9 nucleases, Cell Rep 14 (9) (2016) 2263–2272. [DOI] [PubMed] [Google Scholar]
- [23].Heppert JK, Dickinson DJ, Pani AM, Higgins CD, Steward A, Ahringer J, Kuhn JR, Goldstein B, Comparative assessment of fluorescent proteins for in vivo imaging in an animal model system, Mol. Biol. Cell 27 (22) (2016) 3385–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.