Abstract
Background
Gut bacterial diversity is decreased in a proportion of patients with septic shock. We attempted to validate the hypothesis that low bacterial diversity increases the risk of mortality.
Material/Methods
All patients with septic shock seen at 2 medical center from 2016 through 2019 were included in this cohort study. Total DNA was isolated from stool, and high-throughput sequencing was performed. Clinical data were extracted from patient medical records and hospital databases. Patients were grouped by gut microbiota bacterial diversity (measured by Shannon diversity index) on presentation. We used logistic regression analysis to evaluate the risk of 28-day mortality in septic patients with low Shannon diversity index.
Results
Of the 150 patients enrolled in this study, low bacterial diversity (Shannon index <3.0) was found in 80 patients and normal diversity (Shannon index ≥3.0) was found in 70 patients. Low diversity was associated with a higher unadjusted mortality risk, compared to those with normal diversity (odds ratio [OR] 2.04, 95% confidence interval [CI] 1.35–2.83). However, this result became non-significant after adjusting the confounding factors such as age, sex, severity of disease, comorbid status, usage of probiotics, enteral nutrition, and antimicrobial drugs (OR 1.93, 95% CI 0.55–2.69).
Conclusions
Our study does not support that low gut bacterial diversity is an independent risk factor for mortality in intensive care unit patients with septic shock.
MeSH Keywords: Critical Illness; RNA, Ribosomal, 16S; Sepsis
Background
Sepsis is a life-threatening organ dysfunction caused by infection. Sepsis is a serious public health problem with startlingly high morbidity and mortality rates. Septic shock is defined as a type of sepsis with circulatory and cellular metabolic disorders, which is severe enough to increase mortality [1]. Septic shock is common in patients admitted to the intensive care unit (ICU). Development of septic shock has been associated with short-term and long-term morbidity, prolonged hospitalization, and high healthcare costs which can be as high as $4651 per day in an ICU in the United States [2]. Given the clinical consequences of septic shock, identifying the risk factors at its early stage is of great importance and a priority in ICU [2].
A break in intestinal microbiota balance can induce or aggravate many types of diseases, such as diabetes mellitus, adiposis, and atherosclerosis [3–5]. In our previous study [6], compared with healthy volunteers, bacterial diversity was found to be significantly reduced in patients with septic shock. A relatively higher mortality was found in low bacterial diversity patients, but it was not statistically significant. This result might have been due to the limited sample size (only 15 patients). Reduced intestinal diversity can lead to many pathogenic conditions, including Clostridium difficile infection, ulcerative colitis, and adiposis [4]. Taur et al. [7] collected fecal specimens from 80 recipients of allogeneic hematopoietic stem cell transplantation at the time of stem cell engraftment. The 3-year overall survival rates in the low, medium, and high diversity groups were 36%, 60%, and 67%, respectively. Low diversity was found to have statistically significant effect on mortality after multivariate regression analysis.
However, only a limited number of clinical studies have investigated the microbiome of ICU patients. Additionally, the available studies did not focus on specific diseases [8,9]. Therefore, the effect of gut bacterial diversity on septic shock patients remains unknown. Exploring the variation of microbiome possible shows a new perspective for the prevention and treatment of septic shock in the future. Herein, based on previous studies, we hypothesized that septic shock patients have low microbial diversity and a higher mortality rate.
Material and Methods
Patients
Our study was a prospective, cohort study on ICU patients with septic shock. The study was conducted in accordance with the protocol and statistical analysis plan, which were authorized by the ethics committee. According to local law, written informed consent was obtained from the patients or their proxies before study inclusion. This study was registered, and the identification number was 17013870 (chictr.org).
The study was conducted in 2 Chinese teaching hospitals from May 2016 to January 2019. Consecutive adults were enrolled who fulfilled the following conditions: infection or suspected infection patients with sequential organ failure assessment (SOFA) score of 2 points or more, which indicates sepsis [1]. In addition, sepsis patients who showed clinically persistent hypotension, and vasoconstrictors were needed after adequate volume resuscitation to maintain mean arterial pressure ≥65 mmHg (1 mmHg=0.133 kPa) and serum lactate concentration >2 mmol/L. The exclusion criteria were: patients <18 years or with a length of stay <2 days, and patients transferred from another acute care facility, whose treatments prior to admission or vital status on discharge could not be ascertained. In addition, if a patient was admitted more than once during the study period, only the first hospitalization details were used.
Study protocol
A trained data extractor checked the ICU patient database to identify patients who have been diagnosed with septic shock within the past 24 hours. Septic shock diagnosis was further verified by a senior physician in our team. Patients that met the inclusion but not the exclusion criteria were enrolled in the study. Baseline data were acquired from patient medical records, including age, gender, admission source, and comorbidities such as diabetes, end-stage renal disease, and disease severity such as SOFA Score, and acute physiology and chronic health evaluation (APACHE) II score. We used SOFA score [10] and Charlson comorbidity index score [11] to assess the disease severity and comorbid conditions, respectively. The study endpoint was 28-day mortality. This cutoff value was selected to avoid a significant increase in the likelihood of mortality associated with factors other than septic shock over time [12]. Microbiota diversity was quantified using Shannon index, which is a popular diversity index in the ecological field. It reflects the richness of bacterial community. We referred to our previous study [6] as well as the cutoff values of other studies [7,8]. The cutoff value for low and normal diversity in this study was set as Shannon index=3.0. The low bacterial diversity group comprised patients with Shannon index <3.0, while normal bacterial diversity group comprised patients with Shannon index ≥3.0.
According to one study [8] and our previous study [6], the ratio of low to normal bacterial diversity in septic patient population was 1: 1. Assuming the 28-day mortality rate of normal bacterial diversity patients was 30% [13], we needed 122 patients (61 in each group) to obtain 80% power for detecting 25% relative increase in the 28-day mortality rate with 1-side alpha risk of 0.05. Since non-randomized studies overestimate the magnitude of the effect, we included 150 patients.
Intestinal microbiota analysis
Fresh feces specimen collection was conducted at the day before antimicrobial drug use, and transferred to liquid nitrogen container immediately. We used enema operation for patients without spontaneous defecation to collect samples. The 16S rRNA gene sequencing of sample DNA was carried out according to the method of Lankelma et al. [8]. Optimal bacterial DNA isolation method using bead-beating technique [14]. Briefly, tissue samples were incubated in lysis buffer and proteinase K at 56°C, before being placed in the DNA purification machine. The resulting DNA was then used in preparing clone libraries and for performing quantitative polymerase chain reaction (PCR). PCR targeting the 16S rRNA-encoding gene was performed as previously described by Young et al. [15]. Illumina libraries were prepared using amplicons according to the manufacturer’s instructions. Primer pair sequences for the V3 and V4 region were used according to the protocol of Illumina Miseq system for 16S metagenomics sequencing library preparation. Gene fragments (V3–V4 region) were sequenced by MiSeq (Illumina, San Diego, CA, USA).
Statistical analysis
Univariate analysis was performed using the Fisher’s exact test for dichotomous variables, and the Student’s t-test and Wilcoxon rank sum test were used for analyzing normal and non-normal continuous variables, respectively. We conducted several logistic regression analyses to examine the association between microbial diversity and 28-day mortality. The adjusted factors in the first model were sex and age. Age was stratified at 65 years point. Sex, age, and severity of illness (SOFA score) were adjusted in the second model. The comorbid illness (Charlson comorbidity index) was added in the third model. The fourth model adjusted all the aforementioned factors and the usage of probiotics, enteral nutrition, and antimicrobial drugs. In the fourth model, analogous structure was evaluated by using different variables. Accordingly, we used the collinearity diagnostics to check for multicollinearity. Kaplan-Meier curves were used to analyze cumulative survival rates in patients with different levels of microbial diversity. SPSS 22.0 for Windows (SPSS Inc., Chicago, IL. USA) was used for all analyses. A P-value <0.05 was considered significant.
Results
Baseline characteristics
We collected a total of 150 stool samples from septic shock patients (mean age 62 years; 63% males). The overall 28-day hospital mortality rate was 42%. There were 80 patients with low bacterial diversity and 70 patients with normal bacterial diversity. The characteristics of the patients are listed in Table 1. A significant difference in age at baseline between low and normal microbial diversity groups was observed. Statistically significant differences also existed in chronic obstructive airways disease and Charlson comorbidity index between low and normal microbial diversity groups.
Table 1.
Characteristics of all included patients.
| Characteristic | Low bacterial diversity group (n= 80) | Normal bacterial diversity group (n=70) | P value |
|---|---|---|---|
| Male gender, % (n) | 61 (49) | 62 (43) | 0.4 |
| Age, yrs median (IQR) | 63 (50–75) | 52 (44–67) | 0.01 |
| Admission source: Emergency Department, % (n) | 50% (40) | 50% (35) | 1.00 |
| Blood stream infection, % (n) | 31% (25) | 39% (27) | 0.84 |
| Pre-existing conditions, % (n) | |||
| Co-existing disease | 94 (75) | 96 (67) | 0.77 |
| Diabetes | 25 (20) | 28 (20) | 0.78 |
| Hypertension | 35 (28) | 29 (20) | 0.30 |
| Ischaemic heart disease | 19 (15) | 14 (10) | 0.43 |
| Morbid obesity | 12 (4) | 14 (4) | 0.78 |
| Renal failure | 12 (4) | 18 (5) | 0.31 |
| Chronic obstructive airways disease | 6 (2) | 17 (5) | 0.01 |
| Immunosuppression | 16 (5) | 7 (2) | 0.08 |
| Charlson comorbidity index (median (IQR)) | 3 (1–4) | 0 (0–2) | <0.01 |
| Indicators of disease severity, % (n) | |||
| APACHE II, mean (SD) | 22.1 (7.7) | 23.5 (7.9) | 0.14 |
| SOFA Score, mean (SD) | 8.3 (3.1) | 8.1 (3.1) | 0.56 |
| Mechanical ventilation | 56 (18) | 61 (17) | 0.47 |
| Renal replacement therapy | 10 (8) | 10 (7) | 0.53 |
| Enteral feeds at study entry | 40 (13) | 46 (13) | 0.36 |
| Received intravenous steroid | 25 (20) | 29 (20) | 0.91 |
| Platelets (mean, SD) | 193 (123) | 232 (142) | 0.38 |
| Mortality, % (n) | |||
| 28 day mortality | 51 (41) | 32 (22) | 0.03 |
Intestinal microbiota distribution characteristics
The majority bacteria were Bacteroidetes and Firmicutes in intestine under phylum level (Figure 1) in normal bacterial diversity group (relative ratio: 45.83% versus 43.51%). Compared to normal bacterial diversity group, the composition of fecal microbiota in patients with low bacterial diversity indicated a significant change. The ratio of Firmicutes and Bacteroidetes decreased (28.27% versus 39.38%). The ratio of proteobacteria in low bacterial diversity group increased significantly (relative ratio: 21.71% versus 9.53%, P<0.05).
Figure 1.
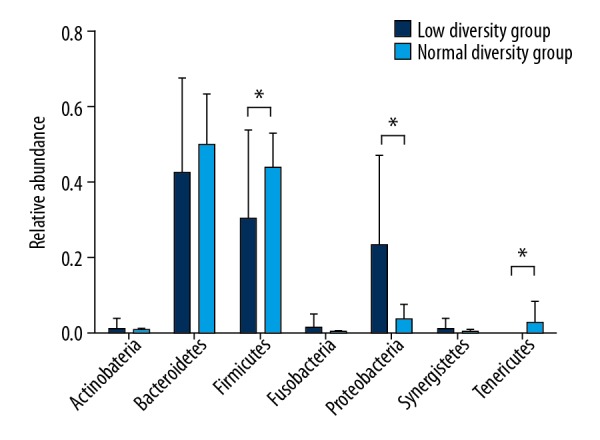
Microbial composition of fecal samples at the phylum level in septic shock patients. Seven main bacteria were compared in this figure. A significantly higher proportion of Firmicutes, and Proteobacteria were found in high bacterial diversity group patients.
Association between microbial diversity and mortality
Based on univariate analysis, 28-day mortality rate was found to be significantly higher among low diversity group (51%), compared to 32% of high diversity patients (odds ratio (OR) 2.04, 95% confidence interval (CI) 1.35–2.83). The adjusted OR for bacterial diversity and mortality in each model are shown in Figure 2. The fourth model which was adjusted with age, sex, SOFA score, Charlson comorbidity index, and usage of probiotics, enteral nutrition, and antimicrobial drugs revealed that OR for low diversity and mortality rate was 1.93 (95% CI 0.55–2.69). The C index of the fourth model was 0.73 (95% CI 0.37–0.82). The Hosmer-Lemeshow test index was 9.03 (P=0.81), indicating that the logistic curve fit into the observed data. Moreover, other models showed similar results, and no significant difference in mortality rate was observed between low and normal microbial diversity in septic shock patients (Figure 2).
Figure 2.
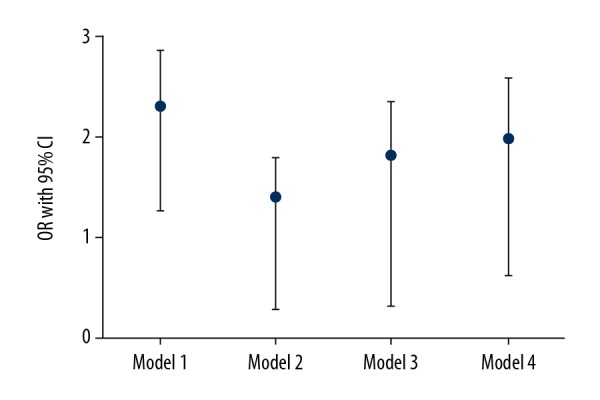
Adjusted odds ratio (OR) and 95% confidence interval (CI) for 28-day mortality in progressively multivariate regression analysis. The adjusted factors in the first model were sex and age. Age was stratified at 65 years old point. Sex, age, and severity of illness (SOFA score) were adjusted in the second model, and comorbid illness (Charlson comorbidity index) was added in the third model. The fourth model adjusted all the aforementioned factors, and usage of probiotics, enteral nutrition, and antimicrobial drugs.
The Kaplan-Meier method was used to study the relationship between microbial diversity and cumulative survival rate. The survival rate in the normal diversity group was higher than that in the low diversity group, and log-rank tests showed significant differences between the 2 groups in terms of cumulative survival rates (hazard ratio 1.88, 95% CI 1.51–3.09, Figure 3). Since the sample sizes in this analysis were small and conducting the analysis without changing the alpha level increased the risk of type 1 error, the findings of the current study should be interpreted cautiously.
Figure 3.
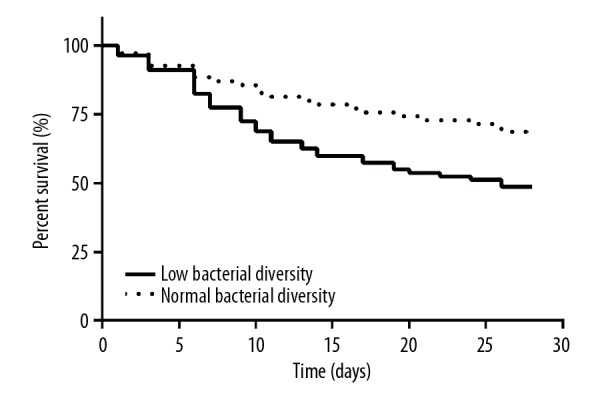
Kaplan-Meier estimation of 2 groups of different levels of bacterial diversity in septic shock patients. The survival rate curve showed that the cumulative survival rate of patients in normal diversity group was higher than that in low diversity group, and the log-rank test showed a significant difference between the 2 groups.
Discussion
We collected 150 stool samples from patients with septic shock for 16S rDNA gene sequencing analysis for 3 years. In our study, approximately 50% of patients had decreased bacterial diversity, and a rough analysis suggest that low bacterial diversity in the intestinal microbiome increased the risk of unadjusted mortality. However, after adjusting for a number of confounding factors, the results showed no independent association between intestinal bacterial diversity and mortality in patients with septic shock.
The break of intestinal microbiota balance can induce or aggravate many types of diseases, such as diabetes mellitus [5], cardiovascular diseases [3], chronic kidney disease [16], and hypertension [17]. The study on the intestinal microecological characteristics of ICU patients is still in its initial stage. Fasting, analgesics, sedatives, vasoactive drugs, stress ulcer prophylaxis drugs, and particularly high-grade multi-drugs in ICU, have all been shown to affect intestinal microbiome in ICU patients [18]. Yeh et al. [19] enrolled 32 ICU patients with trauma or acute surgery in an observational study. Microbial samples were collected from the gut, the oral cavity, and the skin within 48 hours of admission. When compared with healthy volunteers’ microbiome database, the microbiome of ICU patients showed decreased microbial diversity, and reduced symbiotic bacteria. Kelly et al. [20] analyzed 15 patients with ICU respiratory failure. The study showed that, similar to the microbiota in other anatomic sites of the ICU patients, the lower respiratory tract microbiota in patients with tracheal intubation also showed a decrease in alpha diversity, which was further reduced after mechanical ventilation. A study by Dickson et al. [21] illustrated the effects of gut microbiota on the lung microecology. Their study suggested that gut-lung translocation and disorder of the lung microbiome were respectively correlated with the indices of systemic and alveolar inflammation. Nevertheless, all studies consisted of small sample size and were non-randomized controlled trials.
The design of biomarkers based on intestinal microbiome to predict clinical outcome of critically ill patients is a hot topic. In our study, microbiota diversity was quantified using Shannon index. This diversity index is a quantitative indicator of the number of different bacteria that are present in a stool sample, taking into account the uniformity in the distribution of these bacteria in these species [22]. Diversity index value increases both when the number of species increases and when evenness increases. The Shannon index is a well-known diversity index used in microecological studies. The higher the Shannon index value, the higher the community diversity [22]. Simpson index is another indicator, which could also be used to estimate microbial diversity. The higher the Simpson index value, the lower the community diversity. In the low-Shannon index group, the classification of fecal microbial communities is relatively simple, and the number of bacterial species is small. Current research has demonstrated that healthy and intact microbiota can enhance resistance to infection [23]. Loss of microbial diversity might lead to increased mortality. Community diversity decreased in half of the septic shock patients in our study. The reason for the decrease was complex, including the disease itself and the therapy measures. The endotoxin secreted by the pathogenic bacteria could destroy the intestinal environment directly, and intestinal microcirculation ischemia due to prolonged hypotension of septic shock could weaken the intestinal mucosa and change the microenvironment [23]. Several treatments were used for septic shock, including parenteral feeding, gastric acid inhibitory drugs, multiple antibiotics, mechanical ventilation, and rapid liquid resuscitation. The effect of these treatments on gut microbiota is still not completely understood, but might be harmful [24].
The disrupted ecological state of the microbiota subsequently resulted in poor clinical outcome in the later course of hospitalization during septic shock, such as, antibiotic-associated diarrhea, emerging enterogenous infection, and other outcomes, which is not yet known [25]. Reduced mortality rate is currently the primary therapeutic goal for all critically ill patients, especially septic shock patients, whose mortality could be as high as 70%. Previous studies [6,8] and our findings have proven the universality of the decrease in the gut microbiota diversity. The effect of disrupted gut microbiota on the mortality rate in critically ill patients has become a new project. In recipients of allogeneic hematopoietic stem cell transplantation, several studies [7,26] showed that high gut microbiota diversity at the time of stem cell engraftment was associated with high survival rate. In our study, low bacterial diversity was associated with 2 times rude mortality risk in septic shock patients, and the cumulative survival rate of patients in the normal diversity group was higher than that in the low diversity group with Kaplan-Meier method. The mechanism was complex and unclear, and most studies were animal study [27,28]. Some showed that gut microbiota act as a protective factor in host defense during sepsis. Mice with a depleted gut microbiota following infection with Streptococcus pneumoniae have increased bacterial dissemination, inflammation, organ failure, and an accelerated mortality compared to controls [28].
In our study, a considerably higher proportion of Proteobacteria was found in sepsis patients compared to healthy subjects. Proteobacteria is the largest group of gut bacteria. Proteobacteria included many well-known species such as Escherichia coli, Salmonella, Vibrio cholerae, and Helicobacter pylori. Proteobacteria are divided into 5 groups by rRNA sequences, including alpha-, beta-, gamma-, delta-, epsilon-proteobacteria. The well-known hospital-acquired infectious bacteria in the ICU such as Klebsiella belonging to beta-proteobacteria, Escherichia coli, Pseudomonas aeruginosa, and Salmonella to gamma-proteobacteria [29]. The excessive growth of proteobacteria in the gut of septic shock patients in our study might indicate that the rate of enterogenous infection was higher than expected.
The advantage of our study was that the sample size was large. The sample size in most of the previous studies was small, and they were generally concentrated in critically ill patients rather than specific diseases. Additionally, in terms of methodology, this study conducted a systematic multiple regression analysis to determine the relationship between gut microbiota diversity and mortality. This study also had flaws. On one hand, stool sample in each patient was collected only once, thus, the dynamic changes could not be studied. In addition, microbial diversity was estimated only by the Shannon index. Some other indices like Simpson index, and Chao index were not calculated.
Conclusions
This observational study indicated that low bacterial diversity was not associated with worse mortality rate in septic shock patients. More biomarkers of gut microbiota need to be explored.
Footnotes
Source of support: National Natural Science Foundation of China (No. 81370364)
Conflict of interest
None.
References
- 1.Singer M, Deutschman CS, Seymour CW, et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3) JAMA. 2016;315(8):801–10. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arefian H, Heublein S, Scherag A, et al. Hospital-related cost of sepsis: A systematic review. J Infect. 2017;74(2):107–17. doi: 10.1016/j.jinf.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 3.Tang WH, Wang Z, Levison BS, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368(17):1575–84. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen J, Obin MS, Zhao L. The gut microbiota, obesity and insulin resistance. Mol Aspects Med. 2013;34(1):39–58. doi: 10.1016/j.mam.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Giongo A, Gano KA, Crabb DB, et al. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. 2011;5(1):82–91. doi: 10.1038/ismej.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wan YD, Zhu RX, Wu ZQ, et al. Gut microbiota disruption in septic shock patients: A pilot study. Med Sci Monit. 2018;24:8639–46. doi: 10.12659/MSM.911768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taur Y, Jenq RR, Perales MA, et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood. 2014;124(7):1174–82. doi: 10.1182/blood-2014-02-554725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lankelma JM, van Vught LA, Belzer C, et al. Critically ill patients demonstrate large interpersonal variation in intestinal microbiota dysregulation: A pilot study. Intensive Care Med. 2017;43(1):59–68. doi: 10.1007/s00134-016-4613-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ojima M, Motooka D, Shimizu K, et al. Metagenomic analysis reveals dynamic changes of whole gut microbiota in the acute phase of intensive care unit patients. Dig Dis Sci. 2016;61(6):1628–34. doi: 10.1007/s10620-015-4011-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raith EP, Udy AA, Bailey M, et al. Prognostic accuracy of the SOFA score, SIRS criteria, and qSOFA score for in-hospital mortality among adults with suspected infection admitted to the intensive care unit. JAMA. 2017;317(3):290–300. doi: 10.1001/jama.2016.20328. [DOI] [PubMed] [Google Scholar]
- 11.Bannay A, Chaignot C, Blotiere PO, et al. The best use of the Charlson comorbidity index with electronic health care database to predict mortality. Med Care. 2016;54(2):188–94. doi: 10.1097/MLR.0000000000000471. [DOI] [PubMed] [Google Scholar]
- 12.Vincent JL, Nelson DR, Williams MD. Is worsening multiple organ failure the cause of death in patients with severe sepsis? Crit Care Med. 2011;39(5):1050–55. doi: 10.1097/CCM.0b013e31820eda29. [DOI] [PubMed] [Google Scholar]
- 13.Hernandez G, Ospina-Tascon GA, Damiani LP, et al. Effect of a resuscitation strategy targeting peripheral perfusion status vs. serum lactate levels on 28-day mortality among patients with septic shock: The ANDROMEDA-SHOCK randomized clinical trial. JAMA. 2019;321(7):654–64. doi: 10.1001/jama.2019.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salonen A, Nikkila J, Jalanka-Tuovinen J, et al. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: Effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J Microbiol Methods. 2010;81(2):127–34. doi: 10.1016/j.mimet.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 15.Young VB, Schmidt TM. Antibiotic-associated diarrhea accompanied by large-scale alterations in the composition of the fecal microbiota. J Clin Microbiol. 2004;42(3):1203–6. doi: 10.1128/JCM.42.3.1203-1206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hobby GP, Karaduta O, Dusio GF, et al. Chronic kidney disease and the gut microbiome. Am J Physiol Renal Physiol. 2019;316(6):F1211–17. doi: 10.1152/ajprenal.00298.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marques FZ. Missing heritability of hypertension and our microbiome. Circulation. 2018;138(14):1381–83. doi: 10.1161/CIRCULATIONAHA.118.036224. [DOI] [PubMed] [Google Scholar]
- 18.Akrami K, Sweeney DA. The microbiome of the critically ill patient. Curr Opin Crit Care. 2018;24(1):49–54. doi: 10.1097/MCC.0000000000000469. [DOI] [PubMed] [Google Scholar]
- 19.Yeh A, Rogers MB, Firek B, et al. Dysbiosis across multiple body sites in critically ill adult surgical patients. Shock. 2016;46(6):649–54. doi: 10.1097/SHK.0000000000000691. [DOI] [PubMed] [Google Scholar]
- 20.Kelly BJ, Imai I, Bittinger K, et al. Composition and dynamics of the respiratory tract microbiome in intubated patients. Microbiome. 2016;4:7. doi: 10.1186/s40168-016-0151-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dickson RP, Singer BH, Newstead MW, et al. Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat Microbiol. 2016;1(10):16113. doi: 10.1038/nmicrobiol.2016.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim BR, Shin J, Guevarra R, et al. Deciphering diversity indices for a better understanding of microbial communities. J Microbiol Biotechnol. 2017;27(12):2089–93. doi: 10.4014/jmb.1709.09027. [DOI] [PubMed] [Google Scholar]
- 23.Haak BW, Wiersinga WJ. The role of the gut microbiota in sepsis. Lancet Gastroenterol Hepatol. 2017;2(2):135–43. doi: 10.1016/S2468-1253(16)30119-4. [DOI] [PubMed] [Google Scholar]
- 24.Dickson RP. The microbiome and critical illness. Lancet Respir Med. 2016;4(1):59–72. doi: 10.1016/S2213-2600(15)00427-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Sun J, Zhang J, et al. Enzyme inhibitor antibiotics and antibiotic-associated diarrhea in critically ill patients. Med Sci Monit. 2018;24:8781–88. doi: 10.12659/MSM.913739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doki N, Suyama M, Sasajima S, et al. Clinical impact of pre-transplant gut microbial diversity on outcomes of allogeneic hematopoietic stem cell transplantation. Ann Hematol. 2017;96(9):1517–23. doi: 10.1007/s00277-017-3069-8. [DOI] [PubMed] [Google Scholar]
- 27.Deshmukh HS, Liu Y, Menkiti OR, et al. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat Med. 2014;20(5):524–30. doi: 10.1038/nm.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schuijt TJ, Lankelma JM, Scicluna BP, et al. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut. 2016;65(4):575–83. doi: 10.1136/gutjnl-2015-309728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Zhou KH, Chen W, et al. Epidemiology and risk factors for nosocomial infection in the respiratory intensive care unit of a teaching hospital in China: A prospective surveillance during 2013 and 2015. BMC Infect Dis. 2019;19(1):145. doi: 10.1186/s12879-019-3772-2. [DOI] [PMC free article] [PubMed] [Google Scholar]


