Abstract
Terahertz (THz) demethylation is a photomedical technique applied to dissociate methyl-DNA bonds and reduce global DNA methylation using resonant THz radiation. We evaluated the performance of THz demethylation and investigated the DNA damage caused by THz irradiation. The demethylation rate in M-293T DNA increased linearly with the irradiation power up to 48%. The degree of demethylation increased with exposure to THz radiation, saturating after 10 min. Although THz demethylation occurred globally, most of the demethylation occurred within the partial genes in the CpG islands. Subsequently, we performed THz demethylation of melanoma cells. The degree of methylation in the melanoma cell pellets decreased by approximately 10–15%, inducing ∼5–8 abasic sites per 105 bp; this was considerably less than the damaged DNA irradiated by the high-power infrared laser beam used for generating THz pulses. These results provide initial data for THz demethylation and demonstrate the applicability of this technique in advanced cancer cell research. THz demethylation has the potential to develop into a therapeutic procedure for cancer, similar to that involving chemical demethylating agents.
1. Introduction
Terahertz (THz) radiation consists of electromagnetic waves in the 0.1–10 THz (3.3–330 cm−1) frequency range, whose spectrum lies between the microwave and the infrared regions. THz energy is correlated with collective vibration, rotational motion, or torsional motion from intramolecular domains and weak hydrogen bonds or van der Waals forces from intermolecular interactions [1–4]. Because these low-energy motions are closely associated with the chemical and structural characteristics of biomolecules, a number of THz biomedical studies have been performed utilizing the relatively lower photon energy of THz radiation (1 THz = 4.14 meV) [5–10]. In particular, the resonance of the chemical bonds between the methyl group and DNA in cancer DNA was reported to be within approximately 1.6–1.7 THz. The resonance intensity enabled the differentiation of the types of cancer cells at the common center frequency, and the results indicate that this technique could serve as a cancer biomarker [11,12]. The resonance is believed to originate from DNA methylation, an epigenetic process, through which methyl groups (CH3–) are attached to the cytosine in DNA molecules. In epigenetics, gene expression is regulated and programmed by DNA methylation, which is a chemical process that occurs in the genome without changing the DNA sequence. The resonance in DNA methylation can likewise serve as a clue to manipulate the methyl-DNA bond because the energy accumulation coming from resonant THz photons might lead to bond dissociation. The THz dissociation of methyl-DNA bonds causes a decrease in the global degree of methylation, i.e., demethylation [12]. Recent studies show that high-power THz radiation having sufficient power modifies various biological activities [13–15]. THz demethylation is one of the biological modifiers using high-power terahertz radiation, which removes methyl groups from DNA molecules using resonance energy. This can serve as a novel optical method to manipulate DNA methylation as an epigenetic modifier.
Because DNA methylation plays a key role in genomic functioning, the aberration of DNA methylation is a critical factor associated with carcinogenesis via abnormal gene expression. The abnormal activity in tumor suppressor genes results in cancer growth and metastasis [16–18]; therefore, the manipulation of aberrant DNA methylation is crucial in cancer treatment. If the methyl groups during DNA methylation are properly removed from the silenced tumor suppressor genes that are hypermethylated, demethylation may prevent cancer progression and induce apoptosis of cancerous cells through the reprogrammed genes [19–22].
Various demethylation agents, like decitabine (Decogen TM) or azacitidine (Vidaza TM), approved by the U.S. Food and Drug Administration (FDA), have been developed for cancer treatment over the past decade [23–27]. These agents reduce the degree of DNA methylation by inhibiting DNA methyltransferases during epigenetic inheritance [22,28,29]. However, these drugs can lead to many side effects as they are applied to the non-targeted lesion, and it is not easy to adjust the dosage properly [30]. THz demethylation is targetable, and it is relatively easy to control the radiation energy using optical techniques. Because THz radiation has a penetration depth of a few hundred micrometers, which can reach into the tissues to be treated, THz demethylation might be achieved at the cellular level [4,31].
In this study, we evaluated DNA demethylation performed using THz radiation on extracted DNA in terms of the irradiation power, exposure time, and gene position. The results may offer an applicable standard for THz demethylation at the cellular level. As THz demethylation involves high-power radiation, we also demethylated melanoma cell pellets to verify the effect of THz radiation on the cellular structure and investigated the DNA damage caused by THz radiation through quantifying abasic sites (AP sites). AP sites indicate nucleobase loss or removal, and they are one of the primary indicators of DNA damage. Ionizing radiation, toxicity of medicines, or other chemical-enzyme activities can induce multiple AP sites, and high numbers of unrepaired sites may be mutagenic and can lead to cell death.
2. Experimental methods
2.1. THz demethylation system
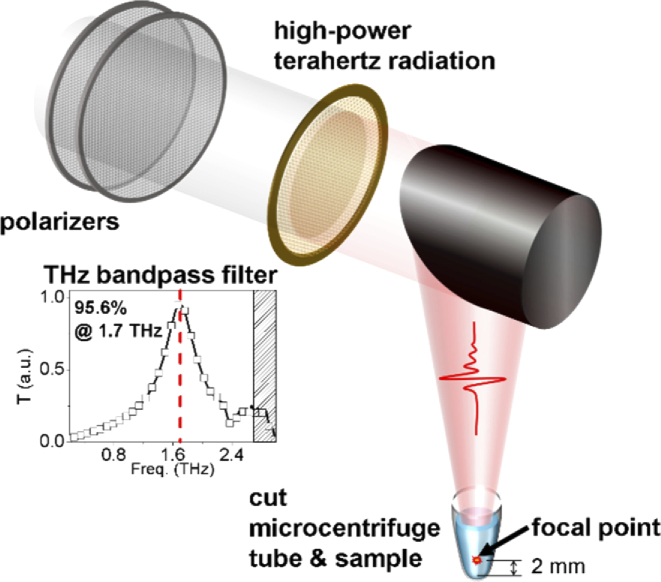
THz radiation was generated using an MgO–LiNbO3 crystal excited by an amplified laser beam with 0.8-mJ energy per pulse from a regenerative amplifier (Spitfire; Spectra-Physics) system, with a 1-kHz repetition rate at an 800-nm wavelength. The amplifier was injected with a seed beam from a Ti:sapphire oscillator (Tsunami; Spectra-Physics, CA, USA) and pumped using a pump laser (Empower; Spectra-Physics) with a pump power of 9 W. The oscillator generated 35-fs laser pulses at an 800-nm wavelength and 80-MHz repetition rate, which were pumped by a 532-nm pump laser (millennia Vs; Spectra-Physics) with a pump power of 5 W. The amplified laser beam was tilted at 62° to the pulse front using a grating with a groove number of 2000 mm−1. The high-power THz pulse of 2.4 mW/cm2 was generated by the LiNbO3 crystal and passed through a silicon window, which blocked the residual laser beam. The THz radiation was collected by a parabolic mirror and passed to a cross-shaped THz bandpass filter; this limited the frequency of the THz radiation to a 1.7-THz center frequency with a 0.57-THz full width at half maximum (Tydex, St. Petersburg, Russia). The 1.7-THz radiation was focused vertically at 2 mm above the top of the cut microcentrifuge tube, which served as a sample container. The THz power was controlled by a pair of grid polarizers and measured using a pyroelectric detector (T-Rad; Gentec-EO, Quebec City, Canada). The cut microcentrifuge tube was held vertically by a sample holder and covered by a thin plastic film to prevent evaporation of the samples (Fig. 1). The temperature of the sample holder was controlled by a pair of thermoelectric coolers, maintaining the samples at a temperature of –4 °C.
Fig. 1.
Schematic showing THz demethylation using resonant THz radiation system. High-power THz radiation was generated using a regenerative amplifier and LiNbO3 crystal. The THz filter limited the THz bandwidth to around the resonance frequency of the methyl-DNA bonds.
2.2. Preparation of extracted DNA samples
Ethical approval for this study was obtained from institutional boards at the Seoul National University Boramae Medical Center. We prepared the human embryonic kidney (HEK) 293T cell line at the central laboratory of the Boramae Medical Center and the SK-MEL-3 melanoma cell lines of the lymph node at the Korean Cell Line Bank (Seoul, Republic of Korea). Cells were cultured in disposable petridishes and plated at a density of 3 × 105 cells per 100-mm2 dish.
Isolated DNA samples were extracted from 293 T cells using the FavorPrep Blood/Cultured Cell Genomic DNA Extraction Mini Kit (FAVORGEN, Ping-Tung, Taiwan) according to the manufacturer’s protocol. After purification, the genomic DNA was dissolved in distilled water to remove the interruption from other residual materials. The 293 T DNA was artificially methylated (methylated 293 T, M-293 T) using the DNA methyltransferase (DNMT) enzyme (New England Biolabs Ltd., MS, USA) according to the manufacturer’s instructions, and the M-293 T DNA sample was purified again. In the experiment, we used M-293 T DNA to verify the response of DNA molecules to THz radiation. To measure the concentration of DNA samples, we used a NanoDrop ND-1000 UV-Vis spectrophotometer (Wilmington, DE, USA). The concentration of the M-293 T DNA samples was 300 ± 7 µg/ml.
2.3. Melanoma cell pellets
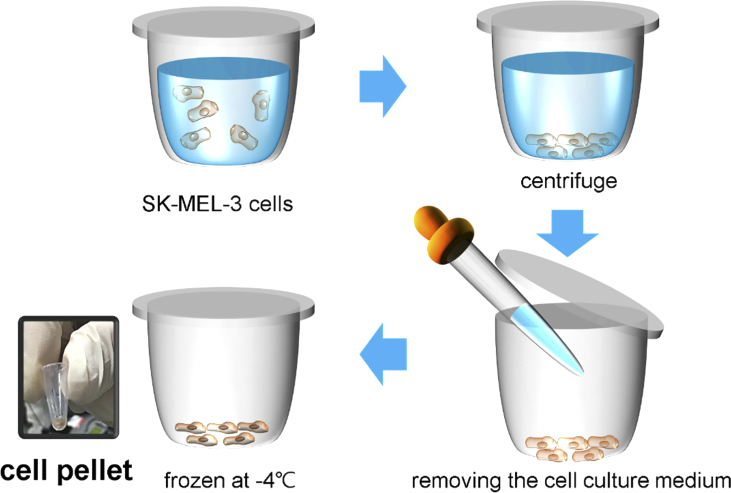
We cultured SK-MEL-3 melanoma cells in a T175 flask and transferred them to four 100-mm2 dishes and obtained 10-µl melanoma cell pellets (cell count: approximately 2 × 105 per cell pellet) from each cultured dish. The cells detached from the dish surface using trypsin (TrypLE™ Express, Gibco, Denmark) were collected into a centrifuge tube. We removed the cell culture medium after centrifugation for 5 min at 1500 rpm using an Allegra X-15R centrifuge (Beckman coulter, CA, USA). The cell pellets were frozen slowly to prevent the damage of cell membranes and stored at –80 °C. The customized temperature controller was composed of a pair of a square-shaped thermoelectric coolers and a centrifuge tube holder made of copper. The wall of the centrifuge tube was in contact with the copper holder, and the temperature of the cell pellet inside the tube was maintained at −4 °C during the THz exposure process (Fig. 2).
Fig. 2.
Collection process of melanoma cell pellets. Absorption of THz radiation by water molecules was reduced in frozen cell pellets.
2.4. Quantification of DNA methylation
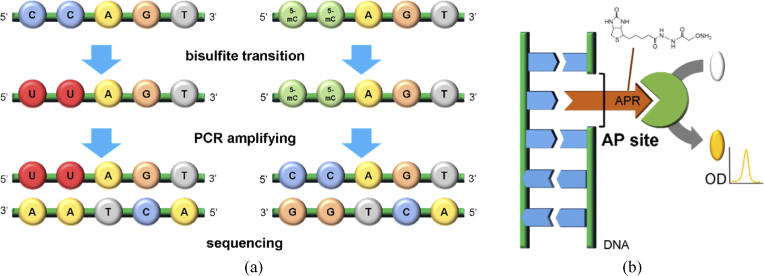
The degree of methylation in genomic DNA was obtained by the enzyme-linked immunosorbent assay (ELISA) method using a Methylamp Global DNA Methylation Quantification Ultra Kit (Epigentek Inc., NY, USA) [32]. The ELISA method is sensitive to global DNA methylation in DNA strands above a 100-bp length. As low as 0.2 ng of methylated parts in 50 ng of the input genomic DNA can be detected using the light-absorbing antibody. The optical density (OD) measured using the ELISA method presents the global degree of DNA methylation through comparison with the positive and negative controls. The OD values were normalized based on the value of the control sample, which was not exposed to THz radiation, with the experimental samples in the same environment. The methylation pattern was determined by whole-genome bisulfite sequencing (Macrogen, Inc., Seoul, KR) (Fig. 3a). We read a total of 131,394,586,424 bases in M-293 T DNA using bisulfite sequencing and analyzed 493,635 genes.
Fig. 3.
Biological methods for verifying THz demethylation. (a) Bisulfite sequencing to quantify the degree of DNA methylation in partial genes. (b) Investigation of DNA damage by measuring AP sites, indicating the locations of missing nucleobases.
2.5. Assessment of DNA damage using AP sites
DNA damage was investigated by quantifying the number of AP sites. The aldehyde reactive probe (APR) reacts on an AP site, which depicts the location of missing bases resulting from DNA damage. The APR was tagged with the biotin/streptavidin-enzyme conjugate of the ELISA substrate to detect the AP sites by measuring the OD (Fig. 3b). We used the OxiSelect™ Oxidative DNA Damage Quantitation Kit (AP sites) (Cell Biolabs Inc., CA, USA) [33,34]. The number of AP sites was obtained through comparison with the value obtained from the standard curve. As a positive control (PC), a DNA sample with an infrared-beam-induced damage containing AP sites was prepared. The 800-nm infrared beam constituted a portion, 160 mW, of the radiation from the regenerative amplifier. It was incident on the SK-MEL-3 melanoma cell pellets at –4 °C for 30 min, the same conditions used during exposure to THz radiation.
2.6. Statistical data processing
We obtained the degree of methylation by averaging two OD values using the ELISA reader, SpectraMax Plus 384 (Molecular Devices, LLC, CA, USA). Each ELISA measurement was repeated three times, and the three datasets with the averaged OD values were averaged again. The fitted lines were obtained using Origin Pro 9 (OriginLab Corporation, MA, USA), a software package for numerical computation and visualization.
3. Results and analysis
3.1. Dependence of THz irradiation power
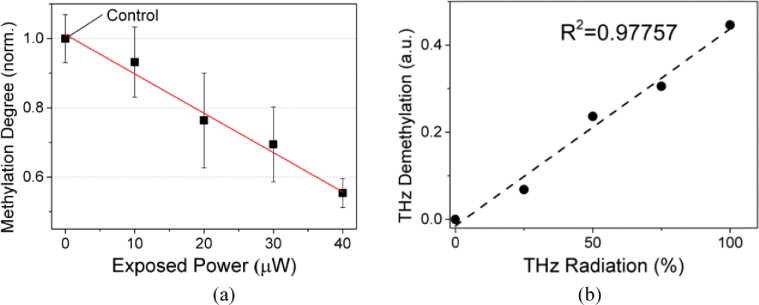
Because THz demethylation is regarded to occur owing to the energy accumulation by low-energy photons similar to that in infrared photodissociation, the demethylation rate might be influenced by the power of the THz radiation (i.e., total THz photon flux) and exposure time, along with other factors [35]. We first measured the power dependency of THz demethylation in M-293T DNA at a constant exposure time of 30 min. The absorption cross section was controlled by the fixed height of the sample holder and a constant sample amount (10 µl). As shown in Fig. 4, the methylation degree decreases with a 10-µW increase in the THz irradiation power. We averaged six OD values obtained through ELISA quantification during triplicate experiments performed under the same condition [12]. The values of the methylation degree were normalized to that of the control sample (non-irradiated DNA).
Fig. 4.
Power dependency of THz demethylation in M-293 T DNA. (a) Degree of methylation decreased with increasing THz irradiation power. THz power was augmented via 10-µW increments, and we obtained each data point by averaging six OD values obtained via ELISA quantification. Error bars represent the standard deviation of each data point. (b) THz demethylation rate exhibiting linear dependency on the irradiation power (R2 = 0.97757).
3.2. Dependence of THz demethylation in the genomic region on THz exposure time
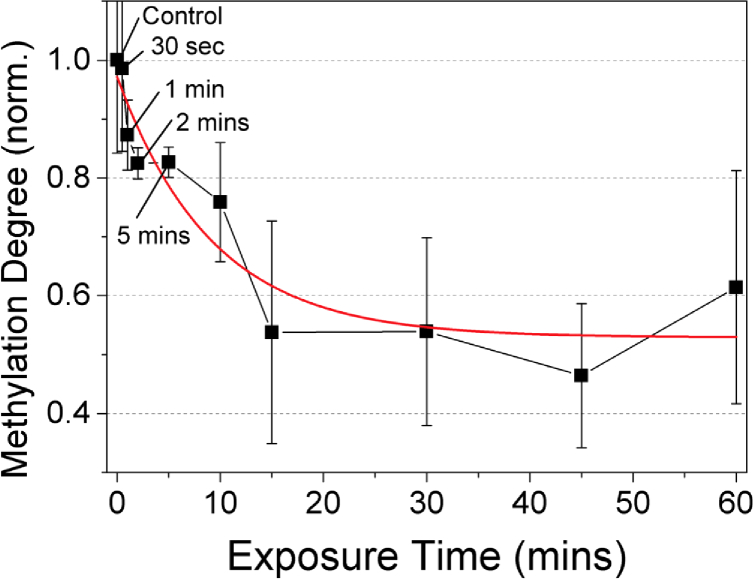
In our previous study, THz demethylation was completed within 15 min, although the power dependence was linear thereto [12]. To verify the relationship between THz demethylation and the exposure time, we applied 40-µW THz radiation to 10-µl DNA solution samples at various exposure times. Figure 5 shows the normalized methylation degree of M-293T DNA at different exposure times. We performed triplicate experiments, measuring the OD values twice. Each data point was obtained by averaging and normalizing the measured OD values. The methylation degree decreased for a longer exposure time; however, it began to saturate after 10 min, similar to the results in our previous research [12]. Photoactivation by absorption of infrared photons is known to be a stepwise process with tiny internal accumulation per step [35]. Because the THz photon has much lower energy than the infrared photon (1.7 THz = 7.03 meV), and demethylation is a large molecular phenomenon, we conjectured that the accumulation of energy over dissociative levels requires a longer time than other infrared multiple photon dissociations.
Fig. 5.
THz demethylation dependence on exposure time in M-293T DNA. Irradiation power was held constant at 40 µW. Below 15 min of exposure time, the degree of methylation decreased with longer exposure times; however, the rate began to saturate after 10 min, as observed in a previous study [6]. The measurements were repeated six times and averaged at each data point. Error bar indicates the standard deviation.
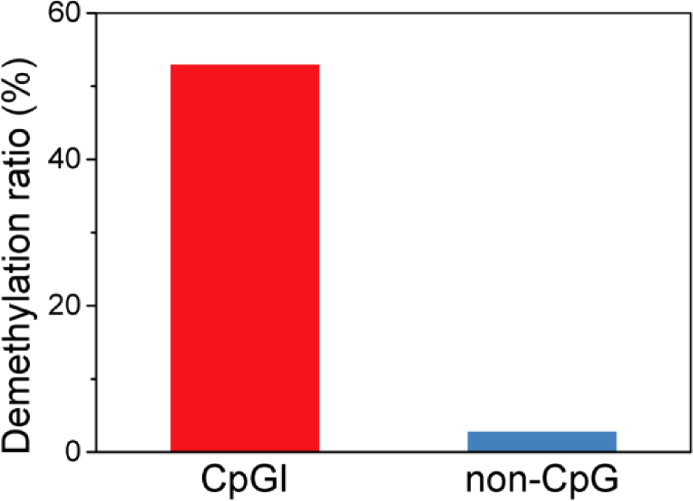
We also investigated the performance of THz demethylation in partial genes. Bisulfite sequencing allows for measuring the degree of methylation in partial genes through ELISA quantification, which is used to quantify DNA methylation in genomic-scale DNA. We compared the quantitative DNA methylation between the control and experimental sample that was exposed to the maximum demethylation conditions (40 µW, 30 min). We obtained the degree of methylation in a total of 493,635 genes. As shown in Fig. 6, THz demethylation mostly occurs in genes at the CpG islands (CpGI), which are genomic regions, with a high frequency of CpG sites (sites of cytosine-phosphate-guanine nucleotides). In the CpGI sites, genes were demethylated by 53%, while genes were demethylated by less than 3% in non-CpG sites. We concluded that the distribution of the methylated sites influenced the efficiency of THz demethylation; however, further investigation is required to confirm this.
Fig. 6.
Asymmetrical pattern of THz demethylation in partial genes. The demethylation ratio of genes at CpGI sites was much higher than that of the genes at non-CpG sites. Data were obtained through bisulfite sequencing of M-293 T DNA.
3.3. Demethylation of the melanoma cells and verification of DNA damage by high-power THz radiation
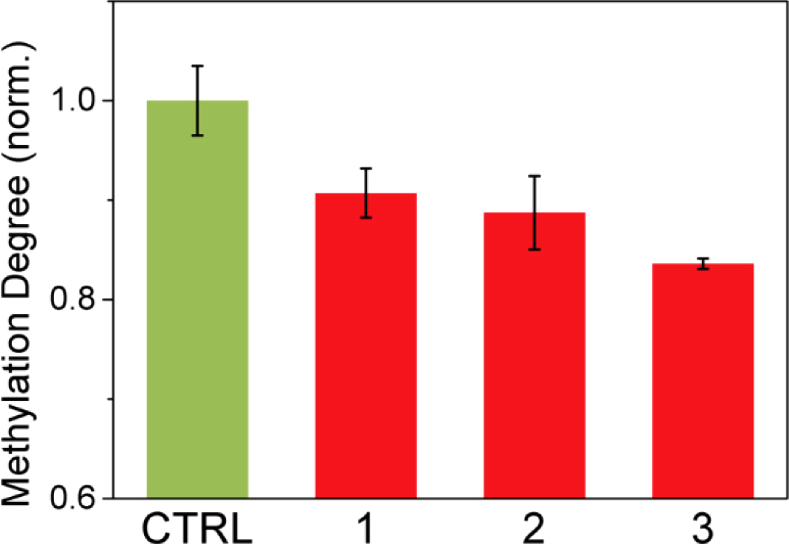
THz radiation was applied to the melanoma cell pellets to assess the effect of THz demethylation at the cell level and verify the damage to the DNA bases due to THz irradiation. DNA was packaged in the cells in contrast with the isolated environment. The experiments using frozen pellets were performed to analyze the effect of demethylation in packed DNA in cells using THz radiation and the influence of other cell constituents. We cultured SK-MEL-3 melanoma cells in a T175 flask and transferred to four 100-mm2 dishes. Two cell pellets of 10 µl were generated from each dish, and we totally prepared eight frozen cell pellets at –4 °C for two experimental sets. Each experimental set consisted of a control and three THz exposure samples. We applied 40-µW THz radiation to three cell pellets (1–3) for 30 min. The control sample (CTRL) was treated accordingly in the absence of THz radiation. The experiments were performed in duplicate, and the methylation degree was obtained by averaging the obtained OD values, which were measured twice through ELISA quantification of extracted DNA from the cell pellets. The methylation degrees of experimental samples were normalized with that of the control sample. The error bar in the control data was from two repeated experiments. The separated irradiated groups showed the similar results unaffected by the cell culture process. THz demethylation decreased the methylation degree of DNA inside melanoma cells by approximately 10–15%, as shown in Fig. 7. This result indicates that high-power THz radiation can affect not only isolated DNA in the aqueous environment, but also natural DNA inside the cell structure.
Fig. 7.
THz demethylation of melanoma cell pellets (1–3). THz demethylation occurred within the natural cell structure. Data was averaged across four measurements. Error bars shows the standard deviation.
Because THz demethylation uses high-power THz radiation and modulated the DNA structure directly by breaking the methyl-DNA bonds, the damage to other DNA structures should be assessed before further developing this method for use in clinical applications. The number of AP sites is a good indicator of DNA damage, with the potential to induce lethal mutations or cell death by blocking DNA replication and transcription. Moreover, AP sites can generate DNA strand breaks [36].
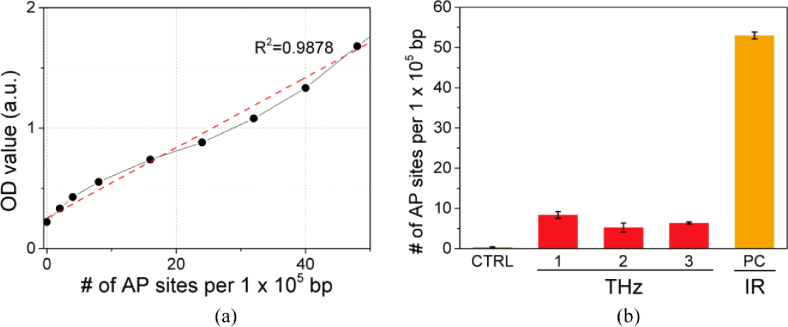
We set up a standard curve with OD values depicting the AP sites per 105 nucleotides before quantifying the number of AP sites in the DNA (Fig. 8a). The number of AP sites was then determined by referring to the standard curve. As shown in Fig. 8b, THz radiation induces a few AP sites (∼5–8 AP sites/105 bp). Nevertheless, these numbers were much smaller compared to the PC, which damaged genomic DNA using an 800-nm infrared beam with a power of 160 mW.
Fig. 8.
DNA damage via THz demethylation in melanoma cell pellets. (a) Standard curve for measurement of AP sites. (b) Number of AP sites produced by THz demethylation. THz demethylation induced a few AP sites; however, there were fewer sites compared to the DNA damaged by infrared radiation. Error bars for each data point represent the standard deviation.
4. Summary
THz demethylation has the potential to become a novel clinical method for cancer treatment as it can manipulate the degree of DNA methylation, which is an epigenetic regulator in cancer biology. Because this is an optical technique, the operating conditions during THz irradiation can be controlled accurately to reduce the risk of side effects.
In this study, we evaluated the effectiveness of THz demethylation in terms of the irradiation power and exposure time. The degree of THz demethylation in M-293T DNA increased with the irradiation power. As for the exposure time, the demethylation began to saturate after 10 min. The global degree of methylation decreased by approximately 48% under the maximum demethylation conditions of our system (40 µW for 15 min). In partial genes, THz demethylation mainly affected cytosine in CpGI (53%), where CpG sites were abundant. Moreover, we investigated the DNA demethylation and damage in melanoma cells induced by THz radiation. THz demethylation reduced the degree of methylation in the melanoma cell pellets by approximately 10–15% and produced five to eight AP sites per 105 bp. Nevertheless, the number of AP sites was much smaller than that in the DNA damaged by infrared radiation.
Our study provides initial data regarding THz demethylation and discusses the potential of this method to develop into a novel procedure for cancer treatment. Although we have demonstrated THz demethylation with frozen cells to reduce the absorption of liquid water [37], this problem at a physiologically relevant temperature can be solved when a higher THz power is applied using state-of-the-art THz sources delivering power that is an order of magnitude higher than that used in this experiment [38–40]. For internal organs, THz waveguides can be utilized to deliver THz radiation although we plan to start with superficial cancers such as skin melanoma [41,42].
Compliance with ethical standards
All procedures involving human cell lines were in accordance with the ethical policies of the institutional boards at the Seoul National University Boramae Medical Center.
Additional Information
All figures were drawn by Hwayeong Cheon, one of the authors.
Acknowledgments
The work was partly supported by the Institute for Information & communications Technology Promotion (IITP) grant funded by the Korean government (MSIT) (No. 2017-0-00422, Cancer DNA demethylation using ultra-high-power terahertz radiation), National Research Foundation of Korea (NRF) grant, funded by the Korean government (MSIT) (NRF-2017R1A2B2007827), and by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (NRF-2017R1A6A3A11033807). We sincerely thank Jungeun Lee for assistance with the biological techniques, and Donggun Lee and Seo-Yeon Jeong for fabricating the experimental sample holder.
Funding
National Research Foundation of Korea10.13039/501100003725 (NRF-2017R1A2B2007827, NRF-2017R1A6A3A11033807); Institute for Information and Communications Technology Promotion10.13039/501100010418 (2017-0-00422).
Disclosures
The University of Seoul, along with H. Y. C. and J. –H. S., has filed provisional patents regarding the technology and intellectual property reported here (patent pending number PCT/KR2018/014489; inventors: H. Y. C. and J. –H. S.; title: Method and apparatus for demethylation and cancer treatment using method and apparatus for demethylation;). This patent covers the method and device used for THz demethylation and cancer treatment method and the device for THz demethylation. H. –J. Y. and M. R. C. declare no potential conflict of interest.
References
- 1.Thrane L., Jacobsen R. H., Uhd Jepsen P., Keiding S. R., “THz reflection spectroscopy of liquid water,” Chem. Phys. Lett. 240(4), 330–333 (1995). 10.1016/0009-2614(95)00543-D [DOI] [Google Scholar]
- 2.Walther M., Plochocka P., Fischer B., Helm H., Uhd Jepsen P., “Collective vibrational modes in biological molecules investigated by terahertz time-domain spectroscopy,” Biopolymers 67(4-5), 310–313 (2002). 10.1002/bip.10106 [DOI] [PubMed] [Google Scholar]
- 3.Andersen J., Heimdal J., Mahler D. W., Nelander B., Wugt Larsen R., “Communication: THz absorption spectrum of the CO2 -H2O complex: Observation and assignment of intermolecular van der Waals vibrations,” J. Chem. Phys. 140(9), 091103 (2014). 10.1063/1.4867901 [DOI] [PubMed] [Google Scholar]
- 4.Cheon H., Yang H.-J., Son J.-H., “Toward clinical cancer imaging using terahertz spectroscopy,” IEEE J. Sel. Top. Quantum Electron. 23(4), 1–9 (2017). 10.1109/JSTQE.2017.2704905 [DOI] [Google Scholar]
- 5.Fischer B. M., Walther M., Uhd Jepsen P., “Far-infrared vibrational modes of DNA components studied by terahertz time-domain spectroscopy,” Phys. Med. Biol. 47(21), 3807–3814 (2002). 10.1088/0031-9155/47/21/319 [DOI] [PubMed] [Google Scholar]
- 6.Kim K. W., Kim H., Park J., Han J. K., Son J. H., “Terahertz tomographic imaging of transdermal drug delivery,” IEEE Trans. Terahertz Sci. Technol. 2(1), 99–106 (2012). 10.1109/TTHZ.2011.2177175 [DOI] [Google Scholar]
- 7.Reid C. B., Reese G., Gibson A. P., Wallace V. P., “Terahertz Time-Domain Spectroscopy of Human Blood,” IEEE J. Biomed. Health Inform. 17(4), 774–778 (2013). 10.1109/JBHI.2013.2255306 [DOI] [PubMed] [Google Scholar]
- 8.Oh S. J., Kim S.-H., Bin Ji Y., Jeong K., Park Y., Yang J., Park D. W., Noh S. K., Kang S.-G., Huh Y.-M., Son J.-H., Suh J.-S., “Study of freshly excised brain tissues using terahertz imaging,” Biomed. Opt. Express 5(8), 2837–2842 (2014). 10.1364/BOE.5.002837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Son J.-H., Terahertz Biomedical Science and Technology (CRC Press, 2014). [Google Scholar]
- 10.Son J.-H., Oh S. J., Cheon H., “Potential clinical applications of terahertz radiation,” J. Appl. Phys. 125(19), 190901 (2019). 10.1063/1.5080205 [DOI] [Google Scholar]
- 11.Cheon H., Yang H. J., Lee S. H., Kim Y. A., Son J.-H., “Terahertz molecular resonance of cancer DNA,” Sci. Rep. 6(1), 37103 (2016). 10.1038/srep37103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheon H., Paik J. H., Choi M., Yang H.-J., Son J.-H., “Detection and manipulation of methylation in blood cancer DNA using terahertz radiation,” Sci. Rep. 9(1), 6413 (2019). 10.1038/s41598-019-42855-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korenstein-Ilan A., Barbul A., Hasin P., Eliran A., Gover A., Korenstein R., “Terahertz Radiation Increases Genomic Instability in Human Lymphocytes,” Radiat. Res. 170(2), 224–234 (2008). 10.1667/RR0944.1 [DOI] [PubMed] [Google Scholar]
- 14.Titova L. V., Ayesheshim A. K., Golubov A., Rodriguez-Juarez R., Woycicki R., Hegmann F. A., Kovalchuk O., “Intense THz pulses down-regulate genes associated with skin cancer and psoriasis: a new therapeutic avenue?” Sci. Rep. 3(1), 2363 (2013). 10.1038/srep02363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franchini V., De Sanctis S., Marinaccio J., De Amicis A., Coluzzi E., Di Cristofaro S., Lista F., Regalbuto E., Doria A., Giovenale E., Gallerano G. P., Bei R., Benvenuto M., Masuelli L., Udroiu I., Sgura A., “Study of the effects of 0.15 terahertz radiation on genome integrity of adult fibroblasts,” Environ. Mol. Mutagen. 59(6), 476–487 (2018). 10.1002/em.22192 [DOI] [PubMed] [Google Scholar]
- 16.Luczak M. W., Jagodzinski P. P., “The role of DNA methylation in cancer development,” Folia Histochem Cytobiol. 44(3), 143–154 (2006). [PubMed] [Google Scholar]
- 17.Szyf M., Pakneshan P., Rabbani S. A., “DNA methylation and breast cancer,” Biochem. Pharmacol. 68(6), 1187–1197 (2004). 10.1016/j.bcp.2004.04.030 [DOI] [PubMed] [Google Scholar]
- 18.Taberlay P. C., Jones P. A., “DNA methylation and cancer,” Prog. Drug Res. 67, 1–23 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Szyf M., “DNA methylation and cancer therapy,” Drug Resist. Updat. 6, 341–353 (2003). 10.1016/j.drup.2003.10.002 [DOI] [PubMed] [Google Scholar]
- 20.Kelly T. K., De Carvalho D. D., Jones P. A., “Epigenetic modifications as therapeutic targets,” Nat. Biotechnol. 28(10), 1069–1078 (2010). 10.1038/nbt.1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hervouet E., Cheray M., Vallette F., Cartron P.-F., “DNA methylation and apoptosis resistance in cancer cells,” Cells 2(3), 545–573 (2013). 10.3390/cells2030545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roulois D., Yau H. L., De Carvalho D. D., “Pharmacological DNA demethylation: Implications for cancer immunotherapy,” OncoImmunology 5(3), e1090077 (2016). 10.1080/2162402X.2015.1090077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hollenbach P. W., Nguyen A. N., Brady H., Williams M., Ning Y., Richard N., Krushel L., Aukerman S. L., Heise C., MacBeth K. J., “A comparison of azacitidine and decitabine activities in acute myeloid leukemia cell lines,” PLoS One 5(2), e9001 (2010). 10.1371/journal.pone.0009001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee Y. G., Kim I., Yoon S. S., Park S., Cheong J. W., Min Y. H., Lee J. O., Bang S. M., Yi H. G., Kim C. S., Park Y., Kim B. S., Mun Y. C., Seong C. M., Park J., Lee J. H., Kim S. Y., Lee H. G., Kim Y. K., Kim H. J., “Comparative analysis between azacitidine and decitabine for the treatment of myelodysplastic syndromes,” Br. J. Haematol. 161(3), 339–347 (2013). 10.1111/bjh.12256 [DOI] [PubMed] [Google Scholar]
- 25.Ren J., Singh B. N., Huang Q., Li Z., Gao Y., Mishra P., Hwa Y. L., Li J., Dowdy S. C., Jiang S. W., “DNA hypermethylation as a chemotherapy target,” Cell. Signalling 23(7), 1082–1093 (2011). 10.1016/j.cellsig.2011.02.003 [DOI] [PubMed] [Google Scholar]
- 26.Linnekamp J. F., Butter R., Spijker R., Medema J. P., van Laarhoven H. W. M., “Clinical and biological effects of demethylating agents on solid tumours – A systematic review,” Cancer Treat. Rev. 54, 10–23 (2017). 10.1016/j.ctrv.2017.01.004 [DOI] [PubMed] [Google Scholar]
- 27.Malik P., Cashen A. F., “Decitabine in the treatment of acute myeloid leukemia in elderly patients,” Cancer Manage. Res. 6, 53–61 (2014). 10.2147/CMAR.S40600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Licht J. D., “DNA methylation inhibitors in cancer therapy: the immunity dimension,” Cell 162(5), 938–939 (2015). 10.1016/j.cell.2015.08.005 [DOI] [PubMed] [Google Scholar]
- 29.Eramo A., Pallini R., Lotti F., Sette G., Patti M., Bartucci M., Ricci-Vitiani L., Signore M., Stassi G., Larocca L. M., Crinò L., Peschle C., De Maria R., “Inhibition of DNA methylation sensitizes glioblastoma for tumor necrosis factor-related apoptosis-inducing ligand-mediated destruction,” Cancer Res. 65(24), 11469–11477 (2005). 10.1158/0008-5472.CAN-05-1724 [DOI] [PubMed] [Google Scholar]
- 30.Christman J. K., “5-Azacytidine and 5-aza-2’-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy,” Oncogene 21(35), 5483–5495 (2002). 10.1038/sj.onc.1205699 [DOI] [PubMed] [Google Scholar]
- 31.Oh S. J., Kim S.-H., Jeong K., Park Y., Huh Y.-M., Son J.-H., Suh J.-S., “Measurement depth enhancement in terahertz imaging of biological tissues,” Opt. Express 21(18), 21299 (2013). 10.1364/OE.21.021299 [DOI] [PubMed] [Google Scholar]
- 32.Kurdyukov S., Bullock M., “DNA methylation analysis: choosing the right method,” Biology (Basel, Switz.) 5(1), 3 (2016). 10.3390/biology5010003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zaika E., Wei J., Yin D., Andl C., Moll U., El-Rifai W., Zaika A. I., “p73 protein regulates DNA damage repair,” FASEB J. 25(12), 4406–4414 (2011). 10.1096/fj.11-192815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garama D. J., Harris T. J., White C. L., Rossello F. J., Abdul-Hay M., Gough D. J., Levy D. E., “A synthetic lethal interaction between glutathione synthesis and mitochondrial reactive oxygen species provides a tumor-specific vulnerability dependent on STAT3,” Mol. Cell. Biol. 35(21), 3646–3656 (2015). 10.1128/MCB.00541-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brodbelt J. S., “Photodissociation mass spectrometry: New tools for characterization of biological molecules,” Chem. Soc. Rev. 43(8), 2757–2783 (2014). 10.1039/C3CS60444F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tomas L., “Instability and decay of the primary structure of DNA,” Nature 362(6422), 709–715 (1993). 10.1038/362709a0 [DOI] [PubMed] [Google Scholar]
- 37.Sim Y. C., Park J. Y., Ahn K.-M., Park C., Son J.-H., “Terahertz imaging of excised oral cancer at frozen temeprature,” Biomed. Opt. Express 4(8), 1413–1421 (2013). 10.1364/BOE.4.001413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nawata K., Tokizane Y., Takida Y., Minamide H., “Tunable backward terahertz-wave parametric oscillation,” Sci. Rep. 9(1), 726 (2019). 10.1038/s41598-018-37068-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nugraha P. S., Krizsan G., Lombosi C., Palfalvi L., Toth G., Almasi G., Fulop J. A., Hebling J., “Demonstration of a tilted-pulse-front pumped plane-parallel slab terahertz source,” Opt. Lett. 44(4), 1023–1026 (2019). 10.1364/OL.44.001023 [DOI] [PubMed] [Google Scholar]
- 40.Bosco L., Franckie M., Scalari G., Beck M., Wacker A., Faist J., “Thermoelectrically cooled THz quantum cascade laser operationg up to 210 K,” Appl. Phys. Lett. 115(1), 010601 (2019). 10.1063/1.5110305 [DOI] [Google Scholar]
- 41.Wang K., Mittleman D. M., “Metal wires for terahertz wave guiding,” Nature 432(7015), 376–379 (2004). 10.1038/nature03040 [DOI] [PubMed] [Google Scholar]
- 42.Ji Y. B., Lee E. S., Kim S.-H., Son J.-H., Jeon T.-I., “A miniaturized fiber-coupled terahertz endoscope system,” Opt. Express 17(19), 17082–17087 (2009). 10.1364/OE.17.017082 [DOI] [PubMed] [Google Scholar]