Abstract
We present for the first time one-to-one correspondence between standard hematoxylin/eosin (H&E) stained tissue sections and stimulated Raman histology (SRH) - a label-free technique in which stimulated Raman scattering (SRS) and second harmonic generation (SHG) are combined to generate virtual H&E images. Experiments were performed on both human thin cryogenic slides from the gastrointestinal tract (GI) and thick freshly excised biopsies from endoscopic surgery. Results on cryogenic slides evidenced an excellent agreement between SRH and H&E images while the ones on biopsies established the relevance of SRH for rapid intraoperative histology to assist in surgical decision making.
1. Introduction
Recently, stimulated Raman histology (SRH) [1] has emerged as a label-free microscopy technique, using stimulated Raman scattering (SRS) [2], that produces virtually stained images that imitate standard eosin and hematoxylin (H&E) staining. SRH was first demonstrated on brain cryogenic sections [3] and later on applied to the gastrointestinal tract (GI) [4]. In the GI, SRS had to be combined with second harmonic generation (SHG) to concurrently extract the collagen distribution whose role is crucial in both cancer development and progression [5,6]. SRH is based on the ability of SRS to highlight cell bodies and cell nuclei distributions. The latter being a key element in cancer diagnoses as cell mitosis, nuclei aspects and sizes are central for pathologists to deliver accurate diagnoses [7]. The distinction between cell bodies and cell nuclei can be performed by investigating solely two wavenumbers by SRS at 2845 cm−1 and 2930 cm−1 corresponding to CH2 and CH3 chemical bonds, respectively [8–10]. This information is sufficient to generate H&E like images [3]. Adding the collagen fibrils organisation using SHG allows to generate HES (hematoxylin, eosin, saffron) images where both cell bodies, cell nuclei and fibrotic surrounding tissue are revealed for cancer stage and aggressiveness assessments [4]. Contrary to standard histology, SRH does not require any labelling and can perform virtual HES sections from unprepared fresh bulk biopsy samples opening the road for fast intra-operative histology to assist in surgical and decision making. So far earlier SRH reports [3,4] evaluated the agreement between SRH and H&E images on the same tissue but at different locations or on adjacent sections, but the one-to-one comparison on identical samples is still missing. In this paper we demonstrate for the first time the quasi-perfect matching between SRH and H&E images on identical cryogenic slides for both normal and cancerous tissues. Unprocessed cryogenic slides from human specimens were first imaged with SRH before their immediate staining using H&E for direct comparison. To further demonstrate the potential of SRH to be used in the intraoperative workflow, we performed SRH directly on fresh bulk (millimetre size) excised biopsies from gastric endoscopy. Here again SRH virtual sections showed comparable features to HES sections from the same patient. These results demonstrate the potential of SRH as a powerful tool for cancer diagnosis in an intraoperative context.
2. Material and methods
The three colour SRS setup used for these experiments follows the one presented in [11]. Briefly, two pump beams were generated by two optical parametric oscillators (OPO1 and OPO2 Emerald APE) pumped at 515 nm by a frequency doubled Ytterbium (Yb) fiber laser (2 ps pulse, repetition rate 80 MHz) (Emerald engine APE). The Yb laser emitting at 1031 nm was concomitantly used as the Stokes beam. Pump beams were tuned to 797.3 nm and 792.2 nm for OPO1 and OPO2 to match the molecular vibrations corresponding to CH2 (2845 cm−1) and CH3 (2930 cm−1) chemical bonds, respectively. OPO1 and OPO2 output were modulated at f1=14.34 MHz and f2=14.74 MHz respectively, using acousto-optic modulators (AA Optoelectronic MT200-AO). The three beams (1031 nm, 797.3 nm and 792.2 nm) were recombined using dichroic mirrors (TBP01-900/11 Semrock; DMSP900R Thorlabs), overlapped in space and time and send into a custom build scanning microscope [12]. A x40 objective (Nikon, PLAN, NA = 1.2 water) was used to focus the beams onto the sample while a x60 objective (Nikon, Fluor, NA = 1, water) was used as a condenser to collect the light in the forward direction. The Stokes beam was filtered (SP980, Semrock) and sent towards a photodiode (APE LIA photodiode) connected to a dual lock-in-amplifier (HF2LI Zurich Instrument) which demodulated the signal at the two distinct frequencies f1 and f2 to provide the two CH2 (2845 cm−1) and CH3 (2930 cm−1) SRS signals simultaneously at each pixel. Meanwhile the SHG signal was detected in the epi direction using a bandpass filter (425/50) and a photomultiplier tube (Perkin Elmer, MP-943). Laser powers used at the sample were 15mW for each beam for thin cryogenic slides and 25mW (for each beam) for thick biopsies. Large scale images were performed by stitching 100µm x 100 µm galvo-scanned images next to each other. The system was fully controlled by a custom made software [13]. For a 100 µm x 100 µm (200 × 200 pixels) galvo scan, a dwell time per pixel of 40 µs with 3 accumulations; a 1 mm x 1 mm stitched image required 25 min.
For cryogenic slides, fresh tissues were immersed into liquid nitrogen before being sectioned to 5 - 10µm thick slices. Tissue slices were deposited onto microscope slides and imaged with SRS and SHG without further preparation before being stained with H&E. Note that no cover glass was used, this is to allow H&E staining after SRS and SHG imaging; cryogenic slides were in direct contact with the water from the objective which sometimes induced tissue surface crinkling. To overcome this problem and to take into account for the thickness of the cryogenic slides several SRH and SHG images were acquired at different z planes, if needed, and summed up in order to catch relevant tissue information with depth in each spatial point.
For thick biopsies imaging, fresh cubic millimetre tissues were extracted from patients during surgery and squeezed between two coverslips to be imaged without further preparation while standard HES was performed in parallel on a different sample from the same tissue area. Data processing was performed using a SRH Matlab homemade software while H&E images were processed using the Calopix software.
The SRH Matlab based custom software used as input the three images (1) SRS2845cm-1, (2) SRSsous = SRS2930cm-1 - SRS2845cm-1 and (3) SHG. Homemade look up tables (LUT) in the pink, dark purple and orange/brown shades were respectively applied to (1), (2) and (3) in order to virtually reproduce eosin, hematoxylin and saffron (HES) staining and then merged together to produce the final SRH images as explained in details in [4].
3. Results and discussion
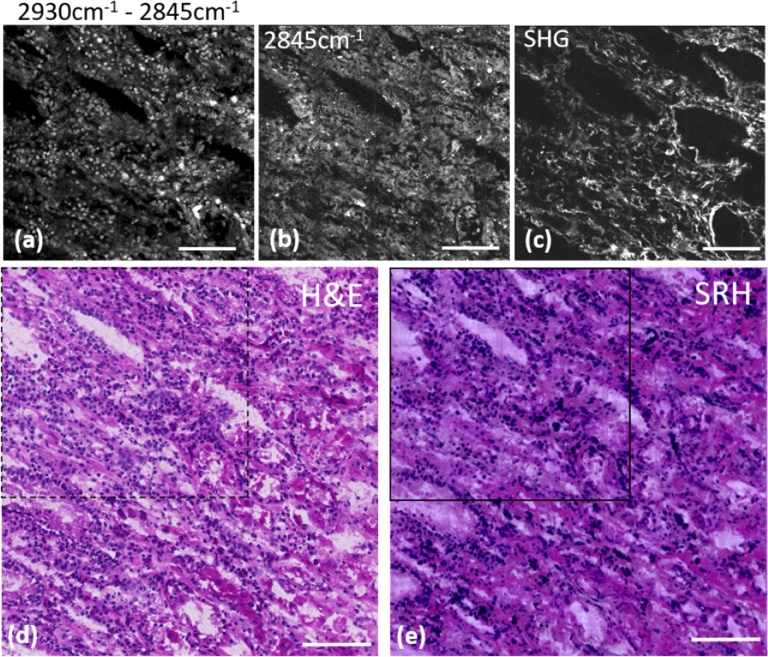
Figures 1 and 2 display one to one comparison of SRH with H&E on the same cryogenic slide for muscosa human stomach and normal human colon respectively. To quantitatively compare the techniques, a pink LUT was used to virtually stain the collagen of the stomach muscosa in virtual eosin so that quantitative correlation could be calculated between the SRH and H&E images. As a proof of concept the sample in Fig. 1 was inflammatory stomach tissue providing a higher concentration of nuclei with both various sizes and shapes as compared to normal stomach tissue. All the relevant features are present in both images: both nuclei distribution and cellular surrounding are equally present in both SRS (a, b), SRH (e) and H&E (d) images and glandular cells which have larger nuclei can be distinguished from the lympho-plasmocytic cells from the inflammatory chorion (Fig. 1(a), (d), (e)). Together with the inflammation, discrete collagen alterations can be seen -collagen fibers being discriminated using SHG (Fig. 1c)- that are indicative of a chronic gastritis. The coloured H&E (d) and SRH (e) images presented a correlation coefficient of 0.80. While high, this coefficient is believed to be mainly limited by the non-perfect matching of the colors assigned via the colouring program with H&E colors from the staining.
Fig. 1.
One to one SRH/H&E comparison on a 8 µm thick cryogenic slide of muscosa from human stomach. (a) Built image of SRS2930cm-1 – SRS2845cm-1 displaying the nuclei distribution of the region highlighted with the black squares in the H&E (dashed square) and SRH (solid square) images; (b) SRS image at 2845cm−1 highlighting the cell body distribution of the same region; (c) SHG image providing the collagen distribution of the same region. (d) H&E staining of a larger region (0.6mmx0.6mm) of the same cryogenic slide. (e) SRH image built using (a, b and c) displaying the same extended region as in (d) and demonstrated similar features. Scale bar is 100 µm.
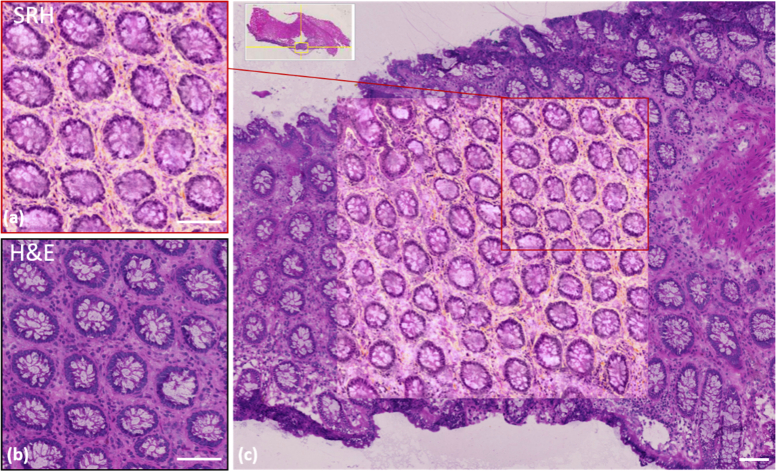
Fig. 2.
One to one SRH/H&E comparison on a 8µm thick cryogenic slide for normal human colon. (a) SRH image and (b) the H&E corresponding image of the same cryogenic region. (c) SRH image of a wider (1 mm x 1 mm) area encapsulated into a larger region of the identical slide stained with H&E. The red square corresponds to (a). Scale bar is 100 µm
On Fig. 2, one can observe the excellent agreement between SRH (a) and H&E (b) images on the same 0.5 mm x 0.5 mm area of normal colon muscosa. All the nuclei present around the crypts as well as the quasi-totality of the nuclei intercalated within the collagen fibrils can be identified. SRH and H&E crypt sizes, number and forms are strictly identical as well as vacuoles aspects, the latter being due to the sectioning process. SRH images provide more information (i.e. collagen fibrils in orange shades) as H&E staining on fresh samples did not include saffron-staining. Figure 2(c) allows better appreciation of the concordance between the techniques especially at the edges of the SRH image where the transitions with the H&E image match for all crypts, and collagen fibres. When a pink LUT was applied to the collagen distribution to build the SRH image of colon (data not shown) quantitative correlation could be calculated and also provided a correlation factor close to 0.80.
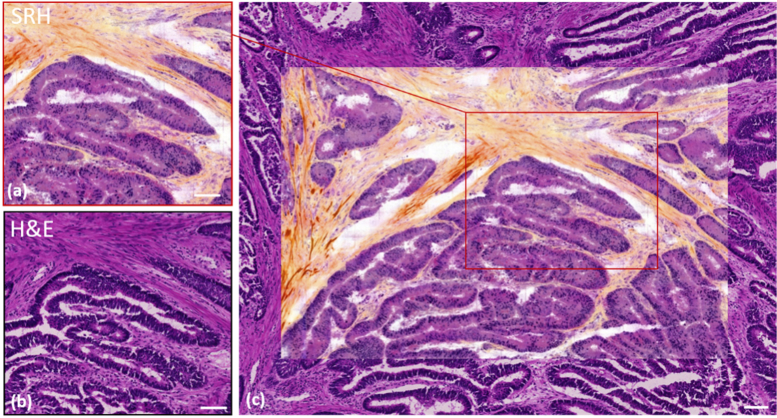
The same conclusions of a close-to-perfect agreement between the SRH and H&E images for a cancerous (adenocarcinoma) colon cryogenic slide is evidenced in Fig. 3. Figure 3(a) and (b) show the same region imaged with both approaches while (c) offers a wider field of view containing both the SRH image (in the middle) superimposed with the H&E image. Close examination of Fig. 3(a) and (b) shows that the detailed tissue architecture is equally revealed with SRH and H&E. For instance, collagen fibres surrounding elongated glands can be seen. These glands lack vacuoles and therefore show a high level of muco secretion loss. A closer examination of the glands shows that accurate details such as nuclei incorporated into collagen fibres are also well defined in both cases.
Fig. 3.
One to one SRH/H&E comparison on a 10 µm thick cryogenic slide for cancerous (adenocarcinoma) human colon. (a) SRH image and (b) the H&E corresponding image of the same cryogenic region. (c) SRH image of a wider area (1.2 mm x 0.8 mm) encapsulated into a larger region of the identical slide stained with H&E. The red rectangle corresponds to (a). Scale bar is 100µm.
Nuclei size differs slightly in the glands between SRH and H&E, this is due to the relatively large thickness of the cryogenic slide (10µm) that somehow degrades the quality of the H&E image while the SRH image provides a sharper sectioning along the z axis due to its multiphoton nature. This intrinsic SRH z sectioning ability is also responsible for the few missing nuclei in both Fig. 2(a) and Fig. 3(a) the SRH acquisition plane ‘missing’ these few out-of-focus nuclei. In Fig. 3(c), the collagen fibres revealed by SHG clearly delineate the frontier between SRH and H&E images. Here again, on Fig. 3(c) we observe an excellent match between collagen fibres, glands and nuclei at the SRH / H&E junction. Both these examples demonstrate the capability of SRH to produce high quality histological images with relevant meaning in comparison with standard H&E staining.
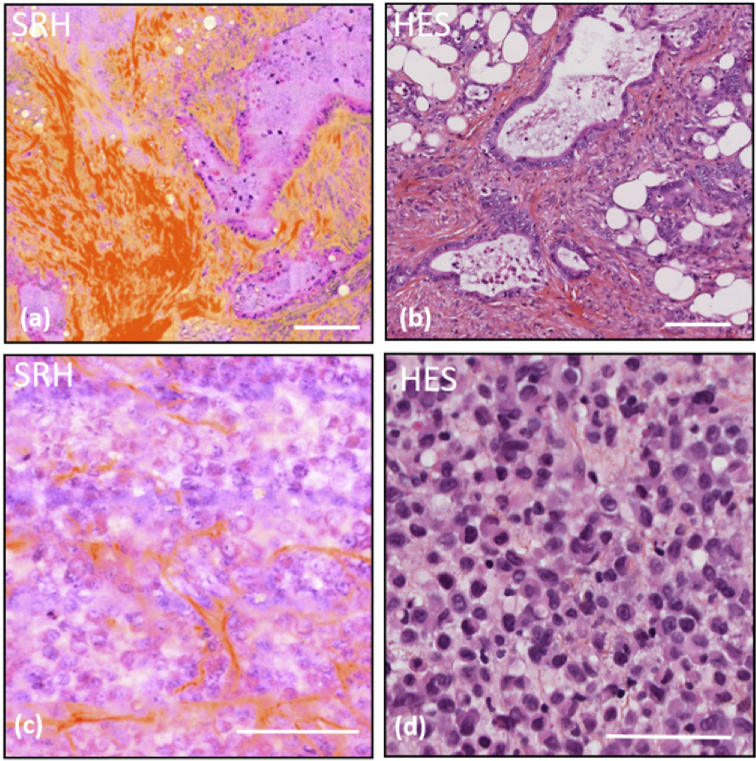
To further establish the potential for SRH to be used in an intraoperative context, experiments were performed in situations where intra-operative histology on biopsy dictates the surgery decision. For instance, in the GI the detection of peritoneal metastasis during a gastric surgery stops immediately the operation and orients the patient toward chemotherapy treatments. Such decisive feature can be revealed using SRH as exemplified in Fig. 4(a) and (b) that compares SRH and H&E images taken from omentum slices. Healthy omemtum is made of adipose tissue solely and the presence of glands and collagen fibres is a sign of malignancy of the tissue. The presence of atypical glands shown in Fig. 4(a) and (b) attest for adenocarcinomatous localisation.
Fig. 4.
SRH/HES comparison for critical pathological situations. (a) SRH image of a cancerous omemtum, (b) standard HES of cancerous omemtum from the same patient, (c) SRH image from a thick biopsy sample, imaged directly after its gastric endoscopic excision, of a poorly differentiated adenoma case, (d) standard HES from the same patient as in (c). Scale bar is 100 µm.
Most importantly such key features can be readily identified by SRH on unprocessed thick biopsy samples directly after their excision in the operatory room. In this case SRH is advantageous as it avoids the burden of work performed during the operatory workflow that requires slicing and H&E staining. Figure 4(c) shows a SRH images performed on a fresh thick colon biopsy directly excised from gastric endoscopy. It can be compared with Fig. 4(d) that presents a conventional HES image taken from the same tissue. Here both SRH and H&E images reveal a poor differentiated adenocarcinoma case. Three elements have to be taken into account to deliver an accurate diagnostic: (I) the number of nucleoli (when DNA is more concentrated within the nuclei), (II) cytosolic to nuclei ratio and (III) nuclei size and shape diversity.
As shown in both Fig. 4(c) (SRH) and (d) (HES), no characteristic gland architecture can be seen, nuclei are numerous within thin collagen fibres attested to a poor differentiated adenocarcinoma. Numerous nucleioli are revealed in both SRH and HES images and some nuclei with both abnormal sizes or shapes are also visible. Cytosolic content in pink shades can be identified with both methods while collagen fibres are better highlighted in SRH. Note that the SRH image quality on this thick biopsy performed 30 µm below the tissue surface is similar to the one obtained on thin cryogenic slides. Furthermore, SRH image (Fig. 4(c)) was obtained in 25 minutes whereas the HES image (Fig. 4(d)) required 24h. SRH speed could be further improved by a factor 2 to 4 using less accumulation and a dual focus strategy [14]. These results demonstrate the viability and relevance of SRH in an intraoperative context to reveal key histological tissue features for surgery guidance and decision making.
4. Conclusions
In this paper, we evidenced for the first time the quasi-faultless similarity between SRH and standard H&E staining via one-to-one comparison over the same cryogenic slides for healthy and cancerous (adenocarcinoma) human colon. We also showed the relevance of SRH images to rapidly identify critical intraoperative situations such as the detection of peritoneal metastasis in omentum that dramatically affects the gastric surgery workflow. Finally, we showed that SRH is suitable to image unprocessed thick biopsy samples directly after their excisions in the operatory room providing similar information than conventional HES images. This study provides a first demonstration of the direct correspondence between SRH with HES images and paves the way for extended studies in human oncology, and more generally for rapid histology. Ultimately SRH could be performed directly in living tissue if implemented in intraoperative microscopes [15] or in flexible endoscopes as recently reported [16].
Acknowledgments
We acknowledge support from the Centre National de la Recherche Scientifique (CNRS) and Aix-Marseille University A*Midex.
Funding
Centre National de la Recherche Scientifique10.13039/501100004794; Aix-Marseille Université10.13039/100007586; Agence Nationale de la Recherche10.13039/501100001665 (A-M-AAP-ID-17-13-1702228-15.22-RIGNEAULT, ANR-11-IDEX-0001-02, ANR-11-INSB-0006); Institut National de la Santé et de la Recherche Médicale10.13039/501100001677 (18CP128-00, PC201508).
Disclosures
The authors declare that there are no conflicts of interest related to this article. All tissue sections were collected in the context of Institut Paoli-Calmettes approved protocol from patients who provided informed consent. Tissues in excess of what was needed for diagnosis were eligible for imaging. All methods were performed in accordance with the European Union guidelines and regulations Commission Directive 2006/17/EC of 8 February 2006 as regards technical requirements for the donation, procurement and testing of human tissues and cells.
References
- 1.Cicerone M. T., Camp C. H., “Histological coherent Raman imaging: a prognostic review,” Analyst 143(1), 33–59 (2018). 10.1039/C7AN01266G [DOI] [PubMed] [Google Scholar]
- 2.Rigneault H., Berto P., “Tutorial: Coherent Raman light matter interaction processes,” APL Photonics 3(9), 091101 (2018). 10.1063/1.5030335 [DOI] [Google Scholar]
- 3.Orringer D. A., Pandian B., Niknafs Y. S., Hollon T. C., Boyle J., Lewis S., Garrard M., Hervey-Jumper S. L., Garton H. J. L., Maher C. O., Heth J. A., Sagher O., Wilkinson D. A., Snuderl M., Venneti S., Ramkissoon S. H., McFadden K. A., Fisher-Hubbard A., Lieberman A. P., Johnson T. D., Xie X. S., Trautman J. K., Freudiger C. W., Camelo-Piragua S., “Rapid intraoperative histology of unprocessed surgical specimens via fibre-laser-based stimulated Raman scattering microscopy,” Nat. Biomed. Eng. 1(2), 0027 (2017). 10.1038/s41551-016-0027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sarri B., Canonge R., Audier X., Simon E., Wojak J., Caillol F., Cador C., Marguet D., Poizat F., Giovannini M., Rigneault H., “Fast stimulated Raman and second Harmonic generation imaging for intraoperative gastro-intestinal cancer detection,” Sci. Rep. 9(1), 10052 (2019). 10.1038/s41598-019-46489-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ueno H., Jones A. M., Wilkinson K. H., Jass J. R., Talbot I. C., “Histological categorisation of fibrotic cancer stroma in advanced rectal cancer,” Gut 53(4), 581–586 (2004). 10.1136/gut.2003.028365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Z. H., Ji C. D., Xiao H. L., Zhao H. B., Cui Y. H., Bian X. W., “Reorganized Collagen in the Tumor Microenvironment of Gastric Cancer and Its Association with Prognosis,” J. Cancer (Wyoming, Aust.) 8(8), 1466–1476 (2017). 10.7150/jca.18466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baba A. I., Câtoi C., Comparative Oncology (The Publishing House of the Romanian Academy, 2007). [PubMed] [Google Scholar]
- 8.Freudiger C. W., Pfannl R., Orringer D. A., Saar B. G., Ji M. B., Zeng Q., Ottoboni L., Ying W., Waeber C., Sims J. R., De Jager P. L., Sagher O., Philbert M. A., Xu X. Y., Kesari S., Xie X. S., Young G. S., “Multicolored stain-free histopathology with coherent Raman imaging,” Lab. Invest. 92(10), 1492–1502 (2012). 10.1038/labinvest.2012.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ji M., Orringer D. A., Freudiger C. W., Ramkissoon S., Liu X., Lau D., Golby A. J., Norton I., Hayashi M., Agar N. Y., Young G. S., Spino C., Santagata S., Camelo-Piragua S., Ligon K. L., Sagher O., Xie X. S., “Rapid, label-free detection of brain tumors with stimulated Raman scattering microscopy,” Sci. Transl. Med. 5(201), 201ra119 (2013). 10.1126/scitranslmed.3005954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He R., Xu Y., Zhang L., Ma S., Wang X., Ye D., Ji M., “Dual-phase stimulated Raman scattering microscopy for real-time two-color imaging,” Optica 4(1), 44–47 (2017). 10.1364/OPTICA.4.000044 [DOI] [Google Scholar]
- 11.Heuke S., Sarri B., Audier X., Rigneault H., “Simultaneous dual-channel stimulated Raman scattering microscopy demultiplexed at distinct modulation frequencies,” Opt. Lett. 43(15), 3582–3585 (2018). 10.1364/OL.43.003582 [DOI] [PubMed] [Google Scholar]
- 12.Brustlein S., Ferrand P., Walther N., Brasselet S., Billaudeau C., Marguet D., Rigneault H., “Optical parametric oscillator-based light source for coherent Raman scattering microscopy: practical overview,” J. Biomed. Opt. 16(2), 021106 (2011). 10.1117/1.3533311 [DOI] [PubMed] [Google Scholar]
- 13.Ferrand P., “GPScan.VI: A general-purpose LabVIEW program for scanning imaging or any application requiring synchronous analog voltage generation and data acquisition,” Comput. Phys. Commun. 192, 342–347 (2015). 10.1016/j.cpc.2015.03.010 [DOI] [Google Scholar]
- 14.Heuke S., Sarri B., Lombardini A., Audier X., Rigneault H., “Dual-focus stimulated Raman scattering microscopy: a concept for multi-focus scaling,” Opt. Lett. 43(19), 4763–4766 (2018). 10.1364/OL.43.004763 [DOI] [PubMed] [Google Scholar]
- 15.Killory B. D., Nakaji P., Gonzales L. F., Ponce F. A., Wait S. D., Spetzler R. F., “Prospective evaluation of surgical microscope-integrated intraoperative near-infrared indocyanine green angiography during cerebral arteriovenous malformation surgery,” Neurosurgery 65(3), 456–462 (2009). 10.1227/01.NEU.0000346649.48114.3A [DOI] [PubMed] [Google Scholar]
- 16.Lombardini A., Mytskaniuk V., Sivankutty S., Andresen E. R., Chen X., Wenger J., Fabert M., Joly N., Louradour F., Kudlinski A., Rigneault H., “High-resolution multimodal flexible coherent Raman endoscope,” Light: Sci. Appl. 7(1), 10 (2018). 10.1038/s41377-018-0003-3 [DOI] [PMC free article] [PubMed] [Google Scholar]