Abstract
γ-Aminobutyric acid (GABA), the major inhibitory neurotransmitter, is challenging to measure using proton spectroscopy due to its relatively low concentration, J-coupling and overlapping signals from other metabolites. Currently, the prevalent methods for detecting GABA at ultrahigh field strengths (≥ 7 T) are GABA-editing and model fitting of non-editing single voxel spectra. These two acquisition approaches have their own advantages: the GABA editing approach directly measures the GABA resonance at 3 ppm, whereas the fitting approach on the non-editing spectrum allows the detection of multiple metabolites, and has an SNR advantage over longer echo time (TE) acquisitions. This study aims to compare these approaches for estimating GABA at 7 T. We use an interleaved sequence of semi-LASER (sLASER: TE = 38 ms) and MEGA-sLASER (TE = 80 ms). This simultaneous interleaved acquisition minimizes the differential effect of extraneous factors, and enables an accurate comparison of the two acquisition methods. Spectra were acquired with an 8 ml isotropic voxel at six different brain regions: anterior-cingulate cortex, dorsolateral-prefrontal cortex, motor cortex, occipital cortex, posterior cingulate cortex, and precuneus. Spectral fitting with LCModel quantified the GABA to total Cr (tCr: Creatine + Phosphocreatine) concentration ratio. After correcting the T2 relaxation time variation, GABA/tCr ratios were similar between the two acquisition approaches. GABA editing showed smaller spectral fitting error according to Cramér–Rao lower bound than the sLASER approach for all regions examined. We conclude that both acquisition methods show similar accuracy but the precision of the MEGA-editing approach is higher for GABA measurement. In addition, the 2.28 ppm GABA resonance was found to be important for estimating GABA concentration without macromolecule contamination in the GABA-edited acquisition, when utilizing spectral fitting with LCModel.
Introduction
γ-Aminobutyric acid (GABA), the major inhibitory neurotransmitter, is present at about one millimolar concentration in the human brain [1]. GABA plays an important role as a potential biomarker in neurological and neuropsychiatric disorders such as cancer [2], multiple sclerosis [3], Alzheimer’s disease [4], epilepsy [5], schizophrenia [6], and autism [7].
Existing in-vivo approaches for measuring GABA include positron emission tomography (PET) [8], single photon emission tomography (SPECT) [9], proton magnetic resonance spectroscopy (1H MRS) [10] and chemical exchange saturation transfer (CEST) [11, 12]. Among these, MRS at ultrahigh filed (UHF ≥ 7 T) is widely used due to high sensitivity and specificity for measuring GABA [13]. Nevertheless, GABA MRS is still challenging by reason of its low concentration and resonances overlapping with higher concentration signals [14]. Previously, various acquisition strategies for proton MRS have been proposed for measuring GABA, such as J-editing [15] (e.g. MEGA-PRESS (MEsher-GArwood Point RESolved Spectroscopy) [16, 17]) and double quantum filters [18].
The MEGA-editing method [14] is the most commonly used approach. A MEGA pulse is used to distinguish the GABA signal at 3 ppm from the overlapping creatine signal, based on the J difference editing approach [16, 17]. MEGA-editing combines conventional localization techniques with one or more additional frequency selective editing pulses, which invert the signal at 1.9 ppm. As the GABA resonance at 3 ppm signal is coupled to the 1.9 ppm GABA resonance, the difference between the signals obtained with and without the editing pulses should give an unambiguous GABA signal at 3 ppm. However, the MEGA-editing method also has disadvantages. It theoretically works best at certain echo times (TEs), when the GABA is refocused in anti-phase to the signal obtained without editing pulses (TE = (2n-1) / 2J, where J is 7.35 Hz, n = 1, 2, 3…). Therefore, the TE for GABA editing is largely standardized at 68 ms as the next possible TE beyond 68 ms (204 ms) would imply a significant T2 decay of the signal. At 7 T, it is quite challenging to implement the MEGA-semi-LASER(MEGA-sLASER) sequence with TE = 68 ms due to the lack of time to insert all pulses within 68ms. Therefore, longer TEs have frequently been used (e.g., 72 ms [19] and 74 ms [6, 20, 21]. However, Edden, et al. [22] reported that a TE of 80 ms showed a reduction in co-edited macromolecule (MM) compared to TE 68 ms even though the GABA SNR is reduced by approximately 7% in comparison with that of TE = 68 ms due to the combined but opposing effects of increased editing efficiency and greater T2-relaxation. In addition, motion, respiration, and frequency drift induce experimental instabilities that may result in subtraction artifacts [23].
Another extensively used GABA measurement method is to obtain a non-edited spectrum, and measure the GABA signals using spectral fitting via LCModel [24, 25]. As these data are normally acquired at a shorter TE, the acquired spectrum is superior to the edited spectrum in terms of sensitivity. In addition, not only GABA but also all metabolites are quantified simultaneously. However, a spectral fitting procedure is required in order to measure metabolite concentrations. Previously, a number of studies have shown the feasibility of GABA detection in combination with LCModel analysis at different field strength: 3 T [26, 27], 4 T [28] and 7 T [29, 30]. Even though GABA is detectable at various field strengths, it has also been reported that spectra that are acquired at higher field strength show superior fitting precision due to the high spectral dispersion, better SNR and simplified spectra [31].
Given these two leading but different GABA measurement techniques: GABA-editing and non-editing acquisitions, there have been various attempts to assess their relative merits. Terpstra, et al. [32] compared STEAM (Stimulated Echo Acquisition Mode [33–35], TE = 5 ms) with MEGA-PRESS (TE = 68 ms) at 4 T in a validation study measuring glutathione (GSH). The concentrations of GSH measured with these two methods in that study were similar, suggesting a similar accuracy of the two techniques for those conditions. Sanaie Nezhad, et al. [36] compared the accuracy and sensitivity of PRESS (Point RESolved Spectroscopy [37, 38], TE = 35 ms) and MEGA-PRESS (TE = 130 ms) at 3 T to measure GSH, and found that PRESS is not an accurate and reliable method to measure GSH in vivo. They also showed that the spectral fitting of the PRESS spectra cannot reliably quantify the concentration of GSH when the concentration is 4 mM or less. In addition, one conference proceeding by Chen, et al. [39] reported that the GABA to total Cr (tCr: creatine (Cr) + phosphocreatine(PCr)) ratios from occipital cortex (OC) and motor cortex (MC), measured by the MEGA sLASER (TE = 72ms) and STEAM (TE = 17ms) approaches, are comparable after T2 relaxation correction. However, after MM correction, GABA editing results were superior to those of the non-editing spectral acquisitions in terms of both reliability and reproducibility. Nevertheless, the authors still suspected that MM contamination and editing efficiency might have had an influence on GABA quantification.
In addition, it is widely known that the 3 ppm signal measured with the GABA-editing technique is typically contaminated by co-edited MM due to the overlapped resonance frequencies of the GABA and MM by the editing pulse, and this is generally considered to be a methodological limitation of this approach [14, 40, 41]. Therefore, this measured 3 ppm signal is commonly referred to as GABA+: GABA ‘plus’ co-edited MM [17, 42].
A standard approach to obtain (relative) GABA concentrations from non-edited single voxel spectroscopy (SVS) spectra is to use LCModel [24, 25] that fits metabolites together with additional MM, and lipid baselines. Furthermore, the GABA signal is fitted to all three methylene groups of GABA, of which the resonance at 2.28 ppm is uncontaminated by the MM signal. This should ensure that the GABA estimated in this way does not include MM. We hypothesize that if the MEGA-edited spectrum also contains a visible signal at 2.28 ppm then application of LCModel will similarly yield estimates of GABA concentration that are free of MM contamination. We can test this hypothesis by obtaining spectra from the same voxel using both acquisition techniques, to test whether the GABA concentrations obtained are the same. This study aims to compare two GABA measurement methods in combination with LCModel fitting by comparing relative GABA concentrations and corresponding spectral fitting quality at various brain regions of healthy individuals. In order to minimize the effect of external factors, a single interleaved sequence, which acquires spectra of the two measurement methods simultaneously was used. As a result, factors such as motion, shim, and B1 inhomogeneity should be the same for both techniques.
Material and methods
Sequence implementation
The MEGA method can be implemented with conventional SVS acquisition methods. It was initially implemented in a PRESS sequence [16, 17, 37, 38]. However, at ultrahigh field, PRESS loses efficiency because of higher B1 field inhomogeneity. In contrast, the sLASER sequence [43] offers improved performance mainly because of the use of adiabatic refocusing pulses that are insensitive to B1 field inhomogeneity and have high bandwidth, leading to a small chemical shift displacement error (CSDE) [44].
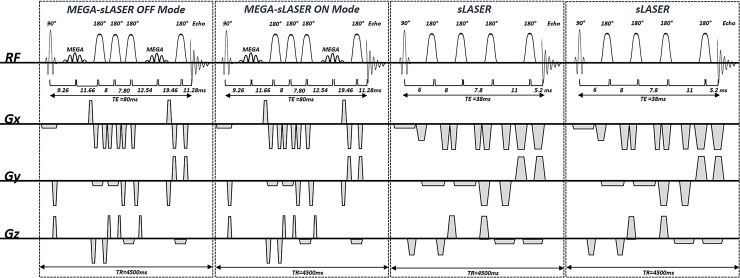
In this study, the sLASER sequence was used to acquire non-editing in-vivo spectra, and MEGA-editing was performed on the basis of the same sLASER sequence, termed MEGA-sLASER [20]. MEGA-sLASER and sLASER SVS spectra were acquired using a single interleaved sequence shown in Fig 1. It consists of the repeated application of four sub sequence blocks: a MEGA-off sLASER, a MEGA-on sLASER, and two sLASER acquisitions. The repetition time (TR) of each sub sequence block was 4500ms.
Fig 1. The pulse sequence diagram of the interleaved sequence.
This figure shows a pulse sequence diagram of the interleaved MEGA-semi-LASER (echo time (TE) = 80 ms) and semi-LASER (TE = 38ms) sequence implemented in the current study. This sequence is composed of four sub-sequence blocks, which have identical repetition times of 4500 ms.
In the implementation of the sLASER sequence with TE = 38 ms, a Shinnar-Le Roux (SLR) 90° excitation pulse (duration = 3.4 ms and bandwidth = 3.5 kHz) was used for slice selection in one direction and two pairs of hyperbolic secant 180° refocusing pulses (duration = 5 ms and bandwidth = 5.3 kHz) were used for slice selection in the two other directions. The spoiler gradients had an amplitude of 25 mT/m and their duration varied between 0.8 and 2 ms. WET (Water suppression Enhanced through T1 effects) water suppression [45] with four RF pulses was used to suppress the water signal. This has a lower sensitivity to B1 variations compared with the three RF pulses WET water suppression. The water suppression block was placed before the localization block of the sequence and is not depicted in the sequence diagram. TE of 38 ms for this sequence is a slightly conservative choice to ensure elimination of unwanted signals and but comparable to those of other sLASER studies at the same field strength; 24 ms [46], 25 ms [47], 28 ms [6], 32 ms [31] and 36ms [48].
MEGA-sLASER was implemented with a TE of 80 ms [22]. Localization components of this sequence were the same as for the sLASER, as described above. A pair of dual-band inversion pulses (duration = 11.52 ms and bandwidth = 133 Hz) was used for both the MEGA-editing and extra water suppression. This editing pulse inverts at 1.9 and 4.7 ppm in on-mode, and at 4.7 and 7.5 ppm in off-mode. Since additional MM suppression was not included in the acquisition sequence except a prolonged TE of 80 ms, our measured GABA signal at 3 ppm is also GABA +.
Ethics statement
The study was conducted at the Erwin L. Hahn Institute for Magnetic Resonance Imaging, University of Duisburg-Essen, Essen, Germany. The experimental protocol was approved by the Ethics Commission of the Medical Faculty of the University Duisburg-Essen (study number 11–4898-BO). All participants provided written informed consent in accordance with the declaration of Helsinki.
Data acquisition
12 healthy volunteers (9 male and 3 female subjects, 29.33 ± 4.49 years old) participated in this study. Experiments were performed on a 7 T whole body system (Magnetom, Siemens, Erlangen, Germany) with a 32-channel transmit/receive (TX/RX) bird-cage coil (Nova Medical, Wilmington, MA). First, T1 weighted images were acquired as an anatomical reference using a three-dimensional magnetization-prepared rapid gradient-echo (3D MPRAGE) [49] with the following scan parameters: 256 slices, slice thickness = 1 mm, TR = 2500 ms, TE = 1.35 ms, inversion time (TI) = 1100 ms, flip angle = 6°, field of view = 256 x 256 x 256 mm3, scan time ≈ 5 min. B0 shimming was performed using FASTESTMAP [50]. Single voxel MRS data were collected from 20 x 20 x 20 mm3 voxels positioned at anterior cingulate cortex (AC), dorsolateral prefrontal cortex (DLPFC), MC, OC, posterior cingulate cortex (PC), and precuneus (PRC) using the interleaved MEGA-sLASER and sLASER sequence (TR = 4500 ms, TEs = 80, 80, 38 and 38 ms, 32 averages of each sub-sequence, bandwidth = 3000 Hz, data points = 1024 and total scan time ≈ 10 min).
Data processing
The signals from each coil element were combined by the signal weighting method [51] that the manufacturer provides by default. MEGA difference spectra were calculated with the jMRUI software package (Version 6.0, http://www.jmrui.eu) [52, 53]. The nominal spectral resolution was increased to 0.27 Hz by zero-filling for a more exact adjustment. The MEGA-edited spectra were acquired by subtracting MEGA-on and MEGA-off spectra after alignment with the 3 ppm peak by the horizontal shift along the frequency axis, to avoid subtraction artifacts [23] caused by frequency drift. After subtraction, 1 Hz line broadening was applied to remove the zero-filling induced high-frequency noise. The same levels of zero-filling and line broadening were applied to the GABA-editing and the non-editing acquisition spectra.
Data analysis
Spectral quality was evaluated on the basis of the SNR and was measured using the 2ppm resonance of total N-acetylaspartate (NAA) and the standard deviation of the white noise area in the 7 to 9 ppm range, for both GABA-editing and non-editing acquisition spectra.
LCModel software (Version 6.3-1L, Stephen Provencher, Ontario, Canada) [24, 25] was used to estimate metabolite concentrations. The Cramér–Rao lower bound (CRLB), was taken as giving the error in the metabolite quantification (expressed in %SD) [24]. For the basis set for the LCModel analysis, parametric spectral models of alanine (Ala), aspartate (Asp), ascorbate (Asc), glycerophosphocholine (GPC), choline (Cho), phosphocholine (PCh), Cr, PCr, GABA, glucose (Glc), glutamine (Gln), glutamate (Glu), glycine (Gly), GSH, myo-inositol (mI), lactate (Lac), NAA, N-acetylaspartylglutamate (NAAG), phosphoethanolamine (PE), scyllo-inositol (Scyllo) and taurine (Tau) were simulated using the NMRSIM module included in TOPSPIN suite (Version 3.6, Bruker, Rheinstetten, Germany) with identical parameters (e.g. RF pulse profile, RF pulse timing, resonance frequency, and acquisition bandwidth) to those used for the in-vivo acquisition. Chemical shift and J-coupling values for each metabolite were taken from [54, 55].
For the sLASER data, the basis set for the LCModel analysis consisted of all twenty-one simulated metabolites. For the GABA editing method, six edited metabolites were modelled: GABA, Glu, Gln, NAA, NAAG, and GSH were included in the basis set. Each edited spectral model was created by subtracting a simulated MEGA-off spectrum from a simulated MEGA-on spectrum. As LCModel performs a phase correction during spectral fitting, additional phase correction steps were not applied. For MM and lipid signals, the non-parametric basis sets that LCModel provides by default were used [56]. However, one singlet peak with Lorentzian lineshape was also included in the GABA-editing basis set model the MM signal at 3 ppm, and thus avoid over-estimation of GABA due to the co-edited MM [57].
We report regional GABA concentrations as a ratio relative to tCr that was normalized across the scans. We used the tCr signal in the MEGA-off spectrum as an internal reference for the MEGA-editing approach. Previous studies used cut-off criteria of 20% [58] or 30% [59] for CRLB, our study included all GABA and Cr results to avoid CRLB thresholding leading to a bias in measuring low concentration metabolites [60]. Spectral fitting quality was compared with CRLB. In addition, absolute uncertainty was calculated by multiplying the relative CRLB by the estimated concentration [60].
As the data were acquired at different TEs, the signal concentration differences were also compensated [61] for T2. We assumed similar T2 relaxation times in GM and WM for each metabolite, and used values taken from the literature of: T2 tCr = 121 ms and T2 GABA = 63 ms [62] at 7 T. T1 weighted images of each spectroscopy voxel of interest (VOI), which were also used for spectroscopy voxel placement, were segmented using SPM12 (Wellcome Trust Centre for Neuroimaging, University College London, UK) unified segmentation routines to determine the relative proportions of GM, WM and cerebrospinal fluid (CSF) within the spectroscopy voxel. We calculated a volume fraction for each tissue component.
Regional GABA concentrations and CRLBs were also compared with a one-way ANOVA was performed using SPSS (Ver. 22, IBM, NY). The null hypothesis was that there is no difference between regions. A p-value of less than 0.05 was considered statistically significant. Bland-Altman analysis [63] as implemented in MedCalc Statistical Software v18.11.6 (MedCalc Software bvba, Ostend, Belgium) was used to compare the raw GABA concentrations (determined by LCModel analysis) acquired by the two acquisition approaches over all voxels.
Potential MM contributions in the edited 3 ppm peak of the MEGA approach were also approximated by amplitude ratios between the fitted GABA line by LCModel and the acquired GABA+ peak at 3 ppm.
Results
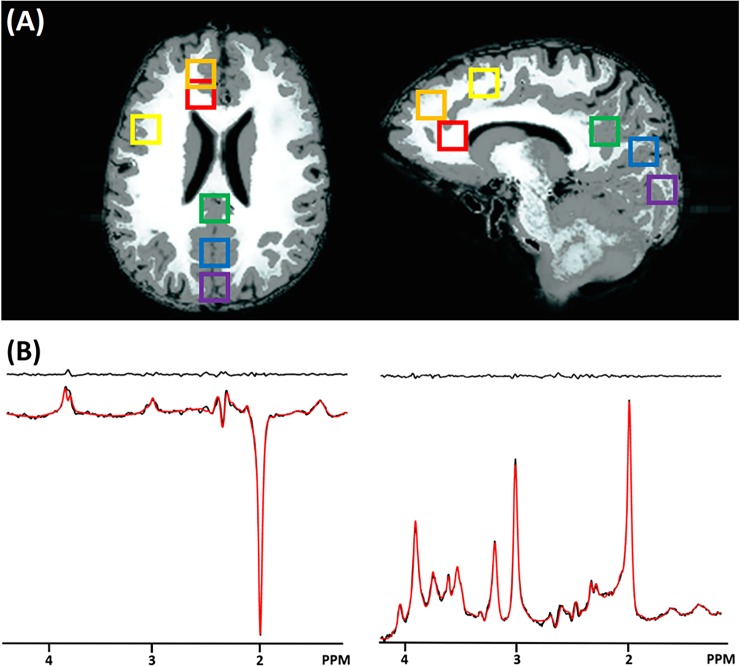
Water linewidths obtained after performing 2–3 iterations with the FASTESTMAP sequence were below 14 Hz for all voxels. Fig 2A depicts locations of VOIs on the GM (gray) and WM (white) maps after T1 weighted image segmentation. Fig 2B shows examples of fitting GABA with LCModel using sLASER (top) and the MEGA-sLASER (bottom) for all six regions. Table 1 summarizes mean metabolic concentration ratios between GABA and tCr, CRLB values and absolute uncertainties for GABA and tCr at six different brain regions for the two different acquisition approaches. Measured spectral SNRs are also summarized in Table 1.
Fig 2. Spectroscopy voxel positions and exemplary results.
(A) Six spectroscopy voxel positions: anterior cingulate cortex (AC: red), dorsolateral prefrontal cortex (DLPFC: orange), motor cortex (MC: yellow), occipital cortex (OC: purple), posterior cingulate cortex (PC: green), and precuneus (PRC: blue) on the GM (gray) and WM (white) map. (B) Example results of the LCModel analysis for the MEGA-sLASER (left) and the sLASER (right) that were acquired with the interleaved acquisition from a single voxel (PRC). The fitted line is plotted in red and residual signal (the difference between in-vivo and fitting lines) is plotted at the top.
Table 1. Averaged SNR and LCModel results from six brain regions by the semi-LASER (sLASER) and the MEGA-sLASER approaches.
| SNR | Concentration ratio | CRLB(%) | Absolute uncertainty | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GABA/tCr | GABA | tCr | GABA | tCr | ||||||||
| Technique | sLASER | MEGA-sLASER | sLASER | MEGA-sLASER | sLASER | MEGA-sLASER | sLASER | MEGA-sLASER | sLASER | MEGA-sLASER | sLASER | MEGA-sLASER |
| TE (ms) | 38 | 80 | 38 | 80 | 38 | 80 | 38 | 80 | 38 | 80 | 38 | 80 |
| AC | 511.80 ± 90.25 | 325.17 ± 62.40 | 0.13 ± 0.06 | 0.07 ± 0.03 | 41.20 ± 8.42 | 21.33 ± 6.60 | 1.00 ± 0.00 | 1.83 ± 0.67 | 0.17 ± 0.07 | 0.05 ± 0.02 | 0.03 ± 0.01 | 0.02 ± 0.01 |
| DLPFC | 435.37 ± 146.20 | 388.87 ± 93.90 | 0.22 ± 0.09 | 0.10 ± 0.02 | 31.33 ± 6.70 | 26.00 ± 6.68 | 1.33 ± 0.47 | 1.33 ± 0.47 | 0.18 ± 0.02 | 0.06 ± 0.02 | 0.05 ± 0.02 | 0.03 ± 0.01 |
| MC | 540.78 ± 155.41 | 294.18 ± 57.73 | 0.16 ± 0.03 | 0.11 ± 0.05 | 36.50 ± 4.11 | 19.50 ± 12.08 | 1.17 ± 0.37 | 1.17 ± 0.37 | 0.17 ± 0.03 | 0.05 ± 0.03 | 0.05 ± 0.03 | 0.04 ± 0.03 |
| OCC | 417.24 ± 37.00 | 410.63 ± 126.73 | 0.28 ± 0.03 | 0.19 ± 0.04 | 29.00 ± 7.09 | 29.33 ± 8.76 | 1.33 ± 0.47 | 1.17 ± 0.37 | 0.30 ± 0.21 | 0.12 ± 0.05 | 0.05 ± 0.03 | 0.03 ± 0.01 |
| PC | 441.11 ± 102.62 | 277.74 ± 78.50 | 0.26 ± 0.09 | 0.18 ± 0.04 | 31.83 ± 4.30 | 28.50±11.72 | 1.17 ± 0.37 | 1.33 ± 0.47 | 0.22 ± 0.09 | 0.11 ± 0.08 | 0.04 ± 0.02 | 0.03 ± 0.02 |
| PRC | 487.66 ± 143.81 | 314.86 ± 102.78 | 0.25 ± 0.05 | 0.18 ± 0.03 | 31.00 ± 6.58 | 27.50 ± 5.19 | 1.17 ± 0.37 | 1.17 ± 0.37 | 0.32 ± 0.08 | 0.11 ± 0.04 | 0.05 ± 0.02 | 0.03 ± 0.01 |
LCModel estimated relative GABA concentrations to total Cr (tCr) with Cramér–Rao lower bound (CRLB) for the sLASER and MEGA-sLASER. SNR was measured using the peak amplitude of the total NAA peak at 2 ppm and the standard deviation of the white noise area in the 7 to 9 ppm range These metabolic concentrations are directly measured values by LCModel before correcting for T2-relaxation. Absolute uncertainties were calculated by multiplying the relative CRLB from the estimated concentration [60]. The measured value was expressed as mean ± SD.
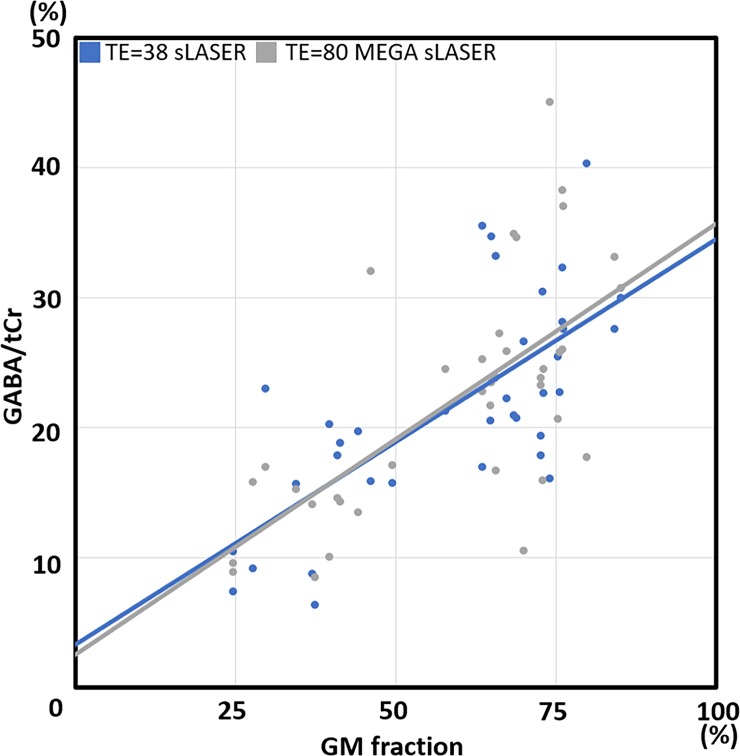
Fig 3 shows the correlation between GABA/tCr and GM volume fraction in the spectroscopy voxel estimated with the two different acquisitions after T2 correction between sLASER (TE = 38 ms) and MEGA-sLASER (TE = 80 ms). We found a linear correlation between the GABA/tCr concentration ratio and GM tissue distribution (linear regression line: yTE = 38 sLASER = 29.27x + 4.68 and yTE = 80 MEGA−sLASER = 33.18x + 2.54, %). In addition, these regression lines were almost identical for the two methods. This result shows a large dependence of measured GABA concentration on GM volume fraction, independent of the two acquisition methods. On the assumption that CSF does not contain significant quantities of metabolites [64], estimated percentage GABA distributions in the GM and WM using the linear regression lines were (GM%: WM%) 86%: 14% for sLASER and 93%: 7% for MEGA-sLASER, very much in line with the literature values for the relative distribution between GM and WM [65].
Fig 3. Correlation between GABA/total Cr(tCR) and gray matter (GM) volume fraction in the spectroscopy voxel.
Spectroscopy signal was measured by two different approaches (blue: echo time (TE) = 38 semi-LASER (sLASER), gray: TE = 80 MEGA-sLASER) across all examined voxels after the T2 correction. GABA concentration was in directly proportional to the GM tissue fraction. Linear regression lines of these two methods are almost identical. Equations of the linear regression lines were yTE = 38 sLASER = 29.27x + 4.68 and yTE = 80 MEGA−sLASER = 33.18x + 2.54, unit (%).
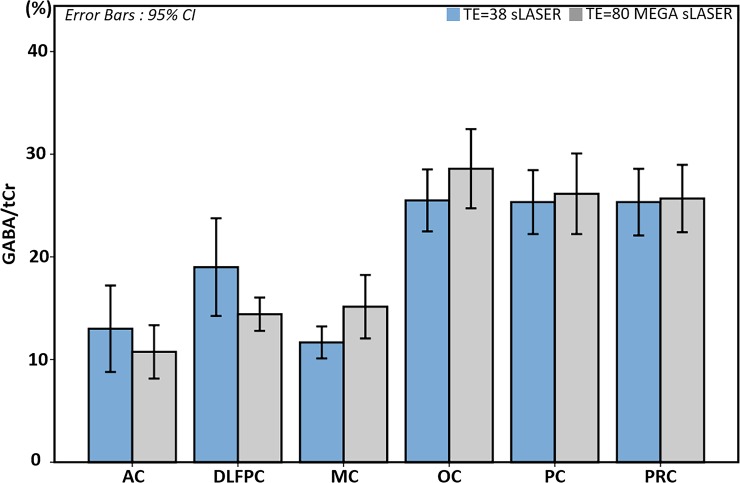
We also found similar GABA to tCr ratios acquired with the two methods for each voxel (Fig 4A). Error bars indicate 95% confidence intervals. Regional GABA concentrations showed statistically significant differences between group means as determined by a one-way ANOVA (F(5,36) = 0.302, p = 0.019) for the sLASER method, and (F(5,36) = 6.015, p < 0.001) for the GABA editing method. OC, PC, and PRC showed similarly high mean GABA/tCr ratios as compared with the other three regions: AC, DLPFC, and MC.
Fig 4. GABA/ total Cr(tCr) comparisons after considering the T2 relaxation times of GABA and tCr.
T2 correction factor for echo time (TE) = 80 MEGA-semi-LASER was 1.488 for GABA/tCr. After applying this factor, we found no statistically significant differences between group means across acquisition methods determined by a one-way ANOVA. Error bars indicate 95% confidence intervals.
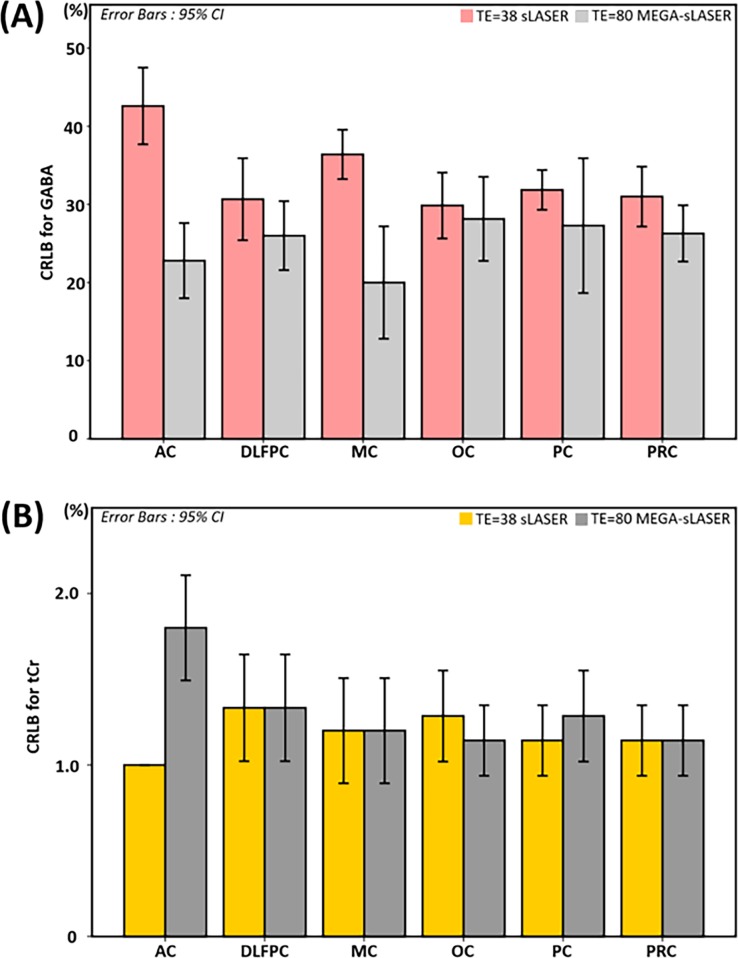
Fig 5 shows CRLB comparisons of GABA (A) and tCr (B), estimated by the two different acquisition strategies. For both metabolites, the MEGA-editing approach showed slightly better spectral fitting quality than that of the non-editing acquisition approach according to CRLB for all regions examined. Error bars also indicate 95% confidence intervals.
Fig 5. Cramér–Rao lower bound (CRLB) comparison.
This figure compares CRLBs of (a) GABA and (b) total Cr (tCr) between semi-LASER (sLASER) and MEGA-sLASER for the six brain regions examined. Error bars indicate 95% confidence intervals, in accordance with Table 1.
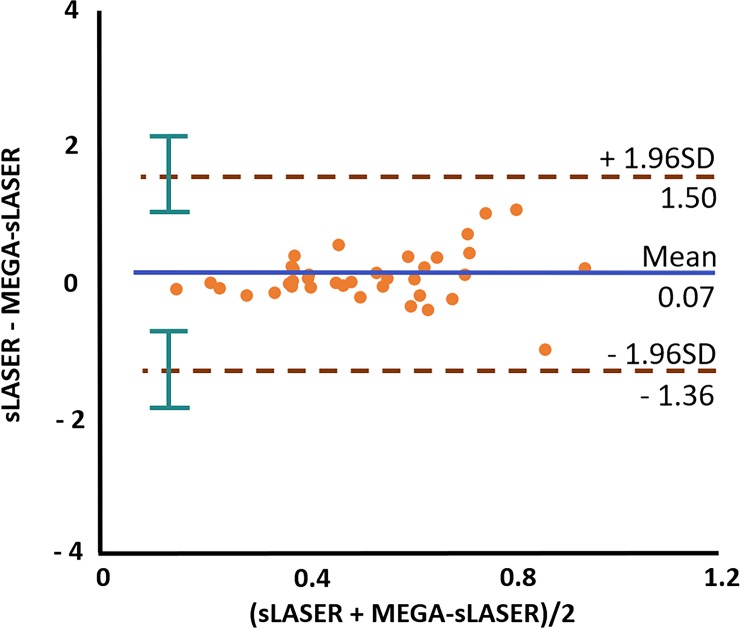
The Bland-Altman plot in Fig 6 shows the mean bias ± SD with 95% confidence intervals between the sLASER and MEGA-sLASER results as 0.06 ± 0.73 (blue line), and the limits of agreement were −1.36 and 1.50 (brown lines).
Fig 6. Bland-Altman plot for GABA.
Bland-Altman plot of the GABA concentration from the interleaved measurements using the semi-LASER (sLASER) and MEGA-sLASER approaches afterthe LCModel analysis. The blue horizontal line shows a mean value and two brown lines show ± 1.96 * SD with 95% confidence intervals (green) across all measurements.
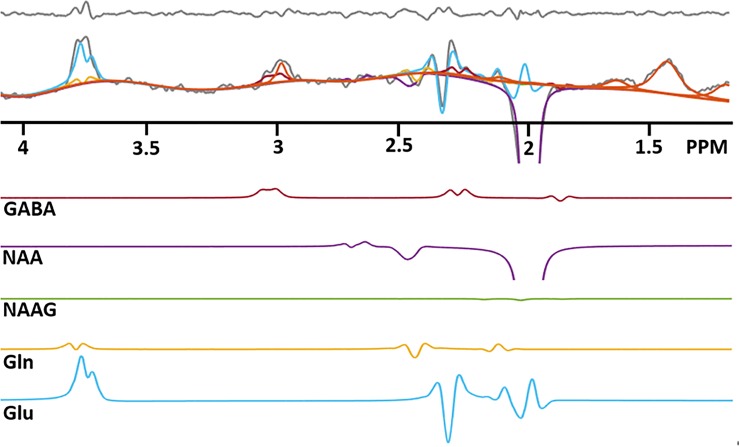
One example of the GABA edited spectrum (gray) with metabolite fitting lines: GABA (red), NAAG (green), NAA (purple), Gln (yellow) and Glu (blue), which were estimated from LCModel is shown in Fig 7. GABA estimation is clearly to a large degree determined by the fit to the 2.28 ppm peak. Pure GABA accounts for 47.21 ± 17.04 percent of the total 3 ppm signal, calculated as the ratio of the LCModel GABA fitted line at 3 ppm to the total measured 3 ppm peak averaged over all subjects.
Fig 7. One example of GABA edited spectrum (gray) with metabolite fitting lines.
GABA (red), NAAG (green), NAA (purple), Gln (yellow), Glu (blue), Glu (blue) and macromolecule (MM: orange), which were estimated from LCModel. Top panel indicates residual, which is a difference between in-vivo and fitting lines. Directly observable GABA-2CH2 resonance at 2.28 ppm plays an important role in the GABA quantification with spectral fitting. GABA estimation is limited by the 2.28 peak, which is the true GABA. It prevents over-estimating GABA by co-edited macromolecule. Note that the bottom part of the NAA signal is truncated in this figure.
Discussion
The interleaved sequence, which we implemented in this study, provides simultaneous MEGA-editing and non-editing acquisitions in a single sequence. By acquiring concurrently, the interleaved acquisition balances any instability across the two techniques. Therefore, we could avoid potential problems of registration, motion, B1-inhomogeneity or different shim quality that may have occurred if we would have acquired sequentially. It is widely known that adiabatic RF pulse based sLASER sequence has the advantage of the less level of CSDE in comparison of the PRESS sequence at ultrahigh field [43] owing to the use of adiabatic refocusing pulses. The sequence used in this study had a maximum CSDE of 8.4% [66].
After applying the T2 correction factor, which compensates the different TEs, the GABA/tCr ratios for the two acquisition techniques did not differ significantly. From the fact, almost all difference values are within the confidence limits, and the p-value is greater than 0.05, we conclude that there was no significant difference between the GABA acquisition methods (See Fig 6). In addition, the uniform distribution of difference values shows that there is no systematic bias between the two methods (calculated bias = 0.01). Judging from results of the linear regression lines (see Fig 3) and the Bland-Altman analysis (see Fig 6), the two different approaches showed similar accuracy in terms of the GABA estimation across the brain regions in combination with proper compensation steps.
Regional GABA concentration
Even though the main purpose of this study was to compare the two methodological approaches in combination with LCModel analysis, we were also able to compare GABA concentrations across different brain regions. As we mentioned in the Introduction, it is widely known that GABA is mostly concentrated in GM (approximately 90% in GM and 10% in WM). Not only spectroscopy studies [30, 65, 67] but also an ex-vivo study [68] obtained similar GABA distribution ratios between GM and WM. Chen, et al. [69] reported that GABA concentration linearly increased with GM volume fraction for one of the brain regions we also studied (AC). In our result, as we can see in Fig 3, GABA concentrations were largely driven by the GM tissue fraction. In addition, estimated GABA distribution ratios in the GM and WM by the linear regression lines are in close agreement with the literature value. This result confirms that GABA is heavily concentrated in GM, and highlights the importance of the tissue volume correction for GABA quantification [70].
In addition to the current study, other studies, which investigated regional GABA concentration differences while accounting for tissue volume variations, found differences in the regional GABA level. However, there is some inconsistency in the literature. For instance, Ganji, et al. [30] reported higher concentrations of GABA in the frontal lobe than in the OC at using PRESS (TE = 92 ms) at 7 T. Öngür, et al. [71] found higher GABA/Cr ratio in a frontal region (AC) than in an occipital region (parieto-occipital cortex) measured with the MEGA-editing method at 4 T. On the other hand, Bhagwagar, et al. [72] found higher GABA/Cr ratio measured with MEGA-editing in the occipital region (OC) than that in AC. Durst, et al. [73] also reported higher GABA in OC than in frontal lobe using 3 T MEGA-PRESS. Van der Veen, et al. [74] similarly found higher GABA concentration in the OC than that of the anterior area (MPFC) with the MEGA-editing method comparing two voxels in frontal and occipital lobes. Van der Veen, et al. [74] selected voxels that had the same GM and WM tissue volume to exclude tissue volume effects. They proposed therefore that the difference should be attributed to factors other than GM volume. The trend in our data is in accordance with that of the latter studies [72–74].
Recent molecular Imaging studies, which quantified GABAA receptor using positron emission tomography (PET) with 11C flumazenil [75, 76], and single-photon emission computed tomography (SPECT) with 123I-iomazenil [77] have visualized GABAA receptor densities in the human brain. These reports have shown not only that the GABAA receptor is more heavily concentrated in GM, but also its density in posterior brain areas is higher than that of anterior areas. This supports our finding that GABA concentration in posterior regions such as OC, PC, and PRC showed higher GABA levels than those of anterior regions: AC, DLPFC, and MC.
MM contamination
We found no statistically significant difference for the quantified GABA concentrations with LCModel between two acquisition techniques. Our finding could potentially be explained by one of two possibilities: The LCModel fitting result of the non-editing spectrum also contains MM contamination, or LCModel efficiently quantifies the GABA signal from the GABA edited spectrum without the interference of the co-edited MM.
LCModel estimates MM and lipid signals at each fixed chemical shift location with non-parametric spectral models including a smoothing B-spline as a baseline [24]. Because these MM and lipid signals are not only measured values but also part of the formulaic model, these quantification approaches still risk over- or under-estimating metabolic concentrations in the co-edited signal. It is quite challenging to determine from our LCModel quantification results of sLASER whether the estimation of GABA is contaminated by MM. However, we can assume the presence of MM contamination in the MEGA-sLASER spectra. In contrast to MM and lipid signals, metabolic signal quantification uses parametric spectral models, which are included in a basis set. LCModel finds metabolic concentrations by changing the amplitudes of simulated metabolic models with their lineshapes unchanged. Therefore, all resonances of GABA also play an important role in solving the optimization problem. In our spectral fitting results, we found that the directly discernible 2.28 ppm GABA signal was essential for distinguishing the GABA signal from GABA+ (See Fig 7). GABA estimation was largely determined by the size of GABA-2CH2 resonance at 2.28 ppm. Because the 2.28 ppm region is not related to the co-edited MM, the visible GABA resonance at 2.28 ppm represents an uncontaminated GABA signal. This result suggests that GABA quantification with spectral fitting is able to separate pure GABA from GABA+ provided that the 2.28 ppm GABA resonance is also recorded. Previous studies by Ganji, et al. [30] and Choi, et al. [78] also highlighted the significance of this GABA-2CH2 resonance in terms of the spectral fitting procedure.
LCModel provides a GABA-editing exclusive control file called ‘mega-press-3’ [56]. This setting excludes quantification of the baseline and MM signals assuming baseline and MM signals are identical between MEGA-on and -off spectra, and that they hence cancel in the subtraction. Since this setting mainly attempts to estimate the 3 ppm peak as GABA+, this necessarily overestimates the GABA concentration as co-edited MM will not be eliminated. Therefore, we did not use this approach.
We corrected for T2 relaxation using the T2-value of 63 ms taken from the study by Intrapiromkul, et al. [62]. After correction, the values obtained were similar between the two methods (See Fig 3 and Fig 4A). This consistency strongly supports the correctness of the T2-value used, and also that the analyses have yielded results without MM contamination. Our estimate that MM constitutes approximately 53% of the signal at 3 ppm is at the upper limit of, but not inconsistent with previous literature values 44% [15], 41–49% [79], and 52–57% [80].
Spectral fitting quality
Since the thresholded CRLB estimation may cause a bias [60], we included all GABA and tCr results. We also compared absolute uncertainties for the GABA estimation for both acquisition methods. As a result, we found that CRLBs and absolute uncertainties of estimated GABA for the MEGA-editing method are lower than those for non-editing acquisition method for all six regions (See Table 1 and Fig 5A).
Compared with the non-editing acquisition, the GABA editing method provides a much simpler spectral pattern with only six metabolic signals present, that are relatively well separated. This explains why the spectral fitting accuracy of the MEGA editing results for estimating GABA is superior to that of the sLASER result, despite the editing method eliminating also a number of lines from the GABA spectrum. MEGA-sLASER and sLASER showed similar CRLB values for tCr (Fig 5B) which is unsurprising given that the tCr concentration of the MEGA-sLASER was estimated from the MEGA-off spectra using a non-editing acquisition. The reasonably good CRLB values of tCr for both approaches confirm that spectral quality and spectral fitting quality with LCModel were good enough for accurate quantification.
Conclusion
This study compared GABA concentration for six different brain regions with the non-editing spectral acquisition approach and the GABA editing approach using a single interleaved sequence acquisition. The simultaneous acquisition of sLASER and MEGA-sLASER minimized differences in confounds which could have occurred in individual acquisitions. Our finding confirms that GABA is heavily concentrated in GM. Therefore, regional GM volume is an important contributor to measured GABA concentration variation. In addition, the ability to reliably measure the GABA resonance at 2.28 ppm would appear to lead to accurate estimates of GABA concentration without MM contamination. A similar concentration of GABA signal measured with the two methods for each voxel was obtained after correction for T2, showing that the two acquisition strategies are both reliable and have a similar accuracy to measure GABA. However, The GABA-editing approach was superior in terms of precision, as assessed by lower CRLB values.
Supporting information
(XLSX)
Acknowledgments
This work was funded by the Helmholtz Alliance ICEMED–Imaging and Curing Environmental Metabolic Diseases, through the Initiative and Networking Fund of the Helmholtz Association.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by the Helmholtz Alliance ICEMED – Imaging and Curing Environmental Metabolic Diseases, through the Initiative and Networking Fund of the Helmholtz Association. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.McCormick DA. GABA as an inhibitory neurotransmitter in human cerebral cortex. J Neurophysiol. 1989;62(5):1018–27. 10.1152/jn.1989.62.5.1018 [DOI] [PubMed] [Google Scholar]
- 2.Bellance N, Pabst L, Allen G, Rossignol R, Nagrath D. Oncosecretomics coupled to bioenergetics identifies α-amino adipic acid, isoleucine and GABA as potential biomarkers of cancer: Differential expression of c-Myc, Oct1 and KLF4 coordinates metabolic changes. Biochimica Et Biophysica Acta (BBA)-Bioenergetics. 2012;1817(11):2060–71. [DOI] [PubMed] [Google Scholar]
- 3.Cawley N, Solanky BS, Muhlert N, Tur C, Edden RA, Wheeler-Kingshott CA, et al. Reduced gamma-aminobutyric acid concentration is associated with physical disability in progressive multiple sclerosis. Brain. 2015;138(9):2584–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bai X, Edden RA, Gao F, Wang G, Wu L, Zhao B, et al. Decreased γ‐aminobutyric acid levels in the parietal region of patients with Alzheimer's disease. J Magn Reson Imaging. 2015;41(5):1326–31. 10.1002/jmri.24665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petroff OA, Rothman DL, Behar KL, Mattson RH. Initial Observations on Effect of Vigabatrin on In Vivo 1H Spectroscopic Measurements of γ‐Aminobutyric Acid, Glutamate, and Glutamine in Human Brain. Epilepsia. 1995;36(5):457–64. 10.1111/j.1528-1157.1995.tb00486.x [DOI] [PubMed] [Google Scholar]
- 6.Marsman A, Mandl RC, Klomp DW, Bohlken MM, Boer VO, Andreychenko A, et al. GABA and glutamate in schizophrenia: A 7 T 1H-MRS study. NeuroImage: Clinical. 2014;6:398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oblak AL, Gibbs TT, Blatt GJ. Reduced GABA A receptors and benzodiazepine binding sites in the posterior cingulate cortex and fusiform gyrus in autism. Brain Res. 2011;1380:218–28. 10.1016/j.brainres.2010.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lingford-Hughes A, Hume SP, Feeney A, Hirani E, Osman S, Cunningham VJ, et al. Imaging the GABA-benzodiazepine receptor subtype containing the α5-subunit in vivo with [11C] Ro15 4513 positron emission tomography. J Cereb Blood Flow Metab. 2002;22(7):878–89. 10.1097/00004647-200207000-00013 [DOI] [PubMed] [Google Scholar]
- 9.Verhoeff NPL, Petroff OA, Hyder F, Zoghbi SS, Fujita M, Rajeevan N, et al. Effects of vigabatrin on the GABAergic system as determined by [123I] iomazenil SPECT and GABA MRS. Epilepsia. 1999;40(10):1433–8. 10.1111/j.1528-1157.1999.tb02016.x [DOI] [PubMed] [Google Scholar]
- 10.Keltner JR, Wald LL, Christensen JD, Maas LC, Moore CM, Cohen BM, et al. A technique for detecting GABA in the human brain with PRESS localization and optimized refocusing spectral editing radiofrequency pulses. Magn Reson Med. 1996;36(3):458–61. 10.1002/mrm.1910360319 [DOI] [PubMed] [Google Scholar]
- 11.Cai K, Haris M, Singh A, Kogan F, Greenberg JH, Hariharan H, et al. Magnetic resonance imaging of glutamate. Nat Med. 2012;18(2):302 10.1038/nm.2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan G, Zhang T, Dai Z, Yi M, Jia Y, Nie T, et al. A Potential Magnetic Resonance Imaging Technique Based on Chemical Exchange Saturation Transfer for In Vivo γ-Aminobutyric Acid Imaging. PLoS One. 2016;11(10):e0163765 10.1371/journal.pone.0163765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Godlewska BR, Clare S, Cowen PJ, Emir UE. Ultra-high-field magnetic resonance spectroscopy in psychiatry. Frontiers in psychiatry. 2017;8:123 10.3389/fpsyt.2017.00123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puts NA, Edden RA. In vivo magnetic resonance spectroscopy of GABA: a methodological review. Progress in nuclear magnetic resonance spectroscopy. 2012;60:29–41. 10.1016/j.pnmrs.2011.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rothman DL, Petroff O, Behar KL, Mattson RH. Localized 1H NMR measurements of gamma-aminobutyric acid in human brain in vivo. Proceedings of the national academy of sciences. 1993;90(12):5662–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mescher M, Tannus A, Johnson MN, Garwood M. Solvent suppression using selective echo dephasing. Journal of Magnetic Resonance, Series A. 1996;123(2):226–9. [Google Scholar]
- 17.Mescher M, Merkle H, Kirsch J, Garwood M, Gruetter R. Simultaneous in vivo spectral editing and water suppression. NMR Biomed. 1998;11(EPFL-ARTICLE-177509):266–72. [DOI] [PubMed] [Google Scholar]
- 18.Shen J, Rothman DL, Brown P. In vivo GABA editing using a novel doubly selective multiple quantum filter. Magn Reson Med. 2002;47(3):447–54. 10.1002/mrm.10104 [DOI] [PubMed] [Google Scholar]
- 19.Chen C, Sigurdsson HP, Pépés SE, Auer DP, Morris PG, Morgan PS, et al. Activation induced changes in GABA: functional MRS at 7 T with MEGA-sLASER. Neuroimage. 2017;156:207–13. 10.1016/j.neuroimage.2017.05.044 [DOI] [PubMed] [Google Scholar]
- 20.Andreychenko A, Boer VO, Arteaga de Castro CS, Luijten PR, Klomp DW. Efficient spectral editing at 7 T: GABA detection with MEGA‐sLASER. Magn Reson Med. 2012;68(4):1018–25. 10.1002/mrm.24131 [DOI] [PubMed] [Google Scholar]
- 21.Houtepen LC, Schür RR, Wijnen JP, Boer VO, Boks M, Kahn RS, et al. Acute stress effects on GABA and glutamate levels in the prefrontal cortex: A 7T 1H magnetic resonance spectroscopy study. NeuroImage: Clinical. 2017;14:195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edden RA, Puts NA, Barker PB. Macromolecule‐suppressed GABA‐edited magnetic resonance spectroscopy at 3T. Magn Reson Med. 2012;68(3):657–61. 10.1002/mrm.24391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris AD, Glaubitz B, Near J, John Evans C, Puts NA, Schmidt‐Wilcke T, et al. Impact of frequency drift on gamma‐aminobutyric acid‐edited MR spectroscopy. Magn Reson Med. 2014;72(4):941–8. 10.1002/mrm.25009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med. 1993;30(6):672–9. 10.1002/mrm.1910300604 [DOI] [PubMed] [Google Scholar]
- 25.Provencher SW. Automatic quantitation of localized in vivo1H spectra with LCModel. NMR Biomed. 2001;14(4):260–4. [DOI] [PubMed] [Google Scholar]
- 26.Napolitano A, Kockenberger W, Auer DP. Reliable gamma aminobutyric acid measurement using optimized PRESS at 3 T. Magn Reson Med. 2013;69(6):1528–33. 10.1002/mrm.24397 [DOI] [PubMed] [Google Scholar]
- 27.Near J, Andersson J, Maron E, Mekle R, Gruetter R, Cowen P, et al. Unedited in vivo detection and quantification of γ‐aminobutyric acid in the occipital cortex using short‐TE MRS at 3 T. NMR Biomed. 2013;26(11):1353–62. 10.1002/nbm.2960 [DOI] [PubMed] [Google Scholar]
- 28.Öz G, Terpstra M, Tkáč I, Aia P, Lowary J, Tuite PJ, et al. Proton MRS of the unilateral substantia nigra in the human brain at 4 tesla: detection of high GABA concentrations. Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine. 2006;55(2):296–301. [DOI] [PubMed] [Google Scholar]
- 29.Wijtenburg SA, Rowland LM, Edden RA, Barker PB. Reproducibility of brain spectroscopy at 7T using conventional localization and spectral editing techniques. J Magn Reson Imaging. 2013;38(2):460–7. 10.1002/jmri.23997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ganji SK, An Z, Banerjee A, Madan A, Hulsey KM, Choi C. Measurement of regional variation of GABA in the human brain by optimized point‐resolved spectroscopy at 7 T in vivo. NMR Biomed. 2014;27(10):1167–75. 10.1002/nbm.3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pradhan S, Bonekamp S, Gillen JS, Rowland LM, Wijtenburg SA, Edden RA, et al. Comparison of single voxel brain MRS AT 3 T and 7 T using 32-channel head coils. Magn Reson Imaging. 2015;33(8):1013–8. 10.1016/j.mri.2015.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terpstra M, Vaughan T, Ugurbil K, Lim KO, Schulz SC, Gruetter R. Validation of glutathione quantitation from STEAM spectra against edited 1H NMR spectroscopy at 4T: application to schizophrenia. Magnetic Resonance Materials in Physics, Biology and Medicine. 2005;18(5):276. [DOI] [PubMed] [Google Scholar]
- 33.Frahm J, Merboldt K-D, Hänicke W. Localized proton spectroscopy using stimulated echoes. Journal of Magnetic Resonance (1969). 1987;72(3):502–8. [Google Scholar]
- 34.Granot J. Selected volume excitation using stimulated echoes (VEST). Applications to spatially localized spectroscopy and imaging. Journal of Magnetic Resonance (1969). 1986;70(3):488–92. [Google Scholar]
- 35.Kimmich R, Hoepfel D. Volume-selective multipulse spin-echo spectroscopy. Journal of Magnetic Resonance (1969). 1987;72(2):379–84. [Google Scholar]
- 36.Sanaei Nezhad F, Anton A, Parkes LM, Deakin B, Williams SR. Quantification of glutathione in the human brain by MR spectroscopy at 3 Tesla: Comparison of PRESS and MEGA‐PRESS. Magn Reson Med. 2017;78(4):1257–66. 10.1002/mrm.26532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ordidge RJ, Connelly A, Lohman JA. Image-selected in vivo spectroscopy (ISIS). A new technique for spatially selective NMR spectroscopy. Journal of Magnetic Resonance (1969). 1986;66(2):283–94. [Google Scholar]
- 38.Bottomley P, Charles H, Roemer P, Flamig D, Engeseth H, Edelstein W, et al. Human in vivo phosphate metabolite imaging with 31P NMR. Magn Reson Med. 1988;7(3):319–36. 10.1002/mrm.1910070309 [DOI] [PubMed] [Google Scholar]
- 39.Chen C, Morris P, Francis S, Gowland P, editors. A comparison of MEGA-sLASER and STEAM for in vivo quantification of GABA at 7T. Proc Int Soc Magn Reson Med; 2015. [Google Scholar]
- 40.Behar KL, Rothman DL, Spencer DD, Petroff OA. Analysis of macromolecule resonances in 1H NMR spectra of human brain. Magn Reson Med. 1994;32(3):294–302. 10.1002/mrm.1910320304 [DOI] [PubMed] [Google Scholar]
- 41.Behar KL, Ogino T. Characterization of macromolecule resonances in the 1H NMR spectrum of rat brain. Magn Reson Med. 1993;30(1):38–44. 10.1002/mrm.1910300107 [DOI] [PubMed] [Google Scholar]
- 42.Mullins PG, McGonigle DJ, O'gorman RL, Puts NA, Vidyasagar R, Evans CJ, et al. Current practice in the use of MEGA-PRESS spectroscopy for the detection of GABA. Neuroimage. 2014;86:43–52. 10.1016/j.neuroimage.2012.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scheenen TW, Heerschap A, Klomp DW. Towards 1H-MRSI of the human brain at 7T with slice-selective adiabatic refocusing pulses. Magnetic Resonance Materials in Physics, Biology and Medicine. 2008;21(1–2):95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scheenen TW, Klomp DW, Wijnen JP, Heerschap A. Short echo time 1H‐MRSI of the human brain at 3T with minimal chemical shift displacement errors using adiabatic refocusing pulses. Magn Reson Med. 2008;59(1):1–6. 10.1002/mrm.21302 [DOI] [PubMed] [Google Scholar]
- 45.Ogg RJ, Kingsley R, Taylor JS. WET, a T1-and B1-insensitive water-suppression method for in vivo localized 1H NMR spectroscopy. Journal of Magnetic Resonance, Series B. 1994;104(1):1–10. [DOI] [PubMed] [Google Scholar]
- 46.Terpstra M, Cheong I, Lyu T, Deelchand DK, Emir UE, Bednařík P, et al. Test‐retest reproducibility of neurochemical profiles with short‐echo, single‐voxel MR spectroscopy at 3T and 7T. Magn Reson Med. 2016;76(4):1083–91. 10.1002/mrm.26022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boer V, van Lier A, Hoogduin J, Wijnen J, Luijten P, Klomp D. 7‐T 1H MRS with adiabatic refocusing at short TE using radiofrequency focusing with a dual‐channel volume transmit coil. NMR Biomed. 2011;24(9):1038–46. 10.1002/nbm.1641 [DOI] [PubMed] [Google Scholar]
- 48.Ip IB, Berrington A, Hess AT, Parker AJ, Emir UE, Bridge H. Combined fMRI-MRS acquires simultaneous glutamate and BOLD-fMRI signals in the human brain. Neuroimage. 2017;155:113–9. 10.1016/j.neuroimage.2017.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mugler JP, Brookeman JR. Three‐dimensional magnetization‐prepared rapid gradient‐echo imaging (3D MP RAGE). Magn Reson Med. 1990;15(1):152–7. 10.1002/mrm.1910150117 [DOI] [PubMed] [Google Scholar]
- 50.Gruetter R, Tkáč I. Field mapping without reference scan using asymmetric echo‐planar techniques. Magn Reson Med. 2000;43(2):319–23. [DOI] [PubMed] [Google Scholar]
- 51.Roemer PB, Edelstein WA, Hayes CE, Souza SP, Mueller O. The NMR phased array. Magn Reson Med. 1990;16(2):192–225. 10.1002/mrm.1910160203 [DOI] [PubMed] [Google Scholar]
- 52.Naressi A, Couturier C, Devos J, Janssen M, Mangeat C, De Beer R, et al. Java-based graphical user interface for the MRUI quantitation package. Magnetic resonance materials in physics, biology and medicine. 2001;12(2–3):141. [DOI] [PubMed] [Google Scholar]
- 53.Naressi A, Couturier C, Castang I, De Beer R, Graveron-Demilly D. Java-based graphical user interface for MRUI, a software package for quantitation of in vivo/medical magnetic resonance spectroscopy signals. Comput Biol Med. 2001;31(4):269–86. 10.1016/s0010-4825(01)00006-3 [DOI] [PubMed] [Google Scholar]
- 54.Govindaraju V, Young K, Maudsley AA. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR Biomed. 2000;13(3):129–53. [DOI] [PubMed] [Google Scholar]
- 55.Govind V, Young K, Maudsley AA. Corrigendum: Proton NMR chemical shifts and coupling constants for brain metabolites. Govindaraju V Young K, Maudsley AA, NMR Biomed. 2000; 13: 129–153. NMR Biomed. 2015;28(7):923–4. [DOI] [PubMed] [Google Scholar]
- 56.Provencher SW. LCModel & LCMgui user’s manual. LCModel Version. 2016:6.3-1L. [Google Scholar]
- 57.Bhagwagar Z, Wylezinska M, Jezzard P, Evans J, Ashworth F, Sule A, et al. Reduction in occipital cortex γ-aminobutyric acid concentrations in medication-free recovered unipolar depressed and bipolar subjects. Biol Psychiatry. 2007;61(6):806–12. 10.1016/j.biopsych.2006.08.048 [DOI] [PubMed] [Google Scholar]
- 58.Raschke F, Fuster‐Garcia E, Opstad K, Howe F. Classification of single‐voxel 1H spectra of brain tumours using LCModel. NMR Biomed. 2012;25(2):322–31. 10.1002/nbm.1753 [DOI] [PubMed] [Google Scholar]
- 59.Chowdhury FA, O'Gorman RL, Nashef L, Elwes RD, RA Edden, JB Murdoch, et al. Investigation of glutamine and GABA levels in patients with idiopathic generalized epilepsy using MEGAPRESS. J Magn Reson Imaging. 2015;41(3):694–9. 10.1002/jmri.24611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kreis R. The trouble with quality filtering based on relative Cramér‐Rao lower bounds. Magn Reson Med. 2016;75(1):15–8. 10.1002/mrm.25568 [DOI] [PubMed] [Google Scholar]
- 61.Edden RA, Intrapiromkul J, Zhu H, Cheng Y, Barker PB. Measuring T2 in vivo with J‐difference editing: Application to GABA at 3 tesla. J Magn Reson Imaging. 2012;35(1):229–34. 10.1002/jmri.22865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Intrapiromkul J, Zhu H, Cheng Y, Barker PB, Edden RA. Determining the in vivo transverse relaxation time of GABA in the human brain at 7T. J Magn Reson Imaging. 2013;38(5):1224–9. 10.1002/jmri.23979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bland JM, Altman D. Statistical methods for assessing agreement between two methods of clinical measurement. The lancet. 1986;327(8476):307–10. [PubMed] [Google Scholar]
- 64.Joanna L, James P, Anthony A, Garnette RS. Nuclear magnetic resonance study of cerebrospinal fluid from patients with multiple sclerosis. Can J Neurol Sci. 1993;20(3):194–8. [PubMed] [Google Scholar]
- 65.Bhattacharyya P, Phillips M, Stone L, Lowe M. In-vivo MRS measurement of gray-matter and white-matter GABA concentration in sensorimotor cortex using a motion-controlled MEGA-PRESS Sequence. Magn Reson Imaging. 2011;29(3):374 10.1016/j.mri.2010.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rohani Rankouhi S, Hong D, Dyvorne H, Balchandani P, Norris DG. MASE‐sLASER, a short‐TE, matched chemical shift displacement error sequence for single‐voxel spectroscopy at ultrahigh field. NMR Biomed. 2018;31(7):e3940 10.1002/nbm.3940 [DOI] [PubMed] [Google Scholar]
- 67.Choi I-Y, Lee S-P, Merkle H, Shen J. In vivo detection of gray and white matter differences in GABA concentration in the human brain. Neuroimage. 2006;33(1):85–93. 10.1016/j.neuroimage.2006.06.016 [DOI] [PubMed] [Google Scholar]
- 68.Perry T, Berry K, Hansen S, Diamond S, Mok C. Regional distribution of amino acids in human brain obtained at autopsy. J Neurochem. 1971;18(3):513–9. 10.1111/j.1471-4159.1971.tb11979.x [DOI] [PubMed] [Google Scholar]
- 69.Chen M, Li G, Zhang Z, Chen L, Pei M, Yan X, et al. The anterior cingulate cortex GABA levels with varied tissue composition measured by in vivo single voxel MRS. Proc Int Soc Magn Reson Med. 2016. [Google Scholar]
- 70.Mikkelsen M, Singh KD, Brealy JA, Linden DE, Evans CJ. Quantification of γ‐aminobutyric acid (GABA) in 1H MRS volumes composed heterogeneously of grey and white matter. NMR Biomed. 2016;29(11):1644–55. 10.1002/nbm.3622 [DOI] [PubMed] [Google Scholar]
- 71.Öngür D, Prescot AP, McCarthy J, Cohen BM, Renshaw PF. Elevated gamma-aminobutyric acid levels in chronic schizophrenia. Biol Psychiatry. 2010;68(7):667–70. 10.1016/j.biopsych.2010.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bhagwagar Z, Wylezinska M, Jezzard P, Evans J, Boorman E, Matthews PM, et al. Low GABA concentrations in occipital cortex and anterior cingulate cortex in medication-free, recovered depressed patients. Int J Neuropsychopharmacol. 2008;11(2):255–60. 10.1017/S1461145707007924 [DOI] [PubMed] [Google Scholar]
- 73.Durst CR, Michael N, Tustison NJ, Patrie JT, Raghavan P, Wintermark M, et al. Noninvasive evaluation of the regional variations of GABA using magnetic resonance spectroscopy at 3 Tesla. Magn Reson Imaging. 2015;33(5):611–7. 10.1016/j.mri.2015.02.015 [DOI] [PubMed] [Google Scholar]
- 74.Veen JWvd, Shen J. Regional difference in GABA levels between medial prefrontal and occipital cortices. J Magn Reson Imaging. 2013;38(3):745–50. 10.1002/jmri.24009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Richardson M, Koepp M, Duncan J, Brooks D, Fish D. Benzodiazepine receptors in focal epilepsy with cortical dysgenesis: An 11C‐flumazenil PET study. Ann Neurol. 1996;40(2):188–98. 10.1002/ana.410400210 [DOI] [PubMed] [Google Scholar]
- 76.D’Hulst C, Heulens I, Van der Aa N, Goffin K, Koole M, Porke K, et al. Positron Emission Tomography (PET) Quantification of GABAA Receptors in the Brain of Fragile X Patients. PLoS One. 2015;10(7):e0131486 10.1371/journal.pone.0131486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nagamitsu S, Sakurai R, Matsuoka M, Chiba H, Ozono S, Tanigawa H, et al. Altered SPECT 123I-iomazenil Binding in the Cingulate Cortex of Children with Anorexia Nervosa. Frontiers in psychiatry. 2016;7 10.3389/fpsyt.2016.00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Choi C, Banerjee A, Ganji S, Dimitrov I, Ghose S, Tamminga C. Contamination-free measurement of GABA in the human brain by optimized PRESS at 7.0 T in vivo Proc Intl Soc Mag Reson Med. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shungu DC, Mao X, Gonzales R, Soones TN, Dyke JP, Veen JW, et al. Brain γ‐aminobutyric acid (GABA) detection in vivo with the J‐editing 1H MRS technique: a comprehensive methodological evaluation of sensitivity enhancement, macromolecule contamination and test–retest reliability. NMR Biomed. 2016;29(7):932–42. 10.1002/nbm.3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mikkelsen M, Singh KD, Sumner P, Evans CJ. Comparison of the repeatability of GABA‐edited magnetic resonance spectroscopy with and without macromolecule suppression. Magn Reson Med. 2016;75(3):946–53. 10.1002/mrm.25699 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.